Abstract
ETHYLENE INSENSITIVE3 (EIN3) is a transcription factor involved in the ethylene signal transduction pathway in Arabidopsis. Two full-length cDNA clones, pVR-EIL1 and pVR-EIL2, encoding EIN3-LIKE proteins were isolated by reverse transcriptase-polymerase chain reaction and by screening the cDNA library of mung bean (Vigna radiata) hypocotyls. VR-EIL1 and VR-EIL2 share 70% identity and display varying degrees of sequence conservation (39%–65%) with previously isolated EIN3 homologs from Arabidopsis, tobacco (Nicotiana tabacum) and tomato (Lycopersicon esculentum) plants. Gel retardation assay revealed that both VR-EILs were able to interact specifically with optimal binding sequence-1, the recently identified optimal binding sequence for tobacco TEIL, with the binding of VR-EIL2 being more efficient than that of VR-EIL1. Transient expression analysis using a VR-EIL::smGFP fusion gene in onion (Allium cepa) epidermal cells indicated that the VR-EIL proteins were effectively targeted to the nucleus. The fusion protein of VR-EIL2 with GAL4 DNA-binding domain strongly activated transcription of a reporter gene in yeast cells, and an essential domain for transcription-stimulating activity was localized to the amino-terminal acidic region that consists of 50 amino acid residues. In contrast with what has been previously found in EIN3- and TEIL-overexpressing Arabidopsis plants, transgenic tobacco seedlings expressing the VR-EIL genes under the control of cauliflower mosaic virus 35S promoter did not exhibit a constitutive triple response. Instead, they displayed a markedly enhanced proliferation of root hairs, one of the typical ethylene response phenotypes, and increased sensitivity to exogenous ethylene. In addition, the pathogenesis-related (PR) genes encoding β-1,3-glucanase, osmotin, and PR1 were constitutively expressed in 35S::VR-EIL lines without added ethylene, and were hyperinduced in response to ethylene treatment. These results indicate that VR-EILs are functional in tobacco cells, thereby effectively transactivating the GCC-box-containing PR genes and enhancing sensitivity to ethylene. The possible physiological role of VR-EILs is discussed in the light of the suggestion that they are active components of the ethylene-signaling pathway and their heterologous expressions constitutively turn on a subset of ethylene responses in tobacco plants.
The gaseous phytohormone ethylene modulates diverse physiological aspects of plant growth and development (Yang and Hoffman, 1984; Abeles et al., 1992). Ethylene also serves as a signaling molecule to elicit specific changes in gene expression at certain stages of a plant life cycle. The production of ethylene in plant tissue is normally low, but can be markedly enhanced by a broad spectrum of developmental and environmental cues, including seed germination, fruit ripening, leaf and flower senescence, and a multitude of biotic and abiotic stresses (Yang and Hoffman, 1984; Abeles et al., 1992).
After synthesis, ethylene must be perceived and its signal transduced into the cells to elicit the physiological responses. Progress in understanding the molecular mechanism of ethylene perception and signal transduction has been recently made by a combination of molecular and genetic approaches using Arabidopsis as a model system (for review, see Kieber, 1997; Chang and Shockey, 1999; Bleecker and Kende, 2000; Stepanova and Ecker, 2000). The triple response is a well-known response of etiolated seedlings to ethylene (Yang and Hoffman, 1984). It comprises three distinct morphological changes: inhibition of hypocotyl elongation, enhancement of radial expansion, and horizontal stem growth. Alteration in the triple response of etiolated Arabidopsis seedlings in response to exogenous ethylene was used to isolate ethylene-related mutants. Among these are the ethylene-insensitive mutants and the recessive mutant that constitutively displays the triple response in the absence of exogenous ethylene (Bleecker et al., 1988; Guzman and Ecker, 1990). Analysis of the mutant genes responsible for these phenotypes has identified a number of components involved in the ethylene signal transduction pathway. Ethylene perception is mediated by a family of membrane receptors, ETR1, ERS1, ETR2, EIN4, and ERS2. Their predicted translation products are reminiscent of bacterial two-component His kinases and function as negative regulators (Chang et al., 1993; Hua et al., 1995, 1998; Schaller and Bleecker, 1995; Hua and Meyerowitz, 1998; Sakai et al., 1998). Loss-of-function mutations at the CTR1 locus result in a constitutive ethylene response, indicating that CTR1 is also a negative regulator of the pathway (Kieber et al., 1993). The CTR1 gene encodes a protein with homology to the Raf family of protein kinases and, hence, a mitogen-activated protein kinase cascade in the ethylene response pathway has been suggested (Kieber et al., 1993). CTR1 acts downstream of the receptor and directly interacts with the receptors in a yeast two-hybrid system (Clark et al., 1998). Thus, plant cells use a unique combination of a prokaryotic-sensing system and a eukaryotic-transducing system for the early step in ethylene signaling. The third gene, ETHYLENE INSENSITIVE2 (EIN2), encodes a novel protein with the amino-terminal 12 integral membrane domains and shows similarity to the disease-related Nramp family of metal-ion transporters of mammalian cells (Alonso et al., 1999).
Epistasis (double-mutant analysis) indicates that EIN3, a positive regulator, acts at the most downstream position of the ethylene-signaling pathway. The EIN3 and three related EIN3-LIKE (EIL1, EIL2, and EIL3) genes were cloned, and their predicted polypeptides were shown to possess common features for nuclear-localized transcription factors (Chao et al., 1997). Solano et al. (1998) have demonstrated that EIN3, as well as EIL1 and EIL2, bind to the ethylene-response element (ERE) present in the upstream region of ETHYLENE-RESPONSE-FACTOR1 (ERF1), an early ethylene-responsive gene encoding the GCC-box-binding protein. The ERF1 protein, in turn, induces various secondary ethylene-responsive pathogenesis-related (PR) genes that contain the GCC-box in their promoters (Solano et al., 1998). These findings indicate that the nuclear proteins EIN3 and ERF1 act sequentially for the transcriptional activation of diverse ethylene-inducible genes in Arabidopsis.
The physiological responses of mung bean (Vigna radiata) seedlings to ethylene have long been studied, such as triple response and root formation (Yang and Hoffman, 1984; Abeles et al., 1992). In addition, mung bean seedlings have been extensively used for the study of regulation of ethylene production in response to auxin and environmental and developmental factors, and ethylene biosynthetic genes are well characterized (Yang and Hoffman, 1984; Abeles et al., 1992; Kim and Yang, 1994; Yi et al., 1999). However, far less has been carried out to investigate the mechanism of the ethylene-signaling pathway and its mode of action in mung bean plants. Application of ethylene in mung bean seedlings caused a great increase in the mRNA level of 1-aminocyclopropane-1-carboxylate (ACC) oxidase (VR-ACO1), the final step of its biosynthetic pathway (Kim and Yang, 1994; Kim et al., 1997; Jung et al., 2000). We are interested in elucidating the molecular mechanism of the mode of action of ethylene in regulating its own production. In this respect, isolation and characterization of mung bean EIN3 homologs would help progress in the understanding of the ethylene action in mung bean seedlings. Here, we report the isolation of two full-length cDNAs encoding mung bean EIN3-LIKE proteins (VR-EIL1 [accession no. AF467784] and VR-EIL2 [accession no. AF467783]) and analysis of their molecular and biochemical properties.
