Abstract
To evaluate the genetic control of stress responses in Arabidopsis, we have analyzed a mutant (uvh6-1) that exhibits increased sensitivity to UV light, a yellow-green leaf coloration, and mild growth defects. We have mapped the uvh6-1 locus to chromosome I and have identified a candidate gene, AtXPD, within the corresponding region. This gene shows sequence similarity to the human (Homo sapiens) XPD and yeast (Saccharomyces cerevisiae) RAD3 genes required for nucleotide excision repair. We propose that UVH6 is equivalent to AtXPD because uvh6-1 mutants carry a mutation in a conserved residue of AtXPD and because transformation of uvh6-1 mutants with wild-type AtXPD DNA suppresses both UV sensitivity and other defective phenotypes. Furthermore, the UVH6/AtXPD protein appears to play a role in repair of UV photoproducts because the uvh6-1 mutant exhibits a moderate defect in the excision of UV photoproducts. This defect is also suppressed by transformation with UVH6/AtXPD DNA. We have further identified a T-DNA insertion in the UVH6/AtXPD gene (uvh6-2). Plants carrying homozygous insertions were not detected in analyses of progeny from plants heterozygous for the insertion. Thus, homozygous insertions appear to be lethal. We conclude that the UVH6/AtXPD gene is required for UV resistance and is an essential gene in Arabidopsis.
DNA damage is a challenge for all organisms exposed to UV irradiation. UV photoproducts consist primarily of cyclobutane pyrimidine dimers and pyrimidine (6-4) pyrimidinone dimers (Mitchell and Nairn, 1989; Pfeifer, 1997). These lesions inhibit DNA replication and transcription and also promote mutagenesis (McGregor, 1999). The effects of UV irradiation are especially detrimental in plants, where sunlight is both a source of damage and a requirement for photosynthesis.
Increasing evidence suggests that plants repair UV-damaged chromosomes using mechanisms similar to those found in humans (Homo sapiens) and yeast (Saccharomyces cerevisiae). These mechanisms include the nucleotide excision repair (NER) pathway, a process which involves recognition of UV lesions, incision of the damaged strand on both sides of the lesion, removal of the damaged fragment, and repair by gap filling and ligation (Batty and Wood, 2000; de Boer and Hoeijmakers, 2000; Prakash and Prakash, 2000). Several potential plant homologs of human and yeast NER genes have been identified. Genetic analyses of these plant genes, including studies of the phenotypes of plants carrying mutations within these genes, provide support for the idea that the NER pathway is conserved in plants.
Lesion recognition during NER involves the homologous heterodimers XPC:HR23B (human) and RAD4:RAD23 (yeast; Balajee and Bohr, 2000; Batty and Wood, 2000; de Boer and Hoeijmakers, 2000; Prakash and Prakash, 2000). The Arabidopsis genome contains potential homologs of both XPC/RAD4 and HR23B/RAD23 (Arabidopsis Genome Initiative, 2000). HR23B expression occurs in Arabidopsis, rice (Oryza sativa), and carrot (Daucus carota), and the carrot gene complements the repair defect in a yeast rad23 mutant (Schultz and Quatrano, 1997; Sturm and Lienhard, 1998).
Damage-induced incision in NER involves two nucleases. The first nuclease makes cuts 5′ to the lesion and consists of homologous heterodimers XPF: ERCC1 (human) and RAD1:RAD10 (yeast). A homologous complex has been implicated in plant DNA repair based on the UV sensitivity of Arabidopsis derivatives carrying either a mutation in the plant XPF homolog (Fidantsef et al., 2000; Liu et al., 2000) or an antisense construct to this gene (Gallego et al., 2000). Arabidopsis XPF mutants are also defective in repair of UV photoproducts (Liu et al., 2000; Li et al., 2002). Homologs of the ERCC1 component of this first nuclease occur in Arabidopsis (Arabidopsis Genome Initiative, 2000) and Lilium longiflorum (Xu et al., 1998), and the L. longiflorum gene complements a repair defect in ERCC1-deficient Chinese hamster (Cricetulus griseus) ovary cells. The nuclease that incises 3′ to DNA lesions is called XPG in humans and RAD2 in yeast. A mutation in the Arabidopsis XPG homolog results in UV sensitivity (Liu et al., 2001b). In addition, a second mutation (uvr1), which fails to complement the AtXPG mutation (Z. Liu and D. Mount, unpublished data) results in defective repair of 6-4 photoproducts (Britt et al., 1993).
Finally, NER requires DNA unwinding by two DNA helicases, called XPB and XPD in humans and RAD25 and RAD3 in yeast. Two potential Arabidopsis homologs of XPB/RAD25 have been identified (Ribeiro et al., 1998; Arabidopsis Genome Initiative, 2000). AtXPB1 complements a repair defect in yeast rad25 mutants, and plants carrying T-DNA insertions in this gene are sensitive to alkylating agents (Costa et al., 2001b). Arabidopsis also encodes a presumed homolog of the XPD/RAD3 genes (Arabidopsis Genome Initiative, 2000).
The AtXPD gene is the subject of the present study. We have examined an Arabidopsis mutation (called uvh6-1) that dramatically increases sensitivity to UV light. We present evidence that this mutation results in a missense change within the Arabidopsis XPD/RAD3 gene. Our findings further support the possibility that the AtXPD/UVH6 gene product functions within the plant NER pathway. In addition, we observe that a T-DNA insertion in AtXPD (uvh6-2) is lethal, suggesting that this gene serves an essential function during plant development.
