Abstract
The SNF1/AMP-activated protein kinase subfamily plays central roles in metabolic and transcriptional responses to nutritional or environmental stresses. In yeast (Saccharomyces cerevisiae) and mammals, activating and anchoring subunits associate with and regulate the activity, substrate specificity, and cellular localization of the kinase subunit in response to changing nutrient sources or energy demands, and homologous SNF1-related kinase (SnRK1) proteins are present in plants. We isolated cDNAs corresponding to the kinase (LeSNF1), regulatory (LeSNF4), and localization (LeSIP1 and LeGAL83) subunits of the SnRK1 complex from tomato (Lycopersicon esculentum Mill.). LeSNF1 and LeSNF4 complemented yeast snf1 and snf4 mutants and physically interacted with each other and with LeSIP1 in a glucose-dependent manner in yeast two-hybrid assays. LeSNF4 mRNA became abundant at maximum dry weight accumulation during seed development and remained high when radicle protrusion was blocked by abscisic acid (ABA), water stress, far-red light, or dormancy, but was low or undetected in seeds that had completed germination or in gibberellin (GA)-deficient seeds stimulated to germinate by GA. In leaves, LeSNF4 was induced in response to ABA or dehydration. In contrast, LeSNF1 and LeGAL83 genes were essentially constitutively expressed in both seeds and leaves regardless of the developmental, hormonal, or environmental conditions. Regulation of LeSNF4 expression by ABA and GA provides a potential link between hormonal and sugar-sensing pathways controlling seed development, dormancy, and germination.
The transition between seed development and germination is accompanied by large-scale changes in gene expression patterns and metabolic pathways. During seed maturation, enzymes and pathways involved in storage reserve accumulation and preparation for desiccation predominate (Girke et al., 2000). Upon imbibition after seed maturation and desiccation, different sets of genes are expressed for reserve mobilization, tissue weakening, and embryo expansion associated with radicle protrusion and seedling growth (Bradford et al., 2000; Gallardo et al., 2002). Abscisic acid (ABA) is an important regulator of seed development and maturation, and several transcription factors determining ABA sensitivity, including ABI3/VP1, ABI4, and ABI5, along with biosynthetic enzymes controlling ABA amount (ABA1, ABA2, and ABA3), are involved in promoting reserve accumulation, developmental arrest, and the imposition of dormancy (Finkelstein et al., 2002; Koornneef et al., 2002). GAs, on the other hand, are essential for germination and subsequent reserve mobilization to support seedling growth (Karssen, 1995; Debeaujon and Koornneef, 2000). Antagonistic interactions between ABA and GA in regulating reserve mobilization in cereal aleurone layers are well documented (Gomez-Cadenas et al., 2001).
Recently, screens for mutants altered in their sensitivity to the presence of sugars during germination and early seedling development have revealed interactions between sugar and hormonal signaling pathways (Finkelstein and Gibson, 2001; Rolland et al., 2002). In particular, a series of mutations have been identified in Arabidopsis based on the ability of mutant seeds to complete germination and of seedlings to develop in the presence of concentrations of Glc, Man, or Suc that are inhibitory to wild-type seeds. These include mig (Man-insensitive germination), sun (Suc-uncoupled), sis (sugar-insensitive), gin (Glc-insensitive), isi (impaired Suc induction), and other mutants exhibiting altered germination or seedling growth responses to sugars (for review, see Finkelstein et al., 2002). Interestingly, a number of mutations identified in this way are allelic to previously known mutations in ABA synthesis (aba) or sensitivity (abi). For example, sun6, sis5, gin6, and isi3 are allelic to abi4; sis4, gin1, and isi4 are allelic to aba2; and some (abi4, abi5, aba1, aba2, and aba3), but not all (abi1, abi2, and abi3), aba and abi mutants exhibit sugar-insensitive phenotypes (Finkelstein et al., 2002). Furthermore, provision of additional sugars partially overcame the inhibition of germination by ABA in both wild-type and abi mutant seeds (Garciarrubio et al., 1997; Finkelstein and Lynch, 2000a). Conversely, ABA increased the expression of a starch biosynthetic gene (ApL3) in response to sugars, suggesting that ABA can enhance the ability of tissues to respond to sugar signals (Rook et al., 2001). ABA also plays important but distinct roles in sugar, osmotic, and cold stress signaling during germination and early seedling growth (Cheng et al., 2002; Kim et al., 2003). Interactions between sugar and hormonal signaling pathways have also been reported for GA, ethylene, and cytokinins (for review, see Gazzarrini and McCourt, 2001; Rolland et al., 2002). Thus, hormonal and sugar regulation of gene expression and plant function are intimately linked, particularly in the case of the transition from seed maturation to germination.
The mechanisms by which plant cells sense sugars and regulate carbohydrate metabolism are complex and multiple pathways have been identified (Gibson, 2000; Smeekens, 2000; Rolland et al., 2002). The SNF1-related kinase (SnRK1) complex is thought to be a central component of the sugar sensing and response mechanism (Halford and Hardie, 1998; Halford et al., 2000, 2003). First identified genetically in yeast (Saccharomyces cerevisiae) as mutants incapable of derepressing Glc-regulated genes and, therefore, unable to grow on sugars other than Glc, the SNF1 (Suc non-fermenting 1) kinase complex and the related mammalian AMP-activated protein kinase (AMPK) complex are metabolic sensors of Glc availability and AMP to ATP ratios, respectively (Hardie et al., 1998). Protein sequence and functional homologies exist between the yeast and mammalian kinase (α) subunits (SNF1/AMPKα), regulatory (γ) subunits (SNF4/AMPKγ), and specification/localization (β) subunits (SIP1/SIP2/GAL83/AMPKβ) that constitute the functional kinase complexes. In yeast, Glc regulates the protein-protein interactions, substrate specificity, and subcellular localization of this complex that modulate SNF1 kinase activity, resulting in the phosphorylation of activators and repressors that control transcription of multiple genes in metabolic pathways required for the utilization of alternative energy sources (Carlson, 1998; Schmidt and McCartney, 2000; Vincent et al., 2001). In mammals, activation of AMPK due to increases in the AMP to ATP ratio during metabolic stress results in the enhancement of ATP-producing pathways and the inhibition of ATP-consuming pathways (Kemp et al., 1999).
Members of several subfamilies of SnRKs have been identified in diverse plants (Halford et al., 2000, 2003). Those most closely related to SNF1 (SnRK1 family) exhibit kinase activity on substrates common to the mammalian and yeast kinases and complement yeast snf1 mutants (e.g. Alderson et al., 1991; Muranaka et al., 1994; Sugden et al., 1999). Antisense suppression experiments in potato (Solanum tuberosum) and wheat (Triticum aestivum) indicated that SnRK1s are involved in regulation of carbon metabolism in planta (Purcell et al., 1998; Laurie et al., 2003). Candidate SNF4-related genes have been identified, including Pv42 in bean (Phaseolus vulgaris; Abe et al., 1995) and AKINγ (Bouly et al., 1999) in Arabidopsis. Plant genes having predicted sequence homology to SnRK1-β subunits are also known, including StubGAL83 from potato (Lakatos et al., 1999) and AKINβ1 and AKINβ2 from Arabidopsis (Bouly et al., 1999). Genes in maize (Zea mays; ZmAKINβγ-1) and Arabidopsis (AtSNF4 or AKINβγ) contain domains homologous to both the β- and γ-subunits of the SnRK1 complex (Lumbreras et al., 2001). Thus, plants contain homologs of all three components common to the yeast and mammalian SNF1/AMPK complexes, and these components interact in a manner consistent with a heterotrimeric structure (or a dimeric structure in the case of the βγ-type proteins) based on yeast two-hybrid and in vivo protein interaction studies (Bouly et al., 1999; Lakatos et al., 1999; Kleinow et al., 2000; Crawford et al., 2001; Ferrando et al., 2001; Lumbreras et al., 2001). Distinct regulatory components such as PRL1 may mediate additional functions of SnRK1 kinases in vivo (Bhalerao et al., 1999; Farrás et al., 2001).
In experiments to identify genes expressed or repressed in association with tomato (Lycopersicon esculentum Mill.) seed germination, we isolated a cDNA having sequence homology to the γ-subunit of the SnRK1 complex (LeSNF4). We report here the functional characterization of this gene and of tomato SnRK1-α and SnRK1-β homolog genes in yeast and their differential expression patterns during tomato seed development and germination. In addition, we show that expression of only LeSNF4 is responsive to GA, ABA, and water stress in tomato seeds and leaves. Our results suggest that hormonal regulation of expression of the γ-subunit of the SnRK1 complex could be one mechanism for cross talk between sugar-sensing pathways and hormonally and environmentally regulated pathways during seed development and germination and plant responses to stress conditions.
