Abstract
Hydroxyproline (Hyp)-rich glycoproteins (HRGPs) participate in all aspects of plant growth and development. HRGPs are generally highly O-glycosylated through the Hyp residues, which means carbohydrates help define the interactive molecular surface and, hence, HRGP function. The Hyp contiguity hypothesis predicts that contiguous Hyp residues are sites of HRGP arabinosylation, whereas clustered noncontiguous Hyp residues are sites of galactosylation, giving rise to the arabinogalactan heteropolysaccharides that characterize the arabinogalactan-proteins. Early tests of the hypothesis using synthetic genes encoding only clustered noncontiguous Hyp in the sequence (serine [Ser]-Hyp-Ser-Hyp)n or contiguous Hyp in the series (Ser-Hyp-Hyp)n and (Ser-Hyp-Hyp-Hyp-Hyp)n confirmed that arabinogalactan polysaccharide was added only to noncontiguous Hyp, whereas arabinosylation occurred on contiguous Hyp. Here, we extended our tests of the codes that direct arabinogalactan polysaccharide addition to Hyp by building genes encoding the repetitive sequences (alanine [Ala]-proline [Pro]-Ala-Pro)n, (threonine [Thr]-Pro-Thr-Pro)n, and (valine [Val]-Pro-Val-Pro)n, and expressing them in tobacco (Nicotiana tabacum) Bright-Yellow 2 cells as fusion proteins with green fluorescent protein. All of the Pro residues in the (Ala-Pro-Ala-Pro)n fusion protein were hydroxylated and consistent with the hypothesis that every Hyp residue was glycosylated with arabinogalactan polysaccharide. In contrast, 20% to 30% of Pro residues remained non-hydroxylated in the (Thr-Pro-Thr-Pro)n, and (Val-Pro-Val-Pro)n fusion proteins. Furthermore, although 50% to 60% of the Hyp residues were glycosylated with arabinogalactan polysaccharide, some remained non-glycosylated or were arabinosylated. These results suggest that the amino acid side chains of flanking residues influence the extent of Pro hydroxylation and Hyp glycosylation and may explain why isolated noncontiguous Hyp in extensins do not acquire an arabinogalactan polysaccharide but are arabinosylated or remain non-glycosylated.
Glycoproteins play diverse roles in the eukaryotic extracellular matrices of both plants and animals. However, in the Hyp-rich glycoprotein (HRGP) superfamily, which is restricted to plants and green algae, Hyp-O-glycosylation forms unique glycomodules that contribute to molecular shape, size, rigidity, stability, solubility, and charge. Such glycomodules often create the bulk of the interactive molecular surface.
Peptide periodicity together with the extent and type of O-glycosylation define three major HRGP families (Kieliszewski and Lamport, 1994). Repetitive Pro-rich proteins (RPRPs) are the most highly periodic and lightly glycosylated, often composed of simple pentapeptide or hexapeptide repeats containing arabinosylated and/or non-glycosylated Hyp.
Extensins exhibit more complex periodicity and glycosylation where repetitive motifs typically of 5, 10, and 16 amino acid residues include monogalactosylated Ser residues (Lamport et al., 1973) and extensively arabinosylated Hyp residues (Smith et al., 1984). On alkaline hydrolysis, both RPRPs and extensins yield a Hyp glycoside profile consisting exclusively of small neutral Hyp arabinooligosaccharides (Lamport and Miller, 1971).
Arabinogalactan-proteins (AGPs) are the least periodic and most highly glycosylated HRGPs. However, AGPs do generally contain repetitive motifs like Xaa-Hyp-Xaa-Hyp and Xaa-Hyp-Hyp (where Xaa is often Ser, Thr, or Ala; Chen et al., 1994; Nothnagel, 1997; Gao et al., 1999) with the Hyp residues frequently glycosylated by short arabinooligosaccharides or significantly larger arabinogalactans (Pope, 1977; Qi et al., 1991). Thus, the Hyp glycoside profile of AGPs often shows a mixture of Hyp-arabinooligosaccharides and Hyp arabinogalactan polysaccharides.
RPRPs, extensions, and AGPs also contain members that are chimeras of an HRGP fused to a non-HRGP. For example, extensin chimeras, such as potato (Solanum tuberosum) lectin (Kieliszewski et al., 1994), and RPRP chimeras (Sheng et al., 1991; Bernhardt and Tierney, 1999) are well known, and recent work indicates that we can include AGPs as well (Cheung et al., 1993; Borner et al., 2002).
AGPs are of considerable current interest because of their apparent role in virtually all aspects of plant development and their location as glycoproteins tethered to the outer leaflet of the plasma membrane through glycosylphosphatidyl inositol (GPI) anchors (Youl et al., 1998; Sherrier et al., 1999; Svetek et al., 1999). Earlier work (Larkin, 1977, 1978; Herman and Lamb, 1992) showed that AGPs are major components of the plasma membrane, whereas more recent work indicates that a significant proportion of the plasma membrane proteome also exists as chimeric proteins containing small AGP domains characterized by their putative AGP glycomodule content (Sherrier et al., 1999; Borner et al., 2002). A high density of bound AGPs may form a periplasmic glycocalyx arranged as a meshwork or “plasmalemmal reticulum” (Gens et al., 2000) that imparts overall mechanical properties to the membrane, with the arabinogalactan polysaccharide substituents imparting its hydrophilic properties. Defining the glycosylation codes that determine Hyp glycosylation, therefore, are essential to interpret the genomic information and associated functions that define the membrane proteome. Inclusion of the substantial carbohydrate contribution to the surface sculpture may help refine our concept of the plasma membrane.