RESULTS
Identification of the UVH6 Gene and the uvh6-1 Mutation
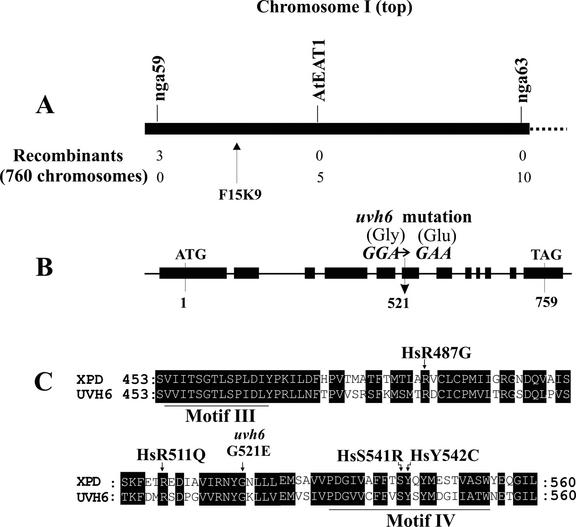
We previously identified a UV-sensitive mutant of Arabidopsis (originally called uvh6 but renamed uvh6-1 in this study) and used it to map the UVH6 locus to the top 6 centimorgans on chromosome I (Jenkins et al., 1995). As shown in Figure 1A, we have further refined the location this gene to the region between physical markers nga59 and AtEAT1. Within this region, we have also identified a candidate gene at locus At1g03190 (Bac locus F15K9). We have designated this gene as AtXPD because it exhibits strong similarity to ERCC2 (XPD) from Chinese hamster and RAD3 (yeast; The Arabidopsis Information Resource, http://www.arabidopsis.org).
Figure 1.
Mapping of the UVH6 locus and organization of the corresponding UVH6(AtXPD) gene. A, Top 3.3 megabases of chromosome I are shown with corresponding physical markers nga59, AtEAT1, and nga63. The UVH6 locus was localized between markers nga59 and AtEAT1, as described in “Materials and Methods,” based on an analysis of DNA from 380 UV-sensitive F2 progeny (760 chromosomes) obtained from a cross between uvh6-1 (Columbia ecotype) and wild-type (Landsberg ecotype) plants. Three UV-sensitive recombinants carrying the Landsberg nga59 polymorphism were identified, and all three lacked both the Landsberg nga63 and AtEAT1 markers. Ten recombinants carrying Landsberg nga63 were also obtained. Five of these carried the Landsberg AtEAT1 marker, whereas none carried the Landsberg nga59 marker. The position of Bac clone F15K9, which carries the candidate repair gene AtXPD (gene F15K9.20, locus At1g03190), is indicated. B, The UVH6/AtXPD gene is diagrammed, based on a comparison between the known genomic and cDNA sequences. The diagram depicts exons (thick bars), introns (thin lines), initiation and termination codons, and the uvh6-1 mutation at codon 521. C, Alignment of the central portion of the human XPD (accession no. CAA36463) and UVH6/AtXPD (accession no. AY090788) protein sequences is shown, encompassing conserved helicase motifs III and IV. The uvh6-1 mutation at amino acid 521 is indicated, and the sites of known human mutations (designated Hs), which produce the diseases xeroderma pigmentosum or trichothiodystrophy, are indicated.
Because mammalian XPD and yeast RAD3 genes are required for NER and because the uvh6-1 mutant is UV sensitive, indicative of a repair defect, we hypothesized that the uvh6-1 mutation maps within the AtXPD gene. To test this possibility, we amplified AtXPD cDNA from wild-type (C10) and uvh6-1 mutant plants (as described in “Materials and Methods”) and determined their corresponding sequences. Compared with the wild-type sequence, we found a single-nucleotide change at codon 521 in the uvh6-1 mutant cDNA sequence. This mutation was also confirmed by sequencing mutant genomic DNA (data not shown). As depicted in Figure 1B, this mutation should result in a Gly to Glu missense substitution. Hence, it seems likely that the UVH6 gene is equivalent to AtXPD and that the sequence change detected in the AtXPD gene is the uvh6-1 mutation.
The UVH6/AtXPD sequence that we obtained from wild-type plants is nearly identical to two unpublished AtXPD mRNA sequences in GenBank (accession nos. AF188623 and AY062471) but contains a few, possibly polymorphic, differences within the coding region. In comparison with AF188623, we observed three differences, one at codon 541 (AGC/AGT), which did not alter the encoded Ser amino acid, and two at codon 554 (GAA/AAG), which changed the encoded Glu to a Lys in our sequence. In comparison with AY062471, one silent change is observed at codon 282 (CGA/CGG). The residues that we observed at all these positions were identical to those found in the Arabidopsis consensus genome sequence (Arabidopsis Genome Initiative, 2000), which, like our sequence, was derived from plants of the Columbia ecotype. Our cDNA sequence has been submitted to GenBank (accession no. AY090788) and is depicted in Figure 1B in relation to the corresponding genomic sequence.
Complementation of UV Sensitivity in the uvh6-1 Mutant by AtXPD DNA
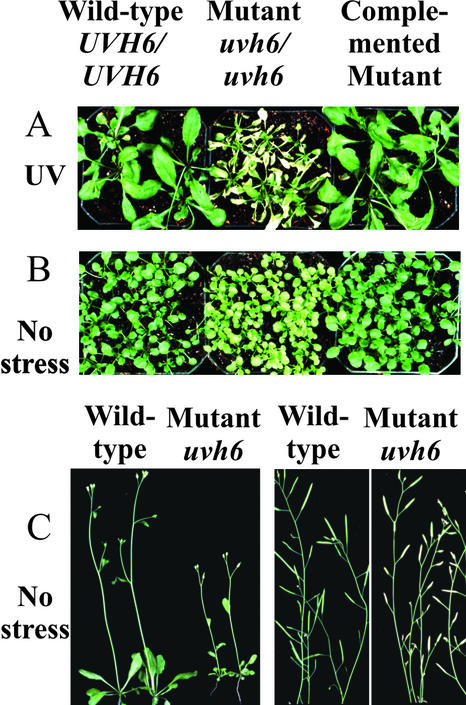
To confirm the identification of the UVH6 gene, we characterized the phenotypes of uvh6-1 mutant plants that had been transformed with wild-type (C10) AtXPD genomic DNA. Primary T1 transformants were examined for UV sensitivity, and 22 of 26 transformants displayed a UV-resistant phenotype. This phenotype is depicted in Figure 2 for T2 generation plants homozygous for the introduced AtXPD transgene. As shown, uvh6-1 mutant plants are abnormally sensitive to UV-C irradiation, compared with the wild-type parent, and exhibit severe browning and death of rosette leaves within 3 d after treatment (compare Fig. 2A, middle with left). This finding confirms our previous report that uvh6-1 plants are hypersensitive to both UV-C and UV-B irradiation (Harlow et al., 1994). In contrast, the transformed plants exhibit the same resistance to irradiation shown by the wild-type parents (right). Thus, the mutant phenotype is complemented by wild-type AtXPD DNA, indicating that the UVH6/AtXPD gene is required for resistance to UV irradiation.
