Abstract
Transcript levels of the Arabidopsis R2R3-AtMYB102 transcription factor gene, previously named AtM4, are rapidly induced by osmotic stress or abscisic acid (ABA) treatment. Reporter gene expression studies revealed that in addition, wounding is required for full induction of the gene. Histochemical analysis showed a local β-glucuronidase induction around the wounding site, especially in veins. In ABA-treated plants, wounding-induced β-glucuronidase activity could be mimicked by the wound signaling compound methyl jasmonate. In silico studies of the AtMYB102 promoter sequence and its close homolog AtMYB74 demonstrated several conserved putative stress regulatory elements such as an ABA-responsive element, its coupling element 1 (CE1), and a W box. Interestingly, further studies showed that the 5′-untranslated region is essential for the osmotic stress and wounding induced expression of the AtMYB102 gene. This 5′-untranslated region contains putative conserved regulatory elements such as a second W box and an overlapping MYB-binding element. These studies suggest that AtMYB102 expression depends on and integrates signals derived from both wounding and osmotic stress.
Abscisic acid (ABA) is the major plant hormone in water stress signaling. ABA regulates plant water balance and osmotic stress tolerance. The role of ABA in the osmotic stress responses has been studied extensively, and the availability of ABA-deficient and -insensitive mutants especially has been most helpful in these studies (Koornneef et al., 1998). Several of these mutants show severe wilting even upon a mild water stress treatments, supporting the role of ABA under stress conditions. ABA levels increase in wilting leaves due to the de novo synthesis of the carotenoid cleavage enzyme 9-cis-epoxycarotenoid dioxygenase (NCED), VP14 in maize (Zea mays). This enzyme catalyzes what appears to be the rate-limiting step in ABA biosynthesis (Tan et al., 1997). A clear correlation in Phaseolus vulgaris was found between NCED (PvNCED1) mRNA expression, NCED protein levels, and ABA levels in dehydrated leaves and roots, indicating the important role of NCED in stress ABA biosynthesis. Moreover, overexpression of the Arabidopsis AtNCED3 results in the accumulation of ABA (Iuchi et al., 2001).
Two separate groups of stress-responsive genes are induced during osmotic stress: the “early response genes” and the “delayed-response genes” (Kiyosue et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994). The early response genes can be induced within minutes. Expression is often transient and is independent of protein synthesis. These genes typically encode transcription factors that activate downstream delayed-response genes. The latter constitute the vast majority of the stress-responsive genes, which are activated by stress more slowly but often show sustained expression. In osmotically stressed Arabidopsis plants, ABA accumulation starts after approximately 2 h after the treatment (Kiyosue et al., 1994). Several genes respond already within this time period to dehydration stress via a rapid ABA-independent signaling pathway (Yamaguchi-Shinozaki and Shinozaki, 1994; Gosti et al., 1995; Hong et al., 1997). The RPK1 gene of Arabidopsis is induced by dehydration stress and by ABA treatment. The dehydration-induced expression is not impaired in the aba1 ABA-deficient mutant or in the ABA-insensitive mutants abi1, abi2, and abi3, suggesting that this ABA-induced gene also is regulated in an ABA-independent way (Hong et al., 1997). Expression of the stress-responsive RD29A gene is rapidly induced in an ABA-independent way followed by a strong ABA-dependent induction phase. These observations can be explained by separate cis-acting elements in the promoter, such as a dehydration-responsive element (DRE) and an ABA-responsive element (ABRE; Yamaguchi-Shinozaki and Shinozaki, 1993, 1994). The DRE element was necessary and sufficient for the first rapid response to osmotic stress induction without the involvement of ABA through the DREB2A and related transcription factors. Full activation of RD29A transcription depends upon the synergy between DRE and ABRE elements in its promoter. Transcription factors with ERF/AP2 domains have been shown to bind to DRE elements, whereas several ABRE-interacting proteins are classified as basic/Leu zipper transcription factors (Carles et al., 2002; Sakuma et al., 2002).
Wounding is another stress factor that often is closely related to osmotic stress. Tissue damage is usually associated with decompartmentalization, release of cellular contents, and a loss of water. Such damage induces local osmotic stress responses similar to those occurring in water-stressed intact plants (Reymond et al., 2000). The central role of jasmonic acid (JA) in plant responses to wounding is well established, but other signaling molecules have been proposed to play important roles in wound signaling as well. These include ABA, ethylene, oligosaccharides, and the oligopeptide systemin. Studies on the wound signal transduction cascades showed involvement of Ca2+ ions (Knight et al., 1993), fatty acids (Ryu and Wang, 1998), systemin (Ryan, 2000), hydrogen peroxide (Orozco-Cardenas and Ryan, 1999), and poly-GalUA in triggering wound signaling. Many of the wound-responsive genes are induced through jasmonate-dependent signal transduction cascades, but jasmonate-independent cascades also exist. For example, in Arabidopsis, oligogalacturonides activate gene expression via a JA-independent pathway (Titarenko et al., 1997).