Earlier, we proposed a sequence-based glycosylation code based on Hyp contiguity. This Hyp contiguity hypothesis postulates arabinosylation of contiguous Hyp residues and galactosylation of noncontiguous Hyp residues (Kieliszewski et al., 1992a, 1995; Kieliszewski and Lamport, 1994). Primary amino acid sequences of known HRGPs show frequent repetitive Ser-Hyp, Ser-Hyp2, and Ser-Hyp4 motifs; therefore, we devised a strategy to test the Hyp contiguity hypothesis based on targeted expression of synthetic genes encoding those repetitive motifs, in effect presenting the cell with an endogenous substrate both for hydroxylation and subsequent glycosylation. An initial test (Shpak et al., 1999) involved the ancient Ser-Hyp repetitive motif that occurs in cell walls of green algae (Woessner and Goodenough, 1992; Ferris et al., 2001), higher plants (de Blank et al., 1993; Vijn et al., 1995), and also in the AGP component of Acacia senegal gum exudates, gum arabic (Goodrum et al., 2000). The noncontiguous Pro construct (Ser-Pro)n targeted for secretion in tobacco (Nicotiana tabacum) Bright-Yellow 2 (BY2) cells gave an expression product (Ser-Hyp)n where all the Hyp residues gained an arabinogalactan (Shpak et al., 1999) but with no evidence of arabinogalactan addition to Ser residues. In contrast, repetitive Ser-Hyp2 and Ser-Hyp4 showed exclusive arabinooligosaccharide addition to contiguous Hyp residues (Shpak et al., 2001). Most recently, the major AGP of tomato (Lycopersicon esculentum), LeAGP-1, purified from transgenic tobacco, yielded a Hyp glycoside profile entirely consistent with predictions based on Hyp contiguity (Zhao et al., 2002). LeAGP-1 is rich in Ser, Thr, and especially Ala, which suggest that repetitive motifs such as Ala-Hyp and Thr-Hyp motifs also serve as arabinogalactan addition sites. On the other hand, examination of other HRGPs shows that not all single Hyp motifs are glycosylated by arabinogalactan addition; this includes the lone Hyp residue (underlined) in the Ser-Hyp4-Ser-Hyp-Ser-Hyp4-Tyr-Tyr-Tyr-Lys canonical repeat of P3-type extensins (Lamport, 1969; Epstein and Lamport, 1984; M. Held and M. Kieliszewski, unpublished data) and the Hyp-Val-Hyp motifs of some PRPs (Kieliszewski et al., 1995). Therefore, here we have continued exploring a range of simple repetitive putative AGP Hyp-glycosylation motifs, specifically repetitive Ala-Hyp, Thr-Hyp, and Val-Hyp, to determine the influence of flanking residues on Hyp-O-glycosylation.
RESULTS
Gene Synthesis, Plasmid Construction, and Tobacco Cell Transformation
Head-to-tail polymerization of the oligonucleotide sets (Fig. 1; Shpak et al., 1999) produced synthetic genes that ranged in size from 609 bp for the TP (Thr-Pro) gene and 309 bp for gene AP (Ala-Pro) to only 69 bp for gene VP (Val-Pro). Several attempts to make VP longer than two internal repeats failed; therefore, we decided to express the shorter construction encoding only 23 amino acids. DNA sequence analysis confirmed the synthetic genes were properly inserted between DNA encoding the tomato AGP signal sequence and the EGFP gene in plasmids that were designated pUC-SSTom-AP-EGFP, pUC-SSTom-TP-EGFP, and pUC-SSTom-VP-EGFP (Fig. 2). Agrobacterium tumefaciens (strain LBA 4404) transformation of tobacco BY2 cells yielded several cell lines from each gene. We chose lines producing the greatest green fluorescence in the growth medium for further biochemical characterization. Two cell lines of each fusion protein were selected for Hyp-O-glycosylation profiling; one cell line of each construction was selected for neutral sugar analysis, amino acid analysis, and peptide sequence analysis.
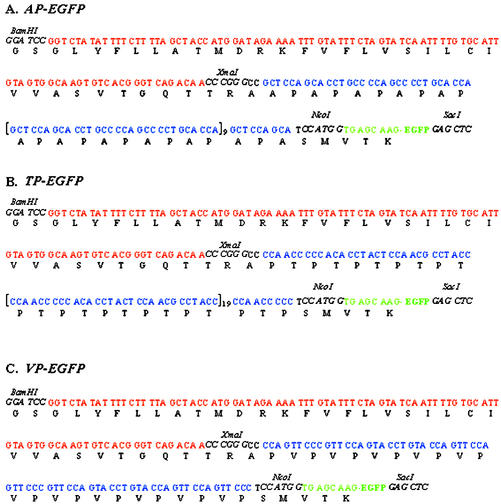
Figure 1.
Oligonucleotides sets used to construct the synthetic genes encoding: A, AP-enhanced green fluorescent protein (EGFP); B, TP-EGFP; and C, VP-EGFP. Internal repeat oligonucleotide sets were polymerized head-to-tail in the presence of the 5′-linker sets. After ligation, the 3′ linker sets were added. The restriction sites (underlined) were used to subclone the genes into pUC18 (BamHI-EcoRI), then into pUC-SStom-SP-EGFP in place of the SP gene (XmaI-NcoI).
Figure 2.
DNA sequencing of pUC-SStom-AP-EGFP (A), pUC-SStom-TP-EGFP (B), and pUC-SStom-VP-EGFP (C) confirmed the synthetic genes sequences and that they were in frame. The genes were then subcloned into pBI121 as BamHI-SacI fragments.