Figure 2.
Recovery of UV resistance and normal growth properties after complementation of a uvh6-1/uvh6-1 homozygous mutant with UVH6/AtXPD DNA. Mutant uvh6-1 plants were transformed with UVH6/AtXPD genomic DNA (from C10 wild-type plants) as described in “Materials and Methods.” C10 wild-type (UVH6/UVH6), uvh6-1 mutant (uvh6/uvh6), and T2 generation-complemented mutant plants were grown for 2 to 3 weeks and subsequently exposed to UV-C irradiation (300 J m2) as described in “Materials and Methods” (A) or maintained under normal growth conditions for 3 weeks (B), 4.5 weeks (c, left), or 6.5 weeks (c, right).
Growth Defects in the uvh6-1 Mutant
In addition to radiation sensitivity, uvh6-1 mutant plants exhibit growth defects that are also suppressed in plants transformed with wild-type UVH6/AtXPD genomic DNA. As shown in Figure 2B (middle), the leaves of uvh6-1 mutant plants are yellow-green, as previously reported (Jenkins et al., 1997). This phenotype is observed when mutant plants are grown under normal (non-stress) conditions and may result from the low chlorophyll content in these plants described previously (Jenkins et al., 1997). In addition, mutant uvh6-1 plants are also substantially smaller when compared with wild-type plants of the same age, as seen in Figure 2C (left). In contrast, uvh6-1 plants that have been complemented with AtXPD genomic DNA are dark-green and exhibit normal growth rates (Fig. 2B, right). Thus, these additional mutant phenotypes appear to result from the same mutation that causes UV sensitivity.
Relationship of AtXPD to Presumed Human and Yeast Homologs
We originally identified the AtXPD gene based on the similarity of its encoded protein sequence to human XPD and yeast RAD3 proteins, which have wellestablished roles in NER (Batty and Wood, 2000; de Boer and Hoeijmakers, 2000; Prakash and Prakash, 2000). We have performed further protein sequence comparisons to assess this similarity and to gain insight into the function of the Arabidopsis protein. Pair-wise alignments (data not shown) indicate that the Arabidopsis sequence (accession no. AAF14582) is 56% and 50% identical to the human and yeast sequences (accession nos. CAA36463 and AAB64698, respectively). The Arabidopsis sequence exhibits strong similarity to both homologs along its entire length, and the residue mutated in the uvh6-1 mutant is conserved in both comparisons. Figure 1C depicts part of the alignment to the human sequence, including the uvh6-1 mutation site. Additional alignments (not shown) with XPD homologs from Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila melanogaster (accession nos. CAA93221, AAK95892, and AAD33587, respectively) also reveal strong sequence similarity and conservation of the G residue at the position of the uvh6-1 mutation. This strong conservation of the uvh6-1 residue in several homologs further suggests that this mutation disrupts the function of the UVH6/AtXPD protein.
Human XPD and yeast RAD3 genes encode DNA helicases essential for repair (Winkler et al., 2000). Seven highly conserved motifs in these sequences are required for helicase activity (Gorbalenya et al., 1989). Our comparison of the AtXPD sequence with the human and yeast homologs revealed especially strong similarity in the regions of all seven motifs (data not shown). Sequence conservation involving motifs III and IV is illustrated in Figure 1C. Thus, it appears likely that the AtXPD plant gene acts as a helicase and performs the same role in DNA repair previously demonstrated in humans and yeast.
Repair of 6-4 Photoproducts in the uvh6-1 Mutant
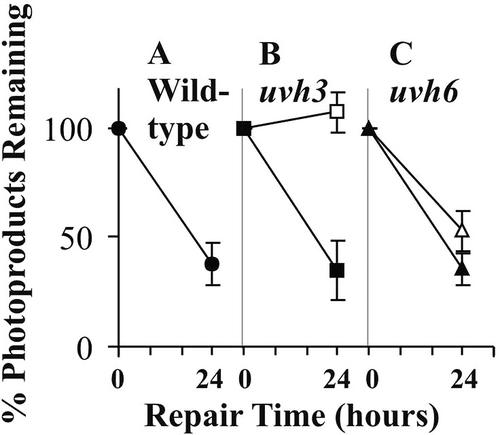
The UV sensitivity of our uvh6-1 mutant suggests that this mutant might be defective in repair of UV photoproducts. To test this possibility, we examined the repair of 6-4 photoproducts in DNA from irradiated plants, comparing wild-type, uvh6-1 mutant, and uvh3 mutant plants. The uvh3 mutant is also UV sensitive and carries a nonsense mutation in the UVH3/AtXPG gene, encoding the homolog of the human XPG 3′-incision endonuclease (Jenkins et al., 1995; Liu et al., 2001b). As shown in Figure 3A, wild-type plants exhibited significant removal (62%) of 6-4 products from their DNA by 24 h after irradiation. In comparison, the uvh6-1 mutant (Fig. 3C) exhibited reduced levels of repair after 24 h (47% photoproduct removal), suggesting that it has a moderate repair defect. No photoproduct removal was detected in the uvh3 mutant (Fig. 3B). This latter result confirms a previous report (Britt et al., 1993) of defective repair in a mutant (uvr1) that fails to complement our uvh3 mutant in genetic tests. Finally, for both the uvh6-1 and uvh3 mutants, complementation by the appropriate wild-type genes resulted in restoration of the wild-type level of repair. The complemented uvh3 mutant showed 66% photoproduct removal (Fig. 3B) and the complemented uvh6-1 mutant showed 65% removal (Fig. 3C).
Figure 3.
Repair of 6-4 photoproducts in DNA from UV-irradiated plants. DNA extracted from irradiated plants was assayed for photoproducts as described in “Materials and Methods.” The plants studied were C10 wild-type (•), mutant uvh3 (□), complemented mutant uvh3 (▪), mutant uvh6-1 (▵), and complemented mutant uvh6-1 (▴). Each point represents the mean of four separate assays, and sds of the means are shown. The difference between the 24-h data points for uvh6-1 and the complemented uvh6-1 plants is statistically significant (P < 0.025).