RESULTS
Isolation of Tomato SnRK1 Complex Genes
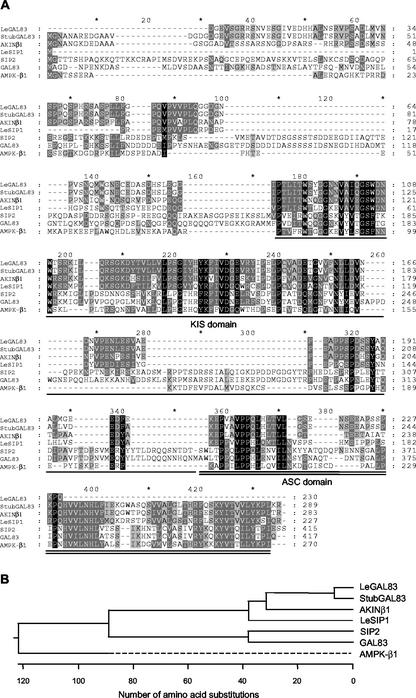
Differential cDNA display using GA-deficient mutant (gib-1) tomato seeds imbibed in the presence or absence of GA was employed to identify candidate gene(s) involved in the regulation or mechanism of seed germination (Cooley et al., 1999). An mRNA identified by one of the cDNAs was present in mature gib-1 seeds and remained abundant in gib-1 seeds imbibed in water (which do not complete germination), but declined within 12 h in the presence of GA, which stimulated germination (expression data presented later). A full-length cDNA corresponding to this mRNA was isolated from a tomato seed cDNA library prepared from gib-1 seeds imbibed for 24 h in water. The complete cDNA (GenBank accession no. AF143742) encoded a predicted protein of 373 amino acids (41,319 kD) having highest sequence homology to a predicted protein from bean (Pv42; 56% identity, 71% similarity) and a predicted protein from the Arabidopsis genome (AAD39660; 55% identity, 70% similarity; Fig. 1A). Regions of homology were also present in yeast SNF4 protein (21% identity, 40% similarity), in other Arabidopsis SnRK1-γ proteins (AKINγ and AtSNF4), and in the γ-domain of the maize ZmAKINβγ-1 protein. The LeSNF4, Pv42, and AAD39660 sequences clustered together, as did the AtSNF4, ZmAKINβγ-1, and SNF4 sequences, whereas the AKINγ sequence was divergent from the rest (Fig. 1B). A motif initially identified in the cystathionine-β-synthase protein (CBS motif) that is repeated four times in the yeast SNF4 and mammalian AMPKγ proteins (Bateman, 1997; Kemp et al., 1999) also appeared four times in the tomato sequence (Fig. 1A). Sequence conservation was greatest in the C-terminal regions of the proteins, coinciding with two of the CBS domains. Based on the sequence homology and functional complementation (see below), we termed the tomato gene LeSNF4.
Figure 1.
LeSNF4 encodes a γ-subunit homolog of the SnRK1 complex. A, Predicted amino acid sequences of tomato LeSNF4 (AF143742), bean Pv42 (U40713), Arabidopsis predicted protein (AAD39660), AKINγ (P80385), AtSNF4 (AF250335), the γ-domain (amino acids 125–497) of maize ZmAKINβγ-1 (AF276085), and yeast SNF4 (172636) were aligned using the Clustal method with PAM250 residue weight (DNASTAR Inc., Madison, WI). Shading indicates extent of conservation/similarity. In addition to direct sequence homology, CBS motifs based upon protein structure (http://www.sanger.ac.uk/cgi-bin/Pfam/) are shared between SNF4 (underlined) and LeSNF4 (overlined). B, Phylogenetic tree of protein sequences shown in A based upon Clustal analysis (DNAStar Inc.).
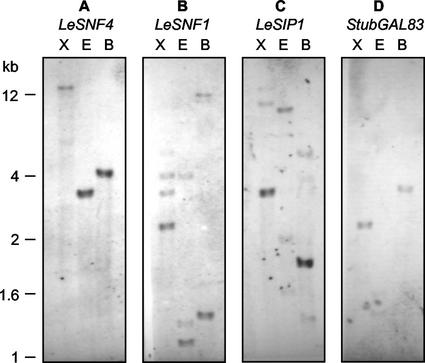
Hybridization analysis of tomato genomic DNA using LeSNF4 as probe revealed only single strong bands, indicating that the mRNA is likely to be encoded by a single gene (Fig. 2A). Genetic mapping supported this conclusion because Chen and Foolad (1999) used LeSNF4 as a DNA hybridization marker and mapped it to a single locus (CG43) on chromosome 6 of the tomato genome. A genomic DNA fragment was isolated encoding LeSNF4 (termed gLeSNF4; AF419320) that contained 1,234 bp 5′ to the translation start site and two introns of 323 bp (starting at bp 43 from the initial ATG of the open reading frame) and 923 bp (starting at bp 793; data not shown).
Figure 2.
DNA gel-blot analysis of tomato genomic DNA. A, Tomato genomic DNA (10 μg) was restriction digested with XbaI (X), EcoRI (E), or BamHI (B), separated on an agarose gel, and hybridized with LeSNF4 cDNA. B, As above, hybridized with LeSNF1 cDNA. C, As above, hybridized with LeSIP1 cDNA. D, As above, hybridized with StubGAL83 cDNA. In a separate experiment using the same restriction enzymes, hybridization with StubGAL83 or LeGAL83 cDNAs resulted in identical banding patterns (data not shown).
A cDNA encoding the tobacco (Nicotiana tabacum) SnRK1-α NPK5 (D26602; Muranaka et al., 1994) was used to isolate a homologous cDNA (termed LeSNF1; AF143743) from a tomato seed cDNA library (data not shown). The predicted amino acid sequence encoded a protein of 514 amino acids (58,824 kD) that is closely related to potato StubSNF1 (U83797; 95% similarity; Lakatos et al., 1999), NPK5 (87% similarity), and kinases from other species in the SnRK1a group (>80% amino acid similarity; Halford et al., 2000). The entire protein sequence of LeSNF1 shared 38% and 43% similarity with yeast SNF1 (M13971) and rat (Rattus norvegicus) AMPKα (U40819), respectively. The kinase domains comprising the N-terminal one-half of the protein shared 97% similarity among these plant proteins and >60% similarity among the plant, yeast, and mammalian proteins. In the more variable C-terminal region, LeSNF1 shared 92% and 76% similarity with StubSNF1 and NPK5, but only 12% to 16% similarity with SNF1 and AMPKα. Pien et al. (2001) reported a partial tomato cDNA encoding 161 C-terminal amino acids having 92% similarity with the corresponding region of StubSNF1, which they also termed LeSNF1. The reported sequence is identical with our sequence over this region. Based on DNA gel-blot analysis, LeSNF1 may be a member of a small gene family in the tomato genome (Fig. 2B).
A tomato expressed sequence tag (EST; AI486580) having significant homology to known SnRK1-β subunits, or SNF1-interacting proteins (SIPs), was used to isolate a full-length cDNA (termed LeSIP1; AF322108) from a tomato seed cDNA library. The predicted amino acid sequence of LeSIP1 shared 50% and 53% amino acid sequence similarity with AKINβ1 and StubGAL83 (Fig. 3A) and 70% with AKINβ2 (not shown). LeSIP1, StubGAL83, and AKINβ1 fell into a phylogenetic group distinct from the yeast (SIP2 and GAL83) and the mammalian (AMPKβ1) proteins (Fig. 3B). The predicted LeSIP1 amino acid sequence contained a conserved internal kinase-interacting sequence (KIS) domain and a C-terminal association with the SNF1 kinase complex (ASC) domain (Fig. 3A). These domains have been shown in yeast to interact with SNF1 (α-subunit) and with SNF4 (γ-subunit), respectively (Jiang and Carlson, 1997). The plant and mammalian proteins are considerably smaller than the yeast GAL83 and SIP2 proteins, with deletions in the region between the KIS and ASC domains and in the N-terminal region (Fig. 3A). DNA gel-blot analysis of tomato genomic DNA using LeSIP1 as a probe detected one major band and several weaker bands, suggesting a small family of related genes in tomato (Fig. 2C). This was confirmed by probing tomato genomic DNA with StubGAL83, which resulted in specific hybridization with a restriction fragment pattern distinct from that of LeSIP1 (Fig. 2D). Little cross hybridization was detected between the two probes at similar DNA fragment sizes, indicating the specificity of the probes for the respective genes. The tomato genome apparently contains at least two SIP-like sequences, LeSIP1 and a tomato homolog of StubGAL83. We subsequently recovered by reverse transcription-PCR a partial tomato cDNA sharing 98% identity in both nucleotide and amino acid sequences with the potato StubGAL83 cDNA (Fig. 3). A Southern blot of tomato genomic DNA using this tomato cDNA as a probe exhibited a hybridization pattern identical to that found when StubGAL83 was used as the probe (data not shown; Fig. 2D). Although not full length (230 amino acids versus 289), this tomato cDNA is evidently the tomato homolog of StubGAL83, so we have termed it LeGAL83 (accession no. AY245177).