RESULTS
Isolation and Characterization of Mung Bean EIN3 Homologs
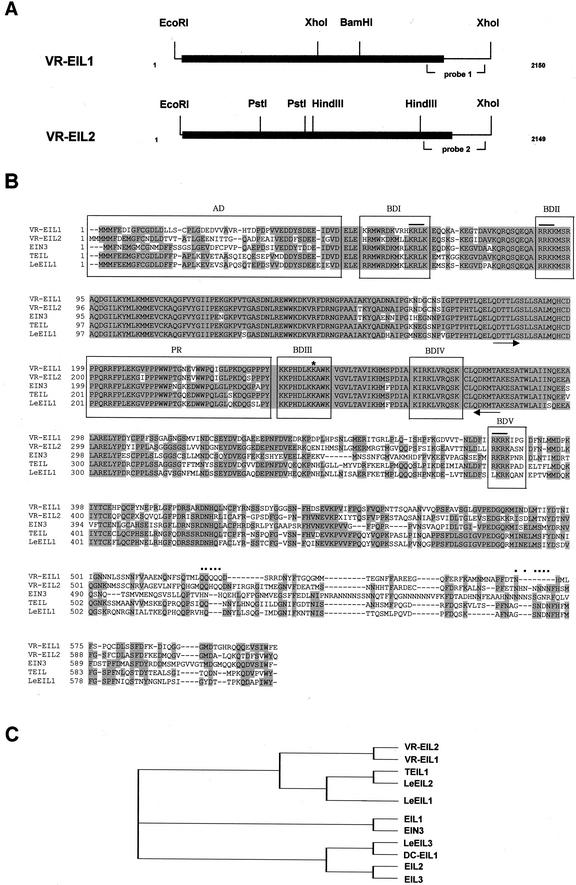
To gain more insight into the mechanism of ethylene action in mung bean vegetative tissues, we proceeded to isolate cDNAs encoding homologs of the Arabidopsis EIN3. Poly(A)+ RNA was isolated from 4-d-old etiolated mung bean hypocotyls. After the synthesis of the first strand cDNA from 1 μg of poly(A)+ RNA, PCR was carried out with mixed oligonucleotides corresponding to the amino acid sequence of QDTTLG as the upstream primer, of QDKMTA as the downstream primer (see “Materials and Methods” for sequences), and the first strand cDNA as the template. These primer amino acid sequences are shown to be highly conserved in Arabidopsis EIN3 and EILs (Chao et al., 1997). Total PCR products of about 300 bp in length were radioactively labeled and used as probes to screen the cDNA library of mung bean hypocotyls under low stringent hybridization and washing conditions (Kim and Yang, 1994). Numerous putative mung bean EIN3 clones were isolated. Subsequent restriction enzyme mapping and DNA sequencing analyses revealed that these clones could be divided into two homology classes. Figure 1A shows the restriction enzyme map analysis of pVR-EIL1 and pVR-EIL2, which contain the longest insert among each class. The pVR-EIL1 clone is 2,150 bp long consisting of a 10-bp 5′-UTR, a 1,827-bp coding region encoding 609 amino acids, and a 313-bp 3′-UTR. pVR-EIL2 is 2,149 bp long comprising a 1-bp 5′-uncoding region, a 1,866-bp coding region of 622 amino acids, and a 282-bp 3′-uncoding region (Fig. 1B). The predicted molecular masses of the polypeptides encoded by pVR-EIL1 and pVR-EIL2 are 67.0 and 68.4 kD, respectively. Complete sequences for pVR-EIL1 and pVR-EIL2 allow comparison of the two full-length VR-EIL genes and analysis of their structural relationship. The overall nucleotide sequence identity between pVR-EIL1 and pVR-EIL2 is 71%, whereas the coding regions are 73% identical at the nucleotide level and 70% at the amino acid level. Both mung bean EILs share 39% to 65% identity at the amino acid level with the Arabidopsis EIN3 and EILs (Chao et al., 1997) and other plant EIN3 homologs, such as tobacco TEIL (Kosugi and Ohashi, 2000), tomato (Lycopersicon esculentum) LeEILs (Tieman et al., 2001), and carnation DC-EIL1 (Waki et al., 2001). Phylogenetic alignment revealed that the VR-EIL1 and VR-EIL2 proteins are the most closely related to TEIL (65% and 64%, respectively), whereas the most distantly related to EIL3 (40%) and EIL2 (39%), respectively (Fig. 1C). As found in other EIN3 homologs, the VR-EIL1 and VR-EIL2 proteins possess an amino-terminal acidic region and a Pro-rich domain (Fig. 1B). In addition, five small clusters of basic amino acids dispersed throughout the protein are also well conserved in both mung bean EILs. The VR-EIL2 polypeptide has a carboxy-terminal Asn-rich region that consists of six Asn residues, as observed in EIN3. However, this poly-Asn repeat is not found in VR-EIL1 and in any of the Arabidopsis EILs and tomato LeEILs. Both mung bean EIL proteins contain a single poly-Gln repeat near the carboxy-terminal portion (Fig. 1B). The Gln-rich region, along with the acidic and Pro-rich regions, has been proposed to be functional as transcriptional activation domains (Chao et al., 1997). Finally, VR-EIL1 and VR-EIL2 have a Lys residue at positions 245 and 246, respectively, which is essential for the function of EIN3 (Fig. 1B, indicated by star). Taken together, these structural conservations suggest that the VR-EIL1 and VR-EIL2 proteins play a role in the ethylene-signaling pathway in mung bean plants.
Figure 1.
Structure of the mung bean EIN3 homologs, VR-EIL1 and VR-EIL2. A, Restriction enzyme map analysis of two mung bean EIL cDNA clones. Solid bars represent the coding regions, whereas solid lines designate the 5′- and 3′-untranslated regions (UTRs). The positions of the gene-specific hybridization probes generated by PCR are indicated. The sequences of pVR-EIL1 and pVR-EIL2 have been deposited in the GenBank data base under accession numbers AF467784 and AF467783, respectively. B, Alignment of the derived polypeptide sequences of VR-EIL1, VR-EIL2, Arabidopsis EIN3 (AtEIN3; Chao et al., 1997), tobacco (Nicotiana tabacum) TEIL (Kosugi and Ohashi, 2000) and tomato (Lycopersicon esculentum) LeEIL1 (Tieman et al., 2001). The conserved amino acid residues are shaded in gray. The amino-terminal acidic domain (AD), Pro-rich region (PR), and five small basic domains (BDI-V) are boxed. The poly-Gln and poly-Asn repeats at the carboxy-terminal portion are marked by dots. Asterisk refers to the Lys residue essential for the function of Arabidopsis EIN3. The putative nuclear-localized signals are indicated by solid lines. The arrows depict the primer amino acid sequences for reverse transcriptase-PCR. Dashes show gaps in the amino acid sequences introduced to optimize alignment. C, Phylogenetic alignment of EIN3 homologs from mung bean (VR-EIL1 and VR-EIL2), Arabidopsis (AtEIN3, AtEIL1, AtEIL2, and AtEIL3; Chao et al., 1997), tobacco (TEIL1; Kosugi and Ohashi, 2000), tomato (LeEIL1, LeEIL2, and LeEIL3; Tieman et al., 2001) and carnation (Dianthus caryophyllus; DC-EIL1; Waki et al., 2001).
VR-EILs Bind Specifically to the Conserved Sequence optimal binding sequence (obs) 1 in Vitro
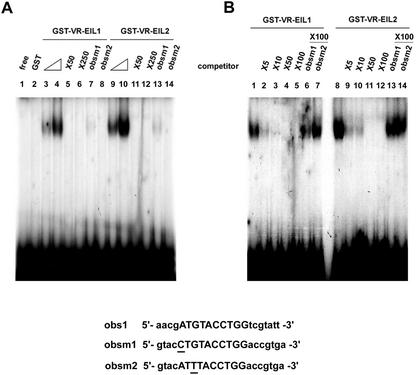
Kosugi and Ohashi (2000) carried out a random binding selection analysis and revealed that the obs for TEIL was A[T/C]G[A/T]A[C/T]CT. This sequence is present in the 5′-upstream region of various ethylene-inducible genes. Among EIN3 homologs, TEIL is the most closely related to VR-EILs (Fig. 1). Thus, we examined whether VR-EILs can bind to the consensus sequence. VR-EIL1 and VR-EIL2 were expressed in Escherichia coli as a fusion protein with glutathione-S-transferase (GST), and purified recombinant proteins were used in a gel retardation assay with a 32P-labeled conserved sequence obs1 (Kosugi and Ohashi, 2000). The full-length 93- to 94-kD GST-VR-EILs gave rise to a single, discrete DNA-protein complex that migrated more slowly than the free probe (Fig. 2A). The intensity of this shifted band increased upon the addition of increasing amounts (50–100 ng) of GST-VR-EILs. The DNA binding specificity of VR-EILs was further confirmed by competition binding experiments, which showed that a 50-fold excess of cold obs1 was enough to displace the labeled probe. The binding capacity of the VR-EIL1 and VR-EIL2 proteins was dramatically reduced when obsm1, which has A→C substitution in the first position of obs1, was used as a probe (Fig. 2A, lanes 7 and 13), whereas we could not detect any interaction between VR-EILs and obsm2 that contains G→T mutation in the third position of the conserved sequence (Fig. 2A, lanes 8 and 14). These results imply that VR-EIL1 and VR-EIL2 bind specifically to obs1 in vitro. The results in Figure 2A also reveal that the intensity of VR-EIL2-DNA complex is stronger than that of VR-EIL1-DNA complex, suggesting that VR-EIL2 interacts with obs1 more effectively that VR-EIL1 does (Fig. 2A, compare lanes 3 and 9, and lanes 4 and 10). To explore whether VR-EIL1 and VR-EIL2 have a different affinity to obs1, we repeated competition binding experiments with varying amounts of cold probe. As shown in Figure 2B, the specific interaction between VR-EIL1 and obs1 was almost completely abolished by a 10-fold excess of cold probe, whereas VR-EIL2 was still able to bind significantly to obs1 in the presence of the same amount of cold competitor (Fig. 2B, lanes 3 and 10). In addition, the ability of VR-EIL1 to bind to obs1 was significantly reduced by a 100-fold molar excess of cold obsm1 (Fig. 2B, lane 6). However, the same amount of obsm1 affects the interaction between VR-EIL2 and obs1 to a much lesser extent (Fig. 2B, lane 13). Overall, these results indicate that VR-EIL1 and VR-EIL2 interact specifically with the conserved sequence obs1, with the binding of VR-EIL2 being more efficient than VR-EIL1.