Characterization of a T-DNA Insertion in UVH6/AtXPD and Its Effect on Plant Viability
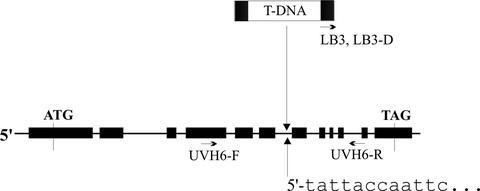
We have identified a line of Arabidopsis plants (called 825 BO5) carrying a T-DNA insertion (which we designate as uvh6-2) within the UVH6/AtXPD gene. The site of insertion was confirmed by DNA sequencing, as depicted in Figure 4. Insertion occurred within the sixth of 11 introns in the UVH6/AtXPD gene and presumably results in a truncated UVH6/AtXPD protein because of altered splicing of UVH6/AtXPD mRNA. Based on the site of insertion, 26% of the coding sequence and four of the highly conserved helicase motifs should be deleted.
Figure 4.
Site of T-DNA insertion (uvh6-2) within the UVH6/AtXPD gene. The position of the insert was confirmed by DNA sequencing, as described in “Materials and Methods.” This site is shown with respect to the UVH6/AtXPD genomic DNA, and the UVH6/AtXPD DNA sequence adjacent to the left border (LB) is shown. The UVH6/AtXPD gene is depicted with exons (thick lines) and introns (thin lines). Positions of the primers (UVH6-F, UVH6-R, LB3, and LB3D) used for PCR reactions and DNA sequencing are indicated. For results shown in Table I, primers UVH6-F and UVH6-R (gene specific) were used to detect wild-type alleles; primers UVH6-R and LB3 (Ga-3 experiment) or LB3D (Ga-2 experiment) were used to detect T-DNA-inserted alleles.
In a preliminary analysis, we asked whether the 825 BO5 line was homozygous or heterozygous for the T-DNA insert. For this analysis, 10 plants were grown from the initial seed stock, and genomic DNA from each plant was subjected to a PCR analysis that distinguishes between wild-type and T-DNA-inserted alleles of UVH6/AtXPD. The locations of primers used in this analysis are shown in Figure 4. Gene-specific primers that anneal to sites flanking the insertion site were used to identify wild-type UVH6/AtXPD alleles. This test detects only wild-type alleles because the presence of the insertion results in an inter-primer distance (greater than 10 kb) that is too long for PCR amplification. To detect alleles carrying the insertion, a second PCR reaction was performed using one of the UVH6-specific primers and a primer that anneals within the T-DNA left border. Thus, DNA from plants heterozygous for the insert should be amplified with both sets of primers, whereas DNA from plants homozygous for either the wild-type or inserted allele should be amplified by only one set of primers. Results of this analysis revealed that five plants were heterozygous and five were homozygous for the wild-type allele. No homozygous T-DNA insertions were detected, suggesting that homozygous insertions are lethal.
To investigate this possibility, we analyzed two of the above heterozygous plants (called Ga-2 and Ga-3) to determine the frequency with which their progeny inherit the T-DNA insert. The heterozygote parents were allowed to produce seeds by self-fertilization, and DNA from individual F1 seedlings was subjected to the same PCR analysis described above. In total, 97 progeny plants were examined. As seen in Table I, 56.7% of these plants were heterozygous and 43.3% carried only wild-type alleles, whereas no plants homozygous for the insert were detected. This result clearly deviates from the theoretical expectation that 25% of the progeny should carry homozygous T-DNA insertions under conditions where the insertion is not lethal (P < 0.01). This result also rules out the possibility that lethality is caused by a second mutation unlinked to the UVH6/AtXPD locus, although the presence of a lethal mutation very closely linked to UVH6 is still possible. Most likely, the results indicate that homozygous uvh6-2 insertions within UVH6/AtXPD are lethal and that the UVH6/AtXPD gene serves an essential function during plant growth.
Table I.
PCR analysis of DNA from progeny of Ga-2 and Ga-3 plants carrying a heterozygous T-DNA insertion (uvh6-2) in the UVH6/AtXPD genea
| Genotype of Progeny
Plantsb
|
No. of Progeny Plants with the Specified Genotype
|
||
|---|---|---|---|
| Progeny from Ga-3 parent | Progeny from Ga-2 parent | Total progenyc | |
| Homozygous wild type | 18 | 24 | 42(43.3%) |
| Heterozygous T-DNA insertion | 29 | 26 | 55(56.7%) |
| Homozygous T-DNA insertion | 0 | 0 | 0(0.0%) |
Genomic DNA was extracted from leaves of F1 seedlings from two plants (Ga-3 and Ga-2) carrying heterozygous T-DNA insertions in UVH6/AtXPD, and the DNA was subjected to PCR analysis, as described in “Materials and Methods.” bGenotypes were scored as homozygous wild type (amplification observed only with gene-specific primers), T-DNA insertion heterozygous (amplification observed with both sets of primers), or T-DNA insertion homozygous (amplification observed only with the T-DNA/gene-specific primer pair). Figure 4 shows the primer annealing sites. cA χ2 analysis of these results generated a value of P < 0.01, indicating that the results are not consistent with the expected Mendelian genetic distribution of 1:2:1.
Expression of the UVH6/AtXPD Gene
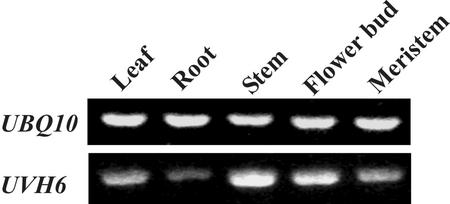
The mRNA expression pattern of the UVH6/AtXPD gene was analyzed as shown in Figure 5. Total RNA was extracted from several plant tissues, specific mRNA sequences were amplified by RT-PCR, and PCR products were visualized after electrophoresis. We previously found that the use of 50 ng of input plant RNA gave a PCR product that was proportional to the input RNA concentration, using 25 to 30 PCR cycles (Liu et al., 2001b). As seen in Figure 5, these conditions gave clearly detectable levels of product with our UBQ10 control, a gene that expresses at a relatively high level. This control also serves to verify that equivalent quantities of RNA were present in each PCR reaction. However, to detect a PCR product for UVH6/AtXPD, which expresses at a relatively low level, we required additional input RNA (200 ng) and 40 PCR cycles. Under these conditions, we observed that UVH6/AtXPD mRNA is expressed to various degrees in leaf, root, stem, flower bud, and meristem tissues. Hence, UVH6/AtXPD appears to be expressed at low levels in all tissues, as expected for a gene required for general transcription.