Figure 3.
LeGAL83 and LeSIP1 have sequence homology with other SnRK1-β subunits. A, Predicted amino acid sequences of tomato LeGAL83 (AY245177), potato StubGAL83 (AJ012215), tomato LeSIP1 (AF322108), Arabidopsis AKINβ2 (AJ132316), rat AMPK-β1 (X95577), yeast SIP2 (L31592), and yeast GAL83 (X72893) aligned using the Clustal method with PAM250 residue weight (DNASTAR Inc.). Shading indicates extent of conservation/similarity. The conserved KIS and ASC domains are marked with single and double underlines, respectively. B, Phylogenetic tree of protein sequences shown in A based upon Clustal analysis (DNAStar Inc.).
LeSNF4 and LeSNF1 Encode Functional Proteins
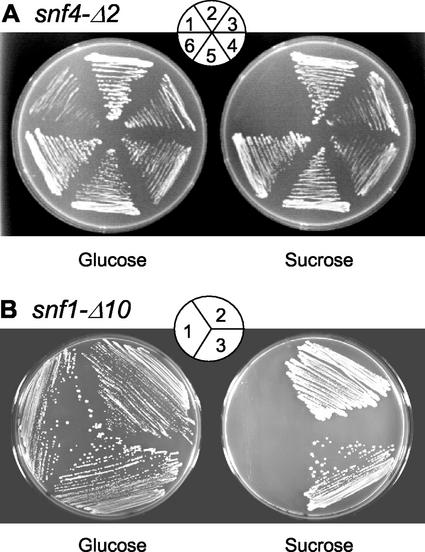
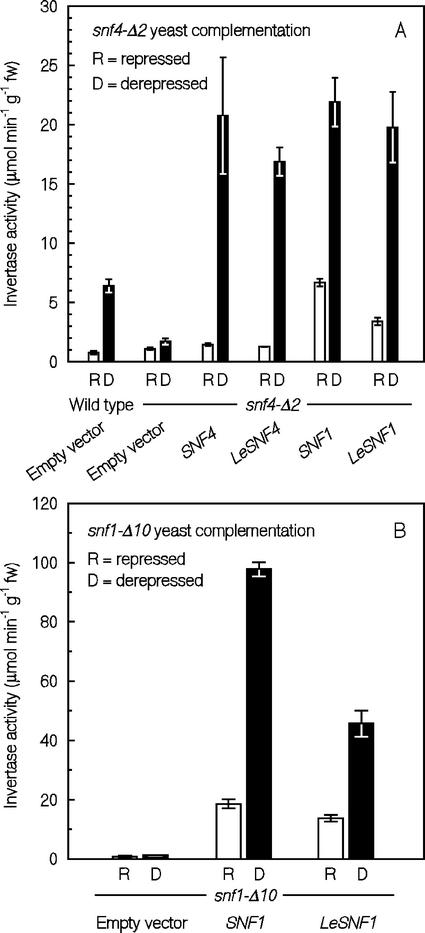
Whether LeSNF4 and LeSNF1 encode functional tomato homologs of the corresponding yeast genes was tested by complementation of mutants deficient in these genes. In yeast, the SNF4 protein is required for activation of SNF1 kinase, which is essential for derepression of sugar-metabolizing enzymes in the absence of Glc. As a consequence, snf4 mutants are able to grow on Glc but not on Suc or other carbohydrate sources (Fig. 4A). Expression of LeSNF4 in a yeast strain having a deletion in the SNF4 gene (snf4-Δ2) restored growth on Suc (or on 2% [w/v] each of Gal, glycerol, and ethanol plus 0.05% [w/v] Glc) essentially equivalent to that in the mutant yeast complemented with the wild-type SNF4 gene (Fig. 4A). Complementation was confirmed by measuring invertase activity from yeast cells in either repressed (2% [w/v] Glc) or derepressed (0.05% [w/v] Glc) conditions (Fig. 5A). Wild-type yeast exhibited an 8-fold increase in invertase activity under derepressed conditions. In contrast, only a 50% increase in invertase activity was detected under derepressed conditions in the snf4-Δ2 mutant transformed with the empty vector. However, when snf4-Δ2 yeast cells were transformed to express either SNF4 or LeSNF4 genes, invertase activity increased by 14- and 13-fold, respectively, under derepressed conditions (Fig. 5A). Thus, LeSNF4 is a functional homolog of SNF4 in regulating derepression of invertase in yeast.
Figure 4.
Complementation of yeast snf4-Δ2 and snf1-Δ10 mutants by LeSNF4 and LeSNF1.A, The snf4-Δ2 mutant was transformed with the empty vector (sector 1), with yeast SNF4 (sector 3), tomato LeSNF4 (sector 4), tomato LeSNF1 (sector 5), or yeast SNF1 (sector 6). Wild-type yeast transformed with the empty vector is shown in sector 2. Transformed strains containing gene inserts were initially identified on selective synthetic complete (SC) medium containing Suc (except those containing empty vector, which were grown on Glc), then were grown on selective SC plates with 2% (w/v) Glc (left) or Suc (right) for 10 d at 29°C. B, The snf1-Δ10 mutant was transformed with the empty vector (sector 1) or with tomato LeSNF1 (sector 2) or yeast SNF1 (sector 3). Transformed strains were selected as above, then were grown on selective SC plates with 2% (w/v) Glc (left) or Suc (right) for 3 d at 29°C. No colonies were recovered on Suc plates after transformation of snf1-Δ10 with vectors containing either LeSNF4 or SNF4 (data not shown). For both A and B, results similar to those shown on Suc medium were obtained when strains were grown on medium containing 2% (w/v) each of Gal, glycerol, and ethanol plus 0.05% (w/v) Glc (data not shown).
Figure 5.
Invertase activity of yeast snf4-Δ2 and snf1-Δ10 mutants expressing LeSNF4 and LeSNF1. A, Invertase activity from wild-type and snf4-Δ2 mutant yeast cells under either repressed (R, 2% [w/v] Glc) or derepressed (D, 0.05% [w/v] Glc) conditions. Mutant cells had been transformed with the empty vector or vectors expressing SNF4, LeSNF4, SNF1, or LeSNF1. Error bars indicate ±95% confidence intervals around the means for assays of three to four independent colonies. B, Invertase activity from snf1-Δ10 mutant yeast cells under either repressed (R, 2% [w/v] Glc) or derepressed (D, 0.05% [w/v] Glc) conditions. Mutant cells had been transformed with the empty vector, SNF1, or LeSNF1. Error bars = ± 95% confidence intervals around the means for assays of three to four independent colonies.
Similar experiments were conducted to test whether LeSNF1 encodes a functional homolog of the yeast SNF1 kinase. The yeast snf1-Δ10 deletion mutant can grow on Glc but not on Suc (Fig. 4B). Expression of LeSNF1 suppressed the mutation and allowed growth on Suc, as did expression of SNF1 (Fig. 4B). In addition, expression of either SNF1 or LeSNF1 stimulated secretion of invertase by snf1-Δ10 cells under both repressed and derepressed conditions, but invertase activity was greater in low Glc (Fig. 5B). When LeSNF4 was expressed in snf1-Δ10 cells, no transformants could be recovered from Suc plates, as expected from the inability of SNF4 to substitute for SNF1 kinase activity (Celenza et al., 1989). On the other hand, expression of either SNF1 or LeSNF1 in snf4-Δ2 mutant yeast cells restored the ability to grow on Suc (Fig. 4A) and stimulated invertase secretion under both repressed and derepressed conditions (Fig. 5A; Jiang and Carlson, 1996). Thus, we conclude that LeSNF1 encodes a functional homolog of SNF1.
