Abstract
In higher plants, etioplast to chloroplast differentiation is characterized by dramatic ultrastructural changes of the plastid and a concomitant increase in chlorophylls and carotenoids. Whereas the formation and function of carotenes and their oxygenated derivatives, the xanthophylls, have been well studied, little is known about the regulation of the genes involved in xanthophyll biosynthesis. Here, we analyze the expression of three xanthophyll biosynthetic genes (i.e. β-carotene hydroxylase [bhy], zeaxanthin epoxidase [zep], and violaxanthin de-epoxidase [vde]) during de-etiolation of seedlings of tobacco (Nicotiana tabacum L. cv Samsun) under different light conditions. White-light illumination caused an increase in the amount of all corresponding mRNAs. The expression profiles of bhy and zep not only resembled each other but were also similar to the pattern of a gene encoding a major light-harvesting protein of photosystem II. This finding indicates a coordinated synthesis during formation of the antenna complex. In contrast, the expression pattern of vde was clearly different. Furthermore, the gene expression of bhy was shown to be modulated after illumination with different white-light intensities. The expression of all xanthophyll biosynthetic genes under examination was up-regulated upon exposure to red, blue, and white light. Gene expression of bhy and vde but not of zep was more pronounced under red-light illumination, pointing at an involvement of the phytochrome system. Expression analysis in the presence of the photosynthetic electron transport inhibitors 3-(3,4-dichlorophenyl)-1,1-dimethyl-urea and 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone indicated a redox control of transcription of two of the xanthophyll biosynthetic genes (bhy and zep).
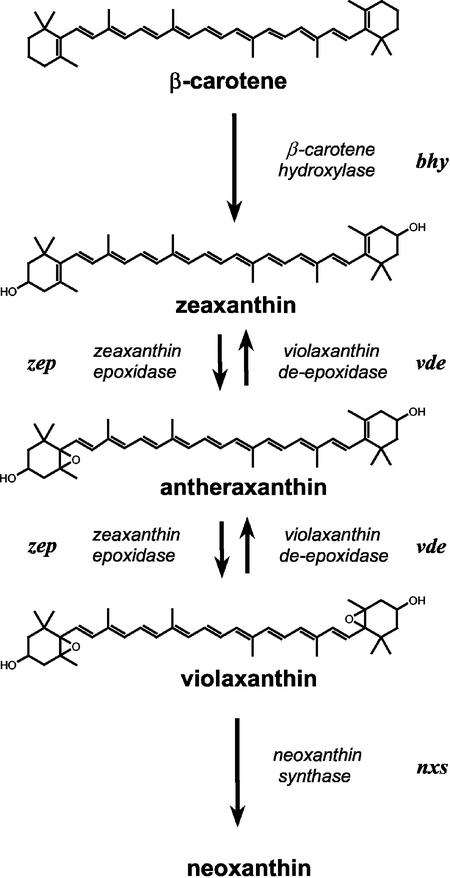
The process of chloroplast differentiation involves dramatic ultrastructural and physiological changes of the plastids (Robertson and Laetsch, 1974; McCormac et al., 1996; Tziveleka and Argyroudi-Akoyunoglou, 1998) and requires the synchronous formation of pigments, lipids, and proteins for the coordinated assembly of a functional photosynthetic apparatus (Vothknecht and Westhoff, 2001). One remarkable feature of de-etiolation is the massive synthesis of chlorophylls and carotenoids. Etioplast to chloroplast transition is therefore a suitable system to study carotenoid gene expression and subsequent product accumulation. In angiosperms, the synthesis of chlorophyll is strictly light dependent (von Wettstein et al., 1995), whereas low levels of carotenoids were shown to be present in etiolated seedlings. In higher plants, carotenoids are essential components of the photosynthetic apparatus. As such, they are involved in light harvesting and protection against excessive light energy (Siefermann-Harms, 1985; Havaux, 1998; Niyogi, 1999). In addition, the presence of the xanthophyll cycle enhances plant photoprotection (Niyogi et al., 1998). This cycle comprises the three xanthophylls zeaxanthin, antheraxanthin, and violaxanthin and their interconversion (Hager, 1975; Demmig et al., 1987). Zeaxanthin can be synthesized in two ways: by hydroxylation of β-carotene or by deepoxidation of violaxanthin (Fig. 1). In the first case, zeaxanthin formation is catalyzed by the enzyme β-carotene hydroxylase using β-carotene as substrate. In the second case, zeaxanthin is converted to violaxanthin by the enzyme zeaxanthin epoxidase under low-light (LL) conditions, whereas high-light conditions lead to the production of zeaxanthin via violaxanthin de-epoxidase due to xanthophyll cycle activity. As members of photosynthetic complexes, the carotenoids are necessary for assembly and stabilization of the corresponding pigment-protein complexes (Plumley and Schmidt, 1987; Herrin et al., 1992). The xanthophylls (lutein, zeaxanthin, antheraxanthin, violaxanthin, and neoxanthin in its 9′-cis rather than the all-trans-form) are predominantly found in the light-harvesting system (Kühlbrandt et al., 1994; Ruban et al., 1994, 2002; Lee and Thornber, 1995; Takaichi and Mimuro, 1998; Croce et al., 2002).
Figure 1.
Biosynthetic pathway of β-carotene-derived xanthophylls in higher plants.
Most of the genes encoding enzymes involved in carotenoid biosynthesis have been cloned, and several xanthophyll biosynthetic genes were identified from higher plants and green algae (Bouvier et al., 1996, 2000; Bugos and Yamamoto, 1996; Marin et al., 1996; Sun et al., 1996; Linden, 1999). Despite the increase in DNA sequence information, our knowledge of the complex regulation of xanthophyll biosynthesis is still limited, especially during light-dependent de-etiolation. Carotenoid enzymes are plastid-located but nuclear-encoded proteins. The preproteins are posttranslationally targeted in the plastids and processed. Thus regulation could take place at transcriptional, posttranscriptional, or translational level. Expression of carotenoid genes depends on the species investigated (Römer et al., 1993) and the developmental stage (Bugos et al., 1999). Moreover, environmental factors have been shown to influence their gene expression (Bouvier et al., 1996; Audran et al., 1998; Steinbrenner and Linden, 2001).
