Abstract
The establishment of the arbuscular mycorrhizal symbiosis results in a modification of the gene expression pattern in both plant and fungus to accomplish the morphological and physiological changes necessary for the bidirectional transfer of nutrients between symbionts. H+-ATPase enzymes play a key role establishing the electrochemical gradient required for the transfer of nutrients across the plasma membrane in both fungi and plants. Molecular analysis of the genetic changes in arbuscular mycorrhizal fungi during symbiosis allowed us to isolate a fungal cDNA clone encoding a H+-ATPase, GmPMA1, from Glomus mosseae (BEG12). Despite the high conservation of the catalytic domain from H+-ATPases, detailed analyses showed that GmPMA1 was strongly related only to a previously identified G. mosseae ATPase gene, GmHA5, and not to the other four ATPase genes known from this fungus. A developmentally regulated expression pattern could be shown for both genes, GmPMA1 and GmHA5. GmPMA1 was highly expressed during asymbiotic development, and its expression did not change when entering into symbiosis, whereas the GmHA5 transcript was induced upon plant recognition at the appressorium stage. Both genes maintained high levels of expression during intraradical development, but their expression was reduced in the extraradical mycelium. Phosphate, a key nutrient to the symbiosis, also induced the expression of GmHA5 during asymbiotic growth, whereas sucrose had a negative effect. Our results indicate that different fungal H+-ATPases isoforms might be recruited at different developmental stages possibly responding to the different requirements of the life in symbiosis.
The arbuscular mycorrhizal (AM) symbiosis formed between the Glomeromycota and the roots of most vascular plants is characterized by reciprocal nutrient exchange between both symbiotic partners. The fungus receives up to 20% of the photoassimilated carbon allocated by the plant to the root. In exchange, the fungus improves plant mineral supply (mainly in phosphate) through the external mycelium extending into the nutrient depletion area surrounding the root (Jakobsen, 1995). A major question in the study of the AM symbiosis is where and how this nutrient exchange takes place. During presymbiosis, the spores of the fungus have poor saprotrophic growth. In the absence of a host plant, limited growth is sustained for up to 4 weeks but then ceases, although the spore remains alive. No further development has been achieved in vitro in the absence of living plant roots. However, some nutrient additives, albeit insufficient to provide continuous growth, have been shown to be perceived (Mosse, 1959; Mosse and Phillips, 1971; Hepper, 1979; Mugnier and Mosse, 1987) and even metabolized (Bago et al., 1999). In the symbiotic interaction, however, the fungus enters the inner cortical root cells to form specialized haustoria called arbuscules. These are branched hyphae with a very thin wall, surrounded by apoplastic space and by the periarbuscular membrane formed by invagination of the plant plasma membrane. There is increasing evidence that phosphate, translocated from the soil through the fungus, is downloaded at the arbuscule interface where it is taken up by plant transporters (Rosewarne et al., 1999; Rausch et al., 2001). In contrast, little is known about where the exchange of carbon takes place. Inter- or intracellular hyphae (i.e. coils) formed in upper cortical cells may be alternative locations for carbon exchange. There is also uncertainty as to what form of carbon is transported at these interfaces. Several approaches have indicated that glucose may be preferred over fructose or sucrose as carbohydrate imported by the fungus from the apoplastic space (Saito, 1995; Shachar-Hill et al., 1995; Solaiman and Saito, 1997; Pfeffer et al., 1999). This is then converted to lipid for transfer within the external mycelium (Pfeffer et al., 1999; Bago et al., 2002). There, it serves to feed this side of the fungal colony that has limitations for the use of externally supplied carbon sources similar to the early developmental phase (Pfeffer et al., 1999). The external mycelium itself is highly active for phosphate uptake, but other mineral nutrients as well such as ammonium or nitrate from the soil solution are also taken up (Jakobsen et al., 1992; Johansen et al., 1992, 1993; Frey and Schüepp, 1993; Tobar et al., 1994; Bago et al., 1996). These nutrients are then transported to the plant through the fungal hyphae to the inner structures of the cortex.
It is known that phosphate and hexoses are usually translocated by means of symporters. These are sustained by an electrochemical gradient in the plasma membrane created by H+-ATPase enzymes. Therefore, an important role has long been proposed for these H+-ATPases at both the plant and the fungal symbiotic interfaces for either phosphate or carbon transport (Smith and Smith, 1990; Gianinazzi-Pearson et al., 1991). In plants, H+-ATPases form a large gene family that is either transcriptionally or/and posttranscriptionally regulated at different stages of plant development. In barley (Hordeum vulgare), Murphy et al. (1997) described the first plant H+-ATPase to be differentially expressed in response to mycorrhizal colonization. More recently, at least two H+-ATPase isoforms have been identified in the interaction of mycorrhizal fungi and tobacco (Nicotiana tabacum; Gianinazzi-Pearson et al., 2000). By using β-glucuronidase-fused promoter constructs, these authors showed induction of these two isoforms in arbuscule-containing cells.
In terms of fungal ATPases, Ferrol et al. (2000) isolated five gene fragments coding for homologs of H+-ATPases in G. mosseae using degenerate primers to the catalytic domain. The authors suggested that a H+-ATPase gene family existed in AM fungi, similar to the situation in plants. There is, however, no information about the conditions that modulate the expression of the genes coding for such isozymes or the developmental stage and location where they are expressed. As a consequence, there is no data about the role that these fungal H+-ATPases might play in the nutrient transfer during symbiosis.
In this paper, we describe the isolation of a new H+-ATPase gene, GmPMA1, that is highly expressed during the asymbiotic stage. We also demonstrate the developmental and nutrient-mediated regulation of this and another H+-ATPase gene, GmHA5, during the mycorrhizal symbiosis.