In several Solanaceous plants, both JA and ABA can activate the expression of wound-responsive genes through the systemin-activated signaling system (Hildmann et al., 1992; Peña-Cortés et al., 1995). This system seems to be absent in species such as barley (Hordeum vulgare), potato (Solanum tuberosum), and Arabidopsis (Leon et al., 2001). In tomato (Lycopersicon esculentum), ABA is necessary for the wound-induced expression of proteinase inhibitor genes (Carrera and Prat, 1998), but ABA does not appear to be a primary signal in wound signaling (Birkenmeier and Ryan, 1998). In Arabidopsis, ethylene acts as a cross talk regulator between the JA-dependent and -independent oligosaccharide-mediated wound signaling pathways (Rojo et al., 1999). The oligosaccharide-mediated repression of the JA-dependent signaling pathway is exerted through the local production and perception of ethylene in damaged tissue.
It is of interest to unravel the regulatory connections between wound-activated signaling pathways and signal transduction pathways triggered by other stress factors such as osmotic stress. Here, the expression analysis of a putative Arabidopsis transcription factor of the R2R3 MYB class is presented. This AtMYB102 gene is responsive to osmotic stress and wounding. Dehydration, osmotic or salinity stress, and ABA application enhance AtMYB102 transcript levels. Also, methyl jasmonate (MeJa) enhances expression synergistically in combination with ABA, whereas MeJa alone has a limited effect. It is proposed that AtMYB102 is a regulatory component, which integrates osmotic stress and wounding signaling pathways in Arabidopsis.
RESULTS
AtMYB102 mRNA Levels Respond to Osmotic Stress and ABA
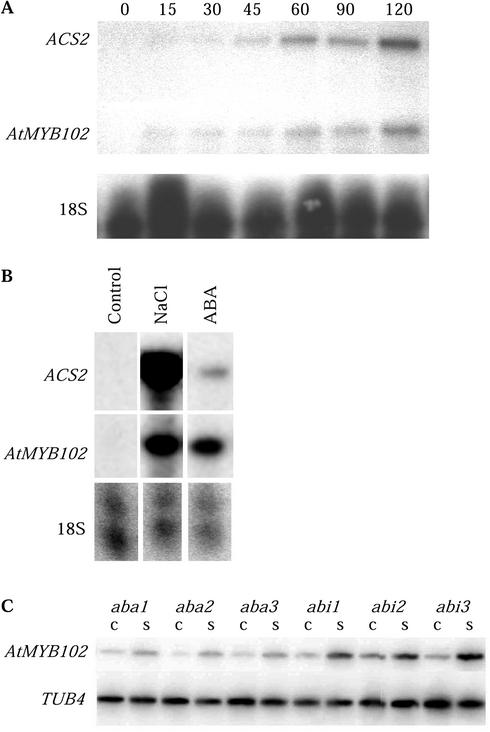
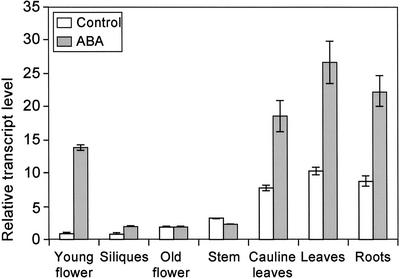
Previously, it was reported that Arabidopsis ecotype Columbia-0 (Col-0) AtMYB102 mRNA levels are induced by dehydration stress in leaves (Kranz et al., 1998). This response of AtMYB102 was further investigated in a time course dehydration experiment. Rosettes were detached from the roots at different time points, and the RNA transcript levels were determined by RNAse protection assays. AtMYB102 transcript levels in leaves increase within 30 min after detachment (Fig. 1A). The stress-responsive ACS2 gene was used as an osmotic stress induction control. ACS2 encodes an enzyme involved in the conversion of 1-aminocyclopropane-1-carboxylic acid (ACC) to ethylene. Salinity stress and ABA effectively induce AtMYB102 mRNA levels (Fig. 1B) as do mannitol- or polyethylene glycol-induced osmotic stress (data not shown). In mutant lines deficient in ABA biosynthesis (aba1-1, aba2-1, and aba3-1) or ABA signal transduction (abi1-1, abi2-1, and abi3-), AtMYB102 mRNA levels are induced (Fig. 1C). Thus, AtMYB102 induction probably does not depend on ABA or ABA-induced signaling per se. AtMYB102 expression and ABA-dependent induction were studied in whole plants. After treatment, plants were dissected and AtMYB102 transcript levels determined by real-time PCR. The gene is prominently expressed in young flowers, leaves, cauline leaves, and roots and is ABA responsive in these tissues (Fig. 2).
Figure 1.
A, Time course dehydration stress experiment. Rosettes were detached and left on a filter paper in a climate room at 70% humidity. RNA was extracted at the time points indicated (in minutes), and RNA protection analyses were performed. The ASC2 gene was used as a control for dehydration stress conditions. B, RNAse protection analyses of the AtMYB102 and the ACS2 control transcripts. Rosettes stage plants were subjected to salinity stress by soil saturation with 200 mm NaCl or were sprayed with 100 μm ABA. The plants were left in a climate room for 4 h before harvest. C, Reverse transcriptase (RT)-PCR analyses of AtMYB102 gene expression. ABA mutant lines were exposed to salinity stress. S, Salinity stress (2 h, 200 mm NaCl soil saturation); C, control.
