Abstract
Using the yeast (Saccharomyces cerevisiae) two-hybrid system and a potato (Solanum tuberosum) KNOX protein, designated POTH1, as bait, we have identified seven distinct interacting proteins from a stolon library of potato. All seven cDNAs are members of the BEL1-like family of transcription factors. Among these proteins, there are at least four regions of high sequence conservation including the homeodomain, the proline-tyrosine-proline three-amino acid loop extension, the SKY box, and a 120-amino acid region upstream from the homeodomain. Through deletion analysis, we identified a protein-binding domain present in the carboxy end of the KNOX domain of POTH1. The protein-binding domain in the BEL1 protein is located in the amino-terminal one-half of the 120-residue conserved region of the BELs. RNA-blot analysis showed differential patterns of RNA accumulation for the BELs in various potato organs. The level of StBEL5 mRNA increased in response to a short-day photoperiod in both leaves and stolons. Similar to sense mutants of POTH1, transgenic lines that overexpressed StBEL5 exhibited enhanced tuber formation even under noninductive conditions. Unlike POTH1 sense lines, however, these BEL lines did not exhibit the extreme leaf and stem morphology characteristic of KNOX overexpressers and displayed a more rapid rate of growth than control plants. Both StBEL5 and POTH1 sense lines exhibited an increase in cytokinin levels in shoot tips. StBEL5 lines also exhibited a decrease in the levels of GA 20-oxidase1 mRNA in stolon tips from long-day plants. Our results demonstrate an interaction between KNOX and BEL1-like transcription factors of potato that may potentially regulate processes of development.
The primary developmental events of plants originate from apical meristems (Clark, 1997; Kerstetter and Hake, 1997). Many of these events are controlled at the molecular level by transcription factors (TFs). TFs are proteins that act as developmental switches by binding to the DNA (or to other proteins that bind to the DNA) of specific target genes to modulate their expression. An important family of TFs involved in regulating the developmental events in apical meristems is the knox (knotted-like homeobox) gene family (Reiser et al., 2000). Knox genes belong to the group of TFs known as the three-amino acid loop extension (TALE) superclass (Bürglin, 1997). These TFs are distinguished by a very high level of sequence conservation in the DNA-binding region, designated the homeodomain, and consisting of three α-helices similar to the bacterial helix-loop-helix motif (Kerstetter et al., 1994). The third helix, the recognition helix, is involved in DNA binding (Mann and Chan, 1996). TALE TFs contain a TALE (Pro-Tyr-Pro) between helices I and II in the homeodomain that has been implicated in protein interactions (Passner et al., 1999). There are numerous TFs from plants and animals in the TALE superclass, and the two main groups in plants are the KNOX and BEL types (Bürglin, 1997). Related genes in animal systems play an important role in regulating gene expression.
In animal developmental systems, members of the TALE superclass physically interact with other TFs to regulate gene expression via a direct effect on transcription of the target gene (Mann and Chan, 1996) or by determining the subcellular location of a key factor (Abu-Shaar et al., 1999; Berthelsen et al., 1999). Specific cooperative DNA binding is facilitated by the tandem protein complex of interacting cofactors (Mann and Chan, 1996; Pinsonneault et al., 1997). Extradenticle (EXD), a TALE TF, functions as a switch that changes homeobox (HOX) proteins from repressors to activators via protein-protein interaction (Pinsonneault et al., 1997). A structural analysis of the protein pairing of EXD and a HOX TF (Ultrabithorax) verified that the P-Y-P loop of EXD binds to a conserved sequence motif in Ultrabithorax to facilitate protein and DNA binding (Passner et al., 1999). EXD and Homothorax (HTH), another TALE TF, interact to facilitate nuclear localization of EXD (Rieckhof et al., 1997). The trimeric interaction of two TALE TFs (EXD and HTH) and a Hox protein facilitates specific binding to the target DNA (Ryoo et al., 1999). Protein interaction in these examples is mediated by specific conserved amino acid sequence motifs (Passner et al., 1999; Ryoo et al., 1999).
Expression patterns and functional analysis of mutations support the involvement of knox genes in specific developmental processes of the shoot apical meristem (SAM). Kn1 from maize (Zea mays), the first plant homeobox gene to be discovered (Vollbrecht et al., 1991), is involved in maintenance of the SAM and is implicated in the switch from indeterminate to determinate cell fates (Clark et al., 1996; Kerstetter et al., 1997; Chan et al., 1998). Transcripts of kn1 in maize (Jackson et al., 1994), OSH1 in rice (Oryza sativa; Sentoku et al., 1999), and NTH15 in tobacco (Nicotiana tabacum; Tamaoki et al., 1997) were localized by in situ hybridization to undifferentiated cells of the corpus and the developing stem but were not detected in the tunica or leaf primordia. Overexpression of kn1 in Arabidopsis (Lincoln et al., 1994) and in tobacco (Sinha et al., 1993) resulted in plants with altered leaf morphologies including lobed, wrinkled, or curved leaves with shortened petioles and decreased elongation of veins. Plants were reduced in size and showed a loss of apical dominance. In plants with a severe phenotype, ectopic meristems formed near the veins of leaves, indicating a reversion of cell fate back to the indeterminate state (Sinha et al., 1993). Overexpression of OSH1 or NTH15 in tobacco resulted in altered morphologies similar to the 35S-kn1 phenotype (Sato et al., 1996; Tamaoki et al., 1997).
Alterations in leaf and flower morphology in 35S-NTH15 or OSH1 transgenic tobacco were accompanied by changes in hormone levels. Although levels of all the hormones measured were changed slightly, both GA and cytokinin levels were dramatically altered (Tamaoki et al., 1997; Kusaba et al., 1998b). RNA-blot analysis revealed that the accumulation of GA 20-oxidase1 mRNA was reduced severalfold in transgenic plants (Kusaba et al., 1998a; Tanaka-Ueguchi et al., 1998). A KNOX protein of tobacco binds to specific elements in regulatory regions of the GA 20-oxidase1 gene of tobacco to repress its activity (Sakamoto et al., 2001). GA 20-oxidase is a key enzyme in the GA biosynthetic pathway necessary for the production of the physiologically inactive GA20 precursor of active GA1 (Hedden and Kamiya, 1997). GA1 and other active GA isoforms are important regulators of stem elongation, the orientation of cell division, the inhibition of tuberization, flowering time, and fruit development (Jackson and Prat, 1996; Hedden and Kamiya, 1997; Rebers et al., 1999).
A homeobox TF of potato (Solanum tuberosum) in the knox family (Reiser et al., 2000), designated POTH1 (potato homeobox; GenBank accession no. U65648) was isolated from an early tuber cDNA library of potato (Rosin et al., 2003a). Sequence analysis indicates that POTH1 is a member of the class I knox gene family (Rosin et al., 2003a) and is also a member of the TALE superclass of homeobox genes (Bürglin, 1997). POTH1 and related knox genes in tobacco, tomato (Lycopersicon esculentum), Arabidopsis, and rice are involved in regulating plant growth by controlling GA synthesis (Tamaoki et al., 1997; Kusaba et al., 1998b; Hay et al., 2002; Rosin et al., 2003a). Overexpression of these knox genes produced plants with altered levels of intermediates in the GA biosynthetic pathway and a reduction in bioactive GAs. These mutants exhibited aberrant leaf formation, dwarfism, and, in the case of POTH1, enhanced tuber formation under both inductive and noninductive conditions (Rosin et al., 2003a).
Another plant homeobox gene family that is closely related to the knox genes is the BEL (BELL) family (Bürglin, 1997; Chan et al., 1998). BEL TFs have been implicated in flower and fruit development (Reiser et al., 1995; Dong et al., 2000). Genetic analysis of BEL1 in Arabidopsis showed that expression of this TF regulated the development of ovule integuments and overlaps the expression of AGAMOUS (Ray et al., 1994; Reiser et al., 1995; Western and Haughn, 1999). In COP1 mutants, the photoinduced expression of ATH1, another BEL TF of Arabidopsis, was elevated, indicating a possible role in the signal transduction pathway downstream of COP1 (Quaedvlieg et al., 1995). Recently, the interaction of BEL1 proteins with KNOX proteins was reported in barley (Hordeum vulgare), Arabidopsis, and maize (Bellaoui et al., 2001; Müller et al., 2001; Smith et al., 2002). Here, we report interactions between a potato KNOX protein involved in the regulation of growth in potato and seven distinct proteins from the BEL1-like family of TFs.