RESULTS
Isolation of a New H+-ATPase Gene from G. mosseae
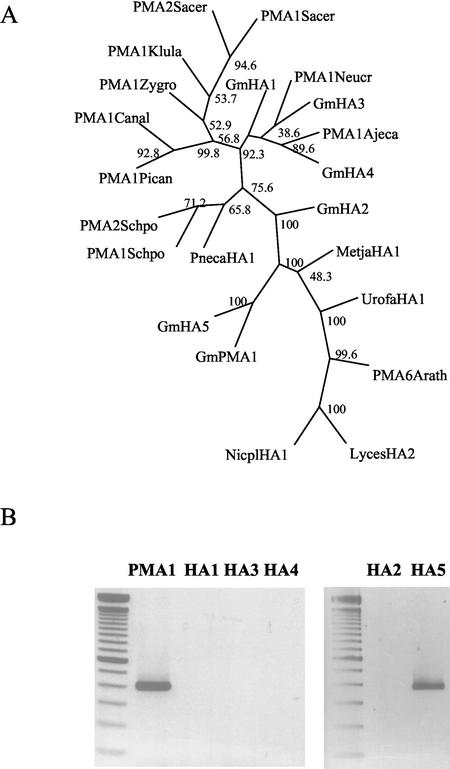
During the screening for genes expressed at early developmental stages of the AM fungal life cycle, a 570-bp cDNA fragment was found whose deduced amino acid sequence displayed high homology to a H+-ATPase from the Archaeon Methanococcus jannaschii, as well as to other H+-ATPases from fungi and plants (Requena et al., 2002). The cDNA sequence also showed homology to the partial gene sequence GmHA5, one of five H+-ATPases described for Glomus mosseae (Ferrol et al., 2000). It was much less similar to the other four H+-ATPase genes described in the same paper. The alignment of the nucleotide and the deduced amino acid sequences showed that the isolated cDNA clone encoded a new isoform that was named GmPMA1. A dendrogram of sequence similarity was created for the catalytic domain of all described G. mosseae H+-ATPase isozymes as well as for several H+-ATPases from other organisms (Fig. 1A). The dendrogram showed that the AM isoforms GmPMA1 and GmHA5 cluster together with other H+-ATPases including the plant H+-ATPases chosen in the analysis, as well as sequences from Archaea, red algae, and the basidiomycete Uromyces fabae. In contrast, the isoforms GmHA1, -2, -3, and -4 cluster with sequences from ascomycete H+-ATPases. The amino acid sequence alignment used to obtain this dendrogram showed that identical gaps existed in the ascomycete sequences as in the four mycorrhizal sequences GmHA1 to -4 (data not shown).
Figure 1.
A, Dendrogram of distances of the catalytic domain from 22 H+-ATPase proteins. GmPMA1 and GmHA5 were aligned using the program ClustalW to protein sequences of H+-ATPases from Brewer's yeast (Saccharomyces cerevisiae; PMA1Sacer, P05030; PMA2Sacer, P19657), Kluyveromyces lactis (PMA1Klula, P49380), Zygosaccharomyces rouxii (PMA1Zygro, P24545), Candida albicans (PMA1 Canal, P28877), Pichia angusta (PMA1 Pican, deduced from AF109913), Neurospora crassa (PMA1 Neucr, P07038), Ajellomyces capsulatus (PMA1 Ajeca, AAB53772); fission yeast (Schizosaccharomyces pombe; PMA1Schpo, P09627; PMA2Schpo, P28876), Pneumocystis carinii (PnecaHA1, AAB06958), M. jannaschii (MetjaHA1, deduced from U67563), U. fabae (UrofaHA1, CAA05841), Arabidopsis (PMA6Arath, Q9SH76), wild tobacco (Nicotiana plumbaginifolia; NicplHA1, A43637), tomato (Lycopersicon esculentum; Lyce-sHA2, AAD55399) and to those deduced from the gene fragments GmHA1 to GmHA5 (AJ133839–AJ133843). Evolutionary distances were calculated based on the Dayhoff PAM matrix using the neighboring-joining method. Bootstrap values of 1,000 data resamplings are indicated. A similar dendrogram was obtained using the program package PUZZLE. B, PCR amplification on genomic DNA from G. mosseae with primers that amplify a similar region in the catalytic domain of H+-ATPases. Primers were designed based on the published sequences for GmHA1 to GmHA5 (AJ133839–AJ133843) and in the sequence of the new isolated gene GmPMA1 (AY149918).
With the aim of isolating the complete genomic sequence coding for the whole H+-ATPase family, a PCR screening was performed on both genomic DNA and on a genomic library from G. mosseae (Hosny et al., 1999). Specific primers for genes GmHA1 to GmHA5 as well as for GmPMA1 were used. Surprisingly, only genes coding for the GmPMA1 and GmHA5 isoforms were present in G. mosseae genomic DNA (Fig. 1B). A low-stringency screening of the genomic library using the GmPMA1 original cDNA clone allowed the isolation of three clones of approximately 11 kb, which, after restriction analysis and partial sequencing, were identical to the gene encoding the isoform GmPMA1 (data not shown).
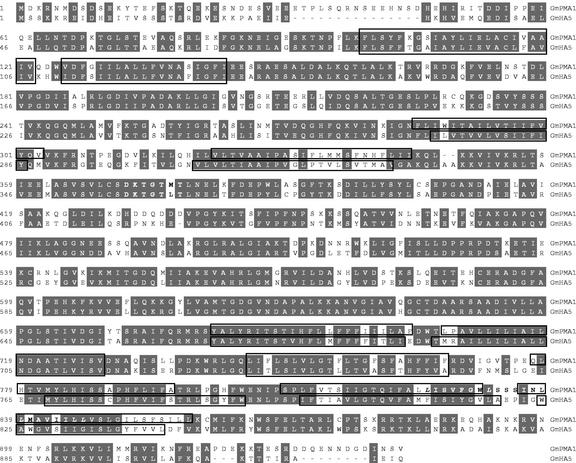
RACE successfully yielded the full-length cDNA clones encoding GmPMA1 and GmHA5, whose sequence were deposited in the National Center for Biotechnology Information database (accession nos. AY149918 and AY193825). An alignment of the deduced amino acid sequences demonstrated that both cDNAs code for different H+-ATPases isoforms with an overall similarity higher than 75% (Fig. 2). The GmPMA1- and GmHA5-deduced proteins have a molecular mass of approximately 105 and 100 kD, respectively, similar to other described H+-ATPases. They contain 10 putative transmembrane domains characteristic of P-type H+-ATPases with the catalytic domain including the E1-E2 phosphorylation site at the beginning of the large cytoplasmic loop (Fig. 2). The Prosite analysis of the GmPMA1-deduced protein showed also the presence of a Leu-zipper pattern with a probability of 7.006e-05 (Falquet et al., 2002) stretching between the middle of the last two transmembrane domains (Fig. 2).
Figure 2.
Amino acid alignment of the deduced proteins GmPMA1 (942 amino acids) and GmHA5 (917 amino acids). Conserved amino acids are marked in gray. The overall similarity between the two sequences is over 75%. The predicted 10 transmembrane helices of both proteins are boxed. The second cytoplasmic loop contains the catalytic domain with the E1-E2 ATPase phosphorylation site (DKTGTMT in GmPMA1; DKTGTLT in GmHA5) in bold. A Leu zipper motif stretching between the last two transmembrane domains and facing outside of the membrane was also found in GmPMA1, and it is also marked in bold.