Figure 2.
Real-time PCR analyses of the AtMYB102 transcript levels in several plant organs. The plants were sprayed with 100 μm ABA and left for 3 h in a climate room before harvest. Adult plants were dissected in the organs indicated.
Osmotic Stress and Wounding Are Both Required for AtMYB102 Expression
The Arabidopsis Col-0 ecotype was transformed with a construct containing a 2-kb AtMYB102 promoter fragment including the mRNA 5′ leader sequence and the first three amino acids of the mature protein fused in frame to the β-glucuronidase (GUS) reporter gene (see “Materials and Methods”). Transformants were generated using the Agrobacterium tumefaciens-mediated floral transformation method and homozygous lines obtained. Surprisingly, neither salt stress nor ABA enhanced GUS reporter enzyme activity in leaf tissues when analyzed histochemically (Fig. 3) or enzymatically (Fig. 4). This was observed in all independent transgenic Col-0 lines tested. Such treatments efficiently induce endogenous AtMYB102 transcript levels (Figs. 1 and 2). These observations were confirmed in all independent transgenic lines tested in the C24 ecotype. These C24 lines were generated with the same construct as the Col-0 ecotype except that a 1-kb promoter element was used. Also, in this ecotype, no induction of GUS activity as determined by histochemistry was found when treated with ABA or salinity stress, whereas in C24 AtMYB102, mRNA levels are induced also (data not shown). With the more sensitive enzymatic GUS assay, an approximately 2-fold induction was observed (Fig. 3B). Interestingly, GUS activity was highly induced in both ecotypes at wounding sites (Fig. 3, A and B; data not shown). This effect of wounding was tested via incisions in leaves and exposure of plants to salt stress by saturating the soil with 200 mm NaCl. Histochemical analysis revealed that GUS activity was prominently induced specifically at the wounding sites in stressed and wounded plants. GUS activity is especially prominent in the vicinity of damaged veins, but a diffuse activity in the adjoining parenchymal tissues was also observed. Staining was absent in the epidermal layer (Fig. 3A).
Figure 3.
A, Histochemical GUS analyses in Col-0 line 2.6 harboring the 2-kb promoter fragment from AtMYB102 translationally fused to the GUS reporter gene. The soil was saturated with 200 mm NaCl, and the plants were left for 4 h in a climate room. Wounding was applied by incision. B, GUS activity assays (line 2.6). The rosettes were exposed for 4 h to 200 mm NaCl via soil saturation. Wounding was applied through cutting of leaf tips. Three leaf slices around the wounding site of each plant were harvested (n = 5).
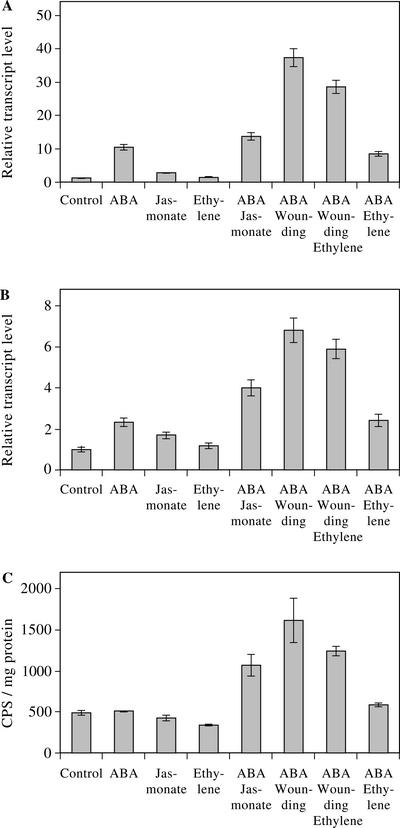
Figure 4.
Real-time PCR analyses of AtMYB102 (A) and GUS fusion transcript levels (B). Ten rosettes of 3-week-old plants were used per treatment and sprayed with 100 μm ABA, 10 μm MeJa, or 10 μm ACC (ethylene precursor). Wounding was applied over the leaf with forceps. Plants were incubated for 4 h in a climate room before processing. C, Representation of the GUS activity in the same samples.
The combination of osmotic stress and wounding and its effect on mRNA levels and GUS enzyme activity was further investigated in the Col-0 ecotype. Pots with 3-week-old transgenic plants were treated with hormones known to be important in osmotic stress and wound signaling. Endogenous AtMYB102 and GUS transcript levels were determined by realtime PCR with Actin2 as the endogenous reference. ABA treatment induces transcript levels as expected. However, GUS mRNA level seems less responsive compared with the endogenous transcript (2- over 10-fold, respectively), as was found for all other treatments (Fig. 4, A and B). Possibly, GUS transcript stability differs from that of the endogenous AtMYB102 transcripts. Alternatively, this might be due to a less efficient PCR with the GUS primers. However, the expression patterns of the GUS and the AtMYB102 transcripts are nearly identical over the treatments as was observed in a number of experiments with similar treatments (data not shown).