Figure 2.
Sequence-specific binding activities of VR-EILs. A, Gel retardation assay showing full-length GST-VR-EILs binding to obs1, the obs for tobacco TEIL (Kosugi and Ohashi, 2000). The radiolabeled probes or varying amounts of cold competitors were incubated in the presence or absence of the purified recombinant GST-VR-EILs (50 and 100 ng). Lane 1, only the free probe; lane 2, the free probe and GST. Lanes 3, 4, 9, and 10, Radiolabeled obs1 probe; lanes 5, 6, 11, and 12, titration with varying amounts of cold obs1 as a competitor; lanes 7 and 13, radiolabeled obsm1 probe; lanes 8 and 14, radiolabeled obsm2 probe. Oligonucleotide probes (obs1, obsm1, and obsm2) were prepared based on the recent results by Kosugi and Ohashi (2000). B, Competition binding analysis. The radiolabeled obs1 probe and purified 100 ng of recombinant VR-EILs were incubated in the presence or absence of varying amounts of cold competitors as indicated. Lanes 1 and 8, Without any competitors; lanes 2 through 5 and 9 through 12, titration with varying amounts of cold obs1 as a competitor; lanes 6 and 13, a 100-fold molar excess of cold obsm1 as a competitor; lanes 7 and 14, a 100-fold molar excess of cold obsm2 as a competitor.
Targeting of VR-EILs to the Nucleus
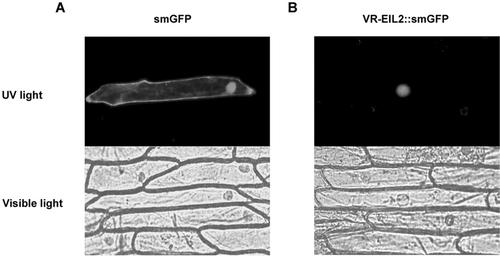
Because VR-EILs specifically bind obs1 in vitro (Fig. 2) and their predicted sequence contains putative nuclear localization signal (Fig. 1B), they are expected to localize to the nucleus. To confirm this, we performed an in vivo targeting experiment that employed a VR-EIL2-fused soluble-modified green fluorescent protein (smGFP) as a fluorescent marker in a transient transfection assay. The smGFP gene was fused to the 3′ end of the pVR-EIL2 coding region in frame under the control of the cauliflower mosaic virus (CaMV) 35S promoter, and the resulting construct was introduced into onion (Allium cepa) epidermal cells by the particle bombardment method. Localization of the fusion protein was then determined by visualization with a fluorescence microscope. As shown in Figure 3, the control smGFP was uniformly distributed throughout the cell (Fig. 3A), whereas the VR-EIL2-smGFP fusion protein was exclusively localized to the nucleus (Fig. 3B). These observations support the notion that the putative nuclear targeting sequence of VR-EIL2 is sufficient and that no additional posttranslational modification may be necessary for the VR-EIL2 protein to be targeted to the nucleus. We obtained identical results with VR-EIL1-smGFP fusion protein (data not shown).
Figure 3.
Nuclear localization of the VR-EIL2 gene product. The smGFP coding region was fused in frame to the 3′ end of the full-length pVR-EIL2-coding region. The construct was introduced into onion epidermal cells by the particle bombardment method and expressed under the control of the CaMV 35S promoter. The expression of the introduced gene was viewed after 12 h by fluorescence microscopy under UV or visible light.
The VR-EIL Proteins Activate Transcription in Yeast
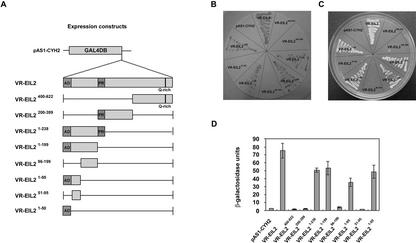
VR-EIL1 and VR-EIL2 proteins possess an amino-terminal acidic region, a Pro-rich domain, and a polyGln repeat at the carboxy terminus that might act as a transcriptional activator motif (Fig. 1). To examine the functional role of these domains, we fused the coding region of VR-EIL2 and its various deletion mutants to the GAL4 DNA-binding domain expression vector and investigated the behavior of each construct as a potential transcriptional activator in yeast cells. In the absence of the GAL4 activation domain, the wild-type VR-EIL2 protein fused to the GAL4 DNA-binding domain, effectively activating transcription of the lacZ reporter gene (Fig. 4). This indicates that VR-EIL2 is able to function as a transcriptional activator in yeast. To identify a minimal transcriptional activation domain, various deletion mutants of VR-EIL2 were also tested by the same method. Figure 4 depicts that deletion of Pro-rich region and carboxy-terminal Gln-rich domain does not result in loss of transcriptional activation. In contrast, deleting the amino-terminal acidic domain almost completely abrogated reporter gene activation. Furthermore, the transcription-stimulating activity was still apparent when the amino-terminal acidic region (1–50 amino acids) was fused to the GAL4 DNA-binding domain; the VR-EIL21–50 mutant protein contained 70% β-galactosidase activity compared with that of the full-length protein (Fig. 4D). Thus, the amino-terminal 50-amino acid residues of VR-EIL2 play a critical role in supporting the ability of VR-EIL2 as a transcriptional activator.
Figure 4.
GAL4 DB-VR-EIL2 fusions and their effect on transcriptional activation of the lacZ reporter gene in yeast cells. A, Schematic overview of the fusion proteins between the GAL4 DNA-binding domain (DB) and various deletion mutants of VR-EIL2 that were investigated for transcription-stimulating activity in yeast. B, The VR-EIL2 and its deletion constructs fused in the GAL4 DB expression vector were transformed into yeast strain CG1945. The transformants were selected by growth on Trp– medium at 30°C for 3 d. Yeast transformants were streaked onto the Glc medium and were grown at 30°C for 48 h, and then β-galactosidase enzyme assay was carried out. C, Yeast transformants were tested for growth in the absence of His. The yeast strain is carrying a modified HIS gene whose transcription is under the control of GAL4 operator. D, β-Galactosidase enzyme activities were quantified after growth of yeast strains in liquid culture using chlorophenol red-β-d-galactopyranoside as substrate. This assay was repeated four times with similar results. The values are means ± sd (n = 4).
Organization and Expression of the VR-EIL Genes
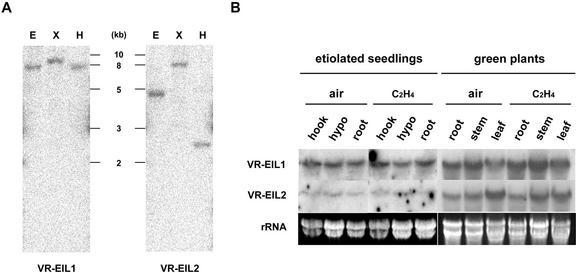
From the results described above, it appears that VR-EILs are nuclear proteins that specifically interact with obs1 in vitro and possess transcriptional activation activity. Therefore, we wanted to characterize the VR-EIL genes in more detail at the molecular level. To assess the exact copy number of the EIL genes in the mung bean genome, we constructed gene-specific probes (probes 1 and 2 for each homology class, Fig. 1A) by PCR based on the divergent 3′-UTRs of pVR-EIL1 and pVR-EIL2. The genomic DNA isolated from mature leaves of mung bean plants was digested with EcoRI, XbaI, or HindIII, and was hybridized with 32P-labeled gene-specific probe 1 or 2 under high stringent conditions. These hybridizations detected only one major band by those enzyme digestions, respectively (Fig. 5A). These results imply that the VR-EIL1 and VR-EIL2 genes are present in a single copy per haploid mung bean genome. With a general probe derived from the conserved coding region and under low stringent hybridization and washing conditions, one to two minor bands were also detected (data not shown). Thus, we could not rule out the possibility of the existence of additional VR-EIL-related genes in the mung bean genome.