Figure 5.
Expression of UBQ10 and UVH6 mRNA in Arabidopsis tissues, as measured by reverse transcriptase (RT)-PCR. Total RNA was isolated from the C10 wild-type plant tissues indicated and assayed, as described in “Materials and Methods.” RT-PCR products were detected after agarose gel electrophoresis, and the product amplified from the fully spliced message is shown in each case. Controls without RT were run in each experiment (not shown) and failed to produce a product, thus ruling out any contribution of genomic DNA to amplification. RNA was prepared from unbolted flower buds, roots, stems, or leaves, using 4-week-old plants or from 2-week-old meristem tissue.
DISCUSSION
We have identified a mutation (uvh6-1) in the Arabidopsis AtXPD gene and have characterized the phenotypes of uvh6-1 mutant plants. AtXPD is predicted to be a homolog of the human XPD and yeast RAD3 genes, which are required for NER of DNA. The uvh6-1 mutation is a missense change from Gly to Glu at position 521 in the UVH6/AtXPD protein. This mutation appears to cause the extreme UV sensitivity exhibited by uvh6-1 plants because mutant plants transformed with wild-type UVH6/AtXPD DNA regain wild-type radiation resistance. Mutant sensitivity to UV light is likely to result from a deficiency in NER, based on our finding that mutant plants exhibit a moderate defect in removal of 6-4 photoproducts from their DNA.
In addition, we have examined a mutant carrying a T-DNA insertion (uvh6-2) within the UVH6/AtXPD gene. This insertion is predicted to produce a truncated UVH6 gene product. Our findings indicate that homozygous insertions within UVH6/AtXPD are lethal, suggesting that this gene encodes an essential function required during plant development.
Role of AtXPD in DNA Repair
Genes involved in NER have been highly conserved from yeast to humans (Prakash and Prakash, 2000), suggesting a common mechanism. This mechanism also appears to be conserved in plants, based on the characterization of several presumed homologs of NER genes in Arabidopsis (Britt et al., 1993; Fidantsef et al., 2000; Gallego et al., 2000; Liu et al., 2000, 2001b; Li et al., 2002). Our findings that the uvh6-1 mutant carries a mutation in the presumed Arabidopsis homolog of XPD (human) and RAD3 (yeast) and that this mutant is both sensitive to UV light and has a moderate repair defect provide additional evidence that NER is conserved in plants.
Human XPD and yeast RAD3 proteins function as helicases during DNA repair, presumably unwinding DNA surrounding target lesions to facilitate incision (Sung et al., 1988; Winkler et al., 2000). We think it highly likely that the Arabidopsis homolog UVH6/AtXPD also acts as a repair helicase. The UVH6/AtXPD protein sequence contains seven highly conserved motifs that are required for helicase function (Gorbalenya et al., 1989). The uvh6-1 mutation lies between motifs III and IV, suggesting that it might impair helicase activity. In addition, the uvh6-1 mutation occurs in a G residue that is highly conserved in humans, yeast, S. pombe, C. elegans, and D. melanogaster, further suggesting that this residue is important for the normal enzymatic function of UVH6/AtXPD.
Potential Role of AtXPD in Regulating Gene Expression and Plant Development
Mutant uvh6-1 plants exhibit phenotypic defects when grown under normal lighting conditions, suggesting that the UVH6/AtXPD protein has a role during plant development in addition to DNA repair. Under normal, non-stress growth conditions, mutant plants are small compared with wild-type plants of the same age. They have a yellow-green appearance, contain subnormal levels of chlorophyll. (Jenkins et al., 1997), and exhibit poorly organized grana stacks within thylakoid membranes (M. Jenkins, unpublished data). Furthermore, the mutants are abnormally sensitive to prolonged exposure to high temperature (37°C; Jenkins et al., 1997). In addition, we have observed that T-DNA insertions within the UVH6 gene are lethal, based on our failure to isolate plants that are homozygous for a T-DNA-insertion (uvh6-2) in this gene. Thus, UVH6/AtXPD appears to serve an essential function during plant growth.
To understand the role of UVH6/AtXPD during plant growth, it is informative to consider the known functions of the human and yeast homologs. Human XPD and the homologous yeast RAD3 proteins have dual roles in repair and transcription initiation (de Boer and Hoeijmakers, 2000; Prakash and Prakash, 2000; Lehmann, 2001). In this latter role, they facilitate initiation of transcription by RNA polymerase II as part of initiation factor TFIIH. Considerable evidence suggests that human XPD mutations can cause developmental abnormalities due to defects in transcription. These mutations produce three different genetic diseases (xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy; de Boer and Hoeijmakers, 2000; Lehmann, 2001) that all result in sensitivity to UV light and repair deficiencies but are distinguished by additional, distinct phenotypes. For example, trichothiodystrophy is associated with brittle hair and Cockayne syndrome with skeletal deformations. Substantial evidence supports the idea that these additional phenotypes result from transcription defects in differentiated tissues, rather than just loss of repair capacity (Bootsma and Hoeijmakers, 1993; de Boer et al., 1998; Lehmann, 2001; Liu et al., 2001a; Viprakasit et al., 2001; Keriel et al., 2002).
Reminiscent of patients with these human diseases, uvh6-1 mutants exhibit developmental and stress-response defects. These phenotypes are consistent with the possibility that the UVH6/AtXPD gene is required for transcription in plants. Hence, we hypothesize that transcription is abnormal in the uvh6-1 mutant, causing defects in expression of genes required for chlorophyll synthesis, normal growth rate, and heat resistance. However, we cannot rule out the possibility that uvh6-1 mutants are not impaired in transcription per se but, due to their repair defect, accumulate DNA damage that blocks transcription.