The wound signaling hormone jasmonate enhances AtMYB102 transcript levels approximately 2-fold compared with a 10-fold induction by ABA (Fig. 4A). Combined application of these compounds induced the transcript abundance 13-fold. Interestingly, GUS activity was not altered compared with the control treatment for both separate treatments, but the activity increased when both ABA and jasmonate were applied (Fig. 4C). Wounding in combination with ABA further enhances GUS enzymatic activity (Fig. 4C). Wounding disrupts tissues and is accompanied by osmotic stress. This combination boosts transcript levels and GUS activity. These results suggest that AtMYB102 functions in a wound response pathway that may involve jasmonate and that is integrated with an osmotic stress and ABA response pathway.
Ethylene is a plant hormone that is induced locally upon wounding. Therefore, the effect of this hormone on the wounding response was tested. ACC, a precursor of ethylene, was applied to 3-week-old wounded and ABA-treated plants. Ethylene in combination with wounding has a small but consistent inhibitory effect on transcript levels and GUS activity (Fig. 4). No effect was found in combination with ABA alone. Ethylene has been proposed as an inhibitory compound of the local jasmonate-dependent wound response pathway (Rojo et al., 1999), and our results suggest a similar but small inhibitory effect on AtMYB102 expression at the wound site.
The 5′-Untranslated Region (UTR) Is Essential for Gene Expression
Experiments with the GUS reporter construct revealed that ABA induces GUS transcript levels, but GUS activity is not enhanced (Fig. 4, B and C). This observation suggests a posttranscriptional regulatory mechanism of the AtMYB102 gene. The AtMYB102 promoter-GUS construct includes the 5′-UTR of 108 nucleotides (see “Materials and Methods”). Such 5′-UTR regions are often involved in translation regulation, and this possibility was investigated using an AtMYB102 promoter-GUS construct lacking the 108-bp 5′-UTR. Six independent hygromycin-resistant Col-0 lines showing different expression levels were analyzed for GUS mRNA levels and GUS enzymatic activity. Surprisingly, the endogenous AtMYB102 transcript levels and the GUS transcript levels were differentially regulated by salt stress (Fig. 5A). This indicates that the 5′-UTR is essential for the stress response. The GUS transcript levels basically are in agreement with GUS enzymatic activity as observed in the independent lines (Fig. 5B). Moreover, in the lines transformed with the construct lacking the 5′-UTR, wounding does not enhance GUS activity as it does in lines with the intact constructs.
Figure 5.
Analysis 5′-UTR-deleted transgenic GUS reporter lines. A, RT-PCR analyses of GUS transcript levels of six independent lines. C, Control; S, salinity stress (4 h, 200 mm NaCl soil saturation). B, Four lines varying in GUS activity were subjected to 4 h of salt treatment by soil saturation with 200 mm NaCl. In a parallel experiment, wounding was applied to salt-stressed plants by cutting (three leaves of five plants per line). The leaf material at the vicinity of the wounding site was harvested and pooled per line. The error bars are based on two independent experiments.
DISCUSSION
AtMYB102 Is Responsive to Osmotic Stress and ABA
The AtMYB102 was identified previously as a light-regulated gene (AtM4) expressed in seedlings (Quaedvlieg et al., 1996). Further analysis revealed that AtMYB102 is responsive to dehydration stress as examined by the reverse northern technique (Kranz et al., 1998). A time course dehydration experiment showed a rapid increase of AtMYB102 mRNA levels in leaves within 30 min after stress application (Fig. 1A). Experiments with salinity stress (Fig. 1B, lane 2) and osmotic stress induced by mannitol or polyethylene glycol (data not shown) showed the same increase in AtMYB102 transcript levels. Similar to many other dehydration-responsive genes, AtMYB102 is ABA responsive but probably does not depend on ABA or ABA signaling. This is suggested by the salinity response of AtMYB102 in several aba and abi mutants (Fig. 1, B, lane 3, and C). Moreover, the dehydration stress response occurs within 30 min, whereas significant stress ABA levels accumulate after a 2-h lag period (Kiyosue et al., 1994; Zhu et al., 1997).
AtMYB102 Is Wound Responsive
Analyzing GUS reporter gene constructs in the Col-0 and C24 ecotypes provided additional information on regulation and location of AtMYB102 gene expression. Some enhancement of GUS activity was observed in plants of both ecotypes exposed to salinity stress or ABA. This enhancement was only observed with sensitive enzymatic assays but not in histochemical experiments. Surprisingly, GUS activity was strongly induced by wounding in combination with either osmotic stress or ABA treatment (Figs. 3 and 4). Thus, wounding is important for full expression of the GUS reporter gene. Jasmonate is a well-known hormone involved in wounding responses. Applying MeJa in combination with ABA to 3-week-old Col-0 transgenic plants tested its possible involvement. ABA, and to a lesser extent MeJa, enhance AtMYB102 transcript levels but fail to induce GUS activity (Fig. 4), suggesting a posttranscriptional regulatory mechanism. Only the combined application of MeJa and ABA significantly induced GUS activity. These results indicate that jasmonate signaling is required for expression of AtMYB102, but this needs further investigation. Wounding is more effective in combination with ABA than with jasmonate in inducing transcript levels and GUS activity (Fig. 4). Perhaps wounding is more effective than jasmonate due to the locally induced osmotic stress at the wounding site.