Development Regulates Expression of the G. mosseae H+-ATPases
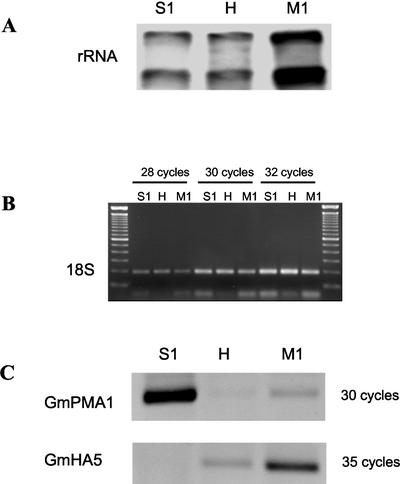
Given that the gene coding for the GmPMA1 isozyme was found in a differential screening of different stages of the fungal life cycle, we were interested in how GmPMA1 and GmHA5 were regulated during development. RNA was extracted from germinated sporocarps (12 d old), from mycorrhizal roots (2-month-old plants), and from extraradical hyphae from those plants. Equal amounts of total RNA from sporocarps and hyphae and five times more of total RNA from mycorrhizal roots were used to synthesize single-stranded (ss) cDNA (Fig. 3A). The primers VAGLO and VANS1, amplifying a fragment from the G. mosseae 18S rRNA, were used to assess that all three ss cDNA samples contained equivalent amounts of fungal cDNA (Fig. 3B). RT-PCR analyses with specific primers for GmPMA1 and GmHA5 on these ss cDNAs showed that the gene GmPMA1 was highly and preferentially expressed during asymbiotic growth but was down-regulated during the symbiotic phase (about 5- to 7-fold reduction; Fig. 3C). However, the gene coding for the isozyme GmHA5 showed a contrasting expression pattern with very low expression during non-symbiotic growth and highly induced expression (50-fold induction) in mycorrhizal roots and extraradical hyphae (8-fold induction).
Figure 3.
Expression of GmPMA1 and GmHA5 H+-ATPases from G. mosseae during different life cycle stages. A, Northern-blot calibration of RNA from the samples used to synthesize the ss cDNA hybridized with a DIG probe containing part of the rRNA genes from Glomus mosseae (H, extraradical hyphae; S1, germinated sporocarps; M1, mycorrhizal roots). B, Calibration by PCR of fungal cDNA present in the ss cDNA synthesized from samples S1, H, and M1. The oligonucleotides VAGLO and VANS1 specific for the 18S rRNA gene from G. mosseae were used as primers. One microliter of a 1:10,000 dilution of the ss cDNA was used as template in 28, 30, and 32 cycles of PCR. C, Reverse transcriptase (RT)-PCR expression pattern of both H+-ATPase genes during presymbiosis (S1), extraradical hyphae (H), and mycorrhizal roots (M1). One microliter of cDNA was used as a template in all cases.
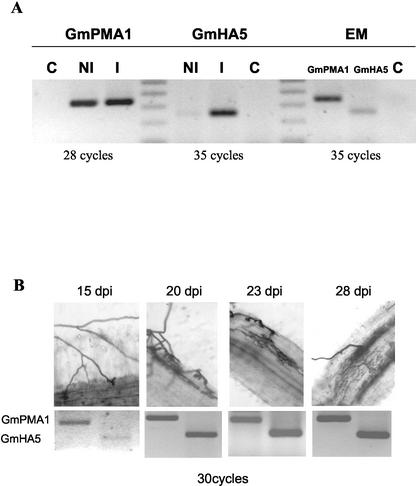
To determine at which point during symbiosis the GmHA5 was induced, we analyzed the expression of both H+-ATPase genes in germinated sporocarps during appressorium formation. Parsley (Petroselinum crispum) seedlings were used as a host, and both symbionts were kept together between two celophane membranes for 10 d. By this time, a large number of appressoria were formed. Plant-triggered sporocarps (induced) were compared versus control sporocarps (noninduced). The expression analysis at this stage showed that although there was no change in the expression level of GmPMA1 after contacting the host plant, strong induction of GmHA5 took place (almost 12-fold; Fig. 4A). Despite the small possible amount of fungal material inside the root at this time, the expression of both genes was already detectable in these roots where appressoria were formed (early mycorrhizal roots). At this stage, GmPMA1 was still more highly expressed than GmHA5 (almost three times).
Figure 4.
A, RT-PCR expression of GmPMA1 and GmHA5 H+-ATPase genes at the appressoria formation stage. C, Water control; NI, noninduced G. mosseae sporocarps; I, induced G. mosseae sporocarps; EM, early mycorrhizal roots). B, RT-PCR expression analysis of GmPMA1 and GmHA5 H+-ATPases during a time course of plant colonization. The top panel shows the trypan blue staining of parsley roots colonized by G. mosseae at 15, 20, 23, and 28 dpi. Although at 15 dpi, only appressoria can be observed on the rhizodermis, further colonization of the cortex can be seen at later time points. The bottom panel shows the expression of both genes at the different time points and the number of PCR cycles used. Relative expression levels were as follows: GmPMA1, 15 dpi = 0.12; 20 dpi = 0.20; 23 dpi = 0.30; 28 dpi = 0.30. GmHA5, 15 dpi = 0.04; 20 dpi = 0.20; 23 dpi = 0.33; 28 dpi = 0.31.
A time-course experiment of mycorrhizal colonization showed how progression of fungal development inside the root cortex affected expression of both H+-ATPase genes. Parsley seedlings collected after 15, 20, 23, and 28 d postinoculation (dpi) with G. mosseae were used for this experiment. Results showed that although very few fungal structures were visible at 15 dpi (basically only multiple appressoria on the epidermal cells; Fig. 4A), expression of GmHA5 is already clearly detectable. As infection progressed and Paris-type development was observed (see pictures in Fig. 4B), the level of GmHA5 expression increased, and both genes were expressed at comparable levels (Fig. 4B).