Figure 5.
Organization and expression of the mung bean EIL genes. A, Genomic Southern-blot analysis of the VR-EIL genes. The mung bean genomic DNA (10 μg per lane) was isolated from leaf tissue, digested with EcoRI (E), XbaI (X), or HindIII (H), and resolved on a 0.7% (w/v) agarose gel. DNA on the gel was transferred to a nylon membrane filter. The filter was hybridized to the 32P-labeled gene-specific probe 1 or probe 2 under high stringent conditions. B, RNA gel-blot analysis of the VR-EIL genes. Total RNAs (20 μg per lane) isolated from different parts of 4-d-old etiolated seedlings or 2-week-old light-grown mung bean plants were resolved on a 1.0% (w/v) formaldehyde-agarose gel. The gel was blotted onto a membrane filter and the blot was hybridized to the 32P-labeled gene-specific probe 1 or probe 2 under high stringent conditions. To examine the effect of ethylene on VR-EILs mRNA expression, intact etiolated seedlings and light-grown plants were enclosed in 3-liter jars containing air or air plus 20 μL L–1 ethylene. After 6 h of treatment, total RNAs were isolated from different parts of plants and were analyzed as described above. The blots were visualized by autoradiography. The equivalence of RNA loading among lanes of the agarose gel was demonstrated by ethidium bromide staining of rRNA on the gel.
To examine the spatial and temporal expression pattern of the VR-EIL1 and VR-EIL2 genes, we monitored the level of corresponding mRNAs in different mung bean vegetative tissues by northern-blot analysis. Total RNAs isolated from apical hooks, hypocotyls, and roots of dark-grown 4-d-old seedlings, or from leaves, stems, and roots of light-grown 2-week-old plants were hybridized with 32P-labeled probe 1 or 2. The substantial level of the transcripts (2.3 kb) for VR-EIL1 and VR-EIL2 was detected in every tissue examined of dark- and light-grown plants (Fig. 5B). However, the relative expression pattern of the two different mRNAs varied in these tissues, with the VR-EIL1 transcript being predominantly present in etiolated seedlings. By contrast, the amount of the two mRNAs was somewhat similar in green plants (Fig. 5B). To investigate whether the activity of the VR-EIL genes is modulated by ethylene, intact mung bean plants were exposed to 20 μL L–1 ethylene for 6 h, and then total RNAs were similarly analyzed. As shown in Figure 5B, the steady-state levels of these mRNAs were not significantly affected by the application of ethylene in dark- and light-grown plants. These results are in line with the previous observations that the expression of Arabidopsis EIN3 and tomato LeEILs is not regulated by exogenous ethylene, and are consistent with the view that the activity of EIN3 and its homologs are possibly subject to control by a posttranslational mechanism (Chao et al., 1997; Kosugi and Ohashi, 2000; Tieman et al., 2001).
Overexpression of VR-EIL1 and VR-EIL2 Enhances Root Hair Formation in Transgenic Tobacco Seedlings
Chao et al. (1997) produced Arabidopsis plants overexpressing the EIN3 and EIL1 genes, respectively, and investigated the effect of overexpression on ethylene responses in transgenic plants. The etiolated T2 generation seedlings exhibited characteristics of a constitutive triple response phenotype in the absence of added ethylene, which were very similar to the ethylene-treated wild-type and ctr1 mutant seedlings. Similarly, when tobacco TEIL cDNA was introduced into Arabidopsis under the control of CaMV 35S promoter, the transgenic seedlings displayed a triple response without ethylene treatment (Kosugi and Ohashi, 2000). These results showed that the enhanced level of EIN3 or EILs resulted in the constitutive activation of the ethylene-signaling pathway. On the other hand, Tieman et al. (2001) constructed tomato plants that expressed antisense RNA for each of the three LeEIL genes. Because LeEILs share significant sequence identities, the antisense expression of one gene conferred the decrease in the expression of other LeEIL genes. As a result, at least one line from each anti-LeEIL-transgenic plant showed a high degree of ethylene insensitivity in many aspects compared with wild-type tomatoes, such as increased seedling length, reduced leaf epinasty, and delayed flower abscission in response to ethylene, and inhibition of fruit ripening (Tieman et al., 2001). The severity of ethylene insensitivity paralleled the level of reduction in the total amount of LeEIL transcripts in antisense plants.
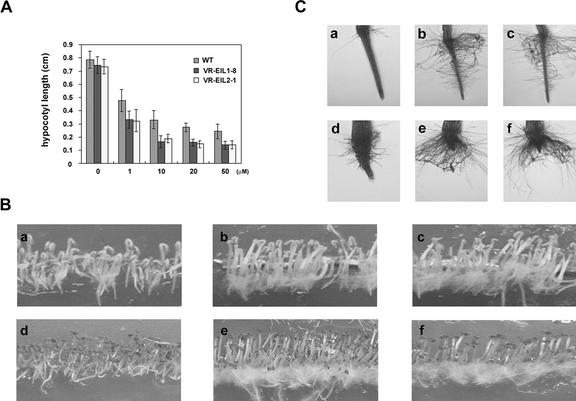
The results described above led us to consider the possibility that the mung bean VR-EILs might be functionally similar to EIN3 or EILs. To examine this possibility, we established transgenic tobacco plants overexpressing VR-EIL1 and VR-EIL2, respectively. Tobacco plants were transformed with binary vectors carrying a fusion of CaMV 35S promoter and pVR-EILs in the sense orientation by means of the Agrobacterium tumefaciens-mediated transformation method. For each gene, several independent primary transformants were obtained based on the resistance against kanamycin, and transgenic plants were subsequently regenerated and used for further analyses. The presence of each of the transgenes was confirmed by PCR analysis (data not shown). In contrast to EIN3- and TEIL-expressing Arabidopsis, 4-d-old etiolated tobacco seedlings of the T2 generation did not display a constitutive triple response; their hypocotyl length was indistinguishable from that of control plants in the absence of added ethylene (Fig. 6A). However, the VR-EIL-overexpressing seedlings showed an increased sensitivity to exogenous ethylene, as indicated by more severely reduced hypocotyl length in comparison with the control seedlings in the presence of 1 to 50 μm ACC, the immediate precursor to ethylene (Fig. 6A). A more unique phenotype specific for both VR-EIL constructs was a marked increase in root hair formation. As shown in Figure 6B, the proliferation of root hairs was pronouncedly elevated in dark- and light-grown transgenic lines without ethylene treatment. A closer inspection revealed that the individual transgenic root had numerous ectopic hairs, and this phenotype was more exaggerated when treated with 10 μm ACC compared with ACC-treated control roots, suggesting that transgenic roots are more sensitive to ethylene (Fig. 6C). Ethylene has long been suggested to play a role in the initiation of root hairs in various plant species, including pea (Pisum sativum), fava bean (Vicia faba), lupine (Lupinus albus), and tulip (Tulipa gesneriana; Abeles et al., 1992). Arabidopsis ctr1 mutation confers production of ectopic root hairs on epidermis cells, which are normally nonhair cells (Dolan et al., 1994). Furthermore, Tanimoto et al. (1995) have demonstrated that Arabidopsis grown with aminoethoxyvinyl-Gly, an effective inhibitor of ethylene biosynthesis, or Ag+, an ethylene action inhibitor, shows a reduced number of root hair, whereas plants grown in the presence of ACC exhibit ectopic root hair, indicating that ethylene is a positive regulator of root hair development. Most recently, Cho and Cosgrove (2002) have shown that ethylene exerts an unambiguous effect on root hair elongation and mediates environment-induced root hair initiation (e.g. auxin and root separation from medium) in Arabidopsis. Thus, the hairy root phenotype of VR-EIL-expressing segregants is consistent with the notion that VR-EIL1 and VR-EIL2 are functioning in tobacco seedlings and activate the ethylene-signaling pathway to turn on a subset of ethylene responses.