Interaction of AtXPD within a Predicted TFIIH Complex
In humans and yeast, the TFIIH transcription initiation complex contains six core components and three additional proteins in an associated kinase CAK complex. Of these nine components, only three core proteins and one CAK protein appear to be conserved in Arabidopsis. Strong matches to Arabidopsis sequences are observed for human core components XPD, XPB (two matched loci), and p44. These matches occur at loci At1g03190, At5g41370, At5g41360, and At1g05050, respectively. The existence of two expressed AtXPB genes (Costa et al., 2001b) suggests that multiple forms of TFIIH may exist in plants. In addition, the CAK catalytic subunit appears to be conserved, based on clustering of an Arabidopsis kinase (locus At1g73690) with human (cdk7) and yeast (KIN28) CAK sequences in a phylogenetic tree of cyclin-dependent protein kinase sequences (http://kinase.ucsf.edu/ksd/). In contrast, sequences that are similar to the other three core and two CAK components are not found in Arabidopsis. Although a recent report (Costa et al., 2001a) claimed to have found two of these components (p52 and cyclinH) in Arabidopsis and sugarcane (Saccharum officinarum and Saccharum spontaneum), our sequence analyses suggest that this report may represent matches to isolated protein domains. Thus, only some of the potential TFIIH core and CAK components can be identified in Arabidopsis, suggesting either that these complexes do not exist or are composed of novel or highly diverged subunits in plants.
Analysis of T-DNA Insertions Suggests That UVH6/AtXPD Is an Essential Gene
In contrast to the subtle growth defects of uvh6-1 mutant plants, we have failed to detect plants carrying homozygous T-DNA insertions in the UVH6/AtXPD gene. The specific insertion that we characterized (uvh6-2) is predicted to cause truncation of the UVH6/AtXPD protein, deleting 26% of its length and four of the seven conserved helicase domains. Thus, the overall structure and presumed helicase activity of this protein should be disrupted. Our failure to isolate plants carrying homozygous insertions in UVH6/AtXPD suggests that this gene is essential for plant growth. Our observation that UVH6/AtXPD is expressed in all plant tissues supports the possibility that this gene might be important for general transcription.
We have observed further an apparent reduction in transmission of the T-DNA insertion within the progeny of plants carrying a heterozygous insertion. These progeny were either heterozygous for the insertion or had lost the insertion and were wild type. However, the observed ratio of heterozygotes:wild type was close to 1:1, rather than the expected 2:1. This finding suggests that transmission of the inserted allele might be impaired in either the female or male gametes (or both). We are currently testing these possibilities by backcrossing heterozygous plants to wild type.
A model to explain the differences observed between the uvh6-1 point mutant and the uvh6-2 T-DNA insertion mutant has been suggested by studies of yeast RAD3 mutations. These mutations exist in two classes. Mutations in the first class are lethal and result in severe defects in RNA polymerase II-dependent transcription under restrictive conditions (Guzder et al., 1994; Prakash and Prakash, 2000), suggesting that the transcription defect causes the lethality. In contrast, the second class of mutations does not impair viability, but most of the mutants are sensitive to UV irradiation and exhibit defects in NER (Prakash and Prakash, 2000). Thus, this second class appears to affect DNA repair, while leaving transcription largely intact. Presumably, XPD mutations that produce diseases in humans are of this second type and cause repair defects with only subtle impacts on transcription. In accord with this model, the uvh6-1 point mutation may also resemble this second class, causing a repair defect with minor effects on transcription, whereas the uvh6-2 T-DNA insertions may resemble the first class, producing a lethal phenotype due to a severe transcription defect.
MATERIALS AND METHODS
Strains and Growth Conditions
The wild-type Columbia (C10) used was derived from a single plant isolate of Arabidopsis and served as the parent of the uvh6-1 mutant, which was isolated as described (Harlow et al., 1994; Jenkins et al., 1995). Plants were normally grown at room temperature (22°C–24°C) under continuous lighting consisting of a combination of 40WT12 Excella and F40 agro bulbs (Philips Lighting Company, Somerset, NJ) positioned approximately 35 cm from the plants (30 μmol photons m–2 s–1).
Seeds carrying a T-DNA insertion (uvh6-2) in the UVH6 gene (garlic line 825 BO5) were obtained from the Torrey Mesa Research Institute (San Diego). Seeds were either surface sterilized and germinated on agar plates (Haughn and Somerville, 1986) or were germinated in soil and grown in a growth chamber at 22°C with 16 h of light (240 μmol photons m–2 s–1) at 22°C and 8 h of dark at 21°C and 75% to 90% humidity.
UV Light Treatment
Two- to 3-week-old plants were irradiated with UV-C light (254 nm) as described by Liu et al. (2001b). They were then incubated for 3 d under F40 GO gold fluorescent lights (Philips Lighting Company, Somerset, NJ) to avoid reversal of UV photoproducts by photoreactivation, a mechanism activated by visible light, and then transferred to standard growth conditions for 7 d. Sensitivity was assessed by the extent of leaf yellowing and tissue death.
Genetic and Physical Mapping of the UVH6 Gene
Recombinant plants were generated by crossing a uvh6-1/uvh6-1 homozygous mutant plant (Columbia ecotype) and a wild-type Landsberg erecta plant. F2 progeny were screened to identify those which exhibited the UV sensitivity of the mutant (Columbia) parent, using protecting foam to shield the meristem during treatment, as described (Harlow et al., 1994). UV-sensitive progeny were then examined for the presence of Landsberg-associated simple sequence length polymorphism physical markers nga59, AtEAT1, and nga63. Polymorphisms were detected by agarose gel analysis of PCR assays (Bell and Ecker, 1994).