Isolation of the Fusion Glycoproteins and the AP, TP, and VP Glycomodules and Sequence Analyses
Fusion proteins AP-EGFP, TP-EGFP, and VP-EGFP eluted from the hydrophobic interaction chromatography column (HIC) in water (not shown). Further fractionation of the green fluorescent HIC fractions by reverse-phase chromatography on a Hamilton polymeric reverse-phase column (PRP-1, Hamilton Co., Reno, NV; Fig. 3) yielded fusion proteins suitable for sequence analyses. Partial protein sequences corroborated the DNA sequences and the amino acid compositions (Tables I and II). After PRP-1 fractionation, typical yields of the fusion glycoproteins from the most productive cell lines were: AP-EGFP, 30 mg L–1 medium; TP-EGFP, 10 mg L–1 medium; and VP-EGFP, 6 mg L–1 medium. Proteolysis of AP-EGFP, TP-EGFP, and VP-EGFP cleaved EGFP from the glycomodules, producing single symmetric peaks during gel filtration chromatography on Superose-12 (Amersham-Pharmacia, Piscataway, NJ; not shown).
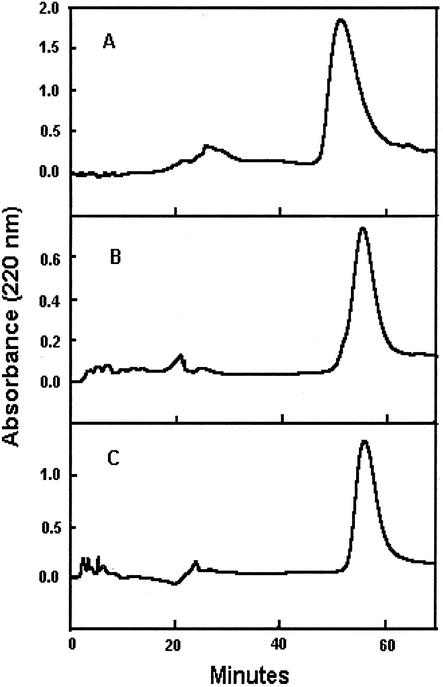
Figure 3.
Reverse-phase fractionation of AP-EGFP (A), TP-EGFP (B), and VP-EGFP (C) on the Hamilton PRP-1 reverse-phase column. Spent media from each cell line were collected and fractionated by hydrophobic interaction chromatography before isolation of the fusion glycoproteins on the PRP-1 column.
Table I.
N-terminal amino acid sequences of the AP and TP modules and of the VP fusion protein
| VP-EGFP | |
|---|---|
| AP module | Ala-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala-Hyp-Ala... |
| TP module | Hyp-Thr-Hyp-Thr-Hyp-Thr-Hyp-Thr-Hyp-Thr-Hyp |
| VP-EGFP | Pro-Val-Hyp-Val-Hyp-Val-Hyp-Val-Hyp |
Table II.
Amino acid molar ratios recovered in the AP, TP, and VP glycomodules
| Amino Acid | AP | TP | VP |
|---|---|---|---|
| nmol | |||
| Hyp | 49 | 37 | 35 |
| Pro | 0 | 16 | 9 |
| Ala | 51 | 0 | 0 |
| Thr | 0 | 47 | 0 |
| Val | 0 | 0 | 56 |
Amino Acid Composition
Amino acid analyses of the isolated glycomodules indicated that 100% of the Pro residues were hydroxylated to form Hyp in AP, 80% in VP, and 70% in TP (Table II).
Carbohydrate Analyses and HF Deglycosylation
All the fusion glycoproteins contained Gal, Ara, Rha, and GlcUA (Table III). Judging by Hyp-glycoside profiles, 100% of the Hyp residues in AP-EGFP contained arabinogalactan polysaccharide adducts, compared with 50% to 60% in TP-EGFP and VP-EGFP, which also contained Hyp-arabinosides and non-glycosylated Hyp (Table IV). TP-EGFP was unique in that it contained an unknown Hyp derivative (Hyp-unknown, Table IV) that eluted later than non-glycosylated Hyp on the Chomobeads C cation exchange column, which meant it was more positively charged than non-glycosylated Hyp.
Table III.
Molar ratios of the monosaccharides in the fusion glycoproteins AP-EGFPa (AP), TP-EGFP (TP), and VP-EGFP (VP)
| Monosaccharide | AP | TP | VP |
|---|---|---|---|
| mol % | |||
| Ara | 31 | 30 | 39 |
| Gal | 41 | 40 | 40 |
| Rha | 13 | 8 | 8 |
| GlcUAb | 15 | 23 | 13 |
EGFP is not glycosylated judging by earlier work with synthetic gene products (Shpak et al., 1999). bDetermined by the colorimetric assay described in reference (Blumenkrantz and Asboe-Hansen, 1973) and by analyses at The Complex Carbohydrate Research Center (University of Georgia, Athens).
Table IV.
Hyp glycoside profiles of AP-EGFP, TP-EGFP, and VP-EGFP
Hyp-PS, Hyp-arabinogalactan polysaccharide; NG-Hyp, non-glycosylated Hyp.
| Hyp Glycoside | AP-EGFP Lines
|
TP-EGFP Lines
|
VP-EGFP Lines
|
|||
|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | 1 | 2 | |
| % total Hyp | ||||||
| Hyp-PS | 100 | 100 | 57 | 64 | 45 | 55 |
| Hyp-Ara-4 | 0 | 0 | 5 | 5 | 0 | 2 |
| Hyp-Ara-3 | 0 | 0 | 10 | 8 | 0 | 10 |
| Hyp-Ara-2 | 0 | 0 | 6 | 6 | 14 | 13 |
| Hyp-Ara | 0 | 0 | 2 | 3 | 8 | 4 |
| Hyp-Unknown | 0 | 0 | 6 | 5 | 0 | 0 |
| NG-Hyp | 0 | 0 | 14 | 10 | 33 | 16 |
β-Elimination of TP-EGFP
The amino acid composition of TP glycomodule after NaOH/sodium sulfite treatment compared with the control (HF deglycosylated TP after the same treatment) showed no loss of Thr or gain of the elimination/sulfite addition product 2-amino-3-sulfonyl butyric acid (Simpson et al., 1972). The amino acid composition of the TP glycomodule after NaOH/sulfite treatment (not shown) was essentially the same as that of the untreated TP glycomodule (Table II).