Phosphate and Sucrose Regulate the Expression of the GmHA5 H+-ATPase Gene during Asymbiosis
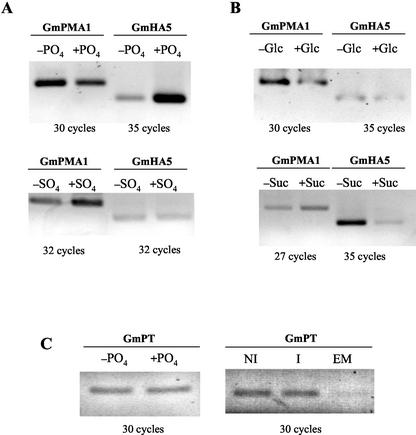
We addressed the question of whether some of the more relevant nutrients in the mycorrhizal symbiosis are able to be perceived and alter the expression of the fungal H+-ATPase genes during asymbiosis. Germinated sporocarps of G. mosseae were exposed to phosphate, Glc, or Suc for 48 h. RT-PCR analyses showed that minute amounts (35 μm) of potassium phosphate induced expression of GmHA5 (about 5-fold) but had no significant effect on expression of GmPMA1 (Fig. 5A). Potassium sulfate did not increase expression of GmHA5, demonstrating a phosphate-specific effect. Glc did not have any noticeable effect on the expression of either H+-ATPase, but Suc had a marked inhibitory effect on expression of GmHA5 (approximately 5-fold reduction; Fig. 5B). The expression analysis of a G. mosseae ortholog of the high-affinity phosphate transporter GvPT (Harrison and van Buuren, 1995) isolated in this work showed that its basal level of expression was not affected by the addition of phosphate to the medium nor was it affected by the presence of the host plant (Fig. 5C).
Figure 5.
A, RT-PCR expression analysis of GmPMA1 and GmHA5 H+-ATPase genes during presymbiosis induced or not with 35 μm K2HPO4 (±PO4) or with K2SO4 (±SO4). B, RT-PCR expression of GmPMA1 and GmHA5 H+-ATPase genes during presymbiosis induced or not with 25 mm Glc (±Glc) or 29 mm Suc (±Suc). C, RT-PCR expression of the phosphate transporter GmPT in sporocarps induced or not with phosphate (±PO4) or with the plant (NI, noninduced with the plant; I, induced with the plant; EM, early mycorrhiza; expression of GmPT inside the roots at the appressoria stage). The number of PCR cycles used is indicated under each picture. Table II shows the relative expression levels for each RT-PCR analysis performed.
DISCUSSION
Plasma membrane H+-ATPases are key enzymes found in plants, fungi, and algae and possibly in archaebacteria and protozoa, although the ion specificity of the latter has not yet been proved (Portillo, 2000). Their function is to generate a proton electrochemical gradient across the plasma membrane required for effective cellular function. Thus, it provides the driving force for the uptake and efflux of ions and metabolites through an interface otherwise impermeable to them and allows the control of intracellular pH. The exchange of metabolites and ions across the plasma membrane is particularly important at the symbiotic mycorrhizal interface because it enables the control of exchanged nutrients between the soil-fungus-plant “compartments.” This maintains the equilibrium that defines the association as mutualistic and not as parasitic. In plants, H+-ATPases are encoded from a large multigene family, and their expression is developmentally regulated in a cell- and tissue-specific manner (Sze et al., 1999). Recently, two of these isoforms have been shown to be specifically regulated during mycorrhiza formation in tobacco (Gianinazzi-Pearson et al., 2000). Isozymes pma2 and pma4 were induced in arbuscule-containing cells showing that the de novo H+-ATPase activity in the periarbuscular membrane resulted from selective induction of these two genes. Similarly, Ferrol et al. (2002) showed changes in plant H+-ATPase expression levels in tomato, not only in roots, but also in leaves of mycorrhizal plants. In contrast, molecular database analysis shows that most fungi contain only one or two genes encoding H+-ATPases, and only one of them usually encodes the enzyme responsible for the main plasma membrane ATPase activity. For example, Brewer's yeast and fission yeast contain two ATPase genes (de Kerchove d'Exaerde et al., 1996). In contrast, the biotrophic rust fungus U. fabae possesses only one gene, although different alleles are sometimes observed in the dikaryotic phase (Struck et al., 1998). In AM fungi, Ferrol et al. (2000) isolated five different fragments corresponding to five putative isoforms of H+-ATPase using a PCR approach with degenerate primers on genomic DNA from G. mosseae. They concluded that in AM fungi, H+-ATPases were encoded by a multigene family as in plants. Using a suppressive subtractive hybridization technique to identify transcriptionally regulated genes during early stages of AM fungal development, we isolated a cDNA fragment from G. mosseae with high homology to different H+-ATPases (Requena et al., 2002). This fragment was different from the five already described, and we therefore isolated the full-length cDNA of this gene. The isolated transcript showed higher homology to the gene coding for the isoform GmHA5 from G. mosseae and to other P-type H+-ATPases from Archaea, fungi, and plants. A bootstrap analysis using the highly conserved catalytic domain from all known G. mosseae isozymes and 16 other related H+-ATPases showed that GmHA5 and the new isolated form, named GmPMA1, formed a separated cluster from the other four G. mosseae isozymes. The latter showed a closer relation with ascomycete H+-ATPase sequences. Specific primers designed to verify the presence of the different isozymes in the genome of G. mosseae showed that only genes coding for GmPMA1 and GmHA5 were present. Expression analysis using cDNA from different fungal developmental stages confirmed this result (data not shown). Therefore, we believe that only GmPMA1 and GmHA5 H+-ATPases belong to the fungus G. mosseae, although we do not exclude that other yet unknown isozymes might exist. GmHA1, -2, -3, and -4 could belong to one of the associated ascomycetes that often cohabit within spores of AM fungi (Redecker et al., 1999). It is interesting that the catalytic domain from the AM isoforms GmPMA1 and GmHA5 appear more closely related to those from plant H+-ATPases than to the catalytic domain of the fungal isoforms. Similar results were observed for U. fabae (Struck et al., 1998). Both the AM fungi and the rust fungus form biotrophic associations with their host plant, and it is tempting to speculate about either horizontal gene transfer or coevolution of the H+-ATPase genes between these fungi and plants.
We showed that symbiosis formation and nutrition seem to be determinants of the differential expression of the both G. mosseae H+-ATPase isoforms, and therefore we can speculate that specific isoforms might be differentially recruited at different developmental or nutritional stages as happens in plants. GmPMA1 is highly expressed during presymbiosis, whereas GmHA5 is expressed at very low levels at this stage. Presymbiotic GmPMA1 expression was not affected by the presence of different nutrients in the growth medium, nor by the presence of the host plant. A lower level of expression occurred during the in planta phase, but the expression dramatically dropped in the extraradical mycelium. In contrast, GmHA5 was strongly induced once appressoria began to form, and it remained highly expressed during the in planta phase. Its expression was higher in the extraradical mycelium than in asymbiotic hyphae, but levels were still low compared with those in planta.