Figure 6.
Overexpression of VR-EIL cDNAs in tobacco plants. A, Ethylene sensitivity of etiolated seedlings of wild type (WT) and VR-EIL-overexpressing transgenic tobacco lines (VR-EIL1-8 and VR-EIL2-1). Wild-type and transgenic seedlings of the T2 generation were germinated in the dark in the absence or presence of different concentrations (1–50 μm) of ACC at 25°C. Data shown indicate the hypocotyl lengths of etiolated 4-d-old seedlings. The values are means ± sd (n = 60). B, Phenotype of the VR-EIL-overexpressing T2 generation seedlings. The seedlings were germinated in the dark for 4 d (a–c) or were grown in a growth chamber under a 16-h daylength for 7 d at 25°C (d–f). a and d, Wild-type plants; b and e, VR-EIL1-8 line; c and f, VR-EIL2-1 line. C, Enhanced formation of root hairs in 35S::VR-EIL lines. The roots were grown in the dark in the absence (a–c) or presence (d–f) of 10 μm ACC for 4 d at 25°C. a and d, Wild-type root; b and e, VR-EIL1-8 root; c and f, VR-EIL2-1 root.
The VR-EIL Genes Effectively Transactivate Various Ethylene-Inducible PR Genes in Transgenic Tobacco Seedlings
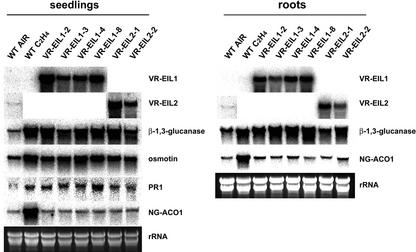
To further investigate the function of VR-EILs in tobacco cells and to define whether the root hair phenotype of VR-EIL constructs was due to the activation of the ethylene signaling pathway, we monitored the expression pattern of various ethylene-responsive genes in transgenic plants. The PR genes encoding β-1,3-glucanase, osmotin, and PR1 were highly induced after ethylene treatment for 6 h in light-grown 7-d-old wild-type tobacco seedlings (Fig. 7). These PR genes are known to have the GCC-box in their promoters (Ohme-Takagi and Shinshi, 1990; Hart et al., 1993; Sessa et al., 1995; Sato et al., 1996). Moreover, as shown in Figure 7, high-level constitutive expression of these PR genes was clearly observed in VR-EIL1- and VR-EIL2-overexpressing transgenic lines without ethylene treatment. The levels of PR mRNAs were approximately correlated with those of the VR-EIL transcripts. Thus, it seems most likely that VR-EILs are highly functional in tobacco cells, thereby effectively transactivating the GCC-box-containing PR genes. As the constitutive root hair formation was the most significant phenotype of VR-EIL-expressing tobacco plants, we next examined the transactivation activities of VR-EILs in transgenic root tissue. VR-EIL1 and VR-EIL2 effectively activated the β-1,3-glucanase gene, as evidenced by the constitutive high level of its mRNA in 35S::VR-EIL roots (Fig. 7). As expected, ethylene caused a marked enhancement of the level of tobacco ACC oxidase mRNA (NG-ACO1, Kim et al., 1998) in control plants (Fig. 7). However, the steady-state level of this transcript was not changed in transgenic lines compared with that of wild-type plants.
Figure 7.
Expression pattern of various ethylene-inducible genes in wild-type (WT) and VR-EIL-overexpressing tobacco plants. Total RNAs (20 μg per lane) were isolated from 7-d-old light-grown wild-type seedlings and roots that had been exposed to air or air plus 20 μL L–1 ethylene for 6 h, and were resolved on a 1.0% (w/v) formaldehyde-agarose gel. The gel was blotted onto a membrane filter and the blot was hybridized to the 32P-labeled cDNA probes for VR-EIL1, VR-EIL2, β-1,3-glucanase (Ohme and Shinshi, 1990), osmotin (Sato et al., 1996), PR1 (Kim et al., 1998), and NG-ACO1 (Kim et al., 1998), respectively, under high stringent conditions. To investigate the transactivation activities of VR-EILs, total RNAs (20 μg per lane) were isolated from 10-d-old green T2 generation plants of various independent 35S::VR-EIL lines (VR-EIL1-2, VR-EIL1-3, VR-EIL1-4, VR-EIL1-8, VR-EIL2-1, and VR-EIL2-2), which had been grown without exogenous ethylene, and were similarly analyzed. The blots were visualized by autoradiography. The equivalence of RNA loading among lanes of the agarose gel was demonstrated by ethidium bromide staining of rRNA on the gel.
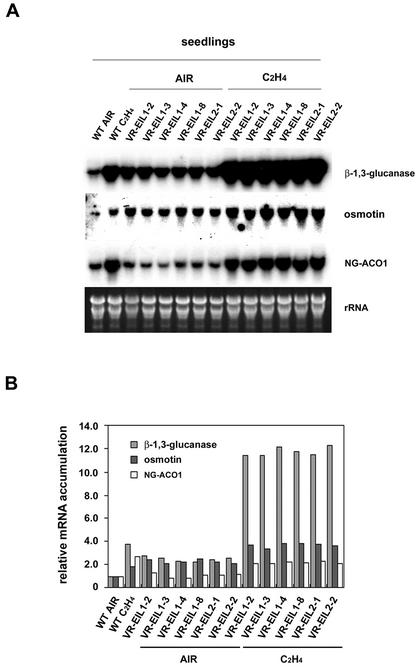
Because 35S::VR-EIL1/2 resulted in hypersensitivity to exogenous ethylene rather than constitutive triple response in tobacco seedlings (Fig. 6, A and C), we considered the possibility that ethylene-induced genes are hyperinducible by ethylene in these transgenic plants. To investigate this possibility, the transgenic seedlings were incubated with exogenous ethylene and the induction patterns of genes were examined. As shown in Figure 8, A and B, the β-1,3-glucanase and osmotin genes were hyperinduced by ethylene treatment; the mRNA level greatly increased in ethylene-treated 35S::VR-EIL1/2 seedlings compared with ethylene-treated wild-type plants, confirming that the induction of β-1,3-glucanase and osmotin is dependent on the function of VR-EIL1/2. Thus, transgenic tobacco plants overexpressing VR-EILs are more sensitive to exogenous ethylene than wild-type plants, indicating that VR-EILs are functionally relevant in tobacco cells. On the other hand, the level of ethylene-inducible NG-ACO1 transcript was almost identical in wild-type and transgenic seedlings in response to exogenous ethylene (Fig. 8).
Figure 8.
Induction pattern of β-1,3-glucanase, osmotin, and NG-ACO1 in response to exogenous ethylene in wild-type (WT) and VR-EIL-overexpressing tobacco plants. A, Total RNAs (20 μg per lane) were isolated from 7-d-old light-grown wild-type and T2 generation plants of various independent 35S::VR-EIL lines that had been incubated with or without 20 μL L–1 ethylene for 6 h, and were resolved on a 1.0% (w/v) formaldehyde-agarose gel. The gel was blotted onto a membrane filter and the blot was hybridized to the 32P-labeled cDNA probes for β-1,3-glucanase, osmotin, and NG-ACO1, respectively, under high stringent conditions. The blots were visualized by autoradiography. The equivalence of RNA loading among lanes of the agarose gel was demonstrated by ethidium bromide staining of rRNA on the gel. B, The relative levels of β-1,3-glucanase, osmotin, and NG-ACO1 transcripts in wild-type and various VR-EIL-overexpressing tobacco plants in the presence or absence of exogenous ethylene. Hybridization signals obtained from the northern-blot analysis of A were quantified with a phosphorimager (Fuji).