Precipitation of the Yariv Reagent
AP-EGFP, TP-EGFP, and VP-EGFP precipitated (β-d-galactosyl)3-Yariv reagent, as did a mixture of AGPs and gum arabic glycoprotein (GAGP) isolated from wild-type tobacco and A. senegal nodules, respectively (Table V).
Table V.
(β-D-galactosyl)3-Yariv reagent coprecipitation of wild-type tobacco AGPs and AGP GAGP (Goodrum et al., 2000) compared with the fusion glycoproteins AP-EGFP, TP-EGFP, and VP-EGFP. The pellet from the precipitated glycoproteins was dissolved in alkali, and the resulting absorbance was measured at 420 nm.
| Weight | Wild-Type Tobacco AGP | GAGP | AP Line 1 | TP Line1 | VP Line 1 |
|---|---|---|---|---|---|
| μg | A420 | ||||
| 50 | 1.24 | 1.27 | 0.56 | 0.42 | 0.23 |
| 100 | 2.43 | 2.53 | 1.26 | 0.88 | 0.55 |
DISCUSSION
Glycoproteins sculpt the surface of all eukaryotic cells. Intriguingly, plants have chosen HRGPs as their major surface glycoproteins; these glycoproteins have no animal homologs and utilize Hyp glycosylation to tailor the molecular properties of both membrane-bound AGPs and cross-linked extensins in the wall. With the Hyp contiguity hypothesis as a guide, further identification of the detailed rules that govern potential Hyp glycosylation, therefore, is essential for interpretation of genomic readouts and functional glycomics. This applies particularly to the plasma membrane proteome because the unusual characteristics of AGPs preclude their facile detection and identification by standard proteomics assays. Based on genomic readouts, the Hyp contiguity hypothesis predicts that arabinogalactan glycomodules are a pervasive feature of the plasma membrane proteome—of 210 GPI-anchored proteins identified in Arabidopsis, 40% contain sites putatively involved in arabinogalactan polysaccharide addition in protein families that include not only classical AGPs but also putative phytocyanins, COBRAs, glycerophosphodiesterases, fasciclins, aspartyl proteases, lipid transfer proteins, receptors, and other unidentified proteins (Borner et al., 2002).
Our data confirm that endogenous plant prolyl hydroxylase(s) recognize common AGP motifs with flanking residues Ala, Val, or Thr when expressed as repetitive VP, TP, or AP and show that the resulting Hyp residues serve as arabinogalactan addition sites with the site occupancy ranging from approximately 50% to 60% (VP and TP) to 100% (AP). Assuming that the relatively low molecular size of VP did not significantly influence hydroxylation or subsequent Hyp-glycosylation, this indicates that the residues flanking Pro/Hyp influence both hydroxylation and the subsequent type and amount of glycosylation, consistent with our earlier results showing that noncontiguous Hyp residues can be arabinosylated or remain non-glycosylated (Kieliszewski et al., 1995). Significantly, contiguous Hyp apparently is never arabinogalactosylated (Shpak et al., 2001). These data also support our earlier putative assignment of approximately 23 arabinogalactan addition sites in LeAGP-1 corresponding to 15 Ala-Hyp, four Thr-Hyp and Ser-Hyp, and one Val-Hyp (Zhao et al., 2002) motifs. Further refinement of the Hyp contiguity code raises the related issues of defining noncontiguous Hyp clusters and the influence of flanking residues on Pro hydroxylation and glycosylation.
What constitutes a noncontiguous Hyp cluster, and can lone occurrences of Hyp be arabinogalactosylated? Although four to five intervening residues permit arabinogalactan addition to noncontiguous Hyp in LeAGP-1 (Zhao et al., 2002), the nine residues between lone Hyp residues of the repetitive extensin motif, Ser-Hyp-Hyp-Hyp-Hyp-Thr-Hyp-Val-Tyr-Lys, do not (Smith et al., 1984). On the other hand, the gymnosperm PRP motif (Kieliszewski et al., 1992a, 1995): Lys-Pro-Hyp-Val-Hyp-Val-Ile-Pro-Pro-Hyp-Val-Val-Lys-Pro-Hyp-Hyp-Val-Tyr contains clustered noncontiguous Hyp motifs, typically Hyp-Val-Hyp-Val (underlined), that are not sites of arabinogalactosylation. Instead, the Hyp-Val-Hyp-Val motif remains non-glycosylated or is arabinosylated (Kieliszewski et al., 1992a, 1995). This suggests either that gymnosperm galactosyltransferase specificity differs from that of dicots, or, alternatively, the galactosyltransferases have a lower affinity for peptide regions with side chains having more bulk than Ala or Ser.