The regulation of phytoene synthase, the first committed step in carotenoid biosynthesis, is the only one that has been examined thoroughly during etioplast to chloroplast transition (von Lintig et al., 1997; Welsch et al., 2000). Later steps in the pathway have been addressed either by investigating one gene at a time or by using one specific light condition only (Bugos and Yamamoto, 1996; Audran et al., 1998). Even the recent application of microarrays for transcriptome analysis did not allow a detailed comparison of xanthophyll biosynthetic gene expression because the arrays contained just a specific selection of genes (Ma et al., 2001; SMD, http://afgc.stanford.edu). Thus conclusions were hampered by the limitations of data availability or by the experimental set up. We therefore reexamined the expression of three xanthophyll biosynthetic genes (β-carotene hydroxylase, zeaxanthin epoxidase, and violaxanthin deepoxidase) in a comparative manner during etioplast to chloroplast differentiation under various light conditions. Our study focused on the following questions: (a) Are the xanthophyll biosynthetic genes under examination coordinately expressed during de-etiolation? (b) How does light quantity and quality influences the steady-state transcript levels of these genes? (c) Is there evidence for a redox and/or photosynthetic control of their expression?
For our investigations, two different low light intensities were chosen to exclude additional effects caused by high-light stress on mRNA transcript levels. Furthermore, the regulation of xanthophyll biosynthesis genes during etioplast to chloroplast differentiation was investigated upon illumination with different light qualities. Application of inhibitors of the photosynthetic electron transport to fully greened seedlings allowed us to examine the impact of the redox status on carotenoid gene transcription levels. Because xanthophylls are constituents of the light-harvesting complexes, a gene encoding a protein of the major light-harvesting system of photosystem (PS) II as well as a chlorophyll biosynthetic gene were included in some of the studies.
RESULTS
Pigment Content and Composition
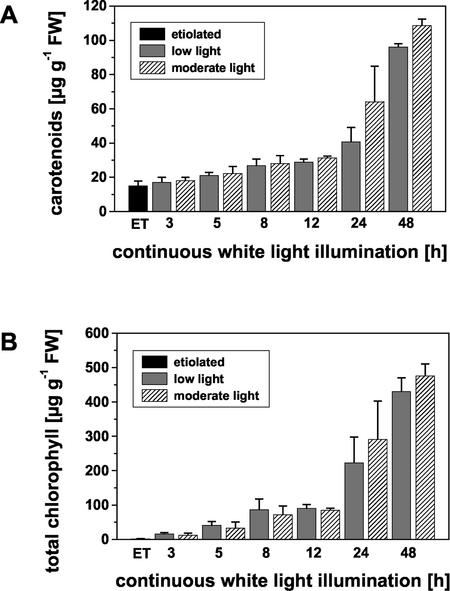
Total pigment content was determined in dark-grown seedlings and in seedlings illuminated with either very low light (LL) or moderate (ML) white-light intensities for various time periods. Dark-grown seedlings had already a carotenoid content of 15.2 μg g–1 fresh weight which increased 3-fold within 24 h of continuous white-light illumination and up to 6-fold within 48 h (Fig. 2A). The carotenoid content was not significantly different between LL- and ML-exposed seedlings, although there was a tendency toward elevated carotenoid levels in seedlings irradiated with ML intensity. In contrast, only trace amounts of chlorophylls could be detected in dark-grown seedlings. Significant accumulation of these pigments occurred after a lag phase at later stages of light-dependent chloroplast differentiation (Fig. 2B). The total chlorophyll content reached values of 476 μg g–1 fresh weight and 431 μg g–1 fresh weight after 48 h of illumination with ML and LL intensity white light, respectively.
Figure 2.
Changes in photosynthetic pigments during chloroplast differentiation of etiolated seedlings upon exposure to continuous white light of different intensities. A, Changes in total carotenoid content. B, Changes in total chlorophyll content. Pigments were extracted from 30 seedlings per time point. Pigment content was determined spectroscopically in acetone. For chlorophylls, the quantification was also done after separation on HPLC. Values are means ± sd of six independent determinations (except for 12 h, n = 4, and 48 h, n = 3). ET, Etiolated seedlings; FW, fresh weight.
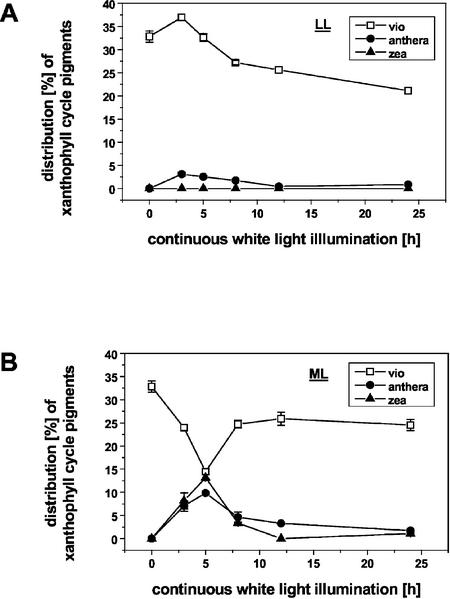
Carotenoid composition was investigated by HPLC. No significant changes in the relative amounts of β-carotene, lutein, and neoxanthin were found concerning the various light regimes (data not shown). However, remarkable differences were detected for the xanthophyll cycle pigments (violaxanthin, antheraxanthin, and zeaxanthin; Fig. 3). The percentage of violaxanthin decreased from 32.8% in etiolated seedlings to 23.9% directly upon ML illumination reaching 14.5% after 5 h of light exposure. Concomitantly, zeaxanthin and the mono-epoxidated intermediate antheraxanthin were transiently accumulated at early stages of chloroplast differentiation (3 and 5 h; Fig. 3B). The highest amount of zeaxanthin was found after 5 h of illumination. Prolongation of the light exposure period resulted in a decrease in the portion of zeaxanthin to nearly undetectable levels and an increase in the relative amount of violaxanthin to 24.6% after 24 h. In contrast to this, LL white light caused an increase in the relative amount of violaxanthin directly after light exposure (Fig. 3A). Seedlings illuminated with this light intensity had a lower violaxanthin to antheraxanthin to zeaxanthin proportion (22%) compared with 27.4% in ML even after 24 h of irradiation.
Figure 3.
Distribution of xanthophyll cycle pigments (percent) during chloroplast differentiation of etiolated seedlings upon exposure to continuous white light of different intensities. A, Illumination with LL (10 μmol m–2 s–1). B, Illumination with ML (100 μmol m–2 s–1). Total pigment extracts were separated by reversed-phase HPLC on a Spherisorb ODS1 column according to Gilmore and Yamamoto (1991). Zeaxanthin was below the detection limit under LL conditions. vio, violaxanthin; anthera, antheraxanthin; and zea, zeaxanthin.
In etioplasts, only chlorophyll a but no chlorophyll b was detectable. However, during light-dependent etioplast to chloroplast transition, the chlorophyll a/b ratio decreased from 5.7 after 3 h of ML illumination to 2.8 after 24 h. The chlorophyll a/b-ratio of seedlings exposed to LL for 3 h reached values of 2.9 and of about 2.6 after 24 h.
