Abstract
The Arabidopsis genome encodes four Dicer-like (DCL) proteins, two of which contain putative nuclear localization signals. This suggests one or more nuclear pathways for processing double-stranded (ds) RNA in plants. To study the subcellular location of processing of nuclear-encoded dsRNA involved in transcriptional silencing, we examined short interfering (si) RNA and micro (mi) RNA accumulation in transgenic Arabidopsis expressing nuclear and cytoplasmic variants of P19, a viral protein that suppresses posttranscriptional gene silencing. P19 binds specifically to DCL-generated 21- to 25-nucleotide (nt) dsRNAs with 2-nt 3′ overhangs and reportedly suppresses the accumulation of all size classes of siRNA. Nuclear P19 resulted in a significant reduction of 21- to 22-nt siRNAs and a 21-nt miRNA, but had a lesser effect on 24-nt siRNAs. Cytoplasmic P19 did not decrease the quantity but resulted in a 2-nt truncation of siRNAs and miRNA. This suggests that the direct products of DCL cleavage of dsRNA precursors of 21- to 22-nt siRNAs and miRNA are present in the nucleus, where their accumulation is partially repressed, and in the cytoplasm, where both normal sized and truncated forms accumulate. DCL1, which contains two putative nuclear localization signals, is required for miRNA production but not siRNA production. DCL1-green fluorescent protein fusion proteins localize to nuclei in transient expression assays, indicating that DCL1 is a nuclear protein. The results are consistent with a model in which dsRNA precursors of miRNAs and at least some 21- to 22-nt siRNAs are processed in the nucleus, the former by nuclear DCL1 and the latter by an unknown nuclear DCL.
“RNA silencing” is the suppression of gene expression through nucleotide (nt) sequence-specific interactions that are mediated by RNA (Voinnet, 2002). RNA silencing is triggered by double-stranded (ds) RNA that is processed by an RNase III activity termed Dicer into short RNAs 21 to 25 nt in length (Hannon, 2002; Zamore, 2002). In plants, RNA silencing can act at the posttranscriptional and transcriptional levels (Cerutti, 2003). The 21- to 22-nt short interfering (si) RNAs and micro (mi) RNAs silence genes posttranscriptionally by targeting cognate mRNAs for degradation by an endonuclease complex (Llave et al., 2002a; Tang et al., 2003). A longer class of siRNAs in plants (24–26 nt) has been implicated in directing homologous DNA methylation and in systemic silencing (Hamilton et al., 2002). RNA-directed DNA methylation (RdDM) can lead to transcriptional gene silencing (TGS) if promoter sequences are targeted by homologous RNA (Mette et al., 1999, 2000; Jones et al., 1999, 2001; Sijen et al., 2001; Aufsatz et al., 2002a, 2002b).
The length and functional diversity of short RNAs in plants are reflected in the multiplicity of Dicer-like (DCL) activities. In contrast to genomes of other organisms, which encode one (human, mouse, fission yeast [Schizosaccharomyces pombe], and Caenorhabditis elegans) or two (fruitfly [Drosophila melanogaster]) Dicer enzymes, the Arabidopsis genome encodes four DCL proteins (Schauer et al., 2002). Two of these, DCL1 and DCL4, are predicted to contain nuclear localization signals (NLS), suggesting that they might be nuclear proteins.
DCL1 is the best characterized member of the DCL gene family in Arabidopsis. Originally identified because weak loss-of-function mutations had dramatic effects on plant development (Schauer et al., 2002), DCL1 has recently been shown to be required for producing miRNAs (Park et al., 2002; Reinhart et al., 2002) but not siRNAs that trigger posttranscriptional gene silencing (PTGS; Finnegan et al., 2003). The existence of multiple DCL enzymes suggests that distinct activities might produce different size classes of short RNA, a possibility that is supported by recent biochemical data obtained using wheat germ extracts (Tang et al., 2003). The existence of potentially nuclear DCL proteins suggests the possibility of one or more pathways for processing dsRNA in the nucleus. To understand RNA-silencing mechanisms, it is important to ascertain whether individual DCL activities produce distinct size classes of short RNA in plants, and where in the cell these processing steps occur.
We have been analyzing RNA-mediated TGS of the moderately active, constitutive nopaline synthase promoter (NOSpro; Mette et al., 1999, 2000; Aufsatz et al., 2002a, 2002b). The system consists of a target locus comprising two NOSpro-driven genes (neomycinphosphotransferase II, NPTII, and nopaline synthase, NOS) and an unlinked silencer locus that encodes a NOSpro dsRNA that is transcribed from an inverted repeat of NOSpro DNA sequences. In the presence of NOSpro dsRNA, which is processed to siRNAs with lengths of 21, 22, and 24 nt (Aufsatz et al., 2002b), the target NOSpro copies become transcriptionally silenced and methylated via RdDM (Mette et al., 2000; Aufsatz et al., 2002a, 2002b).
Because NOSpro dsRNA is synthesized in the nucleus and exerts its effects in that compartment, we have been interested in studying the subcellular location of processing to siRNAs and the identity of the DCL enzyme(s) involved. The presence of different size classes of NOSpro siRNA provides an opportunity to analyze whether these are produced by the same or different DCL enzymes and to assess whether the longer 24-nt class triggers RdDM in our system. Previous work on the NOSpro system using helper component-proteinase (HC-Pro), a suppressor of PTGS encoded by tobacco etch virus (Llave et al., 2000; Mallory et al., 2001), provided indirect evidence for separate nuclear and cytoplasmic pathways for processing dsRNA in plants (Mette et al., 2001). Given the difficulties in isolating clean nuclear and cytoplasmic fractions for analyzing compartmentalization of short RNA populations, we sought to address the question of whether dsRNA processing occurs in the nucleus by exploiting the activity of additional viral suppressors of PTGS.