LeSNF1, LeSNF4, and LeSIP1 Interact in Yeast Two-Hybrid Assays
The presence of conserved KIS and ASC domains predicted that LeSIP1 would bind to LeSNF1 and LeSNF4. Therefore, we tested whether the tomato components of the SnRK1 complex interacted with each other using the yeast two-hybrid assay (Table I). As expected (Jiang and Carlson, 1997), yeast SNF4 and SNF1 proteins showed an interaction that was enhanced under derepressing (0.05% [w/v] Glc) conditions. LeSNF1 and LeSNF4 proteins exhibited little interaction in high (2% [w/v]) Glc, but low Glc enhanced binding by 19-fold. LeSIP1 interacted strongly with both LeSNF1 and LeSNF4 even in high Glc, and this interaction was enhanced approximately 8-fold under low-Glc conditions. LeSNF1 and LeSNF4 showed little binding activity with SNF4 and SNF1, respectively, in high Glc, but strong interactions occurred in low Glc. LeSIP1 showed some binding to SNF1 and SNF4 but much less than with the corresponding tomato proteins. Both LeSNF1 and LeSIP1 exhibited apparent interaction with themselves under low-Glc conditions. We attempted to test the ability of StubGAL83 to interact with LeSNF1 and LeSNF4, but high expression was detected in controls when StubGAL83 was expressed alone in either the binding or activation domains, precluding meaningful tests (data not shown).
Table I.
Two-hybrid analyses of protein-protein interactions among tomato and yeast SnRK1 proteins
| Binding Domain (Bait) | Activation Domain (Prey) | Galactosidase Activity ± SEa | ||
|---|---|---|---|---|
| High Glc (R) | Low Glc (D) | D/Rb | ||
| nmol min-1 g fresh wt-1 | ||||
| SNF4 | SNF1 | 64.2 ± 5.8 | 300 ± 23 | 4.7 |
| LeSNF4 | LeSNF1 | 2.79 ± 0.20 | 52.3 ± 5.9 | 19 |
| LeSIP1 | LeSNF1 | 235 ± 32 | 1,880 ± 220 | 8.0 |
| LeSIP1 | LeSNF4 | 222 ± 11 | 1,670 ± 284 | 7.5 |
| LeSNF1 | SNF4 | 6.25 ± 0.82 | 392 ± 75 | 63 |
| LeSNF4 | SNF1 | 1.94 ± 0.60 | 47.3 ± 5.9 | 24 |
| LeSIP1 | SNF1 | 17.3 ± 1.4 | 46.4 ± 2.0 | 2.7 |
| LeSIP1 | SNF4 | 6.27 ± 1.05 | 21.0 ± 5.0 | 3.4 |
| LeSNF1 | LeSNF1 | 1.59 ± 0.31 | 62.7 ± 28.5 | 39 |
| LeSNF4 | LeSNF4 | NDc | 4.56 ± 1.11 | - |
| LeSIP1 | LeSIP1 | 18.9 ± 4.0 | 84.1 ± 19.1 | 4.4 |
| LeSNF1 | Empty | 6.89 ± 0.82 | NDc | - |
| Empty | LeSNF1 | 5.13 ± 1.57 | NDc | - |
| LeSNF4 | Empty | 5.32 ± 2.23 | NDc | - |
| Empty | LeSNF4 | 5.93 ± 1.54 | NDc | - |
| LeSIP1 | Empty | 3.44 ± 2.29 | NDc | - |
| Empty | LeSIP1 | NDc | NDc | - |
Yeast 190 cells transformed with different coding regions in the binding or activation domains of the yeast two-hybrid vector were grown in SC-Trp-Leu-His plus 2% (w/v) Glc (high Glc [R = repressed]) or 2% (w/v) each of galactose/glycerol/ethanol plus 0.05% (w/v) Glc (low Glc [D = derepressed]) broth containing 25 mM 3-amino-triazole. β-Galactosidase activities in crude protein extracts from three or four yeast colonies were measured as described in “Materials and Methods.” n = 3 or 4. b D/R = value in low Glc (D) divided by value in high Glc (R). c ND, Not detected.
Differential Expression of SnRK1-Related Genes in Tomato
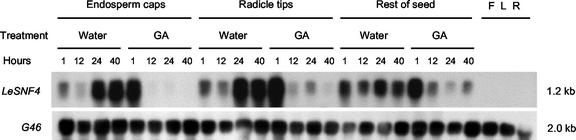
As noted earlier, LeSNF4 was identified as an mRNA that was present in mature gib-1 seeds but declined in the presence of GA (Fig. 6). The expression pattern was actually more complex because LeSNF4 mRNA first declined somewhat in gib-1 seeds imbibed in water before increasing to high abundance after extended imbibition without completing germination (Fig. 6). When imbibed in 100 μm GA4+7, on the other hand, an increased abundance of LeSNF4 mRNA was detected after 1 h, but the amounts then declined to low or undetectable levels before radicle emergence began at approximately 48 h (Fig. 6). The expression pattern was similar in the endosperm cap (endosperm tissue enclosing the radicle tip), the radicle tip, and the rest of the seed (including both the remainder of the embryo and the lateral endosperm surrounding it; Fig. 6; tissue print data not shown). Expression was apparently specific to the seed because the mRNA was not detected in flowers, leaves, or roots of wild-type field-grown plants (Fig. 6).
Figure 6.
Expression of LeSNF4 mRNA in gib-1 tomato seeds and wild-type tissues. Seeds were imbibed for the indicated time in either water or 100 μm GA4+7 at 25°C before dissecting into the endosperm caps (endosperm tissue covering the radicle tip), radicle tips, and the remainder of the seed (for diagram, see Cooley et al., 1999). In addition, flower (F), leaf (L), and root (R) tissues were collected from field-grown wild-type (cv Moneymaker) plants. Extracted total RNAs were hybridized to riboprobes prepared from LeSNF4 (upper) or from a cDNA fragment (G46) having homology to constitutively expressed ribosomal protein sequences to confirm equal RNA loading of each lane (lower).
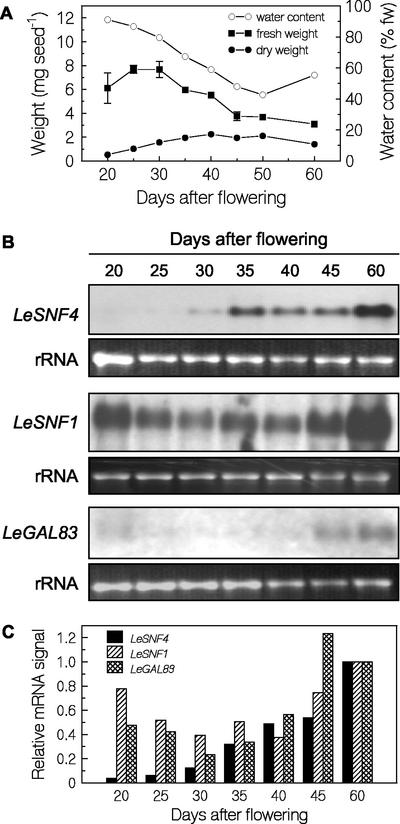
Because LeSNF4 mRNA was present in mature dry seeds and became less abundant during germination in response to GA, we examined the expression of tomato SnRK1-related genes during both seed development and germination. Tomato seeds initially increased in fresh weight as the tissues expanded, followed by dry weight accumulation due to deposition of storage reserves (Fig. 7A). Fresh weight of extracted seeds then fell, apparently associated with collapse of the testa cells (Fig. 7A; Berry and Bewley, 1991). Seed water content decreased during development, reaching values of 40% to 50% (w/w, fresh weight basis) at maturity inside the ripe fruit (Fig. 7A), followed by drying to 6% to 8% once removed from the fruit. LeSNF4 mRNA was barely detectable in tomato seeds at 20 and 25 DAF but increased in abundance after 30 DAF and was most abundant in mature seeds (60 DAF; Fig. 7, B and C). In contrast, LeSNF1 mRNA was abundant at 20 DAP, then declined somewhat before increasing again late in seed development (Fig. 7, B and C). LeSIP1 mRNA could not be detected in northern analyses of developing or germinating seeds (data not shown). However, StubGAL83 hybridized with an mRNA present late in seed development, indicating the expression of the highly homologous LeGAL83 gene (Fig. 7, B and C). Therefore, we used probe prepared from StubGAL83 to assay for the presence of tomato LeGAL83 mRNA. (These experiments were conducted before the isolation of the partial LeGAL83 cDNA described above, which confirmed the virtual identity of their sequences).
Figure 7.
Expression of SnRK1-related genes during tomato seed development. A, Fresh and dry weights and water contents (fresh weight basis) of tomato seeds during development. B, Abundance of LeSNF4, LeSNF1, and LeGAL83 mRNAs during tomato seed development. Total RNA was extracted from freshly harvested seeds at the times indicated and hybridized with probes prepared from the different genes (StubGAL83 probe was used to detect LeGAL83). Lower panels in each pair labeled rRNA are ethidium bromide-stained gels showing loading of RNA in each lane. C, Relative mRNA abundance was quantified by densitometry from the images in B and normalized for RNA loading in each lane. Within each panel, the normalized signal intensity at 60 d after flowering (DAF) was set equal to 1, and the normalized signal intensities in other lanes are shown relative to it.