Thus, design of the repetitive TP and VP construct tested the possibility that a flanking Thr or Val residue might suppress polysaccharide addition to Hyp. Table I does show incomplete hydroxylation of VP and TP motifs with subsequent polysaccharide addition to only 45% to 60% of the Hyp residues (Table III), the rest being arabinosylated, remaining non-glycosylated, or as in the case of TP, with a small amount of an unknown adduct (Table IV). Known flanking residues that influence Pro hydroxylation include Lys-Pro, Tyr-Pro, and Leu-Pro, which are never hydroxylated in contrast to SP and AP motifs that are 100% hydroxylated and undergo 100% polysaccharide addition. Significantly, we have never detected polysaccharide addition to contiguous Hyp motifs (Shpak et al., 2001) where the arabinosides contribute to the stabilization of a poly-Pro II helix and do not react with the (β-d-galactosyl)3-Yariv reagent. However, the more random coil of noncontiguous Hyp polypeptides AP, TP, and VP with their associated arabinogalactan polysaccharides invariably reacted with the (β-d-galactosyl)3-Yariv reagent (Table V), confirming the specificity of this reagent.
We can now deal with several issues that involve the identification of bona fide AGPs. Extreme sequence variability and absence of an obvious single signature motif (Schultz et al., 2000) tends to obscure homologous relationships between AGPs. However, the experimentally based rules of Hyp contiguity identify abundant glycosylation motifs that define AGP glycomodules, AGPs, and AGP domains of many chimeric glycoproteins GPI anchored to the plasma membrane (Borner et al., 2002), although the minimum number of noncontiguous Hyp residues for arabinogalactosylation has not yet been determined. AGP assignments based on other criteria need to be reconsidered. For instance, Arabidopsis SOS5 adhesion protein, identified as an AGP on the basis of glycosylation codes for animal mucins (Shi et al., 2003), has no obvious AGP glycosylation motifs coding for AGP addition. In contrast, LeExt1 of tomato, putatively identified as an extensin (Bucher et al., 2002), contains approximately 21 AGP glycosylation motifs but no extensin cross-linking motifs and, therefore, is more likely a soluble AGP than a cross-linked extensin. Reports of “Hyp-poor” AGPs raise the distinct possibility that arabinogalactan polysaccharide may also be O-linked to Ser or Thr residues in some AGPs such as the chimeric AGP of carrot (Daucus carota), DcAGP1, that apparently lacks Hyp (Baldwin et al., 2001), although it does contain the glycosylation motifs APAPAP, APTPAPAP, APAPAP, and APSHAPTP that would correspond to 13 polysaccharide addition sites based on Hyp contiguity codes. Nevertheless, there is increasing evidence for the β-elimination of arabinogalactan polysaccharides that are O-linked to Ser residues in some AGPs (Qin et al., 2000). This includes purified transgenic LeAGP-1 where biochemical characterization verified both the saccharide and amino acid elimination products, suggesting that another glycosylation code directs polysaccharide addition to specific Ser residues (Z. Zhao, J. Xu, and M. Kieliszewski, unpublished data).
MATERIALS AND METHODS
Creation of Plasmid pUC-SStom-SP-EGFP
We amplified the tomato (Lycopersicon esculentum) LeAGP-1 signal sequence DNA (designated SStom here) as described earlier (Li and Showalter, 1996; Zhao et al., 2002) and then introduced it into the pUC-derived plasmid pUC-SStob-SP-EGFP (Shpak et al., 1999) as a BamHI and XmaI fragment in place of the tobacco (Nicotiana tabacum) extensin signal sequence (SStob; De Loose et al., 1991; Shpak et al., 1999), thus creating the plasmid pUC-SStom-SP-EGFP.
Synthetic Gene and Plasmid Construction
Construction of a given synthetic gene involved three sets of partially overlapping, complementary oligonucleotide pairs (Fig. 1) polymerized as described earlier (Shpak et al., 1999) and inserted into pUC18 as BamHI-EcoRI fragments. The three synthetic genes were designated AP, which encoded 51 AP repeats, TP, which encoded 101 TP repeats, and VP, which encoded 11 VP repeats. The genes were subcloned as XmaI-NcoI fragments into pUC-SStom-SP-EGFP in place of the repetitive gene encoding repeats of Ser-Pro (SP) creating the plasmids pUC-SStom-AP-EGFP, pUC-SStom-TP-EGFP, and pUC-SStom-VP-EGFP (Fig. 2). We sequenced each gene after subcloning into pUC18 and pUC-SStom-SP-EGFP to confirm they were in frame and without error. The genes were then inserted into the plant vector pBI121 (CLONTECH Laboratories, Palo Alto, CA) as BamHI and SacI fragments, replacing the glucuronidase reporter gene and under the control of the 35S cauliflower mosaic virus promoter. DNA sequencing was completed at the Ohio Agricultural Research Center (Ohio State University, Columbus). All primers were designed using Primer Premier Software (PREMIER Biosoft International, Palo Alto, CA) and synthesized by Integrated DNA Technologies, Inc. (Coralville, IA).
Agrobacterium tumefaciens and Tobacco Cell Transformation and Selection of Cell Lines
pUC-SStom-AP-EGFP, pUC-SStom-TP-EGFP, and pUC-SStom-VP-EGFP were introduced into A. tumefaciens strain LBA4404 by the freeze thaw method (McCormick et al., 1986); then, the A. tumefaciens cultures containing the plasmids were used to transform suspension-cultured tobacco cells. The transformed tobacco cells were cultured on solid and liquid Schenk-Hildebrandt medium as described earlier (Shpak et al., 1999). Positive clones of transformed tobacco cells were selected for kanamycin resistance and EGFP fluorescence. EGFP fluorescence was visualized with a Sarastro 2000 confocal laser-scanning fluorescence microscope (Molecular Dynamics, Sunnyvale, CA) using an isothiocyanate filter set with a 488-nm laser wavelength filter, a 510-nm primary beam splitter, and a 510-nm barrier filter.