Expression of Xanthophyll Biosynthetic Genes during Chloroplast Differentiation
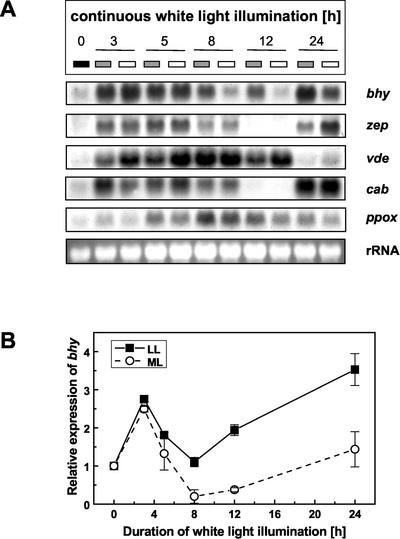
All genes involved in the formation of β-carotene-derived xanthophylls exhibited a low steady-state mRNA-transcript level in etiolated seedlings (Fig. 4A). With the onset of continuous white-light illumination a strong induction in the amount of the corresponding mRNAs was observed. The expression level of the β-carotene hydroxylase gene was found to be modulated by the light intensity (Fig. 4). After 3 or 5 h of continuous white-light illumination, the increase in the transcript level was comparable under ML and LL conditions. However, a prolonged illumination time resulted in a massive reduction in the amount of β-carotene hydroxylase mRNA in ML-treated seedlings with an expression minimum at 12 h, whereas only a moderate decrease was noticeable under LL conditions. After 24 h of continuous white-light irradiation, the expression was again up-regulated under both light regimes (Fig. 4).
Figure 4.
A, Northern-blot analysis of the expression of genes involved in pigment synthesis and light harvesting during chloroplast differentiation of tobacco seedlings under various light intensities. Four-day-old etiolated seedlings were exposed to continuous white light of LL (10 μmol m–2 s–1; gray box) or ML (100 μmol m–2 s–1; white box) intensity for different time periods (0, 3, 5, 8, 12, and 24 h). Gel-blot analysis was performed after separation of total RNA extracted from the individual samples (minimum of 60 seedlings each). For hybridization, nonradioactively labeled DNA fragments of the various xanthophyll biosynthetic genes were used as probes. bhy, β-carotene hydroxylase; zep, zeaxanthin epoxidase; vde, violaxanthin de-epoxidase; cab, chlorophyll a/b binding protein of LHC II; and ET, etiolated seedlings. Bottom, 25S rRNA hybridization for verification of gel loading. B, Densitometric scan of mRNA patterns of the β-carotene gene during chloroplast differentiation of tobacco seedlings. Signals were normalized with respect to the rRNA. The value for etiolated seedlings was set to 1. n = 3.
LL as well as ML illumination caused a strong increase in the expression of zeaxanthin epoxidase gene, and high levels of steady-state mRNA were maintained for the first 5 h. The amount of transcript was reduced subsequently with a minimum reached at 12 h of white-light irradiation. At this time point, the transcript could barely be detected. After 24 h, a very high zeaxanthin epoxidase transcript level was observed again (Fig. 4A).
The expression pattern of the violaxanthin deepoxidase gene (vde) differed from those of the genes mentioned above. Although high amounts of the corresponding transcript were found after short exposure to white light, the transcript level decreased with prolonged illumination periods and remained low at 24 h. All results from northern-blot analyses were verified with RNA from three independent greening series.
Comparative Expression Studies using a Chlorophyll a/b-Binding Protein (cab) and a Chlorophyll Biosynthetic Gene (protoporphyrinogen IX oxidase [ppox])
For comparison, a gene encoding a light-harvesting chlorophyll a/b-binding protein was also included in the expression studies (Fig. 4A). The expression pattern of this gene was similar to those of the xanthophyll biosynthesis genes coding for β-carotene hydroxylase and zeaxanthin epoxidase (Fig. 4). Furthermore, the expression of a chlorophyll biosynthetic gene, namely the plastidal ppox, was examined (Fig. 4A). Low transcript levels in etiolated seedlings increased significantly with the onset of illumination. In contrast to the xanthophyll biosynthetic genes, however, high amounts of steady-state mRNA were only found in later stages of chloroplast differentiation (after 5 h of illumination). The transcript level of ppox was slightly but consistently higher in seedlings exposed to LL conditions.
The Impact of Different Light Qualities on the Expression of Carotenoid Biosynthetic Genes
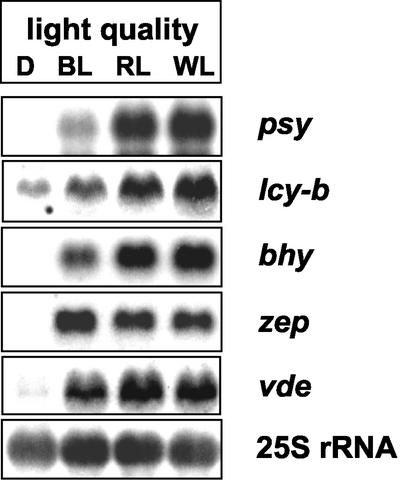
To investigate the effect of light quality on the transcription level of different genes encoding enzymes of the carotenoid biosynthetic pathway, etiolated seedlings were exposed to red, blue, or white light for greening. After 3 h of illumination, seedlings were harvested and subjected to extraction of total RNA. Subsequent northern-blot analysis revealed distinct expression patterns for the different carotenoid genes (Fig. 5). The expression of all genes investigated was induced upon illumination regardless of the light quality used. However, considerable differences in the amount of steady-state transcripts could be observed. The expression of the phytoene synthase gene showed a strong induction after exposure to red light and white light, whereas blue light was less effective. The same phenomenon was observed in the case of β-lycopene cyclase and β-carotene hydroxylase gene expression. The effect of red light on the transcript level of violaxanthin deepoxidase was less pronounced. The amount of zeaxanthin epoxidase mRNA increased upon light exposure, but was not altered by different light qualities.
Figure 5.
Expression of various carotenoid biosynthetic genes during de-etiolation of tobacco seedlings upon illumination with different light qualities. RNA was extracted from seedlings after 3 h of exposure to white light (WL), blue light (BL), and red light (RL) and subjected to northern-blot analysis using DIG-labeled probes for the genes encoding phytoene synthase (psy), β-lycopene cyclase (lcy-b), β-carotene hydroxylase (bhy), zeaxanthin epoxidase (zep), violaxanthin de-epoxidase (vde), and chlorophyll a/b protein (cab) of the major light-harvesting complex. Light intensity was 45 μmol m–2 s–1. Bottom, Hybridization with 25S rRNA for loading verification.