RESULTS
Isolation of Potato KNOX Interactive Proteins
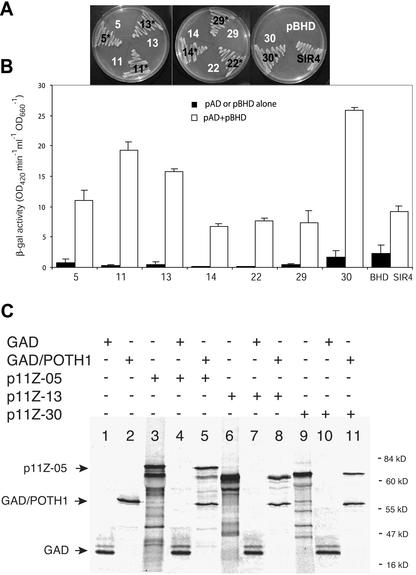
Making use of the two-hybrid selection system in yeast (Saccharomyces cerevisiae), we screened approximately 106 transformants from a stolon cDNA library of potato using POTH1 in the GAL4-binding domain vector, pBridge (CLONTECH Laboratories, Palo Alto, CA), as bait. We identified 38 positive clones that grew on selective media. Of the 38 that were sequenced, 23 clones could be grouped into seven unique genes encoding different members of the TALE superclass of TFs (Chan et al., 1998). All seven, designated StBEL5, -11, -13, -14, -22, -29, and -30 (GenBank accession nos. AF406697, AF406698, AF406699, AF406700, AF406701, AF406702, and AF406703, respectively) showed selective interaction when tested in the yeast system both for nutritional markers and for lacZ activation (Fig. 1, A and B). Interaction occurred also when the prey cDNAs were cloned into pBridge and transformed with POTH1 in a GAL4 activation domain vector (data not shown). As a test for autoactivation, the pAD transformants (5, 11, 13, 14, 22, 29, and 30) did not grow on –His, –adenine, and –Leu medium, and the pBD transformant did not grow on –His, –Trp, and –adenine medium. In vitro-binding experiments verified the results of the two-hybrid selection. POTH1 pulled down three representative StBEL proteins with divergent sequence similarity in the BELL domain (5, 13, and 30) and synthesized by in vitro transcription/translation in immunoprecipitation assays (Fig. 1C).
Figure 1.
Specific interaction of POTH1 with seven BEL1-like proteins of potato. A, Selection on a nutrient carbon medium minus His, Leu, trytophan, and adenine. The pAD plasmid provides Leu selection, the pBD plasmid (pBridge) provides Trp selection, and His and adenine selection are activated from the host strain (AH109) chromosomal DNA. Asterisk, Yeast growth with both plasmids transformed together, whereas the pAD plasmids (designated 5, 11, 13, 14, 22, 29, and 30) are transformed alone (no growth). SIR4, a transcriptional activator of yeast, is used as a positive control and pBHD is POTH1 in pBridge alone. B, POTH1 interacts with all seven BELs as determined by a quantitative yeast two-hybrid assay. LacZ induction in the yeast strain AH109 was assayed in transformed yeast cultures using a quantitative yeast β-galactosidase assay method (Pierce Chemical, Rockford, IL). For each pair, the dark bars on the left represent the pAD or pBHD plasmid alone transformed into yeast. The white bars on the right in each pair represent both plasmids (pAD and pBHD) transformed together. Error bars = se. C, Immunoprecipitates of the in vitro binding of POTH1 to BEL proteins of potato. 35S-labeled GAD:POTH1 fusion protein and the three BEL1 proteins (p11Z-5, -13, and -30) were synthesized in separate in vitro transcription/translation reactions (lanes 2, 3, 6, and 9, respectively). Each of the three BEL1 proteins were incubated with the GAD:POTH1 protein and immunoprecipitated with anti-GAD antibodies (lanes 5, 8, and 11). None of the three BEL proteins bound to the GAD protein alone (lanes 4, 7, and 10). Labeled proteins were visualized by autoradiography after separation by SDS-PAGE. Molecular size markers are shown on the right.
The Proteins That Interact with the Potato KNOX Protein Are Members of the BEL Family of TFs
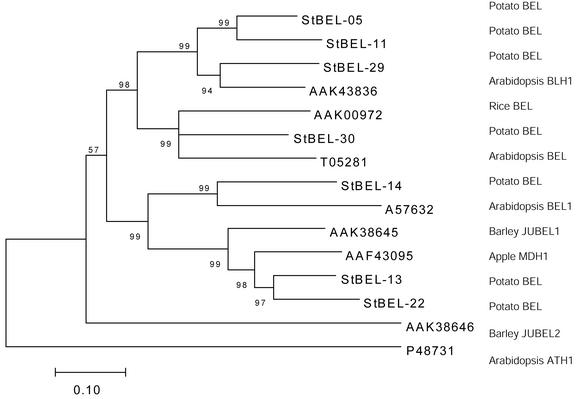
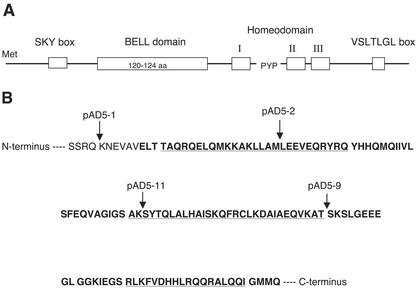
A phylogenetic analysis of the sequences of the seven interacting proteins identified them as members of the BEL1-like family of TFs (Fig. 2). The amino acid sequence of StBEL5 has 56% similarity to BLH1 of Arabidopsis that interacts with KNAT1 (GenBank accession no. AAK43836), StBEL13 matches an apple (Malus domestica) BEL (Dong et al., 2000; GenBank accession no. AAF43095) at 74% similarity, and StBEL30 matches another Arabidopsis BEL (GenBank accession no. T05281) at 59% similarity. The close match of all seven to the conserved homeodomain and the presence of the Pro-Tyr-Pro (P-Y-P) loop between helices I and II (Fig. 3A) distinguish these novel proteins as BEL types in the TALE superclass (Bürglin, 1997). The homeodomain region is nearly identical among these seven (Fig. 3A, encompassing helices I–III). Other regions of conserved sequence identity are shown schematically in Figure 3A. These include the amino-terminal SKY box consisting of 20 amino acids (from Ser-207 to Lys-226 in StBEL5), the 120-amino acid domain starting at Leu-272 of the StBEL5 sequence, and the carboxy-terminal VSLTLGL box beginning at Val-620. Three α-helices were predicted from the conserved 120-amino acid region of the BEL protein StBEL5 (underlined sequence of Fig. 3B). Among the seven BELs, the percent similarity of the amino acid sequence in this conserved 120-amino acid domain ranged from 58% to 90%. Bellaoui et al. (2001) referred to this region as the BELL domain.
Figure 2.
Phylogenetic tree of the BEL1-like proteins of potato. The amino acid sequence of the seven known potato BEL-like proteins was analyzed and compared with BEL proteins of plants. These data were organized into a phylogenetic tree with the ME-Boot program of the MEGA package (version 1.0, Kumar et al., 1993) and the neighbor-joining program (Saitou and Nei, 1987). The numbers listed at the branching points are boot-strapping values that indicate the level of significance (percentage) for the separation of two branches. The length of the branch line indicates the extent of difference according to the scale at the lower left-hand side. Databank accession numbers are listed on the dendrogram, and the common name of the species is listed in the right-hand column.
Figure 3.