A number of viral proteins suppress PTGS (Voinnet et al., 1999) and affect siRNA accumulation (Hamilton et al., 2002). The P19 protein of tomato bushy stunt virus (TBSV) is particularly intriguing because in a recent study, it was shown to be the only viral suppressor of PTGS that repressed the accumulation of all size classes of siRNA produced from transiently expressed dsRNA precursors (Hamilton et al., 2002). In addition, P19 from another virus, Cymbidium ringspot tombusvirus, which shares 74% amino acid identity with TBSV P19, binds in vitro to PTGS-generated 21- to 25-nt dsRNAs with 2-nt 3′ overhangs (Silhavy et al., 2002). Such RNAs are characteristic products of RNase III-like activities, including that of Dicer, which makes two staggered cuts in both strands of a dsRNA (Elbashir et al., 2001a, 2001b). Cymbidium ringspot tombusvirus P19 interacts only weakly with single-stranded RNAs, long dsRNAs, or blunt-ended 21-nt dsRNAs (Silhavy et al., 2002). The ability of P19 to bind the direct products of DCL-catalyzed cleavage and potentially block accumulation of all size classes of short RNA makes it a valuable tool for studying dsRNA processing in plants.
In this paper, we report the effects of cytoplasmic and nuclear variants of P19 protein on accumulation of NOSpro siRNAs and an endogenous miRNA and on TGS in our system. We describe the effects of dcl1 mutations on NOSpro siRNAs, and provide evidence that DCL1 is a nuclear protein. Finally, we report the sequences of 74 cloned NOSpro siRNAs. Our findings support the existence of nuclear pathways for processing dsRNA precursors of miRNAs and at least some 21- to 22-nt siRNAs by distinct DCL activities.
RESULTS
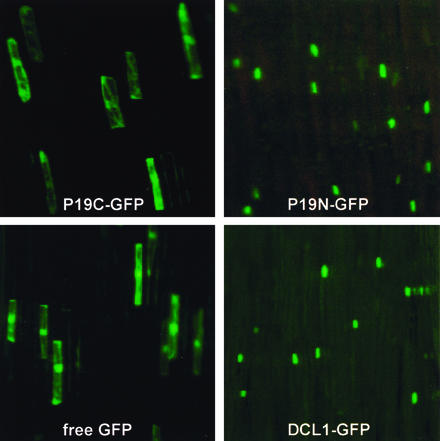
To study the effects of P19 on NOSpro siRNA and endogenous miRNA accumulation, transgene constructs designed to express the TBSV P19 protein were introduced into the Arabidopsis genome. To assay nuclear dsRNA processing steps in the P19 experiments, two transgene constructs were assembled: one with the normal P19 protein (referred to here as P19C), which does not contain a recognizable NLS, and one in which a NLS was added (P19N). In addition, constructs encoding protein fusions between the P19 variants and green fluorescent protein (GFP) were made. When transiently expressed in onion epidermal cells, P19N-GFP was concentrated in nuclei, whereas P19C-GFP was dispersed throughout the cytoplasm (Fig. 1, top). Although we cannot rule out that small amounts of P19C-GFP are in the nucleus as well as the cytoplasm, P19N-GFP appears largely restricted to nuclei.
Figure 1.
Subcellular location of GFP fusion proteins. All proteins are under the control of the 35Spro. Constructs were introduced into onion epidermal cells by particle bombardment. Top left, TBSV P19 protein-GFP (P19C-GFP); top right, TBSV P19 protein-NLS-GFP (P19N-GFP). P19C-GFP appears distributed throughout the cell, probably including the nucleus, whereas P19N-GFP is largely restricted to the nucleus. Bottom left, Free GFP; bottom right, DCL1-GFP.
The P19C and P19N constructs (without GFP) were introduced into plants that were homozygous for both the target locus and the silencer locus. Transgenic plants expressing either the P19C or the P19N protein were recognizable by their small size, serrated leaves, early flowering, and poor fertility. This phenotype was more pronounced in P19C than in P19N plants. The developmental aberrations are consistent with a role for the P19-targeted RNA-silencing pathway in plant development (Silhavy et al., 2002). As described below, the similar phenotype of the P19C and P19N plants probably reflects the fact that both P19 protein variants affect miRNA accumulation, although in different ways. Moreover, the distinct effects of P19C and P19N on short RNA accumulation in transgenic plants (see below) are consistent with relatively strict subcellular locations in cytoplasm and nucleus, respectively, as observed with the transiently expressed P19-GFP protein fusions in onion.
Silencing of NOSpro-driven target genes in P19C and P19N plants was assessed by assaying for the presence of nopaline in leaf extracts. Approximately 30 μg of nopaline (per 100 mg of fresh leaf tissue) can be detected in 70% (v/v) ethanol extracts isolated from the target line. No nopaline can be detected in extracts prepared from plants of the target/silencer line. Additionally, no nopaline was detected in phenotypically affected plants that contained either the P19C or the P19N construct under assay conditions in which the lower limit of detection was 5 μg of nopaline (23 plants, P19N; 10 plants, P19C). Thus, silencing was not alleviated substantially in either P19C or P19N plants. Methylation of target NOSpro copies was also not affected in these plants (supplementary data can be viewed at http://www.plantphysiol.org).