Ethylene might be another factor involved in the observed expression because it is synthesized locally after wounding. Studies on jasmonate-responsive genes like JR1 and JR2 suggest that ethylene inhibits the local jasmonate-dependent signaling pathway (Rojo et al., 1999). Plants treated with ABA and wounding showed a small inhibitory effect of ethylene on both transcript levels and GUS activity. The inhibitory effect of ethylene is restricted to the wound response because the ABA response was not affected (Fig. 4). Several independent experiments confirmed this small inhibitory effect of ethylene (data not shown).
AtMYB102 Integrates Osmotic Stress and Wound Signaling
Full expression of AtMYB102 requires both osmotic stress- and wounding-induced signaling pathways. Most likely, in Arabidopsis the AtMYB102 protein is produced after local wounding with its associated osmotic stress. This allows as of yet unknown target genes to be regulated accordingly in a wounding-dependent manner. Prolonged osmotic stress induces ABA accumulation and a further enhancement of the response in time. In tomato, ABA and jasmonate can activate proteinase inhibitor genes through a common signal transduction pathway (Hildmann et al., 1992; Peña-Cortés et al., 1995). Moreover, the presence of an early common wounding and ABA-mediated signaling pathway was suggested. The dominant Arabidopsis abi1-1 allele reduced the wound-induced expression of the proteinase inhibitor 2 and LAP transcripts (Carrera and Prat, 1998), but ABA does not appear to be a primary signal in wound signaling (Birkenmeier and Ryan, 1998). In these studies, it was observed that the increased ABA levels due to osmotic stress do not induce wound-responsive genes. Similarly, wounding does not enhance osmotic stress-responsive genes except when wounding causes local osmotic stress (Hildmann et al., 1992; Peña-Cortés et al., 1995; Reymond et al., 2000). The Arabidopsis AtMYB102 gene might be a candidate signaling component for such a rapid response to wounding.
The 5′-UTR sequence could be important for a posttranscriptional regulatory mechanism as suggested by experiments where ABA induces GUS mRNA levels but not enzymatic activity in the promoter-GUS transgenic lines. This notion was tested using a leaderless AtMYB102 promoter-GUS construct, but, surprisingly, these experiments showed the presence of essential stress-responsive elements in the 5′-UTR. Steady-state GUS transcript levels in the lines lacking the 5′-UTR remain unchanged during exposure to salt stress alone or combined with wounding (Fig. 5). The GUS transcripts levels were representative for the observed GUS activity, and wounding did not enhance GUS activity in these lines (Fig. 5).
In Silico Promoter Analyses Reveal Conserved Stress-Responsive Elements
The closest homolog of AtMYB102 in the genome of Arabidopsis is AtMYB74. This AtMYB74 revealed considerable sequence conservation with AtMYB102 and has a similar expression pattern (Kranz et al., 1998). Most likely, these genes arose from a single ancestor. The promoter sequences of both genes show conserved cis-elements with homology to previously described sequence motifs (Fig. 6). These include elements like ABRE and its coupling element (CE1), which are spaced as found in other ABA- and dehydration-responsive promoters (Niu et al., 2002). These elements are known to be involved in abiotic stress responses and interact with putative transacting factors. The ABRE can interact with basic domain/Leu zipper-type transcription factors such as ABI5. The CE1 element might interact with AP2-like transcription factors such as ABI4 (Niu et al., 2002). Another conserved element, located 8 bp downstream of the CE1 element, is a W box (Maeo et al., 2001). The W-box is a target of WRKY zinc finger-type transcription factors that are associated with processes like wounding, pathogen defense, and senescence (Hara et al., 2000; Robatzek and Somssich, 2002). The involvement of AtMYB102 in pathogen defense was tested using compatible strains of Peronospora parasitica and Pseudomonas syringae. Neither pathogen was able to induce GUS activity in the AtMYB102 promoter-GUS lines after inoculation (data not shown). In Arabidopsis, wounding- or jasmonate-responsive WRKY proteins might be involved in transcriptional regulation of AtMYB102, possibly interacting with other trans-acting factors mentioned above. The conserved putative promoter elements, like ABRE-CE1 and W box and their interacting factors, likely determine transcription activity of AtMYB102.
Figure 6.
Promoter alignment using the ClustalW program of the homologous AtMYB102 and AtMYB74 genes. The AtMYB102 coding sequence starts at position +108, and the transcript starts at position 0. Transcript initiation site was determined by 5′-RACE (see “Materials and Methods”). Putative conserved boxes are underlined. General gene regulatory elements like the TATA box and CCAAT box are present at positions –30 and –92, respectively. The latter is located between the ABRE motif (GACACGTA) and its Coupling Element (CE1) with the consensus CACCG (Niu et al., 2002). A W box is present with the consensus sequence TTGACT, just 8 bp downstream of the ABRE-CE1 elements (Eulgem et al., 1999). A second W-box consensus sequence is found in the 5′-UTR and overlaps with a MYB-binding consensus sequence, CAGTT (Urao et al., 1993). A 5′-UTR pyrimidine-rich track is present just after the transcription initiation site (Sickinger and Schweizer, 1999).