Isolation of AP-EGFP, TP-EGFP, and VP-EGFP Fusion Proteins
We maintained transformed BY2 cells in liquid culture for 20 d at room temperature on an Innova gyrotary shaker (New Brunswick Scientific, Edison, NJ) rotating at 90 rpm. The culture medium was filtered from cells, concentrated via rotary evaporation at 28°C, and dialyzed against distilled water overnight. The dialyzed medium was further concentrated by rotary evaporation, and NaCl was added to a final concentration of 2 m. Insoluble material was pelleted by centrifugation (13,000g, 20 min, SS-34 rotor), then loaded on an HIC (Phenyl Sepharose 6 Fast Flow, 16 × 700 mm, Amersham-Pharmacia Biotech) equilibrated in 2 m NaCl. A decreasing step gradient was used to elute the fusion glycoproteins, starting with 2 m NaCl and followed by 1 m NaCl, then deionized distilled water. The flow rate was 1 mL min–1, and the fractions were monitored by eye for green fluorescence. We combined the fluorescent fractions and loaded them on a semipreparative polymeric C-18 reverse-phase column (10 μm, PRP-1, 7 × 305 mm, Hamilton) equilibrated with buffer A (0.1% [v/v] aqueous trifluoroacetic acid). Samples were eluted with a gradient of 100% (v/v) buffer A increasing to 70% (v/v) buffer B (80% [v/v] acetonitrile in 0.1% [v/v] aqueous trifluoroacetic acid) for 105 min. The flow rate was 1 mL min–1, and the eluate was monitored at 220 nm on a 1050 HPLC system (Hewlett-Packard, Novi, MI). The fusion glycoproteins eluted in 50% (v/v) buffer B and native AGPs in 30% (v/v) buffer B.
Isolation of AP, TP, and VP Glycomodules
We dissolved 10 mg of AP-EGFP, TP-EGFP, and VP-EGFP in 0.5 mL of deionized, distilled water and heated the samples at 100°C for 5 min. After cooling, an equal volume of freshly made 4% (w/v) ammonium bicarbonate containing 2.5 mm CaCl2 and either pronase or trypsin (1:100 [w/w] final enzyme:substrate ratio). The mixture was incubated at room temperature for 24 h and then freeze dried, redissolved in 0.5 mL of Superose buffer (0.2 m aqueous sodium phosphate [pH 7] containing 0.05% [w/v] sodium azide), and further separated on a semipreparative Superose-12 gel filtration column equilibrated in Superose buffer (see below). The enzymes used were pronase with AP-EGFP and trypsin with TP-EGFP and VP-EGFP.
Superose-12 Gel Filtration Chromatography
Dialyzed and concentrated tobacco cell medium or proteolyzed fusion glycoproteins were loaded on a semipreparative Superose-12 column (16 × 500 mm, Amersham-Pharmacia Biotech), equilibrated, and eluted with Superose buffer at a flow rate of 0.3 mL min–1 and monitored at 220 nm. Peaks absorbing at 220 nm were collected, dialyzed against deionized, distilled water, freeze dried, and then analyzed (see below) to determine which peaks corresponded to the TP, VP, and AP glycomodules.
Amino Acid Composition Analysis and Sequencing of Glycomodules
Amino acid compositions were determined either at the Michigan State University Macromolecular Facility (East Lansing) or in our laboratory on a Beckman HPLC (Beckman Instruments, Fullerton, CA) using methods described earlier (Bergman et al., 1986). Amino acid sequence analyses were carried out at the Michigan State University Macromolecular Facility on a 477-A gas phase sequencer (PE-Applied Biosystems, Foster City, CA). Although VP-EGFP was sequenced without prior deglycosylation or protease treatment, the N termini of AP-EGFP and TP-EGFP were blocked. Therefore, we deglycosylated AP-EGFP and TP-EGFP (see method below) and treated the deglycosylated fusion proteins with trypsin and pyro-Glu amino peptidase to remove both EGFP and the extreme N terminus of the proteins. We then isolated the deglycosylated VP and AP modules by reverse-phase HPLC before sequencing.
Carbohydrate Analysis of AP-EGFP, TP-EGFP, and VP-EGP Fusion Glycoproteins
Hyp-glycoside profiles were determined on 10 mg of base-hydrolyzed fusion glycoproteins as described earlier (Weathers et al., 1977; Shpak et al., 1999). Neutral sugars (60-μg samples of fusion glycoprotein) were analyzed as alditol acetate derivatives by gas chromatography using a 6-foot × 2-mm polyethylene glycol succinate 224 column programmed from 130°C to 180°C at 2°C min–1 (Bhatti et al., 1970; Shpak et al., 1999). We also sent samples for composition analyses to the Complex Carbohydrate Research Center (University of Georgia). Data were captured by Hewlett-Packard Chem Station software. Uronic acids (100 μg of each fusion glycoprotein) were assayed by the colorimetric method based on reaction with m-hydroxydiphenyl (Blumenkrantz and Asboe-Hansen, 1973) using GlcUA as the standard.
Coprecipitation with (β-d-Galactosyl)3-Yariv Reagent
One hundred micrograms of each fusion glycoprotein was dissolved in 300 μL of distilled water. An equal volume of (β-d-galactosyl)3-Yariv reagent (1 mg mL–1 in 2% [w/v] NaCl aqueous solution) was added, and the solutions were allowed to stand for 1 h at room temperature before pelleting the resulting precipitate in a microfuge. The pellets were washed, then dissolved in 0.1 n NaOH, and the absorbance measured at 420 nm, all as described earlier (Jermyn and Yeow, 1975). GAGP was a gift from Dr. Jianfeng Xu (Ohio University Biochemistry Department).