A, Schematic of the amino acid sequence of the BEL1-like proteins of potato. Boxed regions represent conserved sequences identified by aligning all seven BELs. Helices I to III of the homeodomain are designated. The Pro-Tyr-Pro (PYP) loop extension is located between helices I and II. For clarity in labeling, the sequence is not drawn to scale. B, Predicted helices of the putative protein-binding region (BELL domain) of the BEL1 protein StBEL5. The bold letters represent amino acids conserved in other plant BEL1 proteins based on a BLAST analysis of StBEL5. The underlined portion of the sequence represents a predicted α-helix. A consensus for the prediction of the sequence structure was derived by using three software programs for amino acid sequence analysis: sspal, ssp, and nnssp (http://www.softberry.com/berry.phtml?topic=protein). Four deletion constructs from Figure 5B are designated with arrows. Construct pAD5-1 contains amino acids 230 through 653 of pAD-05 (interaction with POTH1), and pAD5-2 contains amino acids 257 through 653 of pAD-05 (no interaction). Construct pAD5-11 consists of amino acids 1 through 286 of pAD-05 (no interaction), and pAD5-9 consists of amino acids 1 through 315 (interaction with POTH1).
The deduced lengths of the seven original cDNAs are 688 amino acids for StBEL5, 535 amino acids for StBEL11, 586 amino acids for StBEL13, 589 amino acids for StBEL14, 612 amino acids for StBEL22, 511 amino acids for StBEL29, and 645 amino acids for StBEL30. 5′-RACE was used to verify the full length of StBEL5, -13, -14, and -30. For blot hybridizations, we used a representative clone from each of the four subgroups (StBEL5, -13, -14, and -30). Southern-blot analysis revealed that these genes are unique and belong to small gene subfamilies, based on the complexity of bands detected by gene-specific probes from each of the cDNAs (data not shown).
Patterns of mRNA Accumulation for the Potato BELs
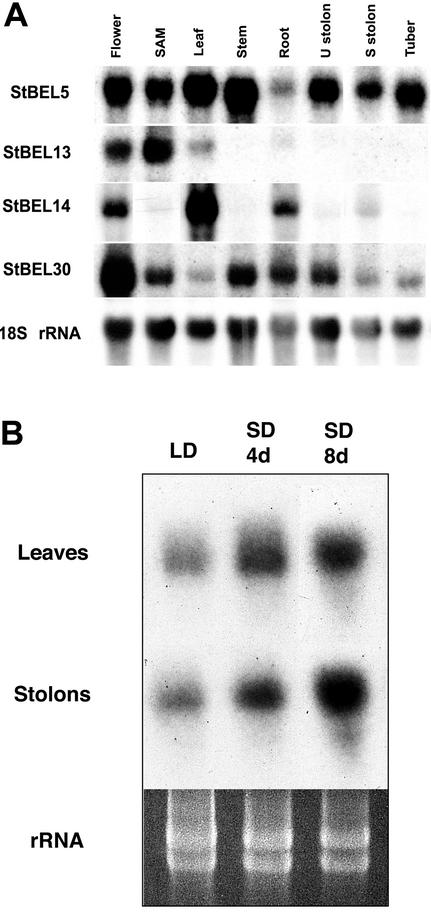
The BEL1-like gene represented by StBEL5 exhibited mRNA accumulation in all organs examined with the greatest levels in leaves and stems (Fig. 4A). Transcript accumulation of StBEL11 and StBEL29 was similar to the pattern of StBEL5 (data not shown). Transcripts for StBEL13 accumulated to the highest levels in the SAM and in fully formed flowers but were barely detectable in other organs (Fig. 4A). The autoradiographs for StBEL13 were exposed 2 times longer than the other StBELs. For StBEL14, transcripts were detected in flowers, leaves, roots, and stolons. The greatest accumulation of StBEL30 was in flowers with detectable levels in all organs examined. To examine more closely the dynamics of StBEL expression during tuber induction, a temporal study was undertaken for the accumulation of StBEL5 transcripts in leaves and stolons of the photoperiod-sensitive potato subsp. andigena grown under SD conditions. Steady-state levels of StBEL5 mRNA increased in both leaves and stolons after exposing the plants to SD conditions (Fig. 4B). Visible tuber formation for the plants grown under SD conditions was observed between 10 and 14 d. In this study, the accumulation of mRNA for the BEL cDNA, StBEL5, was linked to the induction of tuber formation in the leaves and stolons of a potato species responsive to a SD photoperiod.
Figure 4.
A, Northern-blot analysis of the accumulation of mRNA for four BEL1-like cDNAs (StBEL5, -13, -14, and -30) in potato organs. Ten micrograms of total RNA from flowers, shoot tips (SAM), leaves, stems, roots, unswollen stolons (U stolon), swollen stolons (S stolon), and tubers was loaded per lane. Swollen stolons represent an early stage of tuber formation. A probe for the 18S ribosomal RNA was used to verify equal loading of RNA samples (bottom). B, Northern-blot analysis of the accumulation of the mRNA of StBEL5 in leaves and stolons of wild-type (WT) plants grown under long days (LDs, 16 h of light, 8 h of dark) and short days (SDs, 8 h of light, 16 h of dark). Ten micrograms of total RNA was loaded per lane. Leaves and stolons were harvested from the photoperiod-responsive potato subsp. andigena 4 and 8 d after the plants were transferred to SD conditions. Samples were harvested 1 h after the end of the dark period. A gene-specific probe for each BEL cDNA was used. Ethidium bromide-stained ribosomal RNA is visualized as a loading control.
Determining the Protein-Binding Regions in POTH1 and the BEL1-Like Proteins
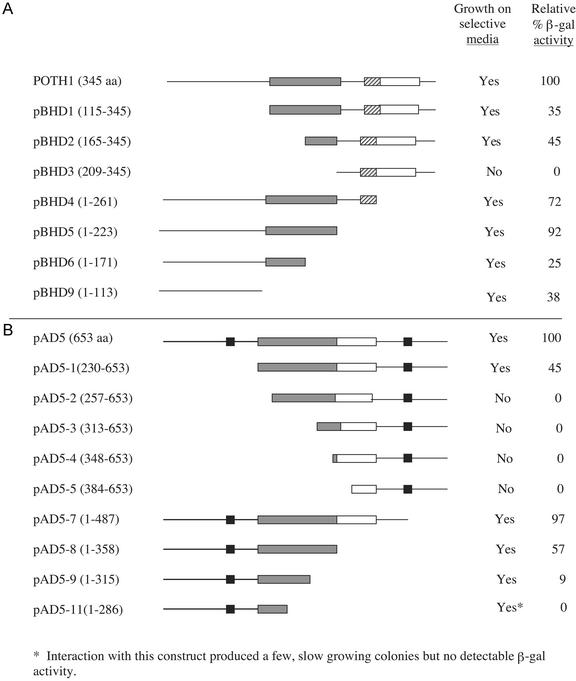
Interaction with StBEL5 was observed with all deletions outside the KNOX domain, with pBHD2 (missing the amino terminus and the first 48 amino acids of the KNOX domain, Fig. 5A), with pBHD6 (missing the carboxy terminus and 52 amino acids of the carboxy end of the KNOX domain), and with pBHD-9 (first amino-terminal 114 amino acids but no KNOX domain sequence). No interaction was observed with pBHD3 (missing all of the KNOX domain and the first 114 amino acids). Control experiments identified the first 114 amino acids of the N terminus (pBHD9) as a transcriptional activator. This construct transformed alone into AH109 exhibited nutrient selection on –His, –Trp, and –adenine medium. Cotransformation of pBHD9 with an empty pGAD cassette produced transformants capable of growth on –His, –Trp, –adenine, and –Leu medium and induction of lacZ (data not shown). None of the other constructs containing this domain were capable of growing on selection media without StBEL5. Using the in vitro-binding protocol, both the pBHD6 construct, containing the amino-terminal one-half of the KNOX domain, and the pBHD9 construct were unable to pull down StBEL5 (data not shown). When the pBHD9 construct was cloned into the pGAD vector, no interaction was observed with StBEL5 in pBridge.
Figure 5.