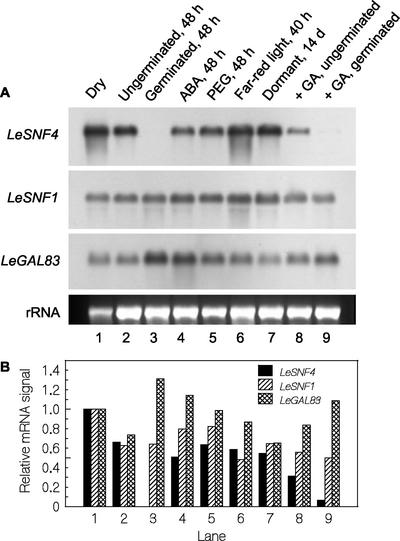
LeSNF4 mRNA was abundant in mature dry wild-type seeds but declined in seeds that had not completed germination by 48 h and was not detected in seeds that had completed germination (Fig. 8, lanes 1–3). LeSNF1 and LeGAL83 transcripts were also present in dry seeds and in imbibed germinating seeds but persisted even after the radicle had protruded (Fig. 8, lanes 1–3). Imbibing seeds in ABA (100 μm), in an osmotic solution (–1.3 MPa PEG), or under FR, all of which inhibited radicle emergence, maintained LeSNF4 mRNA abundance (Fig. 8, lanes 4–6). LeSNF4 mRNA remained abundant in naturally dormant seeds that had not completed germination after 14 d in water (Fig. 8, lane 7). When these dormant seeds were transferred to GA, LeSNF4 mRNA decreased but was still present in seeds that had not completed germination after 48 h and was absent in germinated seeds (Fig. 8, lanes 8 and 9). In contrast, ABA, osmoticum, dormancy, GA, or germination status had relatively little effect on the abundance of LeSNF1 or LeGAL83 mRNAs (Fig. 8, lanes 1–9).
Figure 8.
Expression of SnRK1-related genes in dry and imbibed tomato seeds under different conditions. A, Total RNA was extracted from mature dry wild-type (cv Moneymaker) seeds (lane 1) and seeds imbibed at 25°C under various conditions (lanes 2–9) and hybridized with probes from LeSNF4, LeSNF1, and StubGAL83 (to detect LeGAL83 mRNA). After imbibition in water for 48 h, seeds were separated into ungerminated (lane 2) and germinated (lane 3) seeds. Additional seeds were imbibed in 100 μm ABA (lane 4) or in –1.3 MPa polyethylene glycol (PEG) solution (lane 5) or in water under far-red (FR) irradiation (lane 6); these conditions prevented the completion of germination. Naturally dormant seeds were sampled after failing to complete germination after 14 d of imbibition on water (lane 7). These dormant seeds were transferred to 100 μm GA4+7 to stimulate germination, and after 48 h (approximately 50% germination), seeds were separated into ungerminated (lane 8) and germinated (lane 9) fractions. rRNA, Ethidium bromide-stained gel showing loading of RNA in each lane. Replicate gels were run with identical samples and hybridized with RNA probes for LeSNF4, LeSNF1, or StubGAL83. B, Relative mRNA abundance was quantified by densitometry from the images in A and normalized for RNA loading in each lane. Within each panel, the normalized signal intensity of dry seeds (lane 1) was set equal to 1, and the normalized signal intensities in other lanes are shown relative to it.
Because LeSNF4 mRNA was present in mature dry seeds, its continued presence under conditions preventing germination could be due to increased stability or reduced turnover rather than continued synthesis. Because its abundance declined in imbibed seeds before radicle emergence, we tested whether the transcript would accumulate again if germination were blocked. When wild-type seeds were imbibed for 36 h in water, LeSNF4 mRNA amounts decreased before radicle protrusion (e.g. Fig. 8, lanes 1 and 2). When these seeds were then transferred to –1.3 MPa PEG or to 4°C, LeSNF4 mRNA abundance increased, but LeSNF1 and LeGAL83 mRNA amounts were relatively unaffected (data not shown). Thus, conditions inhibitory to germination re-induced accumulation after LeSNF4 mRNA amounts had initially declined.
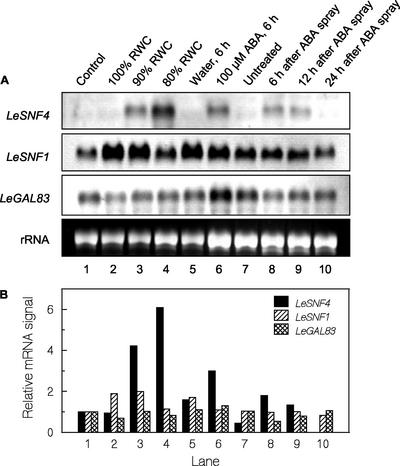
Although LeSNF4 mRNA was not detected in leaves or roots from plants grown under normal field conditions (Fig. 6), we examined the expression of LeSNF4, LeSNF1, and LeGAL83 in leaves exposed to ABA or subjected to water stress (Fig. 9). LeSNF4 mRNA was initially low in leaves from well-watered intact plants (Fig. 9, lanes 1 and 7), in agreement with earlier results (Fig. 6). Excising leaves and maintaining them at 100% RWC for 6 h also failed to induce LeSNF4 expression, but LeSNF4 mRNA accumulated in leaves allowed to dehydrate after excision, with lower RWC resulting in greater accumulation (Fig. 9, lanes 2–4). Similarly, excised shoots maintained with their stems in water did not accumulate LeSNF4 mRNA, but when ABA solution was fed through the stems, LeSNF4 mRNA increased within 6 h (Fig. 9, lanes 5 and 6). A single foliar spray of ABA applied to intact plants also induced transient accumulation of LeSNF4 mRNA within 6 h, followed by its decline to undetectable amounts within 24 h (Fig. 9, lanes 7–10). In contrast, comparatively minor changes in the abundance of LeSNF1 or LeGAL83 mRNAs occurred in shoot tissues in response to these treatments, although LeSNF1 mRNA abundance doubled under mild water stress (Fig. 9, lanes 1–10).
Figure 9.
Expression of SnRK1-related genes in tomato seedling leaves in response to water stress or ABA. A, Leaves were sampled from well-watered plants and frozen immediately (lane 1) or were excised and placed in plastic bags containing moist filter paper either immediately (lane 2) or after different durations of initial water loss resulting in 90% (lane 3) or 80% (lane 4) relative water content (RWC). RNA was extracted from the leaves 6 h after excision. Excised shoots were placed with their cut ends in water (lane 5) or in 100 μm ABA (lane 6) for 6 h before the leaves were sampled for RNA. Intact seedlings (lane 7 = initial leaf samples) were sprayed once with 100 μm ABA and leaves were sampled at 6 (lane 8), 12 (lane 9), and 24 h (lane 10) after treatment. rRNA, Ethidium bromide-stained gel showing loading of RNA in each lane. Replicate gels were run with identical samples and hybridized with RNA probes for LeSNF4, LeSNF1, or StubGAL83 (to detect LeGAL83 mRNA). B, Relative mRNA abundance was quantified by densitometry from the images in A and normalized for RNA loading in each lane. Within each panel, the normalized signal intensity of control leaves (lane 1) was set equal to 1, and the normalized signal intensities in other lanes are shown relative to it.
DISCUSSION
Functional SnRK1-Related Genes Are Present in Tomato
Plant enzymatic or α-subunits of SnRKs have been grouped into three subfamilies (SnRK1, SnRK2, and SnRK3) based upon amino acid sequence homologies (Halford et al., 2000, 2003). We isolated an SnRK1-type cDNA, termed LeSNF1, from germinating tomato seeds that exhibits high predicted amino acid sequence homology to NPK5 of tobacco and StubSNF1 of potato, as would be expected from their close taxonomic relationship. In addition, LeSNF1 complemented an snf1 mutant of yeast, restoring both growth on Suc and derepression of invertase (Figs. 4B and 5B) and indicating that it encodes a functional kinase that can respond to Glc repression signals in yeast. Strong binding of LeSNF1 to yeast SNF4 in the two-hybrid assay only under low-Glc conditions further supports this conclusion (Table I). Like yeast SNF1 (Celenza and Carlson, 1989) and Arabidopsis AKIN10 and AKIN11 (Bhalerao et al., 1999), but unlike tobacco NPK5 (Muranaka et al., 1994), LeSNF1 also suppressed an snf4 mutation in yeast (Figs. 4A and 5A). Suppression of snf4 is likely due to overexpression of LeSNF1 or SNF1 kinase activity, resulting in induction of some invertase activity even in the presence of high Glc concentrations (Fig. 5). However, invertase expression was stimulated severalfold under low-Glc conditions in yeast snf4 mutants expressing LeSNF1 (Fig. 5A), indicating that like SNF1 (Jiang and Carlson, 1996), LeSNF1 remains responsive to signals regulating the Glc repression system in yeast even in the absence of SNF4. Interestingly, interaction of LeSNF1 protein with itself in the two-hybrid assay was enhanced under low-Glc conditions (Table I). In contrast, internal binding of the SNF1 regulatory and kinase domains in yeast is enhanced in high Glc (Jiang and Carlson, 1996). Apparent interaction of LeSNF1 with itself also could be due to the presence of endogenous yeast proteins that could bridge between two LeSNF1 proteins in the two-hybrid system under low-Glc conditions.