The 5′-UTR is essential for stress-induced expression of AtMYB102. This region contains overlapping putative MYB and W-box elements that are conserved in the 5′-UTRs of AtMYB102 and AtMYB74 (Fig. 6). Another conserved element in the 5′-UTR is a pyrimidine-rich tract (Fig. 6). The RNA-binding protein, polypyrimidine tract-binding protein, is known to interact with such tracts and regulates the utilization of premessenger and mRNA in association with other proteins involved in splicing and translation initiation (Shav-Tal and Zipori, 2002). Such a protein might affect translation efficiency of the AtMYB102 mRNA.
In conclusion, osmotic stress in combination with wounding is needed for full expression of AtMYB102. Thus, AtMYB102 seems to integrate signals from these separate signal transduction pathways. It will be interesting to identify the expression mechanisms involved and the downstream processes controlled by AtMYB102. Moreover, overexpression of the gene in combination with phenotypic and metabolic analyses are underway and hopefully will shed light on its function.
MATERIALS AND METHODS
Plant Material
Arabidopsis ecotype Col-0 seeds were sown on soil in 5-cm pots, moistened, and placed for 3 to 5 d in a cold room at 4°C in the dark to synchronize germination. The pots were transferred to a climate room to allow growth to rosette stage with an 8-h photoperiod at 22°C. The plants were sprayed with solutions containing 100 μm ABA (mixed isomers, Sigma-Aldrich Chemie, Steinheim, Germany), 10 μm MeJa (Duchefa, Haarlem, The Netherlands), or 10 μm ACC (Sigma-Aldrich Chemie) and water was used as control. All solutions contained 0.01% (v/v) Silwet L77 (Van Meeuwen Chemicals, Weesp, The Netherlands). About 5 mL of the solution was sprayed on five rosette stage plants. The plants were returned to the climate room at 22°C, 70% relative humidity until harvesting. For salinity stress, the soil was saturated with 200 mm NaCl solution, and about 1 L of the solution was poured on the soil surface of 10 plants and allowed to drain. Wounding of the plants was performed using scalpel or forceps.
RNA Analyses
RNAse Protection and RT-PCR Analyses
Total RNA was prepared from leaves of rosette stage plants using the PUREscript RNA isolation Kit (Gentra Systems, Minneapolis) as described by the manufacturer. RNAse protection analyses were performed on 10 μg of total RNA using the RPAIII Kit (Ambion, Austin, TX) as described by the manufacturer. The 18S endogenous control probe and the RNA ladder were obtained from Ambion and used according to the protocols provided. The AtMYB102 probe was prepared as described before (Quaedvlieg et al., 1996). For preparing an ACS2 probe, a PCR fragment was generated using primers F1213ACS2 (5′-CGAGATCTACGCCGCACAGTC-3′) and R1719ACS2 (5′-CAACGAAGGAAGAGCCAGGAGA-3′), cloned in a pGEM-T vector (Promega, Madison, WI), and linearized with XhoI. For generating the RNA probe, the Riboprobe Combination system T7/T6 (Promega) was used according to the protocol provided.
The first strand cDNA for RT-PCR analysis was synthesized with oligo(dT) primers on 1 to 2.5 μg of total RNA using SUPERSCRIPT II (Invitrogen, Carlsbad, CA) as described by the manufacturer. All RNA was pretreated with DNAse I using the DNA free kit (Ambion) according to the protocol. Amplification reactions contained 2 μL of the 2-fold diluted cDNA pool, 1× polymerase buffer (Roche Diagnostics), 2.5 mm of each dNTP, 10 pmol of each primer, and 2.5 units of Taq DNA polymerase. PCR was performed with denaturation at 94°C for 3 min, followed by 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. A final extension at 72°C for 10 min was performed to complete the reaction.
The primers used were: AtMYB102, FS102cDNA 5′-TAAACAATCAATGGCAAGGTCAC-3′ and 5RACE102C 5′-CTGTGAGTCACTGGATCAATCC-3′; and Tubuline 4, F-B-TUBULIN4 5′-GCTTACGAATCCGAGGGTGCC-3′ and R-B-TUBULIN4 5′-GTCCAGTGTCTGTGATATTGCACC-3′.