Deletion analysis of the binding regions of POTH1 and a potato BEL1-like protein using the yeast two-hybrid system. A, Deletion constructs of POTH1 in pBridge were tested for expression in the yeast strain AH109 and cotransformed with the full-length BEL cDNA, StBEL5, in pGAL4 to test for interaction. B, Deletion constructs of StBEL5 in pGAL4 were cotransformed with the full-length cDNA of POTH1 in pBridge. Interaction was verified with both nutritional selection and β-galactosidase activity. The white box indicates the homeodomain. The gray box indicates the putative protein-protein interaction region (for POTH1, this is the conserved KNOX domain; for StBEL5, this is the BELL domain). The black boxes are conserved sequences identified in the BEL proteins (see Fig. 3A), and the diagonal hatched boxes in POTH1 represent the ELK domain. The numbers in parentheses represent the amino acids of the full-length sequence included in each construct.
Fusion constructs of StBEL5 that dissected the 120-amino acid domain (pAD5-2, -3, -4, -9, and -11) were tested because this is one of the regions that is conserved in BEL TFs from other plant species (Bellaoui et al., 2001; Fig. 3B). Interaction with POTH1 was observed with all constructs that had deletions exclusively outside of the conserved 120-amino acid box (Fig. 5B). The only exception to this was with pAD5-9, which demonstrated an interaction and included a 43-amino acid deletion from the carboxy end of the 120-amino acid domain. Even with as little as a 27-amino acid deletion from the amino end of the 120-amino acid domain, interaction did not occur (Figs. 3B and 5B, pAD5-2). Similar to the results of Bellaoui et al. (2001), deletion of the SKY box (construct pAD5-1) resulted in a 55% decrease in the induction of the lacZ marker as measured by β-galactosidase activity relative to the full-length construct, StBEL5 (Fig. 5B).
Enhanced Tuber Formation in Transgenic Plants That Overexpress the BEL cDNA, StBEL5
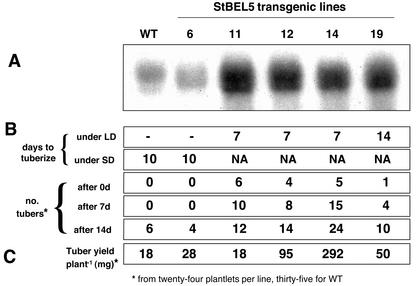
To examine the function of the potato BELs, we analyzed transformed potato subsp. andigena plants that overexpressed StBEL5 from a constitutive promoter. This BEL gene was selected because of its moderate level of activity in stolons and tubers and its increase in RNA levels in response to inductive conditions for tuber formation (Fig. 4). For these experiments, we used a 2,000-bp fragment of the coding sequence of StBEL5 in a sense orientation driven by the cauliflower mosaic virus-35S promoter in the binary vector pCB201 (Xiang et al., 1999). Transformants were identified by PCR analysis of genomic DNA and by detection of the accumulation of sense transcripts of StBEL5 in RNA samples from vegetative meristems. From among approximately 25 positives, four independent lines with the highest levels of StBEL5 mRNA accumulation (Fig. 6A) were selected for evaluation of tuber formation in vitro under both inductive (SD) and noninductive (LD) conditions. The highest expressers of StBEL5 sense transcripts (lines 11, 12, 14, and 19) exhibited tuber formation under LD conditions (Fig. 6B). Control plants (WT and line 6) produced tubers only under SD conditions. The highest overexpressers of StBEL5 also produced more tubers than control plants over the course of this experiment and were more responsive to inductive conditions. After 7 d under SD conditions, the control plants had produced no tubers, whereas the overexpression mutants (lines 11, 12, 14, and 19) had produced 10, eight, 15, and four tubers, respectively (Fig. 6B). After 14 d under SD, controls had increased to six and four tubers, whereas the overexpression lines had increased to 12, 14, 24, and 10 tubers, respectively. Tuber yields (fresh weight) also increased in overexpression lines 12, 14, and 19 (Fig. 6C). The greatest tuber production was exhibited by lines 12 and 14 with a 5- and 16-fold increase, respectively, relative to WT plants. Tubers from the overexpression lines grew larger than controls. Select tubers from line 14 reached fresh weights of almost 700 mg, whereas the largest control tuber reached only 140 mg.
Figure 6.
In vitro tuberization of transgenic plants that overexpress sense transcripts of StBEL5. Northern-blot analysis for the accumulation of mRNA for StBEL5 was performed by using 10 μg total RNA lane–1 from vegetative meristems of in vitro plantlets and gene-specific probes for StBEL5 (A). Equal loading of RNA samples was verified by visualizing ethidium bromide-stained rRNA bands with UV light (not shown). The rate of tuberization (days to tuberize) was determined by the first appearance of tubers from among 24 replicates (B). The number of tubers was scored after 2 weeks of LD conditions (0 d) and after 7 (7 d) and 14 d (14 d) of SD conditions (B). Tubers were harvested and weighed after 21 d (C) from the StBEL5 overexpression (24 plants each) and WT lines (35 plants). Cultured transgenic plants of potato subsp. andigena were grown on a Murashige and Skoog medium with 6% (w/v) Suc under an LD photoperiod (16 h of light, 8 h of dark) in a growth chamber for 2 weeks. For tuber induction, plants were transferred to a Murashige and Skoog medium supplemented with 6% (w/v) Suc and evaluated daily for tuber formation under an SD photoperiod (8 h of light, 16 h of dark) in the growth chamber until tubers formed. All numbered lines were verified as transgenic by using PCR with transgene-specific primers. Control plants were both nontransgenic (WT) and transgenic (StBEL5 line 6).
With whole plants grown in soil under SD conditions for 14 d, StBEL5 overexpression lines produced an average of 3- to 5-fold more tubers per plant and more than a 3-fold greater tuber yield per plant than controls (Table I). Increased yields (as high as 50%) were maintained for these lines even after 6 weeks of growth in soil (data not shown). Seven overexpressing sense lines (lines 7, 11, 12, 14, 16, 19, and 20) also exhibited tuber activity (swollen stolons or tuber formation) on soil-grown plants under LD greenhouse conditions. Five of these plants produced tubers, whereas control plants exhibited no tuber activity. Similar to POTH1 overexpressers (Rosin et al., 2003a), these results show that the accumulation of StBEL5 mRNA is correlated with an increased rate of tuber formation. Other than this enhanced capacity for tuberization, the StBEL5 overexpression lines in Table I did not exhibit the phenotype characteristic of KNOX gene overexpressers, including extreme dwarfism and abnormal leaf morphology (Fig. 7). The abnormal phenotype of KNOX overexpressers is mediated by changes in hormone levels, specifically, a reduction in GAs and an increase in cytokinins (Sato et al., 1996; Tamaoki et al., 1997; Frugis et al., 2001; Rosin et al., 2003a). With the exception of two StBEL5 sense mutants (lines 11 and 20), the leaf and stem growth of the StBEL5 overexpression lines was similar to WT plants (Fig. 7). All five StBEL5 lines exhibited an enhanced rate of growth comparable with control plants (Table II). The average height of line 19 plants was 13.5 cm greater than control plants after 45 d. Fresh weights of leaves and stems of lines 12, 14, and 19 were 29% to 62% greater than control plants. Lines 11 and 20 exhibited a more rapid rate of growth early (10 d), and then growth rate dropped off by 45 d (Table II). Accumulation of StBEL5 transgenic mRNA in line 20 was equivalent to line 11 (data not shown). Three-month-old plants from lines 11 and 20 exhibited a slight reduction in leaf size and stem height as a result of decreased apical dominance. To examine the mechanism for this reduced leaf morphology, cytokinin analysis was performed on shoot apices down to the fourth visible true leaf. Similar to POTH1 overexpressers, shoot tips of both StBEL5 lines 11 and 20 exhibited a 2- to 5-fold increase in the bioactive forms of cytokinin (Table III). The overall magnitude increases in the cytokinin types among the four StBEL and POTH1 mutant lines were remarkably consistent.
Table I.