In contrast to the many known plant SnRK1-α homologs (Halford et al., 2000), relatively few candidate genes for the β- and γ-subunits of the complex have been identified. The first plant γ-subunit candidate, termed Pv42, was isolated serendipitously from bean (Abe et al., 1995), and no further information on this gene has been reported. Bouly et al. (1999) used sequence homology with SNF4 and AMPKγ to isolate an Arabidopsis gene (AKINγ) whose protein product interacted with AKINα1 in two-hybrid assays. However, the sequence of AKINγ is quite divergent from other plant γ-subunit candidates (Fig. 1B), and it did not interact with yeast SNF1 protein or complement a yeast snf4 mutant (Bouly et al., 1999), so whether it functions in vivo as part of an SnRK1 complex is uncertain. Similarly, Slocombe et al. (2002) identified an SnRK1-interacting protein (SnIP1) having slight similarity to other plant SnRK1-γ candidates that also was unable to complement a yeast snf4 mutant. Another candidate γ-subunit gene (AtSNF4) was identified from Arabidopsis as a “weak” suppressor of the yeast snf4 mutation (Kleinow et al., 2000). It was subsequently shown that expression of a longer transcript of this gene containing a 5′ KIS domain in addition to the γ-subunit domain effectively complemented the yeast snf4 mutation (Lumbreras et al., 2001). Similarly, both the full-length ZmAKINβγ-1 protein from maize and the γ-related region alone complemented the yeast snf4 mutation and interacted with AKIN11, an SnRK1-α protein (Lumbreras et al., 2001). Because the only functional plant homologs of SNF4 known at that time were of the βγ-type, these authors proposed that plant SnRK1 complexes may consist of dimers of α- and fused βγ-type subunits (Lumbreras et al., 2001).
We identified a putative SNF4 homolog from tomato (LeSNF4) that exhibited sequence homology to both SNF4 and AMPK-γ and to other potential plant SnRK1-γ proteins (Fig. 1). The tomato protein complemented a yeast snf4 deletion mutant and restored derepression of invertase during Glc starvation (Figs. 4A and 5A). The expressed protein interacted strongly with both yeast SNF1 and LeSNF1 proteins only under low-Glc conditions (Table I). The LeSNF4 gene encodes only a γ-type subunit and does not contain β-related sequences in the 1,234-bp 5′ to the translation start site or in the expressed mRNA. We also identified two tomato cDNAs (LeSIP1 and LeGAL83) containing conserved KIS and ASC domains characteristic of β-subunits (Fig. 3), and LeSIP1 exhibited Glc-sensitive binding to LeSNF4 and LeSNF1 and to itself (Table I). We are not aware of any other reports testing whether β-subunits can self-associate under low-Glc conditions. We have not tested LeGAL83 in the two-hybrid assay and were unable to achieve meaningful results with StubGAL83 in that assay, but StubGAL3 was previously reported to interact with StubSNF1 and SNF4 (Lakatos et al., 1999). Thus, although plants possess genes for chimeric βγ-type proteins that can interact with SnRK1-α proteins (Lumbreras et al., 2001), they also contain genes encoding separate functional γ- and β-type subunits analogous to those in yeast and mammals.
Halford et al. (2000, 2003) have discussed some of the potential biochemical differences between the plant SnRK1 complexes and those of yeast and mammals, such as the likelihood that Suc, rather than Glc, may be the primary sugar regulating the activity of the kinase in plants. Plant SnRK1 kinases are involved in multiple signal transduction and/or regulation pathways, and a number of proteins in addition to SnRK1-β and -γ subunits have been identified that could potentially interact with SnRK1 kinases to influence their activity or substrate specificity (e.g. Bhalerao et al., 1999; Farrás et al., 2001; Fordham-Skelton et al., 2002; Slocombe et al., 2002). Biochemical studies are needed to identify the in vivo substrates for LeSNF1 and whether its activity, localization, and/or substrate specificity are altered by the presence of LeSNF4, SIPs, or sugars. However, the high sequence conservation among eukaryotic SnRK1 components, the ability of LeSNF1 and LeSNF4 to interact with each other and with their yeast counterparts, and their ability to respond to Glc derepression signals in yeast all suggest that these proteins are likely to be involved in regulating carbon metabolism in planta. We are developing transgenic tomato plants in which LeSNF4 expression can be experimentally modified to test this possibility.
LeSNF4 Is Differentially Expressed during Seed Development and Germination and in Response to Hormones and Water Stress
Relatively limited information is available about the expression of genes of the SnRK1 complex in plants. Different SnRK1-α-subunit genes are expressed in vegetative and seed tissues of barley (Hordeum vulgare), and a specific subfamily (SnRK1b) is uniquely expressed in cereal seeds (Hannappel et al., 1995; Takano et al., 1998; Halford et al., 2000). Potato PKIN1 showed somewhat higher expression in stolons than in other tissues (Man et al., 1997). In Arabidopsis, AKINα1, AKINγ, AKINβ1, and AKINβ2 all exhibited low expression in siliques, whereas mRNA abundance of AKINβ1 in leaves may be regulated by light (Bouly et al., 1999). Abundance of StubSNF1 mRNA was low in various plant tissues except flowers, where expression was high, whereas StubGAL83 mRNA was detected in all tissues tested, with lower expression in roots and fruits (Lakatos et al., 1999). AtSNF4 mRNA was most abundant in cell suspension cultures, flower buds, and shoot apices (Kleinow et al., 2000). ZmAKINβγ-1 mRNA was most abundant during early embryogenesis, and its expression in seedlings was not affected by ABA or temperature stress (4°C or 37°C; Lumbreras et al., 2001). Tobacco NPK5 and potato PKIN1 exhibited some variation in mRNA abundance among tissues, but their mRNAs were detected in all tissues tested (Muranaka et al., 1994; Man et al., 1997). Thus, studies of the expression of genes of the plant SnRK1 complex have been limited largely to tissue surveys and some alterations of environmental conditions, and in only one study were putative members of all three SnRK1 components examined (Bouly et al., 1999).
The dramatic changes in carbon fluxes associated with the deposition of reserves during seed development and their mobilization during germination provide an opportunity to test whether expression of the components of the SnRK1 complex are altered during these metabolic transitions. During tomato seed development, LeSNF4 mRNA was initially absent but appeared at 30 d after flowering, which coincided with maximum dry weight accumulation and the first appearance of planteose, a Suc-galactosyl oligosaccharide that accumulates late in tomato seed development (Fig. 7; Downie et al., 2003). The increase in LeSNF4 mRNA coincided with increasing ABA concentrations in tomato seeds between 30 and 40 DAF, and both ABA content and LeSNF4 mRNA remained high thereafter (Fig. 7, B and C; Hocher et al., 1991; Berry and Bewley, 1992). LeSNF4 mRNA was present in imbibed seeds under various conditions where germination was inhibited and was absent from seeds after completion of germination (Figs. 6 and 8). In contrast, LeSNF1 mRNA was present throughout seed development, being the most abundant in mature seeds (Fig. 7, B and C), and LeSNF1 mRNA amounts were essentially constant after imbibition regardless of germination status (Fig. 8). Comparable results were found with true (botanical) potato seeds when LeSNF4 and LeSNF1 probes were used to detect homologous potato mRNAs in similar types of experiments (Alvarado et al., 2000). Although represented in an imbibed seed cDNA library and in a tomato ovary EST library, suggesting that it is not a pseudogene, we were unable to detect LeSIP1 mRNA in developing or germinating seeds or unstressed leaves by hybridization, whereas LeGAL83 mRNA was detected during late seed development and throughout germination (Figs. 7, 8, 9). Different β-subunits of the SnRK1 complex apparently are differentially expressed in a tissue-specific manner, as shown previously for human and Arabidopsis genes (Thornton et al., 1998; Bouly et al., 1999). LeGAL83 mRNA abundance was low during early seed development but increased as seeds approached maturity (Fig. 7, B and C), and like LeSNF1, remained relatively constant in imbibed seeds, regardless of the germination status (Fig. 8). We conclude that at least one SnRK1-α gene (LeSNF1) and one SnRK1-β gene (LeGAL83) are expressed in developing and germinating tomato seeds and that their mRNA abundance is relatively unaffected by conditions that markedly alter expression of LeSNF4.