DISCUSSION
As ethylene exerts profound effects on such diverse aspects during plant growth and in the pathogenesis response, its signal transduction pathway and molecular action mechanism have attracted much interest. The expression of various development- and defense-related genes is controlled by this simple gaseous hormone at the transcriptional level. Ethylene-responsive cis-acting elements and nuclear proteins that specifically interact with these regulatory regions have been identified in several genes such as the tomato fruit E4 and E8 genes (Cordes et al., 1989; Montgomery et al., 1993; Coupe and Deikman, 1997; Deikman et al., 1998), the carnation GST1 gene (Itzhaki et al., 1994; Maxson and Woodson, 1996), and the tobacco PRB-1b and osmotin-like PR-5 genes (Meller et al., 1993; Sato et al., 1996). These studies have found two different types of EREs. The study with the aid of stable deletion analysis of promoters from tobacco chitinase and PRB-1b genes has defined the GCC-box as an ERE, composed of an 11-bp sequence (TAAGAGCCGCC) that is highly conserved in the 5′-upstream region of ethylene-inducible PR protein genes (Sessa et al., 1995; Shinshi et al., 1995). Subsequently, Ohme-Takagi and Shinshi (1995) have isolated four different cDNA clones encoding the GCC-box-binding proteins (EREBPs) and have demonstrated that the accumulation of their transcripts increased in response to ethylene in various tissues of tobacco plants. More recently, these ERFs have been found to modulate the GCC-box-mediated gene expression, as transcriptional activators or repressors in tobacco and Arabidopsis plants, indicating that ERFs respond to extracellular signals in distinct ways (Fujimoto et al., 2000; Ohta et al., 2000a, 2000b). The other kind of ERE was identified to be responsible for the developmental expression of tomato E4 and carnation GST1 genes that are induced during fruit ripening and flower senescence, respectively (Montgomery et al., 1993; Itzhaki et al., 1994). Interestingly, a similar E4- and GST1-like cis-acting element was also found in the promoter of Arabidopsis ERF1 that encodes the GCC-box-binding protein (Solano et al., 1998). Based on these results, Solano et al. (1998) renamed the E4- and GST1-like element as a primary ERE. Furthermore, they have shown that the Arabidopsis EIN3/EILs family, an essential component participating in the ethylene-signaling pathway, directly binds to a primary ERE present in the ERF1 gene. Thus, it seems most likely that there exists an ethylene signaling sequence in the nucleus from EIN3 to ERF1 to a variety of ethylene-responsive genes in Arabidopsis. Using a random binding site selection analysis, Kosugi and Ohashi (2000) have revealed that the consensus binding sequence for tobacco TEIL is A[T/C]G[A/T]A[C/T]CT. This conserved sequence is found to be present in the 5′-upstream regions of E4, GST1, ERF1, and tobacco NsERF2 (Montgomery et al., 1993; Itzhaki et al., 1994; Solano et al., 1998; Kitajima et al., 2000).
In the present study, we have isolated and characterized two full-length cDNA clones, pVR-EIL1 and pVR-EIL2, that encode mung bean homologs of the Arabidopsis EIN3. Both VR-EILs share a high degree of sequence conservation with previously identified EIN3-LIKE proteins from Arabidopsis, tobacco, and tomato plants (Fig. 1). Gel retardation analysis shows that mung bean EILs are able to interact specifically with obs1, the obs for tobacco TEIL (Kosugi and Ohashi, 2000). Competition binding assay suggests that two mung bean proteins have different binding capacity to obs1, with the interaction of VR-EIL2 being more efficient than that of VR-EIL1 (Fig. 2B). Although VR-EIL1 and VR-EIL2 are highly homologous and share various conserved domains throughout the coding region (see below), VR-EIL2 uniquely possesses an Asn-rich region at the carboxy terminus that consists of six Asn residues (Fig. 1B). This domain is also present in Arabidopsis EIN3, but not in VR-EIL1 and other Arabidopsis EILs and tomato LeEILs. Thus, it would be intriguing to identify whether a carboxy-terminal Asn-rich region is functioning in the binding mechanism between VR-EIL2 and the consensus sequence obs1.
Beside an Asn-rich region, VR-EILs contain several distinct architectural properties such as an amino-terminal acidic domain, a Pro-rich region, five small clusters of basic amino acids, and a single poly-Gln repeat near the carboxy terminus (Fig. 1B). Acidic, Pro-rich, and Gln-rich domains have been indicated to function as transcriptional activation motifs (Mitchell and Tjian, 1989; Chao et al., 1997). To unravel an essential domain for transcriptional activation, we performed yeast one-hybrid assay using various deletion mutants of VR-EIL2. The results demonstrate that the amino-terminal 50-amino acid residues are required to activate transcription of a reporter gene in yeast cells; the VR-EIL21–50 mutant protein shows 70% transcription-stimulating activity compared with that of the wild-type protein (Fig. 4). In our experimental conditions, the Pro-rich region and poly-Gln repeat, respectively, contain only the background level of activity for transcriptional activation in yeast, suggesting that they are not essential components for the function of a transcriptional activator. However, we could not rule out the possibility that those domains may be necessary for full activity of VR-EIL2.
To investigate the physiological role of VR-EILs in more detail, we constructed transgenic tobacco plants expressing each of these genes under the control of CaMV 35S promoter. In contrast with what has been previously found in EIN3- and TEIL-overexpressing Arabidopsis plants, we observed that 35S::VR-EIL1/2 tobacco lines did not exhibit a constitutive triple response in the absence of exogenous ethylene (Fig. 6A). Instead, both VR-EIL constructs displayed great increase in root hair formation, and this phenotype was further enhanced in the presence of ACC (Fig. 6, B and C). In addition, various GCC-box-containing PR genes, including β-1,3-glucanase, osmotin, and PR1, were constitutively activated in 35S::VR-EIL1/2 plants without ethylene treatment (Fig. 7). The VR-EIL-overexpressing lines also show an enhanced sensitivity to exogenous ethylene as evidenced by following observations: more severely reduced hypocotyl length and exaggerated root hair formation in comparison with the control seedlings in response to ACC (Fig. 6, A and C) and the hyperethylene-induction of the β-1,3-glucanase and osmotin genes compared with ethylene-treated wild type plants (Fig. 8). Taken together, these results support the notion that VR-EILs are functionally relevant in tobacco cells, which interact with upstream and downstream partners of the tobacco EIN3 homologs and turn on a subset of ethylene responses.
A wealth of information has been documented that ethylene is closely associated with the induction of the ACC oxidase gene in climacteric fruits, resulting in a surge in ethylene production during the ripening process (Yang and Hoffman, 1984; Abeles et al., 1992). Previously, we showed that ethylene markedly stimulated the accumulation of ACC oxidase mRNA in vegetative tissues of mung bean (Kim and Yang, 1994; Jung et al., 2000) and tobacco (Kim et al., 1998) plants. Based on these results, it has been proposed that ethylene plays a key role in regulating ACC oxidase gene expression in vegetative tissues, as it does in fruit tissue, and that the low basal level of ACC oxidase transcript constitutively expressed in vegetative tissues is regulated by the endogenous ethylene present in these tissues (Kim et al., 1997; Jin et al., 1999). This autocatalysis reaction by ethylene results from the transcriptional activation of the ACC oxidase gene (Park et al., 2001). However, the results in Figures 7 and 8 reveal that although the 35S::VR-EIL constructs display the phenotype of ectopic root hair formation, one of the typical ethylene responses, and several GCC-box-containing PR genes are effectively transactivated in these transgenic lines, the expression pattern of ACC oxidase transcript (NG-ACO1) was very similar in transgenic and wild-type tobacco plants. The NG-ACO1 transcript was not constitutively expressed or hyperinduced by ethylene in transgenic plants. The reason for this unexpected result is not clear at this moment, but it could be due to the fact that VR-EILs may not be fully functional in heterologous tobacco cells and, hence, turn on a subset of ethylene responses. In this regard, it is worth noting that the VR-EIL lines do not exhibit a constitutive triple response (Fig. 6A). Alternatively, it is possible that the EIL family might possess slightly different DNA-binding specificity (e.g. distinct target sequences and different binding activities) or differential signal thresholds and sensitivities, resulting in the activation of different sets of gene expression. For instance, NG-ACO1 could be more sensitive to TEIL than to VR-EIL1/2 so that overexpression of VR-EILs did not significantly affect the level of NG-ACO1 mRNA in transgenic tobacco seedlings. Interestingly, our computer database analysis suggests that the promoter region of ACC oxidase from different species, including tomato LeACO1 and LeACO2 (Barry et al., 1996; Blume and Grierson, 1997), apple (Malus domestica) GSAO (Atkinson et al., 1998) and MD-ACO2 (GenBank accession no. AF015787), mung bean VR-ACO1 (Park et al., 2001), and Arabidopsis EAT1 (GenBank accession no. X66719) do not seem to have the consensus sequences for EIN3-binding site or GCC-box. Thus, further experiments are required to define the ERE present in the 5′-upstream region of ACC oxidase and the role of ethylene-responsive trans-acting factors for the regulation of the last step in ethylene production by this simple signaling hydrocarbon in higher plants.