GmHA5 expression was not only triggered by the host plant, but also by micromolar amounts of phosphate in the medium. Lei et al. (1991) described stimulation of plasmalemma ATPase activity in Gigaspora margarita by the exogenous addition of root factors. With the help of cytochemical localization techniques, they showed an increase in H+-ATPase activity at the hyphal root tip of stimulated hyphae and correlated this enhanced ATPase activity to a stimulation in phosphate uptake. However, these authors performed the plant induction experiment on a medium containing 35 μm KH2PO4, which we noted is sufficient to produce an increase in GmHA5 expression even in the absence of the plant. We tested whether induction of GmHA5 runs parallel to an increase in the expression of a homolog of the high-affinity phosphate transporter described for Glomus versiforme and for Glomus intraradices (Harrison and van Buuren, 1995; Maldonado-Mendoza et al., 2001). We showed that neither phosphate nor the presence of the host plant triggered induction of the G. mosseae phosphate transporter during the presymbiotic stage, but similar concentrations of phosphate induced its expression in extraradical hyphae of both G. intraradices (Maldonado-Mendoza et al., 2001). A possible explanation is that during asymbiotic growth, the basal expression level of the high-affinity phosphate transporter could be enough to supply the limited fungal biomass with phosphate at this stage. This is in agreement with results of Maldonado-Mendoza et al. (2001) who showed a tight regulation of the fungal high-affinity phosphate transporter related to the existing phosphate needs of the symbiont. In contrast, the proton gradient necessary for the cotransport of phosphate during presymbiosis would be insufficient and hence the increased GmHA5 expression. In fact, proton pumps have a very slow ion transport rate (approximately 100 ions s–1), in contrast to cotransporters (approximately 300–1,000 ions s–1), that has to be compensated with protein abundance (Sze et al., 1999).
Glc did not have any effect on the expression of either H+-ATPase gene during presymbiosis, in contrast to what was observed for plants and yeast (Mito et al., 1996; Portillo, 2000). The effect of Glc on H+-ATPase gene expression in other fungi has been found to be variable. Candida albicans and U. fabae respond weakly to Glc, whereas Aspergillus nidulans responds initially with a temporary slight increase but then a strong down-regulation (Monk et al., 1993; Struck et al., 1996; Abdallah et al., 2000). The Glc modulation of H+-ATPase expression has been related to the metabolic state of the cell, with the involvement of the Tuf/Rap1/Gcr1 transcription factors also involved in the control of glycolytic genes (Capieaux et al., 1989; Rao et al., 1993; Scott and Baker, 1993). There is evidence that a small proportion of Glc is taken up into the presymbiotic mycelium of AM fungi after several days of incubation (Shachar-Hill et al., 1995; Bago et al., 1999), but its significance to the growth of the fungus seems to be minor in contrast to the breakdown of internal reserves. This could explain why no increase in H+-ATPase protein is required, and therefore there is no change in expression level.
In contrast, Suc down-regulated the expression of GmHA5 during presymbiosis. This could be correlated with the negative effect of Suc on G. mosseae growth reported both at presymbiosis (Mosse, 1959) and in the extraradical mycelium phase (Mugnier and Mosse, 1987). Suc levels higher than 29 mm inhibit the attachment of mycorrhizal hyphae to root surface and the formation of symbiosis. However, it is possible that the amount of Suc used in these experiments exceeded the physiological levels to which the fungus usually meets in soil. The same could be true for the amount of Suc that the fungus faces in the apoplast, given that all isolated plant high-affinity Suc transporters have a Km in the range of 0.2 to 2 mm (Lemoine, 2000; Weise et al., 2000). This would explain why GmHA5 is not down-regulated during the in planta phase but is up-regulated. The next challenge will be to ascertain the localization and activity of both GmPMA1 and GmHA5 in the in planta phase and their role in the carbon transport.
MATERIALS AND METHODS
Strain and Culture Conditions
Isolation of Fungal Asymbiotic Material
Sporocarps from the AM fungus Glomus mosseae (BEG12) were purchased from Biorize (Dijon, France) and surface-sterilized according to Budi et al. (1999). Sporocarps were germinated on MES-buffered water-agar plates as described elsewhere (Requena et al., 1999) and grown in an incubator at 25°C in the dark. After 12 d, the germinated sporocarps were harvested using fine forceps to recover all hyphae and immediately frozen in liquid nitrogen. RNA from germinated sporocarps was extracted as described below and used for expression experiments.
Isolation of Fungal Symbiotic Material
Parsley (Petroselinum crispum) plants were inoculated with G. mosseae (BEG12) and grown in pots divided into compartments separated by a thin nylon mesh (50 μm), which allowed the passage of extraradical hyphae but not of roots (Redecker et al., 1995). The hyphal compartment contained 2-mm glass beads from which the extraradical mycelium could be easily recovered. Plants were grown for 2 months in a phytochamber under controlled conditions of light (150 μE m–2 s–1), temperature (20°C), and photoperiod (14 h of light). Extraradical hyphae were then isolated from the glass bead compartment, washed several times in chilled sterile distilled water, and frozen in liquid nitrogen. Colonized roots free of extraradical hyphae were also collected, washed, and immediately frozen in liquid nitrogen. RNA from both extraradical hyphae and mycorrhizal roots was extracted as described below. The experiment was conducted twice.
Developmental Expression of GmPMA1 and GmHA5
Expression of both H+-ATPase genes GmPMA1 and GmHA5 was studied during different stages of mycorrhizal development. It is then important that RNA from germinated sporocarps, mycorrhizal roots, and extraradical hyphae contain equal amounts of fungal material. To balance the amount of fungal RNA in the mycorrhizal root sample with the other two samples, ss cDNA was synthesized using approximately five times more total RNA in the mycorrhizal samples. Thus, 50 ng of total RNA from spores and hyphae and approximately 250 ng of total RNA from mycorrhizal roots was used to synthesize ss cDNA using hexamer oligonucleotides according to the protocol of the Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). cDNA amounts were estimated by PCR using the primers VAGLO and VANS1 that amplify a 200-bp fragment from the small ribosomal subunit (Simon et al., 1992, 1993). One microliter from a dilution 1:10,000 was used for the PCR, which was performed for 28, 30, and 32 cycles. Expression analyses with specific primers for the H+-ATPase genes were then performed as described below.