PCR products (5 μL) were fractionated on a 1% (w/v) agarose gel, transferred onto Hybond N+ membranes (Amersham, Buckinghamshire, UK), and hybridized with 32P-labeled fragments generated with the PCR primers specified. Hybridization signals were visualized using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Real-Time PCR
RNA was extracted from 100 mg of plant material with the RNeasy Plant Mini Kit (Qiagen Sciences, Germantown, MD), subsequently pretreated with DNAse I using the DNA free kit (Ambion), and cDNA was synthesized with Moloney murine leukemia virus RNase H– (Promega) using 2 μg of total RNA. The cDNA was diluted to 100 μL with Tris-EDTA buffer, and 5 μL was used in a 25-μL PCR reaction. The mixture was set up with 12.5 μL of Taq-Man 2x Universal PCR Master Mix (Applied Biosystems, Warrington, UK), 5 μL of cDNA, 2.5 μL of probe (250 nm final), and 2.5 μL of each primer (900 nm final). PCR was performed with denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing, and extension at 60°C for 1 min. The comparative treshold cycle (Ct) method was used to determine relative RNA levels (User Bulletin no. 2, Applied Biosystems). Actin2 is used as the internal reference, and expression levels are relative to the control treatment.
The primer probe combination of AtMYB102 was: probe, FAM 5′-AGAACTCCAATCTTCCATGCCACCATTCC-′3 Tamra; forward primer, 5′-AACCAATATTTCGAGAACACGATTACT-′3; and reverse primer, 5′-TGTTGTTAAACTGACGAGCTTCATT-′3. The primer probe combination of the Promoter::GUS fusion construct was: probe, FAM 5′-CGGCCGCCTGCAGCAGCCAA-′3 Tamra; forward primer, 5′-CAATGGCAAGGTCACAATCACT-′3; and reverse primer, 5′-TAAACTAGTCAGATCTACAGCGCTAAG-′3. The primer probe combination of Actin2 was: probe, FAM 5′-AAGTCTTGTTCCAGCCCTCGTTTGTGG-′3 Tamra; forward primer, 5′-GCTGAGAGATTCAGATGCCCA-′3; and reverse primer, 5′-GTGGATTCCAGCAGCTTCCAT-′3.
Determination of the Transcription Initiation Site of AtMYB102 Using 5′-RACE
Total RNA was prepared as mentioned for RT-PCR analyses from leaves of rosette stage plants exposed to dehydration stress for 4 h and reverse transcribed as above using 2.5 μg of total RNA and 1.5 pmol gene-specific primer 5′RACE102A (5′-GCTGATGACGACGATTAGTATCC-3′) in a 12-μL reaction mix. The reaction was stopped by heating at 70°C for 15 min and treated with 1 μL of RNAse H (Promega) at 37°C for 30 min. The gene-specific cDNA was purified over a spin column Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and eluted in 19 μL of 10 mm Tris buffer (pH 8.0). The cDNA was C tailed by adding 2.5 μL of One-Phor-All buffer (Amersham-Pharmacia Biotech, Uppsala) and 2.5 μL of 2 mm dCTP. The mixture was heated at 94°C for 3 min, transferred to ice for 5 min, 25 units of terminal deoxynucleotidyl transferase (Amersham-Pharmacia Biotech) was added, and C tailing was performed at 37°C for 10 min and terminated at 70°C for 5 min. PCR was performed on 5 μL of the C-tailed gene-specific cDNA using 20 pmol 5RACE-AP (5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and 20 pmol 5RACE102B (5′-GACATGTTCATGTGATGTGAAG-3′) under RT-PCR conditions as described before with a melting temperature of 55°C and for 35 cycles. The PCR products were separated on a 1% (w/v) agarose gel along a size marker. Visible bands were isolated from the gel using Qiaex II gel extraction kit (Qiagen) and eluted in 20 μL of 10 mm Tris buffer (pH 8.0). A second identical PCR was performed on 5 μL of 1:250 (v/v) diluted gel-isolated fragments with 20 pmol 5RACE-AUAP (5′-GGC-CACGCGTCGACTAGTAC-3′) and 20 pmol 5RACE102C (5′-CTGT-GAGTCACTGGATCAATCC-3′). The obtained fragment was cloned in a pGEM-T vector (Promega) according to the protocol and sequenced. The transcription initiation site could be located 108 bp upstream of the ATG codon.
Histochemistry, Microscopy, and Quantitative GUS Assay
For GUS histochemistry, plant material was stained overnight at 37°C in a 1 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide (Biosynth AG, Staad, Switzerland) solution in 50 mm sodium phosphate (pH 7.0) buffer, 0.1% (v/v) Triton X-100, 10 mm EDTA, 0.5 mm K3Fe(CN)6, and 0.5 mm K4Fe(CN)6. The plant material was cleared in 70% and 95% (v/v) ethanol.
Plant material for microscopy was infiltrated and embedded in a Technovit 7100 (Kulzer, Heraus, Germany) as instructed by the manufacturer. Sections of 4 μm were made on a Reichert-Jung 1140 rotary microtome (Hanau, Germany) carrying a disposable Adams steel knife. Sections were stained with 0.1% (w/v) Ruthenium red (Sigma-Aldrich Chemie) in distilled water for 2 to 5 min at room temperature and fixed under a glass slide cover.