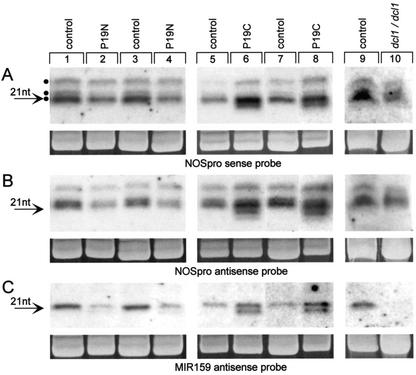
NOSpro siRNAs and the microRNA MIR159 were analyzed in leaves of P19C and P19N plants exhibiting a clear phenotype. MIR159 was chosen for analysis because it is the most abundant miRNA in Arabidopsis leaves, producing a signal on northern blots that is comparable in strength to that produced by NOSpro siRNAs (Mette et al., 2002b). We assume that the amounts of the respective precursor dsRNAs are likewise roughly the same. Therefore, any differences observed in siRNA and miRNA accumulation are probably not due to large disparities in precursor concentration. Although NOSpro siRNAs and MIR159 behaved identically in P19-expressing plants (which was not the case with dcl1 mutations, as described below), differences in accumulation were observed for both types of short RNA in the presence of either P19N or P19C.
In P19N plants, there was an approximately 30% to 40% reduction of both sense and antisense NOSpro siRNAs of the 21- to 22-nt class (Fig. 2, A and B, left) and an approximately 60% reduction of MIR159, which accumulates in one orientation only (Mette et al., 2002b; Fig. 2C, left). The 24-nt NOSpro siRNAs, which were less abundant in control plants than the 21- to 22-nt class, were on average reduced <10% in P19N plants (Fig. 2, A and B, left). The decrease of primarily 21- to 22-nt siRNAs and MIR159 in P19N plants suggests that the direct products of DCL cleavage (to which P19 binds) of the respective dsRNA precursors are present in the nucleus.
Figure 2.
Northern-blot analysis of short RNAs. A, NOSpro sense probe; B, NOSpro antisense probe; C, MIR159 antisense probe (previously termed “40” short RNA, present in sense orientation only [Mette et al., 2002b]). Left panels, Control and P19N plants, two representatives of each; middle panels, control and P19C plants, two representatives of each. Right panels, Control (wild type) and dcl1-9 mutant. Similar results were obtained for dc1-7 and dcl1-8 mutants (data not shown). Arrow indicates 21-nt RNA size marker. The three dots in A left indicate 21, 22, and 24 nt. Ethidium bromide staining of the major RNA species in the samples is shown as a loading control (Mallory et al., 2002).
The overall reduction of the primarily 21- to 22-nt short RNAs seen in P19N plants contrasted with the changes observed in P19C plants, in which novel shorter fragments appeared. For MIR159, one new band of equal intensity that was an estimated 2 nt shorter than the original 21-mer was observed (Fig. 2C, middle). The existence of a single new band suggested a controlled cleavage event occurring predominantly in the cytoplasm, because there was little or no MIR159 19-mer in P19N plants (Fig. 2C, left). For NOSpro siRNAs, up to two new bands were observed just below the 21-mer in both sense and antisense orientations (Fig. 2, A and B, middle). These new fragments are likely 19- and 20-mers, suggesting that 2 nt was truncated, respectively, from the 21- and 22-nt NOSpro siRNAs. Because a 2-nt reduction of the 24-mer would produce a fragment comigrating with the 22-nt siRNA, it is not possible to determine whether the 24-nt NOSpro siRNAs were similarly affected. The observation that P19C had an effect on either the production or the stability of 21- to 22-nt siRNAs and MIR159 suggests that the direct products of DCL cleavage of these dsRNA precursors are present in the cytoplasm.
To study the DCL activity required to produce NOSpro siRNAs, we examined the effects of weak loss-of-function alleles of DCL1: dcl1-7 and dcl1-8, which contain point mutations in the helicase domain (Golden et al., 2002), and dcl1-9, which has a T-DNA insertion in the second of two dsRNA-binding domains (Jacobsen et al., 1999). The two putative NLSs in the 1,909 residue DCL1 protein suggest that it might localize to the nucleus and be active in that compartment.
We tested whether the NLSs in DCL1 were functional by making protein fusions between the DCL1 protein and GFP. These constructs were introduced into onion epidermal cells by particle bombardment. GFP fusion proteins containing the N-terminal half (1,097 amino acids) of the DCL1 protein localized to nuclei of onion epidermal cells (Fig. 1). We were unable to successfully express a full-length DCL1-GFP fusion protein either in Escherichia coli or in bombarded onion cells. This is likely due to the relatively large size of the DCL1 protein because the C-terminal RNase III domains can be expressed as glutathione S-transferase fusions in E. coli (D.S. Merchant, S.E. Schauer, and A. Ray, unpublished data). Nevertheless, the results with the partial DLC1-GFP fusions demonstrate that one or both NLSs are functional, supporting the idea that DCL1 is a nuclear protein.
To assess whether NOSpro dsRNA processing was impaired in dcl1 mutants, NOSpro siRNAs were examined by northern blotting. In dcl1 mutants, all three sizes of siRNA (21, 22, and 24 nt) were detected in both sense and antisense polarities (Fig. 2, A and B, right; only dcl1-9 is shown). The reduced amount of NOSpro siRNAs in mutant plants can be explained by the hemizygous state of the silencer locus, compared with the homozygous state in control plants. In contrast, MIR159 was not detectable in dcl1-9 mutants (Fig. 2C, right). This result is consistent with a requirement for DCL1 in miRNA accumulation (Park et al., 2002; Reinhart et al., 2002). The absence of MIR159 confirmed that the dcl1-9 mutation affected short RNA production in the plants tested. Therefore, DCL1, which is likely localized in the nucleus, is required for generating miRNA but not NOSpro siRNAs.