Rate of tuberization for overexpression lines of StBEL5 under soil-grown, SD conditions
Plants were grown in 10-cm pots under LDs (16 h of light, 8 h of dark) until they reached the 16-leaf stage and then transferred to SDs (8 h of light, 16 h of dark). After 14 d under SDs, four plants per independent line were evaluated for tuber formation. SEs are shown.
| Plant Line | No. Tubers Plant-1 | Tuber Yield Plant-1 |
|---|---|---|
| g | ||
| WT | 2.2 ± 1.4 | 1.4 ± 0.9 |
| StBEL5-12 | 8.0 ± 0.8 | 5.4 ± 1.3 |
| StBEL5-14 | 8.3 ± 0.9 | 4.6 ± 1.3 |
| StBEL5-19 | 11.5 ± 2.1 | 4.7 ± 1.4 |
Figure 7.
Overexpression mutant lines for the potato KNOX gene, POTH1 (lines 15 and 18), and for the BEL1-like protein, StBEL5 (lines 12, 14, and 19). These StBEL5 sense lines had a leaf phenotype similar to WT plants. These are 8-week-old plants grown under LD conditions (16 h of light, 8 h of dark) in the greenhouse supplemented with high-pressure sodium high-intensity discharge lamps. The StBEL5 plants ranged in height from 34 to 39 cm, whereas the POTH1 lines were 7 to 10 cm in height (Rosin et al., 2003a).
Table II.
Plant height and fresh wt of overexpression lines of St-BEL5 under soil-grown, LD conditions
Plants were grown in 10-cm pots under LDs (16 h of light, 8 h of dark), and plant height was measured after 10 and 45 d. Four plants per independent line were evaluated for growth. Fresh wt of leaves and stems was measured after 45 d. SES are shown.
| Plant Line
|
Plant Height
|
Fresh Wt of Stem and Leaves
|
|
|---|---|---|---|
| at 10 d | at 45 d | ||
| cm | g | ||
| WT | 5.3 ± 0.3 | 35.2 ± 2.2 | 18.0 ± 2.6 |
| StBEL5-11 | 7.3 ± 0.4 | 31.9 ± 3.0 | 19.6 ± 1.3 |
| StBEL5-20 | 6.3 ± 0.6 | 32.2 ± 2.0 | 10.8 ± 0.5 |
| StBEL5-12 | 7.1 ± 0.7 | 44.9 ± 0.9 | 23.3 ± 1.2 |
| StBEL5-14 | 6.2 ± 0.2 | 38.2 ± 1.2 | 29.2 ± 1.0 |
| StBEL5-19 | 7.1 ± 0.5 | 48.7 ± 1.9 | 25.5 ± 3.5 |
Table III.
Cytokinin content in shoot tips of POTH1 and StBEL5 overexpression lines grown under LDs (16 h of light, 8 h of dark) in the greenhouse
WT lines are non-transformed potato subsp. andigena. Zeatin types include zeatin, zeatin riboside, dihydrozeatin, and dihyrozeatin riboside. Isopentenyl types include isopentenyl and isopentenyladenine. SE was calculated on three replicates.
| Sample | Zeatin Types | Isopentenyl Types |
|---|---|---|
| picomoles g fresh wt-1 | ||
| WT | 10.5 ± 1.0 | 12.0 ± 1.5 |
| POTH1-15 | 42.5 ± 15 | 35.5 ± 7.0 |
| POTH1-29 | 34.0 ± 12 | 30.0 ± 6.0 |
| StBEL5-11 | 55.5 ± 30 | 31.5 ± 11 |
| StBEL5-20 | 30.5 ± 6.0 | 29.5 ± 6.5 |
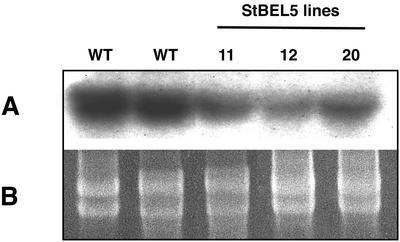
POTH1 sense lines had increased levels of GA53 and GA19 and decreased levels of GA20 and GA1 in shoot tips, implicating a down-regulation of the biosynthetic enzyme GA 20-oxidase1 (Rosin et al., 2003a). Using a probe for the potato GA 20-oxidase1 gene (Carrera et al., 2000), we observed a reduction in GA 20-oxidase1 mRNA in shoots of the most severe mutant phenotypes for POTH1 sense lines (Rosin et al., 2003a; Fig. 7). To determine the effect of overexpression of the POTH1 partner, StBEL5, RNA levels for GA 20-oxidase1 were examined in the stolons of StBEL5 sense lines grown under LD photoperiod conditions. All three of the StBEL5 lines examined (lines 11, 12, and 20) exhibited a reduction in GA 20-oxidase1 mRNA in stolon tips comparable with controls (Fig. 8). No such reduction in the levels of GA 20-oxidase1 mRNA was observed in shoot tips of StBEL5 lines grown under LDs (data not shown).
Figure 8.
Northern-blot analysis of the accumulation of the mRNA of the GA 20-oxidase1 gene of potato (Carrera et al., 2000) in WT plants and sense lines 11, 12, and 20 of StBEL5 (A). Total RNA was extracted from the 2.0-mm distal tip of stolons from plants grown under LD conditions (16 h of light, 8 h of dark). WT RNA was extracted from two separate pools. Ten micrograms of total RNA was loaded per lane. A gene-specific probe for GA 20-oxidase1 was used for hybridization. All three StBEL5 lines exhibited enhanced tuber formation. Ethidium bromide-stained rRNA is visualized as a loading control (B).
To determine the effect of up-regulating StBEL5 mRNA levels on POTH1 RNA accumulation, northerns were performed on total RNA extracted from StBEL5 sense lines 12, 14, 19, and 20 using POTH1 as a probe. There were no changes in the levels of POTH1 mRNA in both shoot tips and stolon tips of these StBEL5 lines relative to WT plants (data not shown). These results indicate that the enhancement of tuber formation in StBEL5 overexpression lines is not mediated by an indirect increase in POTH1 expression.
DISCUSSION
Seven BEL1 Proteins Interact with a KNOX Protein of Potato
Using a yeast two-hybrid library screen, we have identified seven unique proteins from potato stolons that interact with the knotted-like protein, POTH1. Sequence analysis revealed that these interacting proteins are from the BEL1-like family in the TALE superclass of homeodomain proteins. These proteins have conserved regions in common with other TALE proteins, including the homeodomain (comprised of three α-helices) and the Pro-Tyr-Pro “TALE” (Bürglin, 1997). These sequences have been implicated in DNA-binding and protein-protein interactions, respectively (Mann and Chan, 1996; Passner et al., 1999). A second conserved region of 120 amino acids just upstream from the homeodomain (designated the BELL domain by Bellaoui et al., 2001) was identified among BEL proteins by using a BLAST analysis (Fig. 3B, bold letters). Sequence analysis of the predicted secondary structure of this domain reveals the presence of three putative α-helices within the 120 residues (Fig. 3B, underlined sequence). Not all BEL proteins conserve the third helix, however, including the Arabidopsis BEL, ATH1 (Quaedvlieg et al., 1995) and the barley BEL, JUBEL2 (Müller et al., 2001). Protein interaction using the two-hybrid system demonstrated that the first 80 amino acids of this domain (up to QVKAT of the STBEL-5 sequence and comprising the first two predicted helices of this region) are necessary to mediate interaction with POTH1 (interaction of construct pAD5-9 with POTH1). Deletion of as little as the first 20 amino acids of this domain (comprising a major portion of the first predicted helix) interfered with the interaction with POTH1 (Figs. 3B and 5B, construct pAD5-2). Our results also showed that deletion of 43 amino acids from the carboxy-end of the 120-amino acid domain (see Fig. 5B, construct pAD5-9, comprising the third helical structure) did not affect protein interaction. Deletion of the two carboxyl-terminal helices in this region (construct pAD5-11) resulted in a loss of interaction. It is conceivable that all three helical structures contribute to specific binding affinity for POTH1 but that only the amino-terminal two-thirds of the 120-amino acid domain are necessary for binding to occur. Müller et al. (2001) identified a coiled-coil region in a BEL protein of barley that was involved in the interaction with KNOX proteins. This coiled-coil domain overlaps with 48 of the 80 amino acids (and comprising the first helix) that we identified as essential for interaction to occur.