The Effect of Photosynthetic Inhibitors on Carotenoid Gene Expression
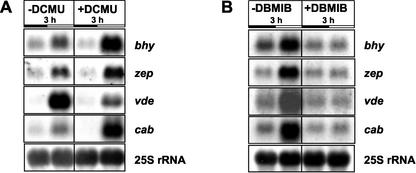
To evaluate the impact of the photosynthetic electron transport on the transcription of carotenoid biosynthetic genes, fully greened 3-d-old seedlings were incubated either with the inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), which blocks the reduction of plastoquinone, or with 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB), which impairs the oxidation of the plastoquinol (Fig. 6). Inhibition of the electron transport by DCMU for 3 h caused an increase in the transcription level of genes encoding β-carotene hydroxylase (bhy) and zeaxanthin epoxidase (zep), whereas the amount of violaxanthin de-epoxidase (vde) transcript was diminished (Fig. 6A). A major light-harvesting gene (cab) used for comparison was also expressed at higher levels upon treatment with DCMU. The opposite effect, namely a down-regulation of the transcript levels of bhy, zep, and cab was observed upon a 3-h incubation with DBMIB (Fig. 6B).
Figure 6.
Effect of the photosynthetic inhibitors DCMU (A) and DBMIB (B) on expression levels of various carotenoid biosynthetic genes. Three-day-old fully greened seedlings were treated with 2 mm DCMU or 50 μm DBMIB for3hinthe dark or in the light, respectively. Untreated seedlings served as control (–DCMU and –DBMIB). bhy, β-Carotene hydroxylase; zep, zeaxanthin epoxidase; and vde, violaxanthin de-epoxidase. Bottom, Hybridization with 25S rRNA for loading verification.
Application of DCMU or DBMIB for 3 h had different effects on the total pigment content of fully greened seedlings. Whereas DCMU treatment caused slightly elevated levels of all pigments (i.e. chlorophyll a and b, carotenoids), the total pigment content of DBMIB-treated seedlings remained unchanged compared with controls (Table I).
Table I.
Effect of photosynthetic inhibitors on total pigment content
Data represent the mean ± sd from three to six independent experiments.
| CONTROL | DCMU 3 h | DBMIB 3 h | |
|---|---|---|---|
| μg g-1 fresh wt | |||
| Chl a | 173.55 ± 10.08 | 197.86 ± 4.99 | 169.82 ± 25.15 |
| Chl b | 50.29 ± 2.75 | 58.70 ± 1.42 | 50.26 ± 6.06 |
| Car | 49.00 ± 2.98 | 56.38 ± 1.48 | 47.61 ± 6.23 |
DISCUSSION
Effect of Different White-Light Intensities on the Steady-State mRNA Levels of Xanthophyll Biosynthetic Genes
White-light illumination led to a general induction of the expression of all three xanthophyll biosynthetic genes (bhy, zep, and vde) under investigation. The increase of the steady-state transcript levels of these genes is likely to be related to the large increase of total carotenoids, which is observed during chloroplast differentiation (compare with von Lintig et al., 1997). Furthermore, our study revealed that illumination with different white-light intensities can also cause a modulation of mRNA transcript levels during de-etiolation of tobacco (Nicotiana tabacum L. cv Samsun) seedlings. The expression of bhy showed the most pronounced light-intensity dependence with highest mRNA levels reached under LL conditions. The regulation of the bhy steady-state transcript level may represent a way to meet the requirement of additional β-carotene derivatives for the assembly of the photosynthetic apparatus (Young, 1991; Herrin et al., 1992) and to adjust the size of the light-harvesting antenna (Horton et al., 1996). The catalytic activity of the enzyme β-carotene hydroxylase leads to the formation of zeaxanthin from β-carotene via the β-branch of the pathway (Sun et al., 1996). Zeaxanthin itself is not only important for the dissipation of excess light energy (Demmig-Adams, 1990; Niyogi, 1999) but is also the substrate for the formation of all other β-carotene-derived xanthophylls (violaxanthin and neoxanthin), which are located in the light-harvesting complexes and are involved in light collection (Kühlbrandt et al., 1994; Lee and Thornber, 1995; Formaggio et al., 2001; Ruban et al., 2002). We therefore assume that the differences in the steady-state transcript level of β-carotene hydroxylase are due to a gene expression-based control mechanism. In contrast to expression analyses comparing just two developmental stages, namely etiolated and fully greened plant material (Bouvier et al., 1996; Bugos and Yamamoto, 1996; http://genome-www4.stanford.edu/Microarray/SMD), we followed the expression levels of the three xanthophyll biosynthetic genes at different illumination intervals. The resulting expression profiles of bhy and zep were similar, but clearly distinct from that of vde. This finding implies a coordination in gene expression of these two xanthophyll biosynthetic genes (bhy and zep), which are necessary to increase the content of β-carotene derivatives with light-harvesting functions (violaxanthin and neoxanthin). High steady-state transcript levels of bhy and zep were found in early stages of chloroplast development after irradiation times of 3 or 5 h, respectively.
In the case of violaxanthin de-epoxidase, a posttranslational control might be a more decisive factor than the regulation of its gene expression because vde transcript levels and the resulting product formation were shown not to be correlated by Rossel et al. (2002). Although we detected higher vde mRNA levels during de-etiolation under ML in comparison with LL, it has to be kept in mind that the enzyme itself requires a low luminal pH and higher light conditions to become active (Hager, 1975). In accordance, we found a transient accumulation of zeaxanthin in early greening stages after 3 and 5 h of ML illumination only, which indicates that this light intensity is sufficient to lead to an acidification of the lumen as a prerequisite for the activation of the enzyme violaxanthin de-epoxidase (Pfündel and Strasser, 1988). Such a transient zeaxanthin accumulation and concomitant reduction in violaxanthin content during chloroplast development were also detected by Krol et al. (1999) performing greening experiments under different temperature conditions in combination with ML and high-light intensities. Moreover, changes in PSII excitation pressure and its release as chloroplast differentiation proceeds might result in the (transient) accumulation of certain xanthophylls (compare with Montané et al., 1998).