The ability of the three dcl1 mutations to alleviate silencing of NOSpro-driven genes was evaluated by testing mutant seedlings for resistance to kanamycin (dcl1-7 and dcl1-8) or the presence of nopaline (dcl1-9). No release of silencing was observed with any of the mutations (data not shown). In addition, methylation of target NOSpro copies was unchanged in mutant plants (supplementary data).
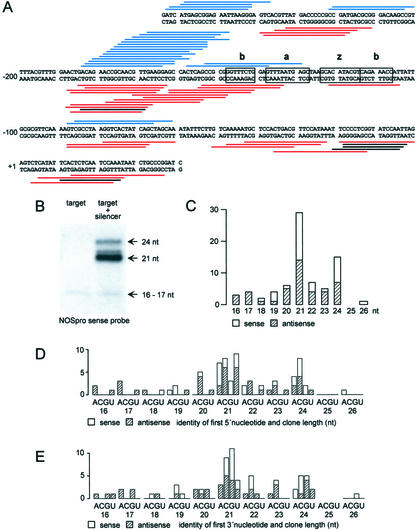
Because dcl1 mutants were not impaired in NOSpro siRNA production, we were interested in determining whether these siRNAs had any special features that might be related to the activity of a different DCL family member. Through an enrichment procedure (Djikeng et al., 2001), we cloned and sequenced 74 unique NOSpro siRNA sequences (Fig. 3A). Consistent with the pattern observed on northern blots, the major size classes of cloned NOSpro siRNAs are 21 and 24 nt (Fig. 3, B and C). Both orientations were recovered in approximately equal amounts, possibly because of stabilization by base pairing to double-stranded DNA. In contrast, miRNAs usually accumulate in only one polarity (Mette et al., 2002b; Park et al., 2002; Reinhart et al., 2002), presumably because only the antisense orientation can by stabilized by base pairing to complementary target mRNA.
Figure 3.
Cloned NOSpro siRNAs. A, NOSpro DNA sequence and positions of sense (blue) and antisense (red) NOSpro siRNAs. B, Those shown in black are 16- to 17-nt NOSpro short RNAs that are also detectable in the target line alone and hence do not result from NOSpro dsRNA processing. Motifs important for promoter activity (a, b, and z) are boxed. C, Histogram showing sizes of cloned NOSpro siRNAs. Consistent with the northern blot, the major sizes cloned were 21 and 24 nt. D and E, Histograms showing the distribution of 5′ and 3′ nts, respectively, of all sizes of NOSpro siRNA.
The 5′ nt for both sense and antisense polarities of the 21 nt class was fairly evenly distributed among adenosine, cytidine, and uridine; guanosine was underrepresented, probably because the T4 RNA ligase used in the cloning procedure discriminates against 5′-phosphorylated guanosine as donor oligonucleotide (Elbashir et al., 2001a). The lack of a preferred 5′ nt is also apparent from the dense group of siRNAs derived from the NOSpro sequence between positions –185 and –165, where nearly all nucleotides over an approximately 20-bp region are represented. For the 24-nt class, the major 5′ nt was cytidine (53%), followed by adenosine (27%). With respect to the 3′ nt for both size classes, cytidine and guanosine were overrepresented (21 nt, 74% CG; 24 nt, 71% CG). These sequence biases do not reflect the sequence composition of the NOSpro, which is composed evenly of A, T, C, and G in both the top and bottom strands.
One size class, 16 to 17 nt, was only cloned in the antisense orientation. Because a 16- to 17-nt band was also observed on Northern blots containing NOSpro siRNA isolated from the target line alone (Fig. 3B), this size class is not due to cleavage of NOSpro dsRNA encoded by the silencer locus. These 16- to 17-nt RNAs do not trigger TGS or RdDM, as evidenced by the fact that target NOSpro copies are active and unmethylated in the absence of the silencer locus (Aufsatz et al., 2002a, 2002b). The center region of the NOSpro from which few siRNAs were recovered contains sequence elements required for promoter activity (Mitra and An, 1989). The NOSpro dsRNA might act as an RNA aptamer for the corresponding transcription factors (Lebruska and Maher, 1999), which could protect from DCL cleavage in this region.
DISCUSSION
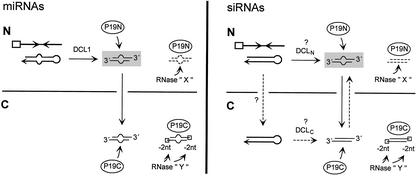
To study the subcellular site where nuclear-encoded dsRNAs are processed, we analyzed siRNA and miRNA accumulation in plants expressing nuclear and cytoplasmic versions of P19, a plant viral suppressor of PTGS. The P19 protein binds in vitro to the direct products of DCL cleavage (21- to 25-nt dsRNAs with 2-nt 3′ overhangs), whereas it interacts poorly with single-stranded RNAs, long dsRNAs, or blunt-ended 21-nt dsRNAs (Silhavy et al., 2002). A separate study reported that P19 suppresses the accumulation of all size classes of siRNA produced from transiently expressed dsRNA precursors (Hamilton et al., 2002). We also tested the function of the putative NLSs of DCL1 and determined the effects of dcl1 mutations on production of siRNAs in a TGS system. The results are consistent with a model in which dsRNA precursors of miRNAs and at least some 21- to 22-nt siRNAs are processed in the nucleus; the former by DCL1 and the latter by a second nuclear DCL that remains to be specified (Fig. 4).