Anhydrous Hydrogen Fluoride Deglycosylation
TP-EGFP, VP-EGFP, or the AP, TP and VP glycomodules (3–5 mg) were dissolved in 500 μL of anhydrous hydrogen fluoride containing 10% (v/v) anhydrous methanol and stirred for 2 h at 0°C. The reaction was quenched with 10 mL of cold, deionized, distilled water, followed by dialysis against deionized distilled water and freeze drying (Sanger and Lamport, 1983). For VP module, the water-quenched solution was dried under nitrogen gas at 0°C, then the small Mr material was removed by size filtration on a Sephadex G-50 column. The deglycosylated fusion proteins or deglycosylated glycomodules were purified on the PRP-1 reverse-phase column using the procedures described above.
β-Elimination of TP Glycomodule
The TP glycomodule (680 μg) was dissolved in 800 μL of 0.1 n HCl (pH 1) and heated for 1 h at 100°C to remove Ara residues. The mixture was dialyzed against deionized distilled water and freeze dried. The dearabinosylated glycomodule (0.30 mg) was heated at 50°C for 5 h in 500 μL of 0.2 n NaOH/1 m sodium sulfite (Whistler and BeMiller, 1958; Simpson et al., 1972; Kieliszewski et al., 1992b); the HF-deglycosylated TP glycomodule was used as a control to test for nonspecific elimination. The reaction mixtures were dialyzed against deionized distilled water and then freeze dried. The eliminated TP modules were hydrolyzed and derivatized with a fluorescein isothiocyanate filter set for amino acid composition analysis as described above.
Preparation of Wild-Type Tobacco AGPs
Endongenous AGPs from non-transformed BY2 cells were prepared as described earlier (Shpak et al., 1999).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Mr. Mick Held for assistance with amino acid analyses, Dr. Elena Shpak for plasmid SStob-SP-EGFP, and Dr. Derek T.A. Lamport for comments on the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021766.
This work was supported by the National Science Foundation (grant no. MCB–9874744), by Ohio University (Molecular and Cellular Biology Program grants), and in part by the National Institutes of Health (grant no. 2–P41–RR05351–06 awarded to the Complex Carbohydrate Research Center).
References
- Baldwin TC, Domingo C, Schindler T, Seetharaman G, Stacey NJ, Roberts K (2001) DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol Biol 45: 421–435 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Tierney ML (1999) Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol 121: 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman T, Carlquist M, Jornvall H (1986) Amino acid analysis by high performance liquid chromatography of phenylthiocarbamyl derivatives. In B Wittmann-Liebold, ed, Advanced Methods in Protein Microsequence Analysis. Springer Verlag, Berlin, pp 45–55
- Bhatti T, Chambers RE, Clamp JR (1970) The gas chromatographic properties of biologically important N-acetylglucosamine derivatives, monosaccharides, disaccharides, trisaccharides, tetrasaccharides and pentasaccharides. Biochim Biophys Acta 222: 339–347 [DOI] [PubMed] [Google Scholar]
- Blumenkrantz N, Asboe-Hansen G (1973) New methods for quantitative determination of uronic acids. Anal Biochem 54: 484–489 [DOI] [PubMed] [Google Scholar]
- Borner GHH, Sherrier DJ, Stevens TJ, Arkins IT, Dupree P (2002) Prediction of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M, Brunner S, Zimmerman P, Zardi GI, Amrhein N, Willmitzer L (2002) The expression of an extensin-like protein correlates with cellular tip growth in tomato. Plant Physiol 128: 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-G, Pu Z-Y, Moritz RL, Simpson RJ, Bacic A, Clarke AE, Mau S-L (1994) Molecular cloning of a gene encoding an arabinogalactan-protein from pear (Pyrus communis) cell suspension culture. Proc Natl Acad Sci USA 91: 10305–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, May B, Kawata EE, Gu Q, Wu H (1993) Characterization of cDNAs for stylar transmitting tissue-specific proline-rich proteins in tobacco. Plant J 3: 151–160 [PubMed] [Google Scholar]
- de Blank C, Mylona P, Yang WC, Katinakis P, Bisseling T, Franssen H (1993) Characterization of the soybean early nodulin cDNA clone Gm-ENOD55. Plant Mol Biol 22: 1167–1171 [DOI] [PubMed] [Google Scholar]
- De Loose M, Gheysen G, Tire C, Gielen J, Villarroel R, Genetello C, Van Montagu M, Depicker A, Inze D (1991) The extensin signal peptide allows secretion of a heterologous protein from protoplasts. Gene 99: 95–100 [DOI] [PubMed] [Google Scholar]
- Epstein L, Lamport DTA (1984) An intramolecular linkage involving isodityrosine in extensin. Phytochemistry 23: 1241–1246 [Google Scholar]
- Ferris PJ, Woessner JP, Waffenschmidt S, Kilz S, Dress J, Goodenough UW (2001) Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40: 2978–2987 [DOI] [PubMed] [Google Scholar]
- Gao M, Kieliszewksi MJ, Lamport DTA, Showalter AM (1999) Isolation, characterization and immunolocalization of a novel modular tomato arabinogalactan-protein corresponding to the LeAGP-1 gene. Plant J 18: 43–55 [DOI] [PubMed] [Google Scholar]
- Gens JS, Fujiki M, Pickard BG (2000) Arabinogalactan protein and wall-associated kinase in a plasmalemmal reticulum with specialized vertices. Protoplasma 212: 115–134 [DOI] [PubMed] [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewksi MJ (2000) Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54: 99–106 [DOI] [PubMed] [Google Scholar]
- Herman EM, Lamb CJ (1992) Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiol 98: 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn MA, Yeow YM (1975) A class of lectins present in the tissues of seed plants. Aust J Plant Physiol 2: 501–531 [Google Scholar]