Effect of Different Light Qualities on the Steady-State mRNA Levels of Xanthophyll Biosynthetic Genes
The effect of different light qualities on gene expression was investigated after exposure of etiolated seedlings to blue, red, or white light of equal quanta. These light qualities led to an increase in the amount of the corresponding mRNAs pointing to an involvement of different photoreceptors (phytochrome and cryptochrome; Lin, 2002). However, the expression levels of the carotenoid genes under examination varied significantly in response to the light quality used. There was a prominent red-light-dependent increase of the steady-state transcript level of the phytoene synthase gene during de-etiolation, indicating a phytochrome control in accordance with data of von Lintig et al. (1997). A larger amount of mRNA in red light in comparison with blue light was also detected for β-lycopene cyclase, for β-carotene hydroxylase, and to a lower extent for violaxanthin de-epoxidase, suggesting that the expression of these genes is also under the control of the phytochrome system. In contrast, zeaxanthin epoxidase responded with a similar induction of gene expression in blue and red light. In this context it is noteworthy that various promoter elements have been identified that exhibit different sensitivities toward blue and red light (Lübberstedt et al., 1994).
Comparative Expression Analysis of Different Components of the Photosynthetic Apparatus
The expression pattern of the genes encoding β-carotene hydroxylase and zeaxanthin epoxidase resembled each other and exhibited high similarity to that of a gene encoding a chlorophyll a/b binding protein of the light-harvesting complex of PSII. These experimental data support the hypothesis that mRNA accumulation is coordinated for individual photosynthetic complexes (Beator and Kloppstech, 1993; Oelmüller et al., 1995). In fact, experiments of Batschauer et al. (1986) demonstrated that the cab mRNAs encoding light-harvesting proteins cannot accumulate in the absence of carotenoids, emphasizing their importance. Using tomato (Lycopersicon esculentum) plants subjected to light/dark cycles, Thompson et al. (2000) could show a high similarity between the expression pattern of the zeaxanthin epoxidase (LeZep 1) and a cab gene. Both exhibited fluctuations of their mRNA levels typical for circadian cycling genes. This finding was recently confirmed by microarray data available for zeaxanthin epoxidase and cab genes (Schaffer et al., 2001; compare with Green et al., 2002; Standard Microarray Database, http://afgc.stanford.edu). On the basis of the observed cycling of the expression level, we postulate that not only the expression of zep but also of bhy is under circadian clock control.
In addition to carotenoids, chlorophylls are essential constituents of the photosynthetic complexes. Therefore, a chlorophyll biosynthetic gene, namely ppox, was included in the expression studies. Analysis of its mRNA pattern revealed a moderate up-regulation upon exposure of etiolated tobacco seedlings to white light (compare with Ma et al., 2001), but higher transcript levels were only detectable in later stages of chloroplast development. Similar to the results observed for bhy gene expression, the transcript level of ppox was slightly but consistently elevated under LL conditions. The increase in transcript levels of ppox and bhy could be causally linked to the augmentation in chlorophyll and carotenoid content (Sisler and Klein, 1963; Akoyunoglou and Argyroudi-Akoyunoglou, 1966). The availability of higher amounts of these light-harvesting pigments might then in turn affect the antenna size, which was previously shown to be enlarged in LL-adapted plants (Thayer and Björkman, 1990).
Effect of Photosynthetic Electron Transport Inhibitors on the mRNA Transcript Levels of Xanthophyll Biosynthetic Genes
Redox control of photosynthetic gene expression has been recently recognized as an important regulatory mechanism (for review, see Pfannschmidt et al., 2001). Within the electron transport chain, the redox status of the plastoquinone pool was shown to specifically control the transcription of chloroplast-encoded genes of the core complexes of PSI and PSII in higher plants or the nuclear-encoded genes of the PSII light-harvesting system in the alga Dunaliella terticolecta (Escoubas et al., 1995; Pfannschmidt et al., 1999). Until now, no data are available that univocally prove the involvement of the plastquinone pool in the control of xanthophyll biosynthetic gene expression. To examine the role of the plastoquinone pool, we applied the site-specific photosynthetic electron transport inhibitors DCMU and DBMIB. DCMU partially inhibits the reduction of plastoquinone thereby decreasing PSII electron pressure and mimicking a transfer to lower light intensities (Teramoto et al., 2002). DCMU treatment of fully greened seedlings for 3 h resulted in an up-regulation of the transcript levels of the β-carotene hydroxylase (bhy) and zeaxanthin epoxidase (zep) gene relative to untreated controls. The expression of a light-harvesting gene (cab) used as control was induced in good agreement with Escoubas et al. (1995). The DCMU treatment also caused an increase in the photosynthetic pigment content (Table I), the same phenomenon was observed after a shift from higher to lower light intensities.
The photosynthetic inhibitor DBMIB partially blocks the oxidation of the plastoquinol therefore mimicking a transfer to higher light conditions. Application of this inhibitor to fully greened tobacco seedlings had opposite effects on the transcript levels of bhy and zep (namely a reduction of the amount of these mRNAs). Interestingly, the expression level of vde was reduced after DCMU as well as DBMIB incubation. Although a down-regulation under LL conditions might have been expected, the reduction of its mRNA level as response to DBMIB application was unforeseen. However, the fact that high-light illumination also led to a reduction in the amount of vde transcript supports our data (Rossel et al., 2002). Using microarrays, Rossel et al. (2002) found an increased transcript level of bhy upon high-light illumination (1,000 μmol m–2 s–1). Application of the photosynthetic electron transport inhibitor DCMU in their study was restricted to a selection of six genes that are not related to the formation of carotenoids. On the basis of our results derived from the inhibitor treatments, we demonstrate for the first time, to our knowledge, that bhy and zep transcript levels are redox controlled by the plastoquinone pool. In contrast, the expression of vde does not seem to be redox controlled but is rather influenced by the photosynthetic electron transport.
MATERIALS AND METHODS
Plant Material and Illumination
All experiments were carried out with tobacco (Nicotiana tabacum L. cv Samsun) seedlings. Seeds were surface sterilized and subsequently germinated and grown for 4 d on Murashige-Skoog agar medium (Murashige and Skoog, 1962) without Suc in complete darkness at a constant temperature of 24°C in a growth room. The seedlings used as experimental material contained only cotyledons. No primary leaves had developed even at the end of the light period. Seedlings were harvested at the same age either in the etiolated stage or after different light periods. Sample collection was carried out at the same time of day for all plant material. Determination of fresh weight revealed no significant changes excluding growth effects (data not shown). Etiolated seedlings were kept in the dark until the illuminated samples reached the end of the light period. To check for developmental influences, etiolated seedlings were also taken at different ages during the experiment. No significant differences were found in the etiolated seedlings, which was additionally verified by pigment analysis. For de-etiolation, the seedlings were transferred to continuous white light and irradiated for 3, 5, 8, 12, 24, or 48 h in the growth chamber. One set of seedlings was illuminated with a very LL intensity (10 μmol m–2 s–1), the other with a 10-fold higher intensity of 100 μmol m–2 s–1 (ML) provided by a combination of l-FLUORA and Universal White fluorescent tubes (Osram, Munich, Germany). After the end of the illumination period, seedlings were harvested and immediately frozen for RNA extraction and pigment analysis. Etiolated seedlings that were kept in darkness served as controls.