MATERIALS AND METHODS
Plant Materials and Ethylene Treatment
Dry seeds of mung bean (Vigna radiata) were soaked overnight in aerated tap water. Seedlings were grown on 0.8% (w/v) agar for 4 d in a dark room at 25°C or for 2 weeks in an environmentally controlled chamber. In the case of tobacco (Nicotiana tabacum cv Samsun NN) plants, wild-type and transgenic seedlings were grown on 0.8% (w/v) agar for 4 d in a dark room or on Murashige and Skoog medium containing 1% (w/v) Suc, B5 vitamin (12 mg L–1), and 0.8% (w/v) agar (pH 5.8) for 7 d in a growth chamber under a 16-h light/8-h dark photoperiod at 25°C. For ethylene treatment, intact plants were enclosed for various periods in 3-liter jars containing air or air plus 20 μL L–1.
PCR
The first strand cDNA, synthesized from 1 μg of poly(A)+ RNA isolated from etiolated mung bean hypocotyls, was amplified by PCR using mixed oligonucleotide primers (CCGGAATTCCA[G/A]GA[T/C]-[G/A]CIAC[G/A/T/C][T/C]T[G/A/T/C] as the upstream primer and CGGGATCCGC[G/A/T/C]GTCAT[T/C]TT[G/A]TC[T/C]TG as the downstream primer). These primer sequences were derived from the conserved amino acid sequences, QDTTLG and QDKMTA, respectively, which were found to be highly conserved in Arabidopsis EIN3 and EILs (Chao et al., 1997). The amplified PCR products were subcloned into pGEM-T Easy vector system I (Promega, Madison, WI). PCR was performed in a total volume of 25 μL containing 1 μL of the first strand cDNA reaction products, 1 μm primers, 10 mm Tris (pH 8.0), 50 mm KCl, 1.5 mm MgCl2, 0.01% (w/v) gelatin, 200 μm deoxynucleotides, and 2.5 units of Taq polymerase (Promega). Thirty-five thermal cycles were carried out, each consisting of 1 min at 94°C, 2 min at 48°C to 52°C, and 2 min at 72°C in an automatic thermal cycler (Perkin-Elmer/Cetus, Norwalk, CT). PCR products were separated on an agarose gel, and the band was eluted and reamplified by PCR to increase the amount of DNA for the subsequent subcloning. The various ethylene-responsive tobacco cDNA clones for expression studies were obtained by reverse transcriptase-PCR using specific primers constructed based on the published DNA sequences as described above.
Screening of cDNA Library and Sequencing of DNA
To isolate mung bean cDNA clones encoding EIN3 homolog, the lambda Zap II mung bean hypocotyl cDNA library (Kim and Yang, 1994) was screened using the 300-bp PCR products as probes by an established procedure (Sambrook et al., 1989). The cDNA inserts containing putative mung bean EILs were subcloned into Bluescript SK plasmid by in vivo excision of pBluescript from Zap II vector as described in the protocols by Stratagene (La Jolla, CA). Sequencing of DNA was performed using the sequenase DNA sequencing kit according to the manufacturer's manual (U.S. Biochemicals, Cleveland).
Cloning and Expression of VR-EILs
The plasmid pGEX4T-1 (Pharmacia, Uppsala, Sweden) was used for the expression of GST-VR-EIL fusion proteins. Escherichia coli BL21 (DE3) strains containing the plasmids were grown at 37°C in 100 mL of 2× Luria-Bertani medium (10 g of trypton, 10 g of yeast extract, and 5 g NaCl L–1) supplemented with 1% (w/v) glycerol as an additional carbon source and 70 μg mL–1 ampicillin. Cells were grown for a further 4 h at 30°C after induction with 1 mm isopropyl β-d-thiogalactoside at 0.6 to 1.0 OD600. The pellet was resuspended in phosphate-buffered saline containing 1 mm phenylmethyl-sulfonyl fluoride. The suspension was sonicated on ice with Vibracell sonicator (Sonics and Materials, Danbury, CT), and Triton X-100 was added to a final concentration of 1% (w/v). Various fusion proteins were purified by affinity chromatography using glutathione Sepharose 4B from GST purification modules (Pharmacia).
Gel Retardation Assay
DNA probes and competitors (obs1, obsm1, and obsm2) for gel retardation assays were produced based on the results of Kosugi and Ohashi (2000). To reduce nonspecific DNA-protein binding, purified GST-VR-EIL proteins (50–100 ng) were preincubated with 0.5 μg of poly (dI-dC) and 0.5 μg of nonspecific DNA oligonucleotide in 20 μL of binding buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 1 mm dithiothreitol, 50 mm NaCl, and 5% [w/v] glycerol) for 10 min on ice. End-labeled DNA probe (0.25 ng) was then added to the reaction mixtures. After incubating for 10 min on ice, the mixtures were loaded onto an 8% (w/v) nondenatured polyacrylamide gel. Before loading, gels were pre-run at 10 V cm–1 for 30 min, and electrophoresis was performed in 0.5× 54 mm Tris-borate, pH 8.3, and 1 mm EDTA for 2.5 h. For competition experiments, varying amounts of cold competitor molecules were preincubated with GST-VR-EILs before addition of the radiolabeled probe. The gel was dried and autoradiographed.
Subcellular Localization of VR-EILs
The smGFP cDNA was fused in frame to the 3′ end of full-length pVR-EIL coding regions. Transient expression of smGFP fusion constructs was then performed by introducing the DNAs into onion (Allium cepa) epidermal cells using the particle bombardment method according to the manufacturer's protocol (Bio-Rad, Richmond, CA). Fluorescence photographs of onion cells were taken using a fluorescence microscope (Axiophot; Zeiss, Jena, Germany) fitted with fluorescein isothiocyanate filters (excitation filter, 450–490 nm; emission filter, 520 nm; dichroic mirror, 510 nm) and color film (Fuji 400; Fuji, Tokyo). The optimal exposure time was 1 s.
Construction of Various VR-EIL2 Deletion Mutants and Transactivation Activity Analysis
The deletion mutant constructions were generated using various restriction sites within the VR-EIL2 cDNA or by PCR. Coding regions for the deleted proteins were fused in frame to the yeast GAL4 DNA-binding domain expression vector pAS1-CYH2. The recombinant plasmids were then transformed into Saccharomyces cerevisiae strain CG1945 as described by the manufacturer (Clontech, Palo Alto, CA). The transformants were selected by growth on Trp– synthetic dropout medium at 30°C for 3 d. The β-galactosidase filter-lift assay was performed according to the manufacturer's protocol (Clontech). β-Galactosidase activities were quantified after growth of yeast strains in liquid culture using chlorophenol red-β-d-galactopyranoside as a substrate.
Isolation of Genomic DNA and Southern-Blot Analysis
The mung bean leaf genomic DNA was isolated as described previously (Park et al., 2001) with modifications. Each gram of mung bean leaf was pulverized under liquid nitrogen and was suspended in 3 mL of extraction buffer (8.0 m urea, 50 mm Tris-Cl, pH 7.5, 20 mm EDTA, 250 mm NaCl, 2% [w/v] sarcosyl, 5% [v/v] phenol, and 20 mm 2-mercaptoethanol). After successive extractions with phenol/chroloform/isoamylalcohol (25:24:1, v/v), the aqueous phase was concentrated by ethanol precipitation. The pellet was resuspended in 10 mm Tris-Cl (pH 7.5) and 1 mm EDTA adjusted to a density of 1.5 g mL–1 by the addition of CsCl and the DNA was centrifuged overnight at 200,000g. The DNA band was collected, extracted with water-saturated 1-butanol, precipitated by ethanol, and then resuspended in 10 mm Tris-Cl (pH 7.5) and 1 mm EDTA. Mung bean genomic DNA (10 μg per lane) was digested with appropriate enzymes, separated by electrophoresis in a 0.7% (w/v) agarose gel, and blotted onto a nylon membrane filter (Amersham, Arlington Heights, IL). The filter was hybridized to 32P-labeled gene-specific probe 1 and probe 2, respectively, under high stringent conditions.