Sequence differences in this putative protein-binding region could contribute to the regulation of POTH1 activity by affecting binding affinity to a shared partner. In the interaction between PIF3, a basic helix-loop-helix factor, and phytochrome A and B, phytochrome B has 10-fold greater binding affinity for the PIF3 partner than phytochrome A (Zhu et al., 2000). A comparison of this 120-amino acid domain in the potato BELs revealed that StBEL5 (expressed ubiquitously) has a 58% similarity match to StBEL13 (expressed predominately in the SAM and flower only) and that StBEL13 has a 63% match to StBEL30 in this conserved region. Such differences in sequence may mediate binding affinities to shared partners and, coupled with expression patterns, could reflect organ-specific differences in function.
Conservation in sequence among these seven proteins can also be identified in two short amino acid sequence motifs, one near the carboxyl-end of the protein (VSLTLGL) and another just upstream of the BELL domain (SKY box, Fig. 3A). Both of these regions are conserved among other plant BELs. Protein interaction studies showed that the VSLTLGL box is not involved in protein interaction with POTH1 and its function remains unknown. Consistent with Bellaoui et al. (2001), we observed that although binding occurred without the 229 amino acids at the amino terminus of StBEL-5 (construct pAD5-1), this 229-amino acid sequence alone, containing the SKY box, was sufficient to mediate an interaction with POTH1 (and other class I KNOX proteins). This 229-amino acid sequence, however, did not interact with a class II KNOX protein (data not shown). Müller et al. (2001) identified the SKY box sequence in the barley BEL protein to be a part of the KNOX-interacting domain. Our deletion analysis indicates that the SKY box enhances the binding affinity of StBEL5 to KNOX partners.
The Protein-Binding Region of POTH1
In addition to the homeodomain, KNOX TFs also contain a conserved region of approximately 100 amino acids, upstream from the homeodomain, known as the KNOX (MEINOX) domain, and postulated to be involved in protein-protein interaction (Bürglin, 1998). Using deletion mutants in the two-hybrid yeast system, we have identified regions of amino acid sequence in the KNOX domain of the class I KNOX protein, POTH1, that are involved in an interaction with the BEL TFs. Binding to the BEL partner is mediated by the KNOX domain but is not dependent on the presence of the first half of the 120-amino acid KNOX region (Fig. 5A). Similar results were obtained by Müller et al. (2001). Sakamoto et al. (1999) showed by using chimeric proteins that the second one-half of the KNOX domain (designated KNOX2) of a tobacco KNOX protein (NTH15, with 63% similarity to POTH1 in the KNOX region) was most important for determining the severity of the mutant phenotype. Their results indicated that this conserved domain was even more important in determining the phenotype than the DNA-binding domain. The deletion analysis for POTH1 in the present study, combined with the results of Sakamoto et al. (1999), suggest that the interaction of the BEL proteins with the KNOX domain may be a prominent control mechanism for mediating KNOX activity and maintaining stable development of the vegetative meristem. KNOX2 contains 18 amino acids that are predicted to form an α-helix and are conserved among all tobacco and potato KNOX proteins. POTH1 has a close sequence match to members of the family of KNOX proteins of tobacco (Nishimura et al., 2000), with an overall sequence similarity ranging from 60% to 73% and an even greater match in the conserved KNOX and homeodomain regions. Using the two-hybrid system, all seven BELs of potato interacted with four other tobacco class I-type KNOX proteins (data not shown). Unlike KNOX proteins of barley (Müller et al., 2001) and rice (Nagasaki et al., 2001), however, POTH1 did not form homodimers in vitro (data not shown). Structural similarities to the MEIS domain of animal homeodomain proteins (Bürglin, 1998) suggest that sequences in the KNOX domain of plants are important for interactions with other proteins (Sakamoto et al., 1999). Our results confirm the function of this domain in an interaction with a BEL1-like protein of potato.
The Function of the BEL/POTH1 Interaction
Through both molecular and genetic analyses, the BEL proteins are known to function in the development of ovules. Reiser et al. (1995) showed that BELL1 of Arabidopsis was involved in the pattern formation of ovule primordium. More specifically, the expression of NOZZLE (a nuclear protein and putative TF) and BELL are spatially linked and interact with other TFs to determine distal-proximal pattern formation during ovule development (Balasubramanian and Schneitz, 2002). Both NOZZLE and BELL are chalazal identity genes that share overlapping functions (Balasubramanian and Schneitz, 2000). In bel1 mutants, the chalazal domain undergoes altered development, and growth of the integuments is replaced by irregular outgrowths (Modrusan et al., 1994). Overexpression of an apple BEL gene (MDH1) in Arabidopsis produced plants that were dwarf, had reduced fertility, and exhibited changes in both carpel and fruit shape (Dong et al., 2000). Overall, these results suggest that BEL proteins function in controlling the formation of carpellate tissues and plant fertility. Overexpression of a cDNA of a barley BEL in tobacco produced plants that were dwarf and exhibited malformed leaves and reduced apical dominance (Müller et al., 2001). This BEL1-like cDNA isolated from floral meristems produced a sense phenotype similar to a class I knox overexpresser (Chan et al., 1998). All seven of the BEL TFs in this study were isolated from stolons, a vegetative organ. Based on these results and the patterns of mRNA accumulation in potato, it is likely that the BEL1 TFs of potato play a diverse role in plant growth by regulating the development of both reproductive and vegetative meristems.
Because the BEL1s of potato and POTH1 interact, the function of one may provide a clue to the function of the other. The KNOX protein of tobacco, NTH15, affects plant growth by regulating GA levels through a direct interaction with a specific motif in regulatory sequences of the GA 20-oxidase1 gene, a key GA biosynthetic enzyme (Sakamoto et al., 2001). NTH15 directly suppresses the expression of GA 20-oxidase1 within specific cells of the SAM to maintain the indeterminate state of corpus cells. The knotted1-like protein of potato, POTH1, is also involved in the regulation of GA synthesis and acts as a developmental switch during tuber formation. Transgenic plants that overexpressed POTH1 had reduced levels of GA 20-oxidase1 mRNA, altered levels of GA intermediates, and exhibited a phenotype that could be partially rescued by GA3 treatment (Rosin et al., 2003a). These plants were dwarf and developed malformed leaves. Under both SD (inductive conditions) and LD (noninductive) photoperiods, POTH1-overexpressing lines produced more tubers than controls (Rosin et al., 2003a). These sense lines exhibited a capacity for enhanced tuber formation. Lines that overexpressed StBEL5 produced tubers even under LD in vitro conditions, whereas control plants produced tubers only after 10 d of SD conditions. Overall, the BEL sense lines produced more tubers at a faster rate than controls even on soil-grown plants. After 14 d of SD conditions, soil-grown StBEL-5 overexpressers exhibited a 3-fold increase in tuber production relative to WT plants (Table I). In addition to enhanced tuber production, select StBEL5 lines exhibited increases in cytokinin levels and a reduction in GA 20-oxidase1 mRNA similar to POTH1 overexpression lines. This increase in cytokinin levels could explain the enhanced rate of growth for the StBEL5 lines, although excessive accumulation may have led to the reduction in growth exhibited by mature plants of lines 11 and 20. GA is involved in regulating cell growth in a tuberizing stolon (Xu et al., 1998) and in contributing to the control of the photoperiodic response of tuber formation (Kumar and Wareing, 1974; Jackson and Prat, 1996, Martínez-García et al., 2001). Low levels of GA in the stolon tip are correlated with tuber induction (Xu et al., 1998). Tuberization is also affected by cytokinin accumulation, with high levels inhibiting and moderate levels promoting tuber formation (Gális et al., 1995; Romanov et al., 2000). Local accumulation of cytokinins in axillary buds of transgenic tobacco produced truncated, tuberizing lateral branches (Guivarc'h et al., 2002). Through an interaction with POTH1, the BEL protein encoded by StBEL5 may also function to regulate hormone levels in stolons or leaves to favor the formation of tubers.