- Kieliszewski M, de Zacks R, Leykam JF, Lamport DTA (1992a) A repetitive proline-rich protein from the gymnosperm Douglas fir is a hydroxyproline-rich glycoprotein. Plant Physiol 98: 919–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Kamyab A, Leykam JF, Lamport DTA (1992b) A histidine-rich extensin from Zea mays is an arabinogalactan protein. Plant Physiol 99: 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA (1994) Extensin: repetitive motifs, functional sites, posttranslational codes and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, O'Neill M, Leykam J, Orlando R (1995) Tandem mass spectrometry and structural elucidation of glycopeptides from a hydroxyproline-rich plant cell wall glycoprotein indicate that contiguous hydroxyproline residues are the major sites of hydroxyproline-O-arabinosylation. J Biol Chem 270: 2541–2549 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Showalter AM, Leykam JF (1994) Potato lectin: a modular protein sharing sequence similarities with the extensin family, the hevein lectin family, and snake venom disintegrins (platelet aggregation inhibitors). Plant J 5: 849–861 [DOI] [PubMed] [Google Scholar]
- Larkin PJ (1977) Plant protoplast agglutination and membrane-bound β-lectins. J Cell Sci 26: 31–46 [DOI] [PubMed] [Google Scholar]
- Larkin PJ (1978) Plant protoplast agglutination by artificial carbohydrate antigens. J Cell Sci 30: 283–292 [DOI] [PubMed] [Google Scholar]
- Lamport DTA (1969) The isolation and partial characterization of hydroxyproline-rich glycopeptides obtained by enzymic degradation of primary cell walls. Biochemistry 8: 1155–1163 [DOI] [PubMed] [Google Scholar]
- Lamport DTA, Katona L, Roerig S (1973) Galactosyl serine in extensin. Biochem J 133: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA, Miller DH (1971) Hydroxyproline arabinosides in the plant kingdom. Plant Physiol 48: 454–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Showalter AM (1996) Cloning and developmental/stress regulated expression of a gene encoding a tomato arabinogalactan protein. Plant Mol Biol 32: 641–652 [DOI] [PubMed] [Google Scholar]
- McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5: 81–84 [DOI] [PubMed] [Google Scholar]
- Nothnagel EA (1997) Proteoglycans and related components in plant cells. Int Rev Cytol 174: 195–291 [DOI] [PubMed] [Google Scholar]
- Pope DG (1977) Relationships between hydroxyproline-containing proteins secreted into the cell wall and medium by suspension-cultured Acer pseudoplatanus cells. Plant Physiol 59: 894–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Fong C, Lamport DTA (1991) Gum arabic glycoprotein is a twisted hairy rope: a new model based on O-galactosylhydroxyproline as the polysaccharide attachment site. Plant Physiol 96: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Yamauchi R, Aizawa K, Inakuma T, Kato K (2000) Isolation and characterization of arabinogalactan-protein from the fruit of Lycium chinese mill. J Appl Glycosci 47: 155–161 [Google Scholar]
- Sanger MP, Lamport (1983) A micro-apparatus for liquid hydrogen fluoride solvolysis: sugar and amino sugar composition of Erisiphe graminus and Triticum aestivum cell walls. Anal Biochem 128: 66–70 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, D'Ovidio R, Mehdy MC (1991) Negative and positive regulation of a novel proline-rich protein mRNA by fungal elicitor and wounding. Plant J 1: 345–354 [DOI] [PubMed] [Google Scholar]
- Sherrier DJ, Prime TA, Dupree P (1999) Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20: 2027–2035 [DOI] [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu J-K (2003) The Arabidopsis SOS5 locus encodes a putative cell surface protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ (2001) Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem 276: 11272–11278 [DOI] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ (1999) Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc Natl Acad Sci USA 96: 14736–14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DL, Hranisavljevic J, Davidson EA (1972) β-elimination and sulfite addition as a means of localization and identification of substituted Seryl and Threonyl residues in proteins and proteoglycans. Biochemistry 11: 1849–1856 [DOI] [PubMed] [Google Scholar]
- Smith JJ, Muldoon EP, Lamport DTA (1984) Isolation of extensin precursors by direct elution of intact tomato cell suspension cultures. Phytochemistry 23: 1233–1239 [Google Scholar]
- Svetek J, Yadav MP, Nothnagel EA (1999) Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem 274: 14724–14733 [DOI] [PubMed] [Google Scholar]
- Vijn I, Yang W-C, Pallisgaard N, Jensen EO, van Kammen A, Bisseling T (1995) VsENOD5, VsENOD12 and VsENOD40 expression during Rhizobium-induced nodule formation of Vicia sativa roots. Plant Mol Biol 28: 1111–1119 [DOI] [PubMed] [Google Scholar]
- Weathers PJ, Jost M, Lamport DTA (1977) The gas vacuole membrane of Microcystis aeruginosa: a partial amino acid sequence. Arch Biochem Biophys 178: 226–244 [DOI] [PubMed] [Google Scholar]
- Whistler RL, BeMiller JN (1958) Alkaline degradation of polysaccharides. Adv Carb Chem 13: 289–329 [DOI] [PubMed] [Google Scholar]
- Woessner JP, Goodenough UW (1992) Zygote and vegetative cell wall proteins in Chlamydomonas reinhardtii share a common epitope, (SerPro)x. Plant Sci 83: 65–76 [Google Scholar]
- Youl JJ, Bacic A, Oxley D (1998) Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci USA 95: 7921–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZD, Tan L, Showalter AM, Lamport DTA, Kieliszewski MJ (2002) Tomato LeAGP-1 arabinogalactan-protein purified from transgenic tobacco corroborates the Hyp contiguity hypothesis. Plant J 31: 431–444 [DOI] [PubMed] [Google Scholar]