Time-Course of Plant Colonization by G. mosseae
One hundred parsley seedlings were grown on 30-mL pots containing inoculum of G. mosseae (BEG12) consisting of terra-green substrate with sporocarps and mycorrhizal roots as propagules. The plants were grown as described above, and the roots were harvested at 15, 20, 23, and 28 dpi. A portion of the root was used for trypan blue staining (Phillips and Hayman, 1970), and the rest was used for RNA isolation and RT-PCR analysis (see below).
Appressorium Induction on Parsley Seedlings
Appressorium formation was triggered by bringing axenically germinated sporocarps of G. mosseae (BEG12) into contact with parsley seedlings between two cellophane membranes on water-agar plates. The plates were kept at room temperature for 240 h, after which the fungal material was carefully separated from the plant under the stereo-microscope. Fungus (induced) and root (early mycorrhiza) were separately frozen in liquid nitrogen. Non-triggered sporocarps maintained under the same growth conditions were used as controls (noninduced). RNA was isolated from the three different samples and used for RT-PCR analysis (see below).
Nutrient Effect on H+-ATPase Expression
Germinated sporocarps were transferred to water-agar plates buffered with MES and containing the different nutrients tested. These were: 25 mm Glc, 29 mm Suc, 35 μm potassium dihydrogen phosphate, or 17.3 μm potassium sulfate. Each of the nutritional treatments had its respective water control consisting of water-agar plates that were also MES buffered.
Isolation of GmPMA1 and GmHA5
A (570-bp) cDNA fragment with homology to several plasma membrane H+-ATPases was isolated in a previous screening by suppressive subtractive hybridization comparing presymbiotic fungal growth versus extraradical hyphae (Requena et al., 2002). Sequence comparison using the National Center for Biotechnology Information database showed that this clone was a new isoform, different from other previously identified from the same fungus (Ferrol et al., 2000). The corresponding full-length cDNA was obtained by RACE using the SMART RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA). Two nested PCR reactions were performed on the SMART cDNA using two pairs of internal primers based on the sequence of the original cDNA fragment (Table I). Total RNA (100 ng) extracted using the RNAeasy kit (Qiagen USA, Valencia, CA) from germinated sporocarps was used as template. The PCR reactions were performed using the proof-reading enzyme Advantage 2 (BD Biosciences Clontech). The obtained fragments were cloned into the pCR2.1 TOPO vector and sequenced (MWG Biotech). The new deduced isozyme was named GmPMA1. A similar procedure was employed to isolate the full-length gene of the previously identified H+-ATPase isozyme GmHA5.
Table I.
List of the primers used in this work
Primers 1 to 8 were used for the RACE experiments with GmPMA1 and GmHA5. Primers 9 to 16 were used to detect the presence of the other four putative H+-ATPase isozymes in the genome of G. mosseae. Expression experiments concerning GmPMA1 were performed with primers 2 and 3, whereas primers 17 and 18 were used for GmHA5. Expression analyses with GmPT were performed with primers 19 and 20.
| Primer No. | Primer Name | Sequence from 5′ to 3′ |
|---|---|---|
| 1 | GmPMA1F2 | tttcccagaaatgtcccgg |
| 2 | GmPMA1F3 | ccagtcaagaaagttcccaag |
| 3 | GmPMA1R1 | cacgttcagctgctgatatcg |
| 4 | GmPMA1R2 | gttggctccaggattatcaacc |
| 5 | GmHA5R1 | gcaaaaccatcggcgcgttcac |
| 6 | GmHA5Rev | tcttcatctgatttctcgggatcaacg |
| 7 | GmHA5F0 | cacaaagatgtcatacgcaacc |
| 8 | GmHA5F1 | gtcttcgtgcccttggtatcgc |
| 9 | GmHA1F1 | actcgtggtttccgttcccttg |
| 10 | GmHA1R1 | tcctccaccgagaccgagc |
| 11 | GmHA2F1 | agtttgcacgccgtggcttcc |
| 12 | GmHA2R1 | cgacagtgtatttgtgtccaggg |
| 13 | GmHA3F1 | cacccgtggtttccgttcgc |
| 14 | GmHA3R1 | accgccaccgagaccaagg |
| 15 | GmHA4F1 | cccgtggtttccgttctcttgg |
| 16 | GmHA4R1 | caccgccaccaaggccgag |
| 17 | GmHA5up1 | aataatgtctccgaggcgagg |
| 18 | GmHA5up3 | ttgaagtagcttgccttttcgc |
| 19 | GmPTF1 | ccaacacgttacagatcaac |
| 20 | GmPTR1 | gtgataaaccttttgtttcagg |
Database Comparison and Phylogenetic Analysis
Using the deduced amino acid sequence of the catalytic domain of the H+-ATPases GmPMA1 and GmHA5, an alignment was carried out using the program ClustalW (Thompson et al., 1994) with data from the SWISSPROT and the GenBank databases. Phylogenetic analysis was conducted with the program packages PHYLIP (Phylogeny Inference Package, v3.573c, Department of Genetics, University of Washington, Seattle) and PUZZLE (v4.02; Strimmer and von Haeseler, 1996). On the basis of the results, a dendrogram was constructed using the program TREEVIEW (Page, 1996). Accession numbers of the sequences used in the alignment are given in the figure legend.
Detection of H+-ATPase Isoforms in G. mosseae Genomic DNA
PCR amplification of a genomic segment located inside the catalytic domain was used to prove the existence of the different H+-ATPase isoforms in the genome of G. mosseae (BEG12; GmPMA1 from this study and GmHA1 to GmHA5 previously identified by Ferrol et al. [2000]). Genomic DNA from 1,000 axenically germinated spores was isolated using the Qiagen DNAeasy kit and PCR amplified with specific primers for each isozyme. The primers were designed to amplify a fragment of similar length in the same area based on the published sequences (GmHA1–GmHA5) and on the new isolated sequence (GmPMA1) Table I. Southern-blot screening of a genomic library (Hosny et al., 1999) was performed using the original GmPMA1 cDNA fragment as a probe under nonastringent conditions.