Although all seven BELs may act to regulate growth during stolon and tuber formation, it is possible that some of the potato BELs are functional only in other organs. The expression patterns of StBEL5, -13, -14, and -30 (Fig. 4A) suggest such a specialization of function. The interaction of POTH1 (and other KNOX proteins of potato) and the various BELs could represent unique complexes with different affinities for DNA-binding motifs. This modification in structure coupled with the regulation of protein accumulation could mediate the activity of POTH1 and determine binding to a select target gene. Third partner protein interaction could also affect the activity of the KNOX/BEL complex via structural modification or subcellular localization. In preliminary studies with two-hybrid screening, we have identified other proteins that interact with StBEL5. Throughout the plant, the various BELs may regulate growth by acting as either activators or repressors of POTH1 activity. The interaction of HOX proteins with MEIS and PBC class proteins produces a complex regulatory network, where even slight changes in protein levels can have profound phenotypic effects (Azpiazu and Morata, 1998). Saleh et al. (2000) suggested a model whereby the complex of two homeodomain proteins, HOX and PBX, can act as a repressor or activator of transcription via interaction with a third partner. In this system, a protein kinase modifies a CREB protein to facilitate its binding to a HOX/PBX complex to activate transcription of the target gene (Saleh et al., 2000).
The results of this study suggest that the physical interaction between the KNOX and BEL1 proteins provides a molecular basis for regulating processes of growth in the potato and that overexpression of each partner alone affects vegetative development and enhances tuber formation.
MATERIALS AND METHODS
Two-Hybrid Selection and Deletion Analysis
The Matchmaker two-hybrid system (CLONTECH) was used for the yeast (Saccharomyces cerevisiae) two-hybrid screen. Yeast transformation and plasmid rescue into DH5-α Escherichia coli cells were according to the manufacturer's instructions. Full-length POTH1 was cloned into the pBridge (CLONTECH) vector and used as bait to screen the potato (Solanum tuberosum cv Desireé) stolon cDNA library in pAD-GAL4-2.1 (Stratagene, La Jolla, CA). Positive interactions were confirmed by cotransforming yeast strain AH109 with each purified pAD plasmid and pBridge:POTH1 and plating on –Leu/–Trp (transformation control) and –Leu/–Trp/–His/–adenine (selection) nutrient medium. Induction of the AH109 reporter gene, lacZ, was measured with a yeast β-galactosidase assay kit (Pierce Chemical). β-Galactosidase activity (Fig. 1B) was determined from a known density of yeast cells and calculated as: 1,000 × OD420/time of color reaction (minutes) × volume of yeast culture (milliliters) × OD600.
The StBEL5 deletion constructs were amplified by PCR, then cloned into the vector, pGAD, in-frame with the GAL4 activation domain. POTH1 deletion constructs were amplified by PCR and cloned into pBridge (CLONTECH) in-frame with the GAL4-binding domain. Sequencing of selected cDNAs and constructs was performed at the Iowa State University DNA Facility (Ames). For deletion analysis, modified constructs of POTH1 were cloned into the pBridge vector for fusion with the DNA-binding domain of GAL4 (Fig. 5A). For StBEL5, constructs were cloned into the pGAD vector for fusion with the activating domain of GAL4 (Fig. 5B). Deletion constructs were made from both the amino and carboxy termini. These mutants were then tested for interaction in the yeast two-hybrid system by cotransforming into yeast strain AH109 with the corresponding full-length partner (StBEL5 in pGAL4 or POTH1 in pBridge). All constructs were sequenced to verify that they were in-frame. Positive interactions were verified for lacZ induction by using a β-galactosidase assay (Pierce Chemical). For POTH1, seven deletion constructs were tested (Fig. 5A). For the BEL TFs, a fusion construct of StBEL5 (653 amino acids of StBEL5 sequence) and nine deletion constructs were tested (Fig. 5B).
GenBank accession numbers for StBEL5, -11, -13, -14, -22, -29, and -30 are AF406697, AF406698, AF406699, AF406700, AF406701, AF406702, and AF406703, respectively.
In Vitro-Binding Assay
In vitro-binding experiments were performed as described by Ni et al. (1998). Full-length sequence for POTH1 was cloned into a pET17b/GAD fusion cassette and transcribed under the control of the T7 promoter. The BEL cDNAs were cloned into pGEM11Z vectors and transcribed under the control of the T7 promoter. 35S-Met-labeled bait and prey proteins were synthesized using the TnT in vitro transcription-translation kit (Promega, Madison, WI) according to the manufacturer's protocols. Each 50-μL TnT reaction contained 2.0 μg of template plasmid DNA and 20 pmol (20 μCi) of labeled 35S-Met. The POTH1:GAD/BEL complex was immunoprecipitated with anti-GAD antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). The proteins from the pellet (one-half the fraction) and for the prey (one-fourth of the reaction volume) were resolved on a 10% (w/v) SDS-PAGE gel and visualized by autoradiography.
Hybridization-Blot Analysis
Total RNA was extracted from various organs of potato subsp. andigena plants grown under a LD photoperiod by using TRI REAGENT according to the manufacturer's manual (Molecular Research Center, Inc., Cincinnati). Swollen stolons (newly formed tubers) and tubers were harvested from SD plants. For Figure 4B, RNA was extracted from leaves and stolons that were harvested from the photoperiod-responsive potato subsp. andigena grown under a SD photoperiod. Total RNA was size fractionated via electrophoresis through a 1.4% (w/v) agarose gel that contained 5.0 mm methyl-mercury hydroxide and transferred onto a MagnaGraph nylon membrane (Micron Separations Inc., Westboro, MA). Hybridization and washing conditions were the same as described by Kolomiets et al. (2001). For autoradiography, membranes were exposed to x-ray film with intensifying screens for 3 to 6 d at –80°C. A 1.2-kb wheat (Triticum aestivum) 18S ribosomal RNA probe was used to confirm uniform loading of RNA for the blots in Figure 4A. Blots presented are representative examples of at least two independent experiments.
Plant Transformation
Transformation and regeneration of plants was undertaken on leaf sections from potato subsp. andigena line 7540 as described by Liu et al. (1995). These autotetraploid andigena plants strictly photoperiodic for tuberization were obtained from the Institut für Pflanzenbau und Pflanzenzüchtung (Braunchsweig, Germany). The sense constructs were made from a 2.0-kb fragment from the StBEL5 cDNA and cloned into the binary vector pCB201 (Xiang et al., 1999) driven by the constitutive cauliflower mosaic virus-35S promoter. Constructs were checked by using PCR with clone-specific primers. Positive recombinants were transferred to the Agrobacterium tumefaciens strain GV2260 by using the procedure of direct transformation (An et al., 1988). Control plants in the tuberization study were andigena plants regenerated in vitro. Functional transformants were identified by PCR analysis of genomic DNA and by detection of the accumulation of sense transcripts of StBEL5 in shoot tip samples. From among these positives, the seven independent transformants (lines 7, 11, 12, 14, 16, 19, and 20 for StBEL5) used in this study were selected on the basis of abundant accumulation of sense mRNA in shoot tips. Quantitative analysis of cytokinins was performed by using liquid chromatography as described previously (Rosin et al., 2003b). Three replicate 200-mg (fresh weight) samples of shoot tips down to the fourth visible expanded leaf were collected, frozen in liquid nitrogen, lyophilized, and analyzed.
Evaluation of Tuber Formation
For in vitro tuberization, cultured transgenic plants were grown on a Murashige and Skoog medium with 6.0% (w/v) Suc under a LD photoperiod (16 h of light, 8 h of dark) in a growth chamber for 2 weeks and then transferred to a SD photoperiod (8 h of light, 16 h of dark) in the same growth chamber. For tuber induction, plants were evaluated daily for tuber formation. Soil-grown plants were grown in 10-cm pots under LDs (16 h of light, 8 h of dark) in the greenhouse supplemented with high-pressure sodium high-intensity discharge lamps until they reached the 16-leaf stage and then transferred to SDs in the growth chamber. After 14 d under SDs, plants were evaluated for tuber formation.
Acknowledgments
We thank Uwe Sonnewald for the stolon two-hybrid library and Makoto Matsuoka for providing us with the NTH cDNAs. We also thank Phil Becraft and Dan Voytas for critical reviews of the manuscript and Harry Van Onckelen for performing the cytokinin analysis.