In light quality experiments, seedlings were exposed to equal amounts of photons (45 μmol m–2 s–1). Blue light was produced by a filter (Schott, Mainz, Germany) permitting light penetration from 320 to 500 nm and exhibiting a maximum at around 400 nm, for red light a cut off filter (Schott, Mainz, Germany) omitting light penetration of wavelengths shorter than 600 nm was used. White light was provided by a xenophot lamp (Osram).
Pigment Determination and Analysis
Pigments were extracted from 30 seedlings per determination and quantified according to Lichtenthaler and Wellburn (1983). For pigment distribution, pigments were separated on a Spherisorb ODS1 5 μ RP 18 column by HPLC as described by Gilmore and Yamamoto (1991). The flow rate was 1.2 mL min–1. Photosynthetic pigments were identified by their retention times, comparison of the retention times and absorption spectra with literature values, and co-chromatography with authentic standards.
Inhibitor Treatments
To investigate the impact of photosynthetic inhibitors on the expression of carotenoid biosynthetic genes and total pigment content, seedlings were germinated under continuous white light (100 μmol m–2 s–1) in water in Erlenmeyer flasks and shaken at 100 rpm. After germination, the 3-d-old seedlings were then subjected to the photosynthetic inhibitors DCMU and DBMIB. All seedlings were harvested after an incubation period of 3 h under continuous white light in the presence of the inhibitor. DCMU was added to a final concentration of 2 mm or 500 μm, respectively. The inhibitory effect on photosynthesis was verified by measuring oxygen evolution with a Clark electrode. Even for the highest DCMU concentration, only a 60% inhibition of photosynthetic activity was observed in good agreement with data of Petracek et al. (1997), thus ensuring that the applied concentrations were not lethal. DBMIB was added to a final concentration of 50 μm. The photosynthetic activity of these seedlings was inhibited by 43% compared with nontreated controls.
DNA Extractions and Manipulations
All DNA extractions were carried out in principle according to Sambrook et al. (1989). The restriction enzymes were purchased from Roche Diagnostics (Mannheim, Germany). Partial cDNAs for zeaxanthin epoxidase and violaxanthin de-epoxidase were obtained after reverse transcription of total RNA from leaves of tobacco (Nicotiana tabacum). Total RNA was extracted as described by Kuntz et al. (1992). Two micrograms of RNA were heated at 65°C for 5 min, chilled on ice, and then reverse transcribed using Moloney murine leukemia reverse transcriptase (MBI Fermentas, Wilna, Lituvia) and oligo(dT) (Pharmacia, Uppsala) as primer according to the protocol provided by the supplier. After heat inactivation of the enzyme for 5 min at 95°C, 2 to 5 μL of the reaction mixture was used either directly or after purification (PCR purification kit, Roche Diagnostics) for amplification with sequence-specific primers. Primers were designed as follows: violaxanthin de-epoxidase, 5′-GGTTTGGATTCAAGAGGTCTGC and 3′-CGGATACTTTGGATCTTGCACG; zeaxanthin epoxidase, 5′-TGGTAYTGYAARTTYGAYACITTYAC and 3′-GCRTCRTCRTCYTCRAACCAITT. The amplified PCR products (690 bp for violaxanthin de-epoxidase and 1,074 bp for zeaxanthin epoxidase) were cloned in a PCR vector pCR 2.1 (Invitrogen, Carlsbad, California) and verified by sequencing. The same primers were also used in PCR labeling reactions (see below).
DNA Labeling
DNA fragments were either labeled with DIG high prime DNA labeling mix according to the recommendations of the supplier (Roche Diagnostics) or by PCR amplification using dioxygenin-11-dUTP. A 548-bp DNA fragment encoding a chlorophyll a/b binding protein (cab) of tomato (Lycopersicon esculentum) was isolated after restriction digest of plasmid pTAB 2.0 with HincII and PvuII (Pichersky et al., 1985). After preparative gel electrophoresis, the DNA insert was eluted with a gel extraction kit from Biozym (Hess. Oldendorf, XXX, Germany). The insert was subsequently labeled as outlined in the manufacturers protocol with DIG high prime (Roche Diagnostics). All other gene probes were labeled by PCR amplification. A 605-bp probe for the plastidic ppox was generated by PCR with the upstream primer GCAGGAATTAGTGGCCTCTGC and downstream primer AGGTAAACGCGGATCGCGGGGCGC from the corresponding tobacco cDNA (Lermontova et al., 1997) as template. A 707-bp-labeled DNA fragment of β-carotene hydroxylase was obtained after PCR amplification using upstream primer AGAGGATCCTGAAATGAAAATTGAGGAGC (includes an inserted BamHI restriction site, underlined) and downstream primer CGAAGCTTCATGATCCCCTGG. A β-carotene hydroxylase cDNA (100 ng; S. Römer, unpublished results) isolated from a tobacco cDNA library (λ Zap II, Stratagene, La Jolla, CA; kindly provided by Dr. Grimm [IPK, Gatersleben, Germany]) was added as a template to the PCR reaction. The 50-μL reaction mix typically contained 50 to 200 ng of DNA, 5 μL of PCR buffer (Qiagen, Hilden, Germany), 10 μL of Q-solution (Qiagen), 4 μL of dNTP-labeling mix (Roche Diagnostics), 20 pmol of upstream and downstream primer, respectively, 1.5 mm MgCl2, and 0.5 μL (2.5 units) of Taq polymerase (Qiagen). PCR was carried out in a thermocycler (PTC-100 Perkin Elmer, Biozym, Hess. Oldendorf). PCR cycle programs were as follows: 25 cycles for amplification of β-carotene hydroxylase (1 min at 94°C, 25 s at 56°C, and 45 s at 72°C), 35 cycles for amplification of zeaxanthin epoxidase (1 min at 94°C, 45 s at 52°C, and 90 s at 72°C), 35 cycles for amplification of violaxanthin de-epoxidase (1 min at 94°C, 45 s at 54°C, and 90 s at 72°C), and 25 cycles for amplification of ppox (1 min at 94°C,25 s at 57°C, and 45 s at 72°C). Cycling was preceded by a 4-min denaturing period at 94°C and followed by a final extension at 72°C for 10 min. For amplification of β-carotene hydroxylase and ppox, 0.5 μL of extra Mg2+ from a 25 mm MgCl2 stock solution was added to the PCR master mix, whereas PCR amplification of zeaxanthin epoxidase and violaxanthin de-epoxidase was done in the presence of 0.8 μL of additional Mg2+.