Mutant Complementation
A 5.6-kb fragment of AtXPD genomic DNA, which carries the UVH6/AtXPD coding region, 3′-untranslated sequences, and a 5′-flanking region (1.2 kb) containing the AtXPD promoter, was amplified by PCR from wild-type (C10) DNA, using the primer pair H6U186 (5′-CAACATTCCGATTTTCCGTCACCT) and XDL2532 (5′-CCTACAGTGAAAATTTGAGCTCCAACAATT). The amplified fragment was first cloned into the pCRII vector, using a TA Cloning Kit (Invitrogen, Carlsbad CA), and then transferred into plasmid pBIN19. The resultant recombinant (pAtXPD) was introduced into Agrobacterium strain GV3101 by electroporation, as described by Mozo and Hooykaas (1991). Transformed bacteria were selected on Luria-Bertani medium (Sambrook et al., 1989) containing 30 mg L–1 gentamycin and 60 mg L–1 kanamycin. DNA from the selected bacteria was transferred into uvh6-1 mutant plants using the floral dip method (Clough and Bent, 1998), and transformed kanamycin-resistant plants were selected from the next generation seed on media containing one-half-strength Murashige and Skoog basal salt mix (Gibco/Life Technologies, Grand Island, NY), 1× B5 vitamins (Bamborg et al., 1968), 1% (w/v) Suc, 0.8% (w/v) Bacto-agar, and 60 mg L–1 kanamycin. Primary transformants were tested, as described by Liu et al. (2001b) to identify UV-resistant, complemented transgenic lines. The UV-resistant phenotypes were further confirmed in T2 generation plants.
DNA Sequencing
To sequence the AtXPD/UVH6 cDNA, partial cDNA fragments were amplified from C10 wild-type and uvh6-1 mutant plants by RT-PCR, as described below, and each fragment was sequenced. A full-length cDNA sequence was assembled, and the uvh6-1 mutation was identified by comparison of wild-type and mutant sequences. The primers used for amplification were: XDU169, 5′-GCCAATTTCGAGATCTAGGTAGGAGGAA; XDU740, 5′-TAAGGGCGTTTGGTAAGAATC; H6U1375, 5′-TACGCCATCTCGGCACCCAGGCT; H6L2699, 5′-AAGTACTGAAGTAATCGGCGT; XDL1740, 5′-ACTTCTCATGTCAAATTTGGTGCT; XDL2382, 5′-CCCATTGTACCCGCCTTATCAT; H6U1534, 5′-AACCGGTGTTTGATCGTTTCCAGT; H6U4835, 5′-CTGGTTGGATACTTTCGCATCTGC; and H6L5357, 5′-AGGAGCCGACGAAGTATTTCTTGA. To sequence the T-DNA insertion within UVH6, DNA was isolated from a plant carrying the insertion, subjected to PCR amplification using primers UVH6-R and LB3 to amplify the region surrounding the insertion site (see Fig. 4) and then sequenced using the LB3 primer (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC).
Repair Assays
Assays were performed as described by Liu et al. (2000). In brief, this procedure involved irradiating seedlings that had been sprouted on agar plates, isolating seedling DNA at 0 and 24 h after irradiation, and quantifying the 6-4 photoproducts present in this DNA by a lesion-specific radioimmunoassay (Mitchell, 1996).
PCR Analysis of UVH6 Alleles
Genomic DNA was isolated from the leaves of seedlings grown in agar or from leaves of older plants grown in soil. To prepare seedlings, seeds were surface sterilized, plated on agar medium (containing 2.5 mm potassium phosphate [pH 5.5], 5 mm KNO3, 2 mm MgSO4, 2 mm Ca[NO3]2, 49 μm ethylenediamine-tetraacetic acid micronutrients, and 5 g L–1 Suc), incubated at 4°C, and then germinated in a growth chamber. DNA was isolated from seedlings as described by Klimyuk et al. (1993) and from older plants as described by Bell and Ecker (1994). PCR reactions were conducted using 42 cycles (30 s of melting at 94°C, 1 min of annealing at 65°C, and 2-min extension at 72°C) for experiments with Ga-3 progeny and 35 cycles (1 min of melting at 95°C, 1 min of annealing at 57°C, and 2.5-min extension at 72°C) for experiments with Ga-2 progeny. Primers used were: UVH6-F, 5′-ATCGTCACTGAATTCTCAGGC; UVH-6-R, 5′-CATGACGGCTGTATCTGCAAG; LB3, see above; and LB3-D, 5′-ATTTCATAACCAATCTCGATACAC. Reaction products were detected in 1.5% (w/v) agarose gels.
RT-PCR Analysis of Gene Expression
Total RNA was isolated from C10 wild-type plant tissues as described by Liu et al. (2001b). RT-PCR was conducted using the OneStep RT-PCR Kit (Qiagen Inc., Valencia, CA) with either 50 (UBQ10) or 200 (UVH6) ng of RNA. Either 26 (UBQ10) or 40 (UVH6) PCR cycles were used for amplification. Eight microliters of each 25-μL reaction was subjected to electrophoresis in 1% to 3% (w/v) agarose, and the identity of each RT-PCR product was confirmed by sequencing. Primers UBQ10U60 (5′-GACTCTCACCGGAAAGAAAT) and UBQ10L573 (5′-TTGTCTTGGATCTTGGCTTTCA) were used to amplify the UBQ10 gene. Primers XDU740 (5′-TAAGGGCGTTTGGTAAGAATC) and XDL2382 (5′-CCCATTGTACCCGCCTTATCAT) were used to amplify the UVH6 gene.
Bioinformatics
Multiple sequence alignments were obtained using the ClustalW program with default parameters (Higgins et al., 1996). The BLAST server (http://www.ncbi.nlm.nih.gov/) was used for database similarity searches (Altschul et al., 1997). Pair-wise alignments and alignment of the AtXPD cDNA and genomic sequences were obtained using LALIGN (Pearson and Miller, 1992), with minor adjustments to make the exon/intron junctions consistent with the consensus sequences for Arabidopsis splice junctions.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
The authors gratefully acknowledge the Torrey Mesa Research Institute (San Diego) for the 825 B05 seeds carrying the UVH6 T-DNA insertion.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.021808.