The expression pattern of LeSNF4 is reminiscent of that of an SnRK2 kinase, PKABA1, which was identified from wheat embryos on the basis of its induction by ABA (Anderberg and Walker-Simmons, 1992). PKABA1 is involved in a signaling pathway by which ABA suppresses the induction of gene expression by GA in cereal aleurone layers (Gomez-Cadenas et al., 2001). Like LeSNF4, PKABA1 mRNA remains abundant in imbibed dormant seeds, but declines soon after imbibition in nondormant seeds (Verhey and Walker-Simmons, 1997). Furthermore, its expression is up-regulated by ABA, dehydration, cold, and osmotic stress in other plant tissues (Holappa and Walker-Simmons, 1995). Similarly, we found that expression of the LeSNF4 gene can be stimulated in leaves by ABA or water stress (Fig. 9). This and the decrease in LeSNF4 mRNA after GA treatment of gib-1 seeds (Fig. 6) are consistent with direct regulation of LeSNF4 expression by these hormones and with the presence of consensus ABA- and GA-responsive elements in the promoter region of the LeSNF4 gene.
Regulation of LeSNF4 Expression Could Link Hormonal and Sugar Response Pathways
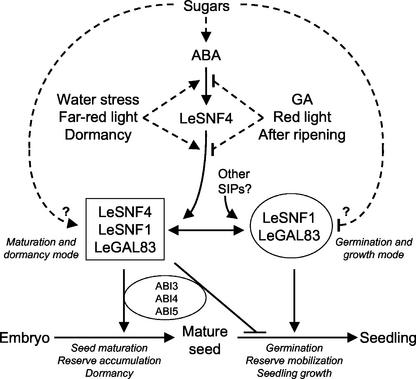
Our discovery that ABA, GA, and other conditions that influence seed germination regulate LeSNF4 expression provides a possible mechanism by which hormonal and sugar-signaling pathways could interact. In yeast and mammals, the SNF1/AMPK kinase complex functions as a metabolic switch, acting on both key enzymes and transcription factors to either induce or repress entire metabolic pathways in response to nutritional or environmental stress or energy demand (Carlson, 1998; Hardie et al., 1998). Rook et al. (2001) proposed that ABA signaling modulates responses to a separate sugar signal, suggesting an explanation for the altered sensitivity of germination to inhibition by sugars in several aba and abi mutants (Cheng et al., 2002; Finkelstein et al., 2002; Rolland et al., 2002). Rook et al. (2001) also proposed that interactions between ABA and sugar-signaling pathways shift development and metabolism between a “storage mode” in which storage reserves are synthesized and their utilization is inhibited and a “mobilization mode” in which storage reserves are catabolized and growth is promoted. Similarly, Gazzarrini and McCourt (2001) proposed that progression from germination to seedling growth involves a transition from “State 1” in which ABA or sugars are capable of blocking reserve mobilization and germination to “State 2” or autotrophic vegetative growth that is much less sensitive to ABA or sugars. The well-known antagonism between ABA and GA in cereal aleurone cells (Gomez-Cadenas et al., 2001) can also be viewed as switching metabolism between the “storage” and “mobilization” modes. Consistent with its role in other organisms and its expression pattern during seed development and germination, we propose that modulation of LeSNF1 kinase activity by the ABA-regulated expression of LeSNF4 could be involved in this metabolic shift (Fig. 10).
Figure 10.
Regulation and hypothetical roles of the SnRK1 complex during seed maturation and germination. This scheme combines recent models (e.g. Gazzarrini and McCourt, 2001; Rook et al., 2001) with data presented here for the tomato SnRK1 complex. ABA synthesis during seed development or in response to high sugar levels during germination results in ABA accumulation, inducing LeSNF4 expression. Additional factors promoting LeSNF4 expression are shown on the left of the ABA signaling pathway, whereas those inhibiting expression are shown on the right. Dashed lines both above and below LeSNF4 are shown because it is unclear where each of these factors acts in influencing expression. When LeSNF4 is present, it binds with LeSNF1/LeGAL83, potentially altering the kinase activity or its interaction with other regulatory factors or substrates, resulting in the maintenance of a “maturation/dormancy” metabolic state and inhibiting reserve mobilization. When LeSNF4 is absent, LeSNF1 may have altered activity, specificity, or interactions with other SIPs, resulting in transition to the “germination/growth” mode required for reserve mobilization and seedling growth. In either of the metabolic modes, the activity of the LeSNF1 complex may be sensitive to regulation by sugars (indicated by the broken lines) and could be involved in the regulation of ABI3, ABI4, and ABI5 and other proteins that are known to influence the transition from seed maturation and dormancy to germination and growth. (Arrows indicate promotion; bars indicate inhibition.)
We hypothesize that when LeSNF4 is expressed during seed maturation (probably in response to ABA), it would bind to LeSNF1/LeGAL83 (or other SIP proteins) and alter the kinase activity of the complex to promote metabolic pathways involved in the accumulation or maintenance of storage reserves and to block those involved in the mobilization or utilization of stored reserves (Fig. 10). For example, LeSNF4 might modulate LeSNF1 activity or substrate specificity to express metabolic pathways that result in the reduction in Glc and accumulation of Suc and oligosaccharides such as raffinose and planteose that occur during seed maturation. After imbibition, expression of LeSNF4 is reduced in seeds that are not dormant or are stimulated by GA, potentially altering LeSNF1 kinase activity to derepress genes encoding enzymes required for reserve mobilization and metabolism (Fig. 10). Conditions that block germination of imbibed seeds, including dormancy, ABA, FR light, or low water potential, maintain LeSNF4 expression (Fig. 10). Provision of sugars can overcome the initial inhibition of germination by ABA (Garciarrubio et al., 1997; Finkelstein and Lynch, 2000a) and induce germination in embryos of the cts (comatose) mutant, which are deeply dormant due to a defect in lipid reserve mobilization (Footitt et al., 2002). Interestingly, ABA did not entirely block lipid mobilization in imbibed Arabidopsis seeds but decreased utilization of Suc, resulting in a doubling of Suc content (Pritchard et al., 2002). ABA action on supply or utilization of metabolic substrates is also consistent with the lack of effect of ABA on the induction of germination-associated cell wall hydrolases and expansins by GA or on the initial weakening of the endosperm cap enclosing the radicle tip of tomato seeds, nonetheless preventing germination (Bradford et al., 2000; Chen and Bradford, 2000).
The developmental arrest associated with embryo quiescence and dormancy that separates the maturation and germination modes requires ABA and is absent in mutants of genes such as ABI3, FUS3, LEC1, and LEC2, which encode transcription factors that influence the expression of a number of genes associated with seed maturation and germination (Nambara et al., 2000; Raz et al., 2001; Koornneef et al., 2002). Additional transcription factors that can bind ABA-responsive elements (ABREs) have been identified in plants, including ABI5, ABRE-binding factors (ABFs), and ABRE-binding proteins (AREBs; Choi et al., 2000; Uno et al., 2000; Carles et al., 2002). In Arabidopsis, ABI5 acts specifically at the postgermination transition in an ABA-dependent manner to maintain the embryo in a quiescent state and block further seedling development (Finkelstein and Lynch, 2000b; Lopez-Molina et al., 2001, 2002). Overexpression of ABF3 exerted an inhibitory effect on germination and early seedling growth and conferred hypersensitivity to ABA (Kang et al., 2002), similar to the effects of ectopic ABI4 expression (Söderman et al., 2000). Genetic and molecular data indicate that ABI3-, ABI4-, and ABI5-related proteins are involved in an interactive network regulating late seed development and germination (Söderman et al., 2000; Nakamura et al., 2001; Bensmihen et al., 2002; Brocard et al., 2002; Niu et al., 2002). Many of these genes are transcriptionally and/or posttranslationally regulated by ABA (e.g. Lopez-Molina et al., 2003), and some of them, including ABI5 (also known as TRAB1), ABF2 (also known as AREB1), and ABF4 (also known as AREB2), are phosphorylated in response to ABA (Uno et al., 2000; Lopez-Molina et al., 2001, 2002; Kagaya et al., 2002; Lu et al., 2002). No plant transcription factors have been demonstrated to be substrates for SnRK1 kinase activity, but the two Ser (S42 and S145) and one Tyr (T201) that are phosphorylated in ABI5 (Lopez-Molina et al., 2002) and Ser-102 in TRAB1 that is phosphorylated in response to ABA and is essential for ABA-induced transcription (Kagaya et al., 2002) are all found in consensus recognition sequences for phosphorylation by SnRK1 (Halford et al., 2003). Interestingly, the same consensus phosphorylation recognition sequence is present in a region of the Mig1 transcriptional repressor in yeast that is required for its regulation by SNF1 in the absence of Glc (Östling et al., 1996). In addition, a wheat transcription factor, TaABF, related to the ABI5 family, was recently identified as a substrate for PKABA1 (Johnson et al., 2002). Although experimental data are as yet lacking, an intriguing possibility is that LeSNF1 is involved in the kinase cascades that phosphorylate the transcription factors regulating late embryogenesis, dormancy, and the transition to germination. Two-hybrid interaction assays with ABI3- and ABI5-related proteins (e.g. Nakamura et al., 2001) conducted in yeast both with and without Glc could be informative in this regard. ABA and/or sugars, acting directly or via expression of LeSNF4, could alter the activity, substrate specificity, or localization of the SnRK1 complex to phosphorylate alternative transcription factors resulting in either the maturation/dormancy or germination/growth metabolic states (Fig. 10).