RNA Extraction and Northern-Blot Hybridization
RNA was extracted from a minimum of 60 seedlings per measurement following the procedure outlined in Kuntz et al. (1992). After separation of total RNA on denaturing agarose/formaldehyde gels, the nucleic acids were transferred onto a positively charged nylon membrane (Biodyne Plus, Pall, Dreieich, Germany) and UV cross-linked. A photo of the ethidium bromide-stained RNA controlled for equal loading before blotting.
Blots were prehybridized in 50% (v/v) deionized formamide, 5 × SSC, 50 mm sodium-phosphate, pH 7.0, 2% (w/v) blocking reagent (Roche Diagnostics), 0.1% (w/v) N-lauroyl sarcosine, and 7% (w/v) SDS for at least 1 h in a hybridization oven (Biometra, Göttingen, Germany) at 42°C. After heat denaturation, 10 to 15 ng of the labeled probe was added per milliliter of hybridization solution. Hybridization was carried out overnight in the hybridization oven at 42°C. Afterward, unbound DIG-labeled DNA was removed by post-hydridization washes. Membranes were washed twice with 2 × SSC containing 0.2% (w/v) SDS for 15 min at room temperature followed by further washing steps with 0.5 × SSC and 0.2% (w/v) SDS and with 0.1 × SSC and 0.2% (w/v) SDS at 65°C. Hybridization signals were detected by chemiluminescence using CDP-Star according to the procedure described by the manufacturer (Roche Diagnostics). After gel-blot analysis and detection, the nylon membranes were incubated twice at 68°C with the northern probe-stripping solution (50% [v/v] formamide, 50 mm Tris-HCl, pH 8, and 1% [w/v] SDS) to remove the probe and could be reused for additional hybridizations.
Quantification of Northern Blots
After detection of the hybridization signals by chemiluminescence, the digitalized images were quantified by positioning a grid over the bands and integrating the individual cells of the grid. For integration the ImageQuant software from Molecular Dynamics (Sunnyvale, CA) was used. Results of the quantification of northern blots were normalized with respect to the total RNA.
Acknowledgments
We are grateful to Prof. B. Piechulla (University of Rostock, Germany) for providing us with the plasmid pTAB 2.0. Furthermore, we thank Prof. B. Grimm (IPK, Gatersleben, Germany) for his gift of the ppox cDNA of tobacco. We are also especially grateful to Prof. P. Böger for advice and support who has enabled us to carry out this work. We thank Dr. B. Lederer for helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019364.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Ro 2047/2–1, 2–2).
References
- Audran C, Borel C, Frey A, Sotta B, Meyer C, Simonneau T, Marion-Poll A (1998) Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiol 118: 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoyunoglou G, Argyroudi-Akoyunoglou JH (1966) Effect of intermittent and continuous light on the chlorophyll formation in etiolated plants at various ages. Physiol Plant 22: 288–295 [Google Scholar]
- Batschauer A, Mösinger E, Kreutz K, Dörr I, Apel K (1986) The implication of a plastid-derived factor in the transcriptional control of nuclear genes encoding the light-harvesting chlorophyll a/b protein. Eur J Biochem 154: 625–634 [DOI] [PubMed] [Google Scholar]
- Beator J, Kloppstech K (1993) The circadian oscillator coordinates the synthesis of apoproteins and their pigments during chloroplast development. Plant Physiol 103: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, d'Harlingue A, Backhaus RA, Kumagai MH, Camara B (2000) Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur J Biochem 267: 6346–6352 [DOI] [PubMed] [Google Scholar]
- Bouvier F, d'Harlingue A, Hugueney P, Marin E, Marion-Poll A, Camara B (1996) Xanthophyll biosynthesis: cloning, expression, functional reconstitution, and regulation of β-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). J Biol Chem 217: 28861–28867 [DOI] [PubMed] [Google Scholar]
- Bugos RC, Chang SH, Yamamoto HY (1999) Developmental expression of violaxanthin de-epoxidase in leaves of tobacco growing under high and low light. Plant Physiol 121: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugos RC, Yamamoto HY (1996) Molecular cloning of violaxanthin deepoxidase from romaine lettuce and expression in Escherichia coli. Proc Natl Acad Sci USA 93: 6320–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce R, Morosinotto T, Castelletti S, Breton J, Bassi R (2002) The Lhca antenna complexes of higher plants photosystem I. Biochim Biophys Acta 1556: 29–40 [DOI] [PubMed] [Google Scholar]
- Demmig B, Winter K, Krüger A, Czygan F-C (1987) Photoinhibition and zeaxanthin formation in intact leaves. Plant Physiol 84: 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B (1990) Carotenoids and photoprotection in plants: a role for the xanthophyll zeaxanthin. Biochim Biophys Acta 1020: 1–24 [Google Scholar]
- Escoubas J-M, Lomas M, LaRoche J, Falkowski PG (1995) Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA 92: 10237–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaggio E, Cinque G, Bassi R (2001) Functional architecture of the major light-harvesting complex from higher plants. J Mol Biol 314: 1157–1166 [DOI] [PubMed] [Google Scholar]
- Gilmore AM, Yamamoto HY (1991) Resolution of lutein and zeaxanthin using a non-endcapped lightly carbon-loaded C18 high-performance liquid chromatographic column. J Chromatogr 543: 137–145 [Google Scholar]
- Green RM, Tingay S, Wang Z-Y, Tobin EM (2002) Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 129: 576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A (1975) Die reversiblen, lichtabhängigen Xanthophyllumwandlungen im Chloroplasten. Ber Deutsch Bot Ges 88: 27–44 [Google Scholar]
- Havaux M (1998) Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci 3: 147–151 [Google Scholar]
- Herrin DL, Battey JF, Crus K, Schmidt GW (1992) Regulation of chlorophyll apoprotein expression and accumulation: requirements of carotenoids and chlorophyll. J Biol Chem 267: 8260–8269 [PubMed] [Google Scholar]
- Horton P, Ruban AV, Walters RG (1996) Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol 47: 655–684 [DOI] [PubMed] [Google Scholar]
- Krol M, Ivanov AG, Jansson S, Kloppstech K, Huner NP (1999) Greening under high light or cold temperature affects the level of xanthophyll-cycle pigments, early light-inducible proteins, and light-harvesting polypeptides in the wild-type barley and the chlorina f2 mutant. Plant Physiol 120: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367: 614–621 [DOI] [PubMed] [Google Scholar]
- Kuntz M, Römer S, Suire C, Hugueney P, Weil JH, Schantz R, Camara B (1992) Identification of a cDNA for the plastid-located geranylgeranyl pyrophosphate synthase from Capsicum annuum: correlative increase in enzyme activity and transcript level during fruit ripening. Plant J 2: 25–34 [DOI] [PubMed] [Google Scholar]