RT-PCR Expression Analyses
Total RNA from germinated sporocarps either induced or not with different nutrients, appressorium stage fungus, extraradical hyphae, or mycorrhizal plants was extracted using the RNAeasy kit (Qiagen USA) and DNase treated using the DNase I enzyme from Promega (Madison, WI). RNA quality and quantity was assessed by northern blot using a DIG probe containing the ribosomal genes from G. mosseae (Franken and Gianinazzi-Pearson, 1996). Identical amounts of RNA were used to synthesize cDNA using oligo(dT) as primer according to the protocol of the Superscript II Reverse Transcriptase (Invitrogen) to be used for the RT-PCR experiments. Transcript levels for both H+-ATPases, GmPMA1 and GmHA5, were estimated by PCR using specific primers spanning an intron, which detect possible genomic DNA contamination (see Table I). The number of cycles and the amount of template used were adjusted for every single experiment to be on the exponential phase of the PCR as noted on each of the figures. A G. mosseae ortholog of the high-affinity phosphate transporter GvPT from Glomus versiforme (Harrison and van Buuren, 1995) was isolated using primers from G. versiforme designed in highly conserved regions. The PCR fragments were cloned and sequenced, and specific primers for G. mosseae were designed (Table I). Expression of the G. mosseae phosphate transporter (GmPT) was studied by RT-PCR in germinated sporocarps induced or not with 35 μm of potassium dihydrogen phosphate for 48 h. GmPT expression was also assayed in germinated sporocarps triggered by the host plant at the appressorium formation stage versus non-triggered sporocarps as well as versus the expression of the GmPT in the early mycorrhiza (see above). All RT-PCR products were quantified by densitometric analysis of ethidium bromide-stained bands using the computer program Quantity One (Bio-Rad Laboratories, Hercules, CA). The values were calculated as a ratio to the amount of RNA in each sample also determined densitometrically. RT-PCR analyses were performed in duplicate on independent samples.
Table II.
Relative expression levels corresponding to Figure 5
NI, Noninduced with the plant; I, induced with the plant; EM, early mycorrhiza.
Acknowledgments
We thank Drs. Uwe Nehls and Reinhard Fischer for critical comments on the manuscript. Special thanks to Prof. P. Jeffries for proofreading our manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019042.
The work was supported in part by structural funds from the University of Tübingen. N.R. was supported by a Margarete von Wrangell Habilitation Stipendium.
References
- Abdallah BM, Simoes T, Fernandes AR, Strauss J, Seiboth B, Sa-Correia I, Kubicek CP (2000) Glucose does not activate the plasma-membrane-bound H+-ATPase but affects pmaA transcript abundance in Aspergillus nidulans. Arch Microbiol 174: 340–345 [DOI] [PubMed] [Google Scholar]
- Bago B, Pfeffer PE, Douds DD Jr, Brouillette J, Becard G, Shachar-Hill Y (1999) Carbon metabolism in spores of the arbuscular mycorrhizal fungus Glomus intraradices as revealed by nuclear magnetic resonance spectroscopy. Plant Physiol 121: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bago B, Vierheilig H, Piche Y, AzconAguilar C (1996) Nitrate depletion and pH changes induced by the extraradical mycelium of the arbuscular mycorrhizal fungus Glomus intraradices grown in monoxenic culture. New Phytol 133: 273–280 [DOI] [PubMed] [Google Scholar]
- Bago B, Zipfel W, Williams RM, Jun J, Arreola R, Lammers PJ, Pfeffer PE, Shachar-Hill Y (2002) Translocation and utilization of fungal storage lipid in the arbuscular mycorrhizal symbiosis. Plant Physiol 128: 108–124 [PMC free article] [PubMed] [Google Scholar]
- Budi SW, van Tuinen D, Martinotti G, Gianinazzi S (1999) Isolation from the Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towards soilborne fungal pathogens. Appl Environ Microbiol 65: 5148–5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capieaux E, Vignais ML, Sentenac A, Goffeau A (1989) The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem 264: 7437–7446 [PubMed] [Google Scholar]
- de Kerchove d'Exaerde AD, Supply P, Goffeau A (1996) Subcellular traffic of the plasma membrane H+-ATPase in Saccharomyces cerevisiae. Yeast 12: 907–916 [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrol N, Barea JM, Azcón-Aguilar C (2000) The plasma membrane H+-ATPase gene family in the arbuscular mycorrhizal fungus Glomus mosseae. Curr Genet 37: 112–118 [DOI] [PubMed] [Google Scholar]
- Ferrol N, Pozo MJ, Antelo M, Azcón-Aguilar C (2002) Arbuscular mycorrhizal symbiosis regulates plasma membrane H+-ATPase gene expression in tomato plants. J Exp Bot 53: 1638–1687 [DOI] [PubMed] [Google Scholar]
- Franken P, Gianinazzi-Pearson V (1996) Construction of genomic phage libraries of the arbuscular mycorrhizal fungi Glomus mosseae and Scutellospora castanea and isolation of ribosomal RNA genes. Mycorrhiza 6: 167–173 [Google Scholar]
- Frey B, Schüepp H (1993) Acquisition of nitrogen by the external hyphae of arbuscular mycorrhizal fungi associated with Zea mays L. New Phytol 124: 221–230 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Arnould C, Oufattole M, Arango M, Gianinazzi S (2000) Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211: 609–613 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Smith SE, Gianinazzi S, Smith FA (1991) Enzymatic studies on the metabolism of vesicular-arbuscular mycorrhizas: V. Is H+-ATPase a component of ATP-hydrolysing enzyme activities in plant-fungus interfaces? New Phytol 117: 61–74 [Google Scholar]
- Harrison MJ, van Buuren ML (1995) A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature 378: 626–629 [DOI] [PubMed] [Google Scholar]
- Hepper C (1979) Germination and growth of Glomus caledonius spores: the effects of inhibitors and nutrients. Soil Biol Biochem 2: 269–277 [Google Scholar]
- Hosny M, van Tuinen D, Jacquin F, Füller P, Zhao B, Gianinazzi-Pearson V, Franken P (1999) Arbuscular mycorrhizal fungi and bacteria: how to construct prokaryotic DNA-free genomic libraries from the Glomales. FEMS Lett 170: 425–430 [Google Scholar]
- Jakobsen I (1995) Transport of phosphorus and carbon in VA mycorrhizas. In A Varma, B Hock, eds, Mycorrhiza. Springer-Verlag, Berlin, pp 297–324
- Jakobsen I, Abbott LK, Robson AD (1992) External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum: 2. Hyphal transport of 32P over defined distances. New Phytol 120: 509–516 [Google Scholar]