Figure 4.
Model for different effects of P19N and P19C on DCL-generated short RNAs (21–25 nt with 2-nt 3′ overhangs). Left, miRNA: The precursor, an imperfect duplex RNA that is transcribed in the nucleus (N) from an imperfect inverted DNA repeat (arrows), is processed by DCL1, a nuclear activity in Arabidopsis. Right, siRNAs: NOSpro dsRNA, a perfect RNA duplex, is transcribed from an inverted DNA repeat downstream of the 35Spro (box). Processing of at least some precursors of 21- to 22-nt siRNAs possibly requires a nuclear Dicer (DCLN) that is not DCL1. If dsRNA precursors are transported to the cytoplasm (C), a cytoplasmic Dicer activity (DCLC) could processes them to siRNAs, which could relocate to the nucleus (dotted arrows). Nuclear products of DCL cleavage could also be transported to the cytoplasm (solid vertical arrow). In their respective compartments, P19N and P19C bind to the immediate products of DCL-catalyzed cleavage. It is hypothesized here that this leads to complete degradation by an unknown RNase (X) in the nucleus, and clipping of the 2-nt 3′ overhangs by a different RNase (Y) in the cytoplasm. Nuclear pathways for processing precursors of miRNA and 21- to 22-nt siRNAs are inferred from the effects of P19N, indicating the presence of DCL cleavage products in the nucleus (shaded gray). Whether all miRNA precursors are processed in the nucleus remains to be determined. Alternative modes of P19 action include inhibiting or deregulating DCL activity.
The ability of P19 to bind specifically to 21- to 25-nt dsRNAs with 2-nt 3′ overhangs makes this protein a unique tool for detecting the primary products of DCL cleavage. In the following discussion, we assume that the observed effects of P19 on siRNA or miRNA accumulation indicate the presence of the respective DCL-generated cleavage products.
The clearest results were obtained for the 21-nt miRNA, MIR159. Loss of MIR159 in dcl1 mutants suggests that DCL1 is required to process the corresponding precursor duplex RNA. Whether DCL1 carries out the processing reaction alone or in conjunction with additional proteins, including other DCLs, remains to be determined. Our results are in agreement with previous findings implicating DCL1 in miRNA production (Park et al., 2002; Reinhart et al., 2002). From our experiments, it can be inferred that DCL1 processes the MIR159 precursor in the nucleus because DCL1-GFP fusion proteins localize to nuclei. Nuclear processing is consistent with the reduction of MIR159 in P19N plants, which indicates that the immediate products of DCL1 cleavage of the MIR159 precursor are in the nucleus (Fig. 4, left). The quantitative decrease of the 21-nt MIR159 in P19N plants is compatible with a previous report that P19 suppresses accumulation of all size classes of short RNA when the dsRNA precursor is supplied transiently (Hamilton et al., 2002). Accumulation of MIR159 is not completely blocked, possibly because of continual synthesis of the precursor dsRNA. Although we have only investigated one miRNA in this study, we presume that other miRNA precursors that require DCL1 for cleavage (Park et al., 2002; Reinhart et al., 2002) are similarly processed in the nucleus. It is conceivable that further miRNA precursors may be exported from the nucleus and processed to their mature form by a cytoplasmic DCL.
The finding that P19C also has an effect on MIR159 suggests that the 21-nt dsRNA with 2-nt 3′ overhangs is transported from nucleus to cytoplasm (Fig. 4, left). It is not known why expression of P19C leads to equal accumulation of two forms of MIR159 (normal sized and truncated) as opposed to simply repressed accumulation of the 21-mer, as observed in the nucleus. The most straightforward explanation for the truncated form is that the 2-nt 3′ overhangs are removed from the 21-nt dsRNA. The postulated clipping reaction takes place predominantly in the cytoplasm, because little or no 19-mer is observed in P19N plants. One possibility is that distinct RNases target P19-bound short dsRNAs in the nucleus and in the cytoplasm (Fig. 4). Until the mode of action of P19 is determined, we cannot rule out alternative models. For example, P19 could inhibit or deregulate DCL activity, resulting in lower accumulation or truncated forms of short RNAs, or it could indirectly influence short RNA production or stability through interactions with other host proteins. These alternatives, however, are not fully consistent with previous arguments that P19 does not suppress Dicer activity and that binding of P19 to short dsRNAs does not require host factors (Silhavy et al., 2002).
The NOSpro siRNAs present a more complex situation. Neither the 21- to 22-nt class nor the 24-nt siRNAs are produced by DCL1, because they are still detectable in dcl1 mutants. This extends recent data showing that DCL1 is not required for siRNAs that trigger PTGS (Finnegan et al., 2003) to siRNAs that are made in an RNA-mediated TGS system. The decrease in the amount of 21- to 22-nt NOSpro siRNAs in P19N plants suggests that at least some of the direct products of DCL cleavage of the NOSpro dsRNA are present in the nucleus. The simplest interpretation of this result is that the dsRNA precursor is cleaved in the nucleus by a nuclear DCL other than DCL1 (Fig. 4, right). DCL4 is the best candidate based on the presence of a single NLS (Schauer et al., 2002). However, we cannot exclude a more complicated scenario in which 21- to 22-nt siRNAs produced in the cytoplasm migrate into the nucleus. The effects of P19C on the 21- to 22-nt siRNAs could be due to either the transport of 21- to 22-nt dsRNA with 2-nt 3′ overhangs from the nucleus into cytoplasm, or to transport of the precursor dsRNA to the cytoplasm and processing by a cytoplasmically located DCL (Fig. 4, right). These issues cannot be resolved until the DCL activity (or activities) required for producing the 21- to 22-nt NOSpro siRNAs are identified. The truncation of 2 nt from 21- to 22-nt siRNAs in P19C plants indicates that these are treated similarly to MIR159 in the cytoplasm.