- Lee AJ, Thornber JP (1995) Analysis of the pigment-protein complexes from barley (Hordeum vulgare): the xanthophyll cycle intermediates occur mainly in the light-harvesting complexes of photosystem I and II. Plant Physiol 107: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Kruse E, Mock H-P, Grimm B (1997) Cloning and characterization of a plastidal and a mitochondrial isoform of tobacco protoporphyrinogen IX oxidase. Proc Natl Acad Sci USA 94: 8895–8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 1111: 591–592 [Google Scholar]
- Lin C (2002) Blue light receptors and signal transduction. Plant Cell Suppl 2002: 207–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H (1999) Carotenoid hydroxylase from Haematococcus pluvialis: cDNA sequence, regulation and functional complementation. Biochim Biophys Acta 1446: 203–212 [DOI] [PubMed] [Google Scholar]
- Lübberstedt T, Bolle CEH, Sopory S, Flieger K, Herrmann RG, Oelmüller R (1994) Promoters from genes for plastid proteins possess regions with different sensitivities toward red and blue light. Plant Physiol 104: 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Queseda A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A (1996) Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J 15: 2331–2342 [PMC free article] [PubMed] [Google Scholar]
- McCormac DJ, Marwood CA, Bruce D, Greenberg BM (1996) Assembly of photosystem I and II during early phases of light-induced development of chloroplasts from proplastids in Spirodelia oligorrhiza. Photochem Photobiol 63: 837–845 [Google Scholar]
- Montané M-H, Tardy F, Kloppstech K, Havaux M (1998) Differential control of xanthophylls and light-induced stress proteins, as opposed to light-harvesting chlorophyll a/b proteins, during photosynthetic acclimation of barley leaves to light irradiance. Plant Physiol 118: 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Niyogi KK (1999) Photoprotection revisited: genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol 50: 333–359 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O (1998) Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R, Schneiderbauer A, Herrmann RG, Kloppstech K (1995) The steady-state mRNA levels for thylakoid proteins exhibit coordinate diurnal regulation. Mol Gen Genet 246: 478–484 [DOI] [PubMed] [Google Scholar]
- Petracek ME, Dickey LF, Nguyen TT, Huber SC, Thompson WF (1997) Light-regulated changes in abundance and polyribosome association of ferredoxin mRNA are dependent on photosynthesis. Plant Cell 9: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Allen JF, Oelmüller R (2001) Principles of redox control in photosynthesis gene expression. Physiol Plant 112: 1–9 [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF (1999) Photosynthetic control of chloroplast gene expression. Nature 397: 625–628 [Google Scholar]
- Pfündel E, Strasser RJ (1988) Violaxanthin de-epoxidase in etiolated leaves. Photosynth Res 15: 67–73 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Bernatzky R, Tanksley SD, Breidenbach RB, Kausch AP, Cashmore AR (1985) Molecular characterisation and genetic mapping of 2 clusters of genes encoding chlorophyll a/b binding proteins in Lycopersicon esculentum (tomato). Gene 40: 247–258 [DOI] [PubMed] [Google Scholar]
- Plumley FG, Schmidt GW (1987) Reconstitution of chlorophyll a/b light-harvesting complexes: xanthophyll dependent assembly and energy transfer. Proc Natl Acad Sci USA 84: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer S, Hugueney P, Bouvier F, Camara B (1993) Expression of the genes encoding the early carotenoid biosynthetic enzymes in Capsicum annuum. Biochem Biophys Res Commun 196: 1414–1421 [DOI] [PubMed] [Google Scholar]
- Robertson D, Laetsch WM (1974) Structure and function of developing barley plastids. Plant Physiol 54: 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV, Pascal A, Lee PJ, Robert B, Horton P (2002) Molecular configuration of xanthophyll cycle carotenoids in photosystem II antenna complexes. J Biol Chem 277: 42937–42942 [DOI] [PubMed] [Google Scholar]
- Ruban AV, Young AJ, Horton P (1994) The effects of illumination on the xanthophyll composition of the photosystem II light-harvesting complexes of spinach thylakoid membranes. Plant Physiol 104: 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefermann-Harms D (1985) Carotenoids in photosynthesis: I. Location in photosynthetic membranes and light-harvesting function. Biochim Biophys Acta 811: 325–355 [Google Scholar]
- Sisler EC, Klein WH (1963) The effect of age and various chemicals on the lag phase of chlorophyll synthesis in dark-grown bean seedlings. Physiol Plant 16: 315–322 [Google Scholar]
- Steinbrenner J, Linden H (2001) Regulation of two carotenoid biosynthesis genes coding for phytoene synthase and carotenoid hydroxylase during stress-induced astaxanthin formation in the green alga Haematococcus pluvialis. Plant Physiol 125: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Gantt E, Cunningham FX Jr (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271: 24349–24352 [DOI] [PubMed] [Google Scholar]
- Takaichi S, Mimuro M (1998) Distribution and geometric isomerism of neoxanthin in oxygenic phototrophs: 9′-cis, a sole molecular form. Plant Cell Physiol 39: 968–977 [Google Scholar]
- Teramoto H, Nakamori A, Minagawa J, Ono T (2002) Light-intensity-dependent expression of Lhc gene family encoding light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Physiol 130: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer SS, Björkman O (1990) Leaf xanthophyll content and composition in sun and shade determined by HPLC. Photosynth Res 23: 331–343 [DOI] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Tziveleka LA, Argyroudi-Akoyunoglou JH (1998) Implications of a developmental-stage dependent thylakoid protease in the stabilization of the light-harvesting pigment-protein complex serving photosystem II during thylakoid biogenesis in red kidney bean. Plant Physiol 117: 961–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Welsch R, Bonk M, Guiliano G, Batschauer A, Kleinig H (1997) Light-dependent regulation of carotenoid biosynthesis occurs at the level of phytoene synthase expression and is mediated by phytochrome in Sinapis alba and Arabidopsis thaliana seedlings. Plant J 12: 625–634 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Westhoff P (2001) Biogenesis and origin of thylakoid membranes. Biochim Biophys Acta 1541: 91–101 [DOI] [PubMed] [Google Scholar]
- Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J (2000) Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211: 846–854 [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Gough S, Kannagara CG (1995) Chlorophyll biosynthesis. Plant Cell 7: 1039–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AJ (1991) The protective role of carotenoids in higher plants. Physiol Plant 83: 702–708 [Google Scholar]