- Johansen A, Jakobsen I, Jensen ES (1992) Hyphal transport of 15N-labelled nitrogen by a vesicular-arbuscular mycorrhizal fungus and its effect on depletion of inorganic soil N. New Phytol 122: 281–288 [DOI] [PubMed] [Google Scholar]
- Johansen A, Jakobsen I, Jensen ES (1993) External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L.: 3. Hyphal transport of 32P and 15N. New Phytol 124: 61–68 [Google Scholar]
- Lei J, Bécard G, Catford JG, Piché Y (1991) Root factors stimulate 32P uptake and plasmalemma ATPase activity in vesicular-arbuscular mycorrhizal fungus, Gigaspora margarita. New Phytol 118: 289–294 [DOI] [PubMed] [Google Scholar]
- Lemoine R (2000) Sucrose transporters in plants: update in function and structure. Biochim Biophys Acta 1465: 246–262 [DOI] [PubMed] [Google Scholar]
- Maldonado-Mendoza IE, Dewbre GR, Harrison M (2001) A phosphate transporter gene from the extra-radical mycelium of anarbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Mol Plant-Microbe Interact 14: 1140–1148 [DOI] [PubMed] [Google Scholar]
- Mito N, Wimmers LE, Bennett AB (1996) Sugar regulates mRNA abundance of H+-ATPase gene family members in tomato. Plant Physiol 112: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk BC, Niimi M, Shepherd MG (1993) The Candida albicans plasma membrane and H+-ATPase during yeast growth and germ tube formation. J Bacteriol 175: 5566–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosse B (1959) The regular germination of resting spores and some observations on the growth requirements of an Endogone sp. causing vesicular-arbuscular mycorrhiza. Trans Br Mycol Soc 42: 273–286 [Google Scholar]
- Mosse B, Phillips JM (1971) The influence of phosphate and other nutrients on the development of vesicular-arbuscular mycorrhiza in culture. J Gen Microbiol 69: 157–166 [Google Scholar]
- Mugnier J, Mosse B (1987) Spore germination and viability of a vesicular arbuscular mycorrhizal fungus Glomus mosseae. Trans Br Mycol Soc 88: 411–413 [Google Scholar]
- Murphy PJ, Langridge P, Smith SE (1997) Cloning plant genes differentially expressed during colonisation of Hordeum vulgare L by the vesicular-arbuscular mycorrhizal fungus Glomus intraradices. New Phytol 135: 291–301 [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pfeffer PE, Douds DD Jr, Becard G, Shachar-Hill Y (1999) Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol 120: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Hayman DS (1970) Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans Br Mycol Soc 55: 158–161 [Google Scholar]
- Portillo F (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim Biophys Acta 1469: 31–42 [DOI] [PubMed] [Google Scholar]
- Rao R, Drummond-Barbosa D, Slayman CW (1993) Transcriptional regulation by glucose of the yeast PMA1 gene encoding the plasma membrane H+-ATPAse. Yeast 9: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Rausch C, Daram P, Brunner S, Jansa J, Laloi M, Leggewie N, Bucher M (2001) A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414: 462–466 [DOI] [PubMed] [Google Scholar]
- Redecker D, Hijri M, Dulieu H, Sanders IR (1999) Phylogenetic analysis of a dataset of fungal 5.8S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet Biol 28: 238–244 [DOI] [PubMed] [Google Scholar]
- Redecker D, Thierfelder H, Werner D (1995) A new cultivation system for arbuscular mycorrhizal fungi on glass beads. Angew Bot 69: 189–191 [Google Scholar]
- Requena N, Fuller P, Franken P (1999) Molecular characterization of Gm-FOX2, an evolutionarily highly conserved gene from the mycorrhizal fungus Glomus mosseae, down-regulated during interaction with rhizobacteria. Mol Plant-Microbe Interact 12: 934–942 [DOI] [PubMed] [Google Scholar]
- Requena N, Mann P, Hampp R, Franken P (2002) Early developmentally regulated genes in the arbuscular mycorrhizal fungus Glomus mosseae: identification of GmGIN1, a novel gene with homology to the C-terminus of metazoan hedgehog proteins. Plant Soil 244: 129–139 [Google Scholar]
- Rosewarne GM, Barker SJ, Smith SE, Smith FA, Schachtman DP (1999) A Lycopersicon esculentum phosphate transporter (LePT1) involved in phosphorus uptake from a vesicular-arbuscular mycorrhizal fungus. New Phytol 144: 507–516 [DOI] [PubMed] [Google Scholar]
- Saito M (1995) Enzyme activities of the internal hyphae and germinated spores of an arbuscular mycorrhizal fungus, Gigaspora margarita Becker & Hall. New Phytol 129: 425–431 [Google Scholar]
- Scott EW, Baker HV (1993) Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol 13: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar-Hill Y, Pfeffer PE, Douds D, Osman SF, Doner LW, Ratcliffe RG (1995) Partitioning of intermediary carbon metabolism in vesicular-arbuscular mycorrhizal leek. Plant Physiol 108: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Lalonde M, Bruns TD (1992) Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl Environ Microbiol 58: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Lévesque RC, Lalonde M (1993) Identification of endomycorrhizal fungi colonizing roots by fluorescent single-strand conformation polymorphism-polymerase chain reaction. Appl Environ Microbiol 59: 4211–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Smith FA (1990) Structure and function of the interfaces in biotrophic symbioses as they relate to nutrient transport. New Phytol 114: 1–38 [DOI] [PubMed] [Google Scholar]
- Solaiman MDZ, Saito M (1997) Use of sugars by intraradical hyphae of arbuscular mycorrhizal fungi revealed by radiorespirometry. New Phytol 136: 533–538 [DOI] [PubMed] [Google Scholar]
- Strimmer K, von Haeseler A (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol 13: 964–969 [Google Scholar]
- Struck C, Hahn M, Mendgen K (1996) Plasma membrane H+-ATPase activity in spores, germ tubes, and haustoria of the rust fungus Uromyces viciae-fabae. Fungal Genet Biol 20: 30–35 [DOI] [PubMed] [Google Scholar]
- Struck C, Siebels C, Rommel O, Wrnitz M, Hann M (1998) The plasma membrane H+-ATPase from the biotrophic rust fungus Uromyces fabae: molecular characterization of the gene (PMA1) and functional expression of the enzyme in yeast. Mol Plant-Microbe Interact 11: 458–465 [DOI] [PubMed] [Google Scholar]
- Sze H, Li X, Palmgren MG (1999) Energization of plant cell membranes by H+-pumping ATPases: regulation and biosynthesis. Plant Cell 11: 677–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobar RM, Azcón R, Barea JM (1994) Improved nitrogen uptake and transport from N-15-labeled nitrate by external hyphae of arbuscular mycorrhiza under water-stressed conditions. New Phytol 126: 119–122 [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]