Article, publication date, and citation information can be found at http://www.plantphysiol.org/cgi/doi/10.1104/pp.103.022434.
This work was supported in part by the Iowa Agriculture and Home Economics Experiment Station. This is journal paper no. J–19520 of the Iowa Agriculture and Home Economics Experiment Station (Ames, IA; project no. 3701).
References
- Abu-Shaar M, Ryoo DH, Mann RS (1999) Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev 13: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, eds, Plant Molecular Biology Manual. Kluwer Academic, Hingham, MA, pp 1–19
- Azpiazu N, Morata G (1998) Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev 12: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Schneitz K (2000) NOZZLE regulates proximal-distal formation, cell proliferation and early sporogenesis during ovule development in Arabidopsis thaliana. Development 127: 4227–4238 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Schneitz K (2002) NOZZLE links proximal-distal and adaxial-abaxial pattern formation during ovule development in Arabidopsis thaliana. Development 129: 4291–4300 [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW (2001) The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13: 2455–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Kilstrup-Nielsen C, Blasi F, Mavilio F, Zappavigna V (1999) The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev 13: 946–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR (1997) Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25: 4173–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR (1998) The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes. Dev Genes Evol 208: 113–116 [DOI] [PubMed] [Google Scholar]
- Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 1–10 [DOI] [PubMed] [Google Scholar]
- Chan RL, Gago GM, Palena CM, Gonzalez DH (1998) Homeoboxes in plant development. Biochim Biophys Acta 1442: 1–19 [DOI] [PubMed] [Google Scholar]
- Clark SE (1997) Organ formation at the vegetative shoot meristem. Plant Cell 9: 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM (1996) The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Dong YH, Yao JL, Atkinson RG, Putterill JJ, Morris BA, Gardner RC (2000) MDH1: an apple homeobox gene belonging to the BEL1 family. Plant Mol Biol 42: 623–633 [DOI] [PubMed] [Google Scholar]
- Frugis G, Giannino D, Nicolodi C, Chiappetta A, Bitonti MB, Innocenti AM, Dewitte W, Van Onckelen HA, Mariotti D (2001) Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol 126: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gális I, Macas J, Vlasák J, Ondrej M, Van Onckelen HA (1995) The effect of an elevated cytokinin level using the ipt gene and N6-benzyladenine on single node and intact potato plant tuberization in vitro. J Plant Growth Regul 14: 143–150 [Google Scholar]
- Guivarc'h A, Rembur J, Goetz M, Roitsch T, Noin M, Schmulling T, Chriqui D (2002) Local expression of the ipt gene in transgenic tobacco (Nicotiana tabacum L. cv. SR1) axillary buds establishes a role for cytokinins in tuberization and sink formation. J Exp Bot 53: 621–629 [DOI] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M (2002) The Gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr Biol 12: 1557–1565 [DOI] [PubMed] [Google Scholar]
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S (1994) Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Jackson SD, Prat S (1996) Control of tuberisation in potato by gibberellins and phytochrome B. Physiol Plant 98: 407–412 [Google Scholar]
- Kerstetter R, Vollbrecht E, Lowe B, Veit B, Yamaguchi J, Hake S (1994) Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 6: 1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Hake S (1997) Shoot meristem formation in vegetative development. Plant Cell 9: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S (1997) Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054 [DOI] [PubMed] [Google Scholar]
- Kolomiets MV, Hannapel DJ, Chen H, Tymeson M, Gladon RJ (2001) Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Wareing PF (1974) Studies on tuberization of Solanum andigena II. Growth hormones and tuberization. New Phytol 73: 833–840 [Google Scholar]
- Kumar S, Tamura K, Nei M (1993) MEGA: Molecular Evolutionary Genetics Analysis, version 1.01. Penn State University, University Park
- Kusaba S, Fukumoto M, Honda C, Yamaguchi I, Sakamoto T, Kano-Murakami Y (1998a) Decreased GA1 content caused by the overexpression of OSH1 is accompanied by suppression of GA 20-oxidase gene expression. Plant Physiol 117: 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba S, Kano-Murakami Y, Matsuoka M, Tamaoki M, Sakamoto T, Yamaguchi I, Fukumoto M (1998b) Alteration of hormone levels in transgenic tobacco plants overexpressing the rice homeobox gene OSH1. Plant Physiol 116: 471–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S (1994) A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ATH, Stephens LC, Hannapel DJ (1995) Transformation of Solanum brevidens using Agrobacterium tumefaciens. Plant Cell Rep 15: 196–199 [DOI] [PubMed] [Google Scholar]
- Mann RS, Chan SK (1996) Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet 12: 258–262 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, García-Martínez JL, Bou J, Prat S (2001) The interaction of gibberellins and photoperiod in the control of potato tuberization. J Plant Growth Regul 20: 377–386 [DOI] [PubMed] [Google Scholar]
- Modrusan Z, Reiser L, Feldmann KA, Fischer RL, Haughn GW (1994) Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W (2001) In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J 27: 13–23 [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Sakamoto T, Sato Y, Matsuoka M (2001) Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 13: 2085–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M (2000) Overexpression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol 41: 583–590 [DOI] [PubMed] [Google Scholar]
- Passner M, Ryoo HD, Shen L, Mann RS, Aggarwal AK (1999) Structure of DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature 397: 714–719 [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Florence B, Vaessin H, McGinnis W (1997) A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J 16: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeek P, Smeekens S (1995) The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. Plant Cell 7: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Robinson-Beers K, Ray S, Baker SC, Lang JD, Preuss D, Milligan SB, Gasser CS (1994) Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc Natl Acad Sci USA 91: 5761–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y (1999) Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J 17: 241–250 [DOI] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL (1995) The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83: 735–742 [DOI] [PubMed] [Google Scholar]
- Reiser L, Sanchez-Baracaldo P, Hake S (2000) Knots in the family tree: evolutionary relationships and functions of knox homeobox genes. Plant Mol Biol 42: 151–166 [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS (1997) Nuclear translocation of Extradenticle requires homothorax, which encodes an Extradenticle-related homeodomain protein. Cell 91: 171–183 [DOI] [PubMed] [Google Scholar]
- Romanov GA, Aksenova NP, Konstantinova TN, Golyanovskaya SA, Kossmann J, Willmitzer L (2000) Effect of indole-3-acetic acid and kinetin on tuberisation parameters of different cultivars and transgenic lines of potato in vitro. Plant Growth Regul 32: 245–251 [Google Scholar]
- Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ (2003a) Overexpression of a knox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol 132: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Hart JK, Van Onckelen H, Hannapel DJ (2003b) Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol 131: 1613–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Marty T, Casares F, Affolter M, Mann RS (1999) Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126: 5137–5148 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguichi-Tanaka M, Iwahori S, Matsuoka M (2001) KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthesis gene in the tobacco shoot apical meristem. Genes Dev 15: 581–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Nishimura A, Tamaoki M, Kuba M, Tanaka H, Iwahori S, Matsuoka M (1999) The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 11: 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M, Rambaldi I, Yang XJ, Featherstone MS (2000) Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltranferases. Mol Cell Biol 20: 8623–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Tamaoki M, Murakami T, Yamamoto N, Kano-Murakami Y, Matsuoka M (1996) Abnormal cell divisions in leaf primordia caused by the expression of the rice homeobox gene OSH1 lead to altered morphology of leaves in transgenic tobacco. Mol Gen Genet 251: 13–22 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, Matsuoka M (1999) Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11: 1651–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7: 787–795 [DOI] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S (2002) Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc Natl Acad Sci USA 99: 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M (1997) Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol 38: 917–927 [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi M, Itoh H, Oyama N, Koshioka M, Matsuoka M (1998) Overexpression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J 15: 391–400 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S (1991) The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 350: 241–243 [DOI] [PubMed] [Google Scholar]
- Western TL, Haughn GW (1999) BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J 18: 329–336 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–718 [DOI] [PubMed] [Google Scholar]
- Xu X, van Lammeren AAM, Vermeer E, Vreugdenhil D (1998) The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol 117: 575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tepperman JM, Fairchild CD, Quail PH (2000) Phytochrome B binds with greater affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci USA 97: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]