Relative GUS-activity was measured using the GUS-Light, Chemiluminescent Reporter Gene Assay (Tropix, Inc., Bedford, MA) as described by the manufacturer. Proteins were extracted from samples containing at most 3 cm2 leaf tissue in a buffer containing 50 mm sodium phosphate (pH 7.0), 10 mm EDTA, 0.1% (w/v) sodium lauryl sarcosine, 0.1% (v/v) Triton X-100, and 10 mm β-mercaptoethanol. Equal amounts of protein as determined by the Bradford assay were used in the reactions.
GUS Reporter Constructs
For the AtMYB102 promoter-GUS construct in C24, the plasmid pM4.1 harboring the promoter was digested with SalI and BstEII and made blunt with Klenow (Amersham-Pharmacia Biotech). The fragment was cloned in the binary vector pBi101.2 using the SalI/SmaI sites, obtaining a translational fusion including the first three gene-specific amino acids. The construct was transformed to C24 ecotype via a root transformation procedure mediated by Agrobacterium tumefaciens (Valvekens et al., 1988). Transformants were selected on Murashige and Skoog medium containing 50 mg L–1 kanamycin.
Using a PCR-based cloning strategy, a 2-kb promoter fragment was fused to the GUS reporter gene in pCAMBIA3381Xa. The primers F102LDR (5′-AATGCGCTCCCCTTTCTC-3′) and R102LDR (5′-GTGACCTTGCCATTGATTGTTTA-3′) were used to amplify the 2-kb promoter region of the AtMYB102 gene on Col-0 ecotype DNA. The proofreading DNA polymerase Pfu was used with Taq DNA polymerase in a 1:80 (v/v) unit ratio. The fragment was cloned in a pGEM-T vector (Promega) and sequenced. A construct with the 5′-end of the AtMYB102 promoter at the vector T7 promoter site was digested with SphI and made blunt with T4 DNA polymerase (Amersham-Pharmacia Biotech) and with PstI. The fragment was cloned into the SmaI and PstI sites of pCAMBIA3381Xa. The translational fusion with the GUS coding region includes three amino acids of the AtMYB102 protein.
The constructs were transformed into Col-0 ecotype by an A. tumefaciens (C58C1)-mediated floral dip method (Clough and Bent, 1998). The putative Basta-resistant transformants (pCAMBIA3381Xa) were sown on pot soil and sprayed once with 75 mg L–1 DL-phosphinothricin after germination (Basta, Duchefa). Hygromycin-containing Murashige and Skoog plates (30 mg L–1) were used for selection of putative transformants containing the binary vector pCAMBIA1381.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019273.
This work was supported by the European Commission (REGIA EU contract no. QLG2–1999–00876).
References
- Birkenmeier GF, Ryan CA (1998) Wound signaling in tomato plants. Evidence that aba is not a primary signal for defense gene activation. Plant Physiol 117: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Carrera E, Prat S (1998) Expression of the Arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J 15: 765–771 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J 18: 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Bertauche N, Vartanian N, Giraudat J (1995) Abscisic acid-dependent and -independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol Gen Genet 246: 10–18 [DOI] [PubMed] [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Hildmann T, Ebneth M, Peña-Cortés H, Sanchez-Serrano JJ, Willmitzer L, Prat S (1992) General roles of abscisic acid and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4: 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Jon JH, Kwak JM, Nam HG (1997) Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol 113: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K (1994) Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol 25: 791–798 [DOI] [PubMed] [Google Scholar]
- Knight MR, Read ND, Campbell AK, Trewavas AJ (1993) Imaging calcium dynamics in living plants using semisynthetic recombinant aequorins. J Cell Biol 121: 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Stam P (1998) Genetic analysis. Methods Mol Biol 82: 105–117 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Leon J, Rojo E, Sanchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem 65: 2428–2436 [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ (2002) Maize ABI4 binds Coupling Element1 in abscisic acid and sugar response genes. Plant Cell 14: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas M, Ryan CA (1999) Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA 96: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA 92: 4106–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Keultjes G, Kock P, Wilmering J, Weisbeek P, Smeekens S (1996) Identification of a light-regulated MYB gene from an Arabidopsis transcription factor gene collection. Plant Mol Biol 32: 987–993 [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20: 135–142 [DOI] [PubMed] [Google Scholar]
- Ryan CA (2000) The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta 1477: 112–121 [DOI] [PubMed] [Google Scholar]
- Ryu SB, Wang X (1998) Increase in free linolenic and linoleic acids associated with phospholipase D-mediated hydrolysis of phospholipids in wounded castor bean leaves. Biochim Biophys Acta 1393: 193–202 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Zipori D (2002) PSF and p54(nrb)/NonO: multi-functional nuclear proteins. FEBS Lett 531: 109–114 [DOI] [PubMed] [Google Scholar]
- Sickinger S, Schweizer M (1999) A high affinity binding site for the polypyrimidine tract binding protein (PTB) is located in the 5′-untranslated region of the rat proteinase alpha1-inhibitor 3 variant I gene. Biol Chem 380: 1217–1223 [DOI] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, Leon J, Sanchez-Serrano JJ (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236: 331–340 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK, Hasegawa PM, Bressan RA (1997) Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci 16: 253–277 [Google Scholar]