The P19 results are less clear for the 24-nt siRNAs. This larger class does not appear to be as affected as the 21- to 22-nt siRNAs or MIR159 in P19N plants. This suggests that there are no primary products of DCL cleavage of this size class (24-nt dsRNAs with 2-nt 3′ overhangs) in the nucleus. Alternatively, binding of P19 to nuclear 24-nt dsRNAs might be precluded if they have blunt ends formed by filling in the 2-nt 3′ overhangs of nuclear 22-nt dsRNAs. A final possibility is that the dsRNA precursor of the 24-nt siRNAs is processed in the cytoplasm. The idea that different DCL activities are required for producing 21- to 22-nt and 24-nt siRNAs, respectively, is consistent with biochemical studies in a wheat germ extract, indicating that each size class requires a distinct DCL (Tang et al., 2003). Moreover, the wheat germ extract, which is essentially cytoplasm, did not properly process pre-miRNAs, consistent with a nuclear location of a wheat (Triticum aestivum) DCL1 homolog. The best candidates for cytoplasmic activities are DCL2 and DCL3, which are not predicted to contain NLSs (Schauer et al., 2002).
The cytoplasmic suppressors of PTGS that have been tested so far appear to affect NOSpro siRNAs and miRNA the same way. As shown here, P19C does not lead to an overall decrease in the amount of NOSpro siRNAs or MIR159. This is consistent with observations made with HC-Pro, a second cytoplasmic suppressor of PTGS. HC-Pro does not reduce NOSpro siRNAs in tobacco (Mette et al., 2001) or in Arabidopsis (M.F. Mette, unpublished data), and it likewise does not reduce miRNA concentration in tobacco (Mallory et al., 2002). The similar effect of cytoplasmic suppressors of PTGS on NOSpro siRNAs and miRNAs might reflect the fact that both are derived from hairpin RNA precursors transcribed in the nucleus.
Although not produced by DCL1, the cloned NOSpro siRNAs generally appear to be typical of other siRNAs that have been cloned and sequenced (Elbashir et al., 2001a, 2001b; Llave et al., 2002b; Tang et al., 2003). Two differences, however, were observed with respect to preferences for 5′ and 3′ nts. In a survey of 423 endogenous small RNA sequences from Arabidopsis (Tang et al., 2003), a 5′-uridine bias for the shorter class (21 nt) was noted, whereas 5′-adenosine was overrepresented in the longer class (24 nt). The 5′-uridine bias was caused by inclusion of miRNAs, which in plants and animals typically begin with uridine. The non-miRNA in the shorter class did not have a 5′ sequence preference (Tang et al., 2003). This was confirmed here for the 21- to 22-nt NOSpro siRNAs. For the 24-nt NOSpro siRNAs, however, 5′-cytidines predominated. Why this differs from the 5′-adenosine bias reported previously (Tang et al., 2003) and whether it reflects the activity of a specific DCL is unknown. At the 3′ ends, cytidine and guanosine predominated for both size classes of NOSpro siRNA. The significance of this is not presently known, because no striking 3′ end bias was seen in short RNAs produced in a fruitfly in vitro system (Elbashir et al., 2001a).
Nuclear processing of dsRNA probably does not occur in all organisms. For example, in humans, the single identified Dicer protein acts in the cytoplasm (Billy et al., 2001; Zeng and Cullen, 2002; Kawasaki and Taira, 2003). This is consistent with the subcellular distribution of processing of miRNA precursors in human cells, which revealed a two-step maturation process: a nuclear step that releases individual approximately 70-nt miRNA precursors from primary polycistronic transcripts and a cytoplasmic step involving Dicer processing of miRNA precursors into mature miRNAs (Lee et al., 2002). Restriction of Dicer activity to the cytoplasm does not rule out the possibility that siRNAs or miRNAs produced in this compartment could migrate to the nucleus and induce genome modifications (Jones et al., 2001; Hall et al., 2002; Volpe et al., 2002). In addition to having possible nuclear pathways for processing dsRNA, plants also differ from mammals in lacking adenosine deaminases acting on RNA, nuclear enzymes that edit long dsRNA in metazoans (Bass, 2002). An emerging view is that plants and animals deal differently with extended duplex RNAs in the nucleus.
Our inability so far to inhibit the production or accumulation of NOSpro siRNAs has prevented resolving which size class of siRNAs triggers RdDM in our system. In addition to the work of Hamilton and coworkers (2002) implicating 24- to 26-nt siRNAs in RdDM, further support for siRNA involvement in epigenetic modifications of the genome is provided by recent findings that mutations in components of the RNAi machinery interfere with heterochromatic silencing and histone methylation in fission yeast (Hall et al., 2002; Volpe et al., 2002) and in Arabidopsis (Zilberman et al., 2003). Although we have studied RNA-mediated TGS of a transgene promoter, several of the Arabidopsis miRNAs identified so far might target promoters of endogenous genes (Park et al., 2002), suggesting a more general role for RNA-mediated TGS in regulating plant gene expression.