RNA Isolation and Northern-Blot Analysis
Total RNAs of mung bean and tobacco plants were obtained by a method adapted from the established protocols reviewed by Lizzradi (1983) with modifications as described previously by Jin et al. (1999). The total RNAs were precipitated overnight at 4°C by the addition of 0.3 vol of 10 m LiCl and then precipitated in ethanol. The RNAs (20 μg) were fractioned by electrophoresis in a 1.0% (w/v) formaldehyde-agarose gel and were blotted onto a nylon membrane filter. Equal loading of RNA was confirmed by visualizing the ethidium bromide-stained ribosomal RNA content under UV light at the end of electrophoresis. The filter was hybridized to various 32P-labeled cDNA probes for mung bean VR-EIL1 and VR-EIL2, and tobacco β-1,3-glucanase, osmotin-like PR-5, PR1, and NG-ACO1. The blot was washed and visualized by autoradiography at –80°C. Hybridization signals were quantified with a phosphorimager (Fuji).
Gene Constructs for Transgenic Plants and Tobacco Transformation
A vector, pBI121, which contained the CaMV 35S promoter and nopaline synthase terminator, was used for tobacco transformation. pVR-EIL cDNA fragments containing the complete open reading frame were generated by PCR using high-fidelity Ex-Tag polymerase (Takara, Kyoto). To facilitate subcloning of PCR products, forward and reverse primers for each cDNA clone were tagged by appropriate restriction enzymes (pVR-EIL1, XbaI/BamHI; pVR-EIL2, BamHI/SacI). The β-glucuronidase reporter gene of pBI121 was eliminated and the tagged pVR-EIL cDNA fragments were inserted into the corresponding sites of pBI121. The fusion gene constructs were transferred to Agrobacterium tumefaciens strain LBA4404 by electroporation according to Shen and Forde (1989). Leaf discs of tobacco cv Samsun NN were transformed essentially as described by Horsch et al. (1985). Transgenic plants were selected on the Murashige and Skoog medium containing 200 μg mL–1 kanamycin and 500 μg mL–1 carbenicillin. The presence of transgene in the transformants was confirmed by PCR and northern-blot analysis with 32P-labeled gene-specific probe 1 and probe 2, respectively. Primary transformants were self-fertilized and seeds were collected. T2 seeds were harvested from individual T1 plants for further studies.
Acknowledgments
We thank Sunglan Chung (Department of Biology, Yonsei University) for her critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022574.
This work was supported by the Plant Diversity Research Center (21st Century Frontier Research Program of Ministry of Science and Technology project no. PF 003105–01) and by Korea Science and Engineering Foundation (Plant Metabolism Research Center, Kyung Hee University, to W.T.K.).
References
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in Plant Biology. Academic Press, San Diego
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress response in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Bolitho KM, Wright MA, Iturriagagoitia-Bueno T, Reid SJ, Ross GS (1998) Apple ACC-oxidase and polygalacturonase: ripening-specific gene expression and promoter analysis in transgenic tomato. Plant Mol Biol 38: 449–460 [DOI] [PubMed] [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D (1996) Differential expression of 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J 9: 525–535 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–8 [DOI] [PubMed] [Google Scholar]
- Blume B, Grierson D (1997) Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental stimuli. Plant J 12: 731–746 [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chang C, Shockey JA (1999) The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol 2: 352–358 [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Cho H-T, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes S, Deikman J, Margossian LJ, Fischer RL (1989) Interaction of a developmentally regulated DNA-binding factor with sites flanking two different fruit-ripening genes from tomato. Plant Cell 1: 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe SA, Deikman J (1997) Characterization of a DNA-binding protein that interacts with 5′-flanking regions of two fruit-ripening genes. Plant J 11: 1207–1218 [DOI] [PubMed] [Google Scholar]
- Deikman J, Xu R, Kneissl ML, Ciardi JA, Kim K-N, Pelah D (1998) Separation of cis elements responsive to ethylene, fruit development, and ripening in the 5′-flanking region of the ripening-related E8 gene. Plant Mol Biol 37: 1001–1011 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett C, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K (1994) Clonal relations and patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins F Jr (1993) A 61-bp enhancer element of the tobacco β-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol Biol 21: 121–131 [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227: 1229–1231 [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM (1995) Ethylene insensitivity conferred by Arabidopsis ERS gene. Science 269: 1712–1714 [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Chang C, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H, Maxson JM, Woodson WR (1994) An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc Natl Acad Sci USA 91: 8925–8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ES, Lee J-H, Park JA, Kim WT (1999) Temporal and spatial regulation of the expression of 1-aminocyclopropane-1-carboxylate oxidase by ethylene in mung bean (Vigna radiata). Physiol Plant 105: 132–140 [Google Scholar]
- Jung T, Lee JH, Cho MH, Kim WT (2000) Induction of 1-aminocyclopropane-1-carboxylate oxidase mRNA by ethylene in mung bean roots: possible involvement of Ca2+ and phosphoinositides in ethylene signaling. Plant Cell Environ 23: 205–213 [Google Scholar]
- Kieber JJ (1997) The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol 48: 277–296 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldman KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim WT, Kang BG, Yang SF (1997) Induction of 1-aminocyclopropane-1-carboxylate oxidase mRNA by ethylene in mung bean hypocotyls: involvement of both protein phosphorylation and dephosphorylation in ethylene signaling. Plant J 11: 399–405 [Google Scholar]
- Kim WT, Yang SF (1994) Structure and expression of cDNAs encoding 1-aminocyclopropane-1-carboxylate oxidase homologs isolated from excised mung bean hypocotyls. Planta 194: 223–229 [PubMed] [Google Scholar]
- Kim YS, Choi D, Lee MM, Lee SH, Kim WT (1998) Biotic and abiotic stress-related expression of 1-aminocyclopropane-1-carboxylate oxidase gene family in Nicotiana glutinosa L. Plant Cell Physiol 39: 565–573 [DOI] [PubMed] [Google Scholar]
- Kitajima S, Koyama T, Ohme-Takagi M, Shinshi H, Sato F (2000) Characterization of gene expression of NsERFs, transcription factors of basic PR genes from Nicotiana sylvestris. Plant Cell Physiol 41: 817–824 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco ethylene-insensitive3 (EIN3) homolog. Nucleic Acids Res 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizzardi PM (1983) Methods for the preparation of messenger RNA. Methods Enzymol 96: 24–48 [DOI] [PubMed] [Google Scholar]
- Maxson JM, Woodson WR (1996) Cloning of a DNA-binding protein that interacts with the ethylene-responsive enhancer element of the carnation GST1 gene. Plant Mol Biol 31: 751–759 [DOI] [PubMed] [Google Scholar]
- Meller Y, Sessa G, Eyal Y, Fluhr R (1993) DNA-protein interactions on a cis-DNA element essential for ethylene regulation. Plant Mol Biol 23: 453–463 [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R (1989) Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245: 371–378 [DOI] [PubMed] [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL (1993) Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc Natl Acad Sci USA 90: 5939–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1990) Structure and expression of a tobacco β-1,3-glucanase gene. Plant Mol Biol 15: 941–946 [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2000a) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H (2000b) Three ethylene-responsive transcriptional factors in tobacco with distinct transactivation functions. Plant J 22: 29–38 [DOI] [PubMed] [Google Scholar]
- Park D, Lee JH, Joo S, Kim WT (2001) Structure and ethylene-induced expression of 1-aminocyclopropane-1-carboxylate oxidase gene in mung bean (Vigna radiata L.). J Plant Biol 44: 1–7 [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM (1998) ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 95: 5812–5817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sato F, Kitajima S, Koyama T, Yamada Y (1996) Ethylene-induced gene expression of osmotin-like protein, a natural isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol 37: 249–255 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Sessa G, Meller Y, Fluhr R (1995) A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol 28: 145–153 [DOI] [PubMed] [Google Scholar]
- Shen WJ, Forde BG (1989) Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucleic Acids Res 17: 8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-tagaki M (1995) Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol 27: 923–932 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR (2000) Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol 3: 353–360 [DOI] [PubMed] [Google Scholar]
- Tanimoto M, Roberts K, Dolan L (1995) Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J 8: 943–948 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26: 47–58 [DOI] [PubMed] [Google Scholar]
- Waki K, Shibuya K, Yoshioka T, Hashiba T, Satoh S (2001) Cloning of a cDNA encoding EIN3-like protein (DC-EIL1) and decrease in its mRNA level during senescence in carnation flower tissues. J Exp Bot 52: 377–379 [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT (1999) Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.). Plant Mol Biol 41: 443–454. [DOI] [PubMed] [Google Scholar]