This work was supported by the National Science Foundation (grant no. MCB–9728125 to D.W.M.) and by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 99–3515100 to E.V.).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Balajee AS, Bohr VA (2000) Genomic heterogeneity of nucleotide excision repair. Gene 250: 15–30 [DOI] [PubMed] [Google Scholar]
- Bamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Batty DP, Wood RD (2000) Damage recognition in nucleotide excision repair of DNA. Gene 241: 193–204 [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bootsma D, Hoeijmakers JH (1993) DNA repair: engagement with transcription. Nature 363: 114–115 [DOI] [PubMed] [Google Scholar]
- Britt AB, Chen JJ, Wykoff D, Mitchell D (1993) A UV-sensitive mutant of Arabidopsis defective in the repair of pyrimidine-pyrimidinone(6-4) dimers. Science 261: 1571–1574 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa RMA, Lima WC, Vogel CIG, Berra CM, Luche DD, Medina-Silva R, Galhardo RS, Menck CFM, Oliveira VR (2001a) DNA repair-related genes in sugarcane expressed sequence tags (ESTs). Genet Mol Biol 24: 131–140 [Google Scholar]
- Costa RMA, Morgante PG, Berra CM, Nakabashi M, Bruneau D, Bouchez D, Sweder KS, Van Sluys MA, Menck CFM (2001b) The participation of AtXPB1, the XPB/RAD25 homologue gene from Arabidopsis thaliana, in DNA repair and plant development. Plant J 28: 385–395 [DOI] [PubMed] [Google Scholar]
- de Boer J, de Wit J, van Steeg H, Berg RJ, Morreau H, Visser P, Lehmann AR, Duran M, Hoeijmakers JH, Weeda G (1998) A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol Cell 1: 981–990 [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenics 21: 453–460 [DOI] [PubMed] [Google Scholar]
- Fidantsef AL, Mitchell DL, Britt AB (2000) The Arabidopsis UVH1 gene is a homolog of the yeast repair endonuclease RAD1. Plant Physiol 124: 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego F, Fleck O, Li A, Wyrzykowska J, Tinland B (2000) AtRAD1, a plant homologue of human and yeast nucleotide excision repair endo-nucleases, is involved in dark repair of UV damages and recombination. Plant J 21: 507–518 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM (1989) Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 26: 4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzder SN, Qiu H, Sommers CH, Sung P, Prakash L, Prakash S (1994) DNA repair gene RAD3 of S. cerevisiae is essential for transcription by RNA polymerase II. Nature 367: 91–94 [DOI] [PubMed] [Google Scholar]
- Harlow GR, Jenkins ME, Pittalwala TS, Mount DW (1994) Isolation of uvh1, an Arabidopsis mutant hypersensitive to ultraviolet light and ionizing radiation. Plant Cell 6: 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Somerville C (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol 266: 383–402 [DOI] [PubMed] [Google Scholar]
- Jenkins ME, Harlow GR, Liu Z, Shotwell MA, Ma J, Mount DW (1995) Radiation-sensitive mutants of Arabidopsis thaliana. Genetics 140: 725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins ME, Suzuki TC, Mount DW (1997) Evidence that heat and ultraviolet radiation activate a common stress-response program in plants that is altered in the uvh6 mutant of Arabidopsis thaliana. Plant Physiol 115: 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keriel A, Stary A, Sarasin A, Rochette-Egly C, Egly JM (2002) XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell 109: 125–135 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JD (1993) Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J 3: 493–494 [DOI] [PubMed] [Google Scholar]
- Lehmann AR (2001) The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev 15: 15–23 [DOI] [PubMed] [Google Scholar]
- Li A, Schuermann D, Gallego F, Kovalchuk I, Tinland B (2002) Repair of damaged DNA by Arabidopsis cell extract. Plant Cell 14: 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Akoulitchev S, Weber A, Ge H, Chuikov S, Libutti D, Wang XW, Conaway JW, Harris CC, Conaway RC et al. (2001a) Defective interplay of activators and repressors with TFIIH in xeroderma pigmentosum. Cell 104: 353–363 [DOI] [PubMed] [Google Scholar]
- Liu Z, Hall JD, Mount DW (2001b) Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J 26: 329–338 [DOI] [PubMed] [Google Scholar]
- Liu Z, Hossain GS, Islas-Osuna MA, Mitchell DL, Mount DW (2000) Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J 21: 519–528 [DOI] [PubMed] [Google Scholar]
- McGregor WG (1999) DNA repair, DNA replication, and UV mutagenesis. J Invest Dermatol Symp Proc 4: 1–5 [DOI] [PubMed] [Google Scholar]
- Mitchell DL (1996) Radioimmunoassay of DNA damaged by ultraviolet light. In GP Pfeifer, ed, Technologies for the Detection of DNA Damage and Mutations. Plenum, New York, pp 73–83
- Mitchell DL, Nairn RS (1989) The biology of the (6-4) photoproduct. Photochem Photobiol 49: 805–819 [DOI] [PubMed] [Google Scholar]
- Mozo T, Hooykaas PJ (1991) Electroporation of megaplasmids into Agrobacterium. Plant Mol Biol 16: 917–918 [DOI] [PubMed] [Google Scholar]
- Pearson WR, Miller W (1992) Dynamic programming algorithms for biological sequence comparison. Methods Enzymol 210: 575–601 [DOI] [PubMed] [Google Scholar]
- Pfeifer GP (1997) Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem Photobiol 65: 270–283 [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L (2000) Nucleotide excision repair in yeast. Mutat Res 451: 13–24 [DOI] [PubMed] [Google Scholar]
- Ribeiro DT, Machado CR, Costa RM, Praekelt UM, Van Sluys MA, Menck CF (1998) Cloning of a cDNA from Arabidopsis thaliana homologous to the human XPB gene. Gene 208: 207–213 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schultz TF, Quatrano RS (1997) Characterization and expression of a rice RAD23 gene. Plant Mol Biol 34: 557–562 [DOI] [PubMed] [Google Scholar]
- Sturm A, Lienhard S (1998) Two isoforms of plant RAD23 complement a UV-sensitive rad23 mutant in yeast. Plant J 13: 815–821 [DOI] [PubMed] [Google Scholar]
- Sung P, Higgins D, Prakash L, Prakash S (1988) Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J 7: 3263–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprakasit V, Gibbons RJ, Broughton BC, Tolmie JL, Brown D, Lunt P, Winter RM, Marinoni S, Stefanini M, Brueton L et al. (2001) Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum Mol Genet 10: 2797–2802 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G (2000) TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem 275: 4258–4266 [DOI] [PubMed] [Google Scholar]
- Xu H, Swoboda I, Bhalla PL, Sijbers AM, Zhao C, Ong EK, Hoeijmakers JH, Singh MB (1998) Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J 13: 823–829 [DOI] [PubMed] [Google Scholar]