The expansion of the DCL gene family in Arabidopsis reflects the diversification of short RNA production, subcellular location, and function in plants. The elaboration of dsRNA processing pathways to produce heterogeneous populations of regulatory short RNAs underscores the importance of RNA-mediated silencing for plant development (Llave et al., 2002a; Rhoades et al., 2002; Tang et al., 2003) and defense to viruses (Voinnet, 2002). The NOSpro siRNA system will be useful for defining the roles of other DCL family members in these silencing pathways.
MATERIALS AND METHODS
Transient Expression of GFP Fusion Proteins
For GFP fusions, soluble-modified red-shifted GFP (smRS-GFP; Davis and Vierstra, 1998) available as CD3-327 from the Arabidopsis Biological Resource Center (Ohio State University, Columbus) was used. To make GFP fusions, two PCR fragments encoding together 1,097 amino acids from the 5′ part of DCL1 were inserted into a modified pDH51, which harbors a cauliflower mosaic virus 35S promoter (35Spro) and terminator separated by a multiple cloning site (Pietrzak et al., 1986) and which contains smRS-GFP in the right reading frame to the DCL1 PCR fragments. For P19C-GFP (normal P19 protein), P19 was amplified by PCR from pBin61-P19 (a gift from D. Baulcombe) and cloned in frame in front of smRS-GFP in the modified pDH51 vector. For P19N-GFP, three SV40 NLSs (amplified from pCFP-NUC [BD Biosciences Clontech, Erembodegem, Belgium]) were inserted in frame in between the P19 coding region and smRS-GFP in the modified pDH51 vector. GFP fusions in the modified pDH51 vectors were introduced into onion epidermal cells by biolistic transformation (Mette et al., 2002a).
Stable Transformation of Arabidopsis and Reporter Gene Assays
For all stable transformations, Arabidopsis ecotype Col-0 was used. For P19C, the pBin61-P19 (P19 under the control of the 35Spro) was mobilized into Agrobacteriun tumefaciens and introduced into plants by the floral dip method (Clough and Bent, 1998). For P19N, P19 was cloned into a modified pDH51, and three SV40 NLSs (amplified as described above) were inserted in frame at the end of the P19 coding region. The construct was cut out of pDH51 and ligated into modified binary vectors containing selection markers for kanamycin (Matzke and Matzke, 1986) or BASTA resistance (Aufsatz et al., 2002b). Wild-type Arabidopsis plants were transformed with pBin61-P19 or the P19N construct in the kanamycin resistance vector as described above. The resulting T1 plants were crossed to the doubly homozygous target/silencer line (Aufsatz et al., 2002a) to produce F1 progeny that were uniformly hemizygous for target and silenced but segregating for P19C or P19N transgenes. P19C- and P19N-expressing plants were clearly distinguishable from normal (control) siblings by their small size, serrated leaves, early flowering, and poor fertility. Alternatively, the doubly homozygous target/silencer line was supertransformed with the P19N construct in the BASTA resistance vector. Target NOSpro reactivation was tested by nopaline assays (Matzke et al., 1989) in 10 P19C-expressing F1 plants and 23 P19N-expressing BASTA-resistant supertransformants. None of these plants was nopaline positive. Short RNA analysis by northern blotting and DNA methylation analysis (procedures described below) were performed on F1 plants hemizygous for target, silencer, and P19C or P19N transgenes, respectively, compared with control F1 plants from the same crosses that were hemizygous for target and silencer only.
Arabidopsis Mutant Crosses and Reporter Gene Assays
Plants homozygous for target and silencer (Aufsatz et al., 2002a) were crossed with plants homozygous for dcl1-7, dcl1-8, and dcl1-9 alleles (Jacobsen et al., 1999; Golden et al., 2002). F1 plants were allowed to self-pollinate. F2 seeds of the dcl1-7 (n = 168) and dcl1-8 (n = 248) crosses were tested for reactivation of the target NOSpro-NPTII gene on Murashige and Skoog agar containing 20 mg L–1 hygromycin (Calbiochem, VWR, Vienna) plus 40 mg L–1 kanamycin (Sigma-Aldrich, Munich). No double-resistant plants were recovered. F2 plants homozygous for the dcl1-9 allele were identified by the characteristic flower phenotype (Jacobsen et al., 1999) and confirmed by the absence of a PCR product diagnostic for the wild-type allele with primers caf1-F, 5′-AATGGGCATCAGCCGTTTAC-3′, and caf1-R, 5′-CTCTTTGCATGAGCCGGTCC-3′. The presence of at least one silencer and target copy was confirmed by selecting plants on Murashige and Skoog agar with 20 mg L–1 hygromycin and by genotyping for the presence of target markers as described previously (Aufsatz et al., 2002a, 2002b). Because the dcl1-9 allele contains a T-DNA insertion that harbors an NPTII gene (Jacobsen et al., 1999), reactivation of NOSpro-driven target genes was restricted to the NOS gene. Ten F2 plants homozygous for dcl1-9 and containing at least one copy of target plus silencer were tested for target reactivation by nopaline assays (Matzke et al., 1989). None of the 10 F2 plants was nopaline-positive. F2 plants homozygous for dcl1-9, hemizygous for the silencer, and homozygous for the target were subjected to short RNA analysis as described previously (Mette et al., 2000).
DNA Methylation Analysis
DNA methylation in the target NOSpro was analyzed by Southern blotting using methylation-sensitive restriction enzymes and Southern blotting as described previously (Matzke et al., 1989). Enzymes and probe used are shown in supplementary data.
Short RNA Analysis
Short RNAs from Arabidopsis leaves were isolated and analyzed by northern blotting as described (Mette et al., 2000). Signals on autoradiograms were quantified using ImageMaster software (Amersham Biosciences, Vienna).
Short RNAs isolated from the doubly homozygous target/silencer lines were cloned following a protocol provided by T. Tuschl and sequenced according to standard procedures (Mette et al., 2002b). To increase the fraction of clones containing NOSpro-sequences, an enrichment-step was included using a published procedure (Djikeng et al., 2001). The PCR products obtained after oligonucleotide-ligations and reverse transcription in the cloning protocol were hybridized to sense or antisense biotin-labeled NOSpro RNA. DNA-RNA hybrids were purified using the Dynabead M-280 Streptavidin system (Dynal Biotech, Hamburg, Germany) and used as the template for a second PCR amplification. Cloning and sequencing of the NOSpro-enriched PCR-products was performed as above. The full length of the biotin-labled NOSpro in vitro transcripts used as molecular bait was confirmed by denaturing agarose gel electrophoresis. The cloned NOSpro siRNAs should thus be representative of the total NOSpro siRNA population.
Supplementary Material
Acknowledgments
We thank David Baulcombe and György Szittya for helpful discussions and two anonymous referees for constructive comments on an earlier form of the manuscript. We are grateful to D. Baulcombe, Sainsbury Laboratory, and Plant Bioscience Limited for supplying the TBSV P19 construct pBin61-P19; Steve Jacobsen for dcl1-9 seeds; and Thomas Tuschl for a short RNA cloning protocol.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021980.
This work was supported by the Austrian Fonds zur Förderung der wissenschaftlichen Forschung (grant no. Z21–MED to M.M. and A.M.) and by the National Science Foundation (grant no. IBN–9982414 to A.R.).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Aufsatz W, Mette MF, van der Winden J, Matzke A, Matzke M (2002a) RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA Suppl 99: 16499–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke A (2002b) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B (2002) RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Müller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H (2003) RNA interference: traveling in the cell and gaining functions? Trends Genet 19: 39–46 [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Davis S, Vierstra R (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E (2001) RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence of retroposon-derived 24–26-nucleotide RNAs. RNA 7: 1522–1530 [PMC free article] [PubMed] [Google Scholar]
- Elbashir S, Lendeckel W, Tuschl T (2001a) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Margis R, Waterhouse P (2003) Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr Biol 13: 236–240 [DOI] [PubMed] [Google Scholar]
- Golden T, Schauer S, Lang J, Pein S, Mushegian A, Grossniklaus U, Meinke D, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130: 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I, Shankaranarayana G, Noma K, Ayoub N, Cohen A, Grewal S (2002) Establishment and maintenance of a heterochromatic domain. Science 297: 2232–2237 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon G (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Jacobsen S, Running M, Meyerowitz E (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones L, Hamilton A, Voinnet O, Thomas C, Maule A, Baulcombe D (1999) RNA-DNA interactions and DNA methylation in posttranscriptional gene silencing. Plant Cell 11: 2291–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe D (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 11: 747–757 [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Taira K (2003) Short hairpin type of dsRNAs that are controlled tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res 31: 700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebruska L, Maher L III (1999) Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry 38: 3168–3174 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee J, Kim S, Kim V (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Kasschau K, Carrington J (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97: 13401–13406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau K, Carrington J (2002a) Cleavage of Scarecrow-like mRNAs targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau K, Rector M, Carrington J (2002b) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A, Ely L, Smith TH, Marathe R, Anandalakshmi R, Fagard M, Vaucheret H, Pruss G, Bowman L, Vance V (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation of the mobile signal. Plant Cell 13: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A, Reinahrt B, Bartel D, Vance V, Bowman L (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA 99: 15228–15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke AJM, Matzke MA (1986) A set of novel Ti plasmid-derived vectors for the production of transgenic plants. Plant Mol Biol 7: 357–365 [DOI] [PubMed] [Google Scholar]
- Matzke M, Primig M, Trnovsky J, Matzke AJM (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J 8: 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke M, Matzke A (2000) Transcriptional gene silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19: 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Matzke A, Matzke M (2001) Resistance of RNA-mediated TGS to HC-Pro, a viral suppressor of PTGS, suggests alternative pathways for dsRNA processing. Curr Biol 11: 1119–1123 [DOI] [PubMed] [Google Scholar]
- Mette MF, van der Winden J, Matzke M, Matzke A (1999) Production of aberrant promoter transcripts contributes to methylation and silencing of unlinked homologous promoters in trans. EMBO J 18: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Kanno T, Aufsatz W, Jakowitsch J, van der Winden J, Matzke M, Matzke A (2002a) Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J 21: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, van der Winden J, Matzke M, Matzke A (2002b) Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol 130: 6–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, An G (1989) Three distinct regulatory elements comprise the upstream promoter region of the nopaline synthase gene. Mol Gen Genet 215: 294–299 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, Shillito R, Hohn T, Potrykus I (1986) Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res 14: 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B, Weinstein E, Rhoades M, Bartel B, Bartel D (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M, Reinahrt B, Lim L, Burge C, Bartel B, Bartel D (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Schauer S, Jacobsen S, Meinke D, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol J, Kooter J (2001) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol 11: 436–440 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart B, Bartel D, Zamore P (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O (2002) RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr Opin Plant Biol 5: 444–451 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Pinto X, Baulcombe D (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc Natl Acad Sci USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Kidner C, Hall I, Teng G, Grewal S, Martienssen R (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Zamore P (2002) Ancient pathways programmed by small RNAs. Science 296: 1265–1269 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen B (2002) RNA interference in human cells is restricted to the cytoplasm. RNA 8: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen S (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.