In conclusion, our results indicate that mRNA abundance of the γ-subunit of the SnRK1 complex in tomato (LeSNF4) is regulated by ABA and GA in a manner consistent with its having a functional role in the maturation to growth transition in seeds and in stress responses in leaves. By adding hormonal regulation to the sugar-, energy-, and stress-sensing functions of a prototypical SnRK1 complex, plants apparently have adapted it for a variety of functions related to source/sink relationships, developmental transitions, and environmental responses.
MATERIALS AND METHODS
Isolation of LeSNF4, LeSNF1, and LeSIP1
Differential cDNA display was performed essentially as described by Liang and Pardee (1992), using mRNA pools from GA-deficient (gib-1) tomato (Lycopersicon esculentum Mill.) seeds (Groot and Karssen, 1987) imbibed in water or in 100 μm GA4+7 for 24 h (Cooley et al., 1999). Anchor primer (dT15MG, where M = G, A, or C) and an arbitrary primer (5′AATCGGGCTG3′) amplified a 300-bp cDNA fragment that was present only in seeds imbibed on water. This fragment was used to retrieve a partial cDNA from a library prepared using mRNA from gib-1 seeds incubated for 24 h on water (Zap Express cDNA Synthesis Kit, Stratagene, La Jolla, CA). The full-length cDNA was obtained using 5′-RACE (Life Technologies, Gaithersburg, MD). The complete cDNA was subsequently used to isolate a single 3.5-kb fragment from a tomato cv VFNT Cherry genomic library in Charon 35 vector (courtesy of R.L. Fischer, University of California, Berkeley). This genomic DNA contained the complete LeSNF4 coding region, including two introns, and 1,234 bp in the promoter region.
LeSNF1
cDNA was isolated from a library prepared using mRNA from gib-1 seeds incubated for 24 h on 100 μm GA4+7 (Zap Express cDNA Synthesis Kit, Stratagene) using tobacco (Nicotiana tabacum) NPK5 cDNA as a probe (courtesy of Dr. Toshiya Muranaka, Sumitomo Chemical Co., Ltd., Takarazuka, Hyogo 665, Japan). LeSIP1 cDNA was isolated from the same library using tomato EST AI486580 (provided by Clemson University Genomics Institute, SC) as a probe. The partial cDNA of LeGAL83 was isolated by reverse transcription-PCR using primers based on the StubGAL83 sequence. cDNAs were sequenced at the Advanced Plant Genetics Facility (University of California, Davis). Sequence comparisons were made using DNAStar software.
Yeast (Saccharomyces cerevisiae) Complementation
LeSNF4 or LeSNF1 cDNAs were fused in frame with pMA424 and pYES and these or empty vectors were transformed into freshly prepared snf4-Δ2 (MCY3915 [MATα snf4-Δ2, His-3Δ200 Leu-2-3, and 112-Trp 1Δ63 Ura-3-52]) or snf1-Δ10 (MCY1846 [MATα snf1-Δ10 Lys-2-801 Ura 3-52]) yeast cells (provided by Dr. Marian Carlson, Columbia University, New York) or wild-type YPH500 competent cells by the lithium acetate method (Rose et al., 1990). The transformed yeast colonies were plated on selective SC medium at 29°C for 3 to 14 d, and growing colonies were characterized by invertase assays and by re-isolation and restriction analysis of the shuttle vector to confirm the presence of the inserted genes.
Invertase Assays
Yeast cells were grown in 3 mL of selective medium at 29°C and harvested at mid-log phase. After centrifuging for 5 min at 15,000g and noting the weight of the pellet, the cells were chilled in ice and washed with 100 μL of chilled sterile water. The cells were resuspended in 100 μL of glass bead disruption buffer and shaken 6 × 1 min each with 80 mg of cooled glass beads (Sigma, St. Louis). The supernatant was spun at 12,000g for 30 min to yield the crude extract. Extract (4 μL) was assayed according to Goldstein and Lampen (1975; except that O-dianisidine was dissolved in methanol) using invertase (Sigma) as the standard.
Yeast Two-Hybrid Protein Interaction Assays
In-frame fusions of members of the SnRK1 complex from tomato and yeast were made by PCR in pGBT9.BS (Trp; binding domain) and pGAD.GH (Leu; activation domain) vectors. Competent Y190 cells were transformed as above with equal quantities (0.25–0.5 μg) of the two vectors. The cells were plated on SC-Trp-Leu-His with 25 mm 3-amino-triazole and grown at 29°C for 3 to 14 d. The transformed colonies were characterized by filter lift assays for β-galactosidase, and colonies showing blue color were grown in 3 mL of selective broth containing 2% (w/v) Glc or 2% (w/v) each of Gal, glycerol, and ethanol plus 0.05% (w/v) Glc with 25 mm 3-aminotriazole. β-d-Galactosidase was assayed spectrophotometrically in extracts (as described above for invertase) using chlorophenyl-red-β-d-galactopyranoside as substrate (Kim et al., 1997). Galactosidase activity was calculated using an extinction coefficient for chlorophenyl red at 570 nm of 75,000 L mol–1cm–1.
Seed Development
Tomato cv Moneymaker plants were grown in the field in the summer of 2000. Flowers were marked at anthesis, and fruits were harvested every 5 d between 20 and 60 DAF. Seeds were separated from the locular tissue by gentle rubbing, and RNA was extracted without drying the seeds.
Northern and Southern Hybridization
Northern hybridizations were conducted as described previously using digoxigenin-labeled RNA probes (Cooley et al., 1999). Hybridization was detected according to the instructions in the Genius System (Boehringer Mannheim Corporation, 1995). All experiments involving mRNA abundance were repeated at least twice with similar results. Southern hybridizations with DNA probes were conducted as described previously (Chen and Bradford, 2000) and detected using enhanced chemiluminescence (ECL kit, Amersham Life Science, Arlington Heights, IL). After hybridization, membranes were washed twice at 42°C with 6 m urea, 0.5% (w/v) SDS, and then washed twice for 5 min each with 2× SSC at room temperature.
Acknowledgments
We thank Dr. Marian Carlson for providing the mutant yeast strains, Dr. Linda Bisson for wild-type yeast YPH500, Dr. Toshiya Muranaka for providing the cDNA of NPK5, Dr. Zsófia Bánfalvi for providing the cDNA of StubGAL83, and the Clemson University Genomics Institute for providing the cDNA of EST AI486580. Dr. Steffen Abel provided valuable advice on the yeast complementation and two-hybrid assays. GA4+7 was a gift from Abbott Laboratories (Chicago).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019141.
This work was supported in part by the National Science Foundation (grant nos. IBN–9407264 and IBN–9722978) and by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 2001–35304 to K.J.B.).
References
- Abe H, Kamiya Y, Sakurai A (1995) A cDNA clone encoding yeast SNF4-like protein (Accession #U40713) from Phaseolus vulgaris L. Plant Physiol 110: 336 [Google Scholar]
- Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreiss M, Shewry PR, Halford NG (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci USA 88: 8602–8605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado V, Nonogaki H, Bradford KJ (2000) Expression of endo-β-mannanase and SNF-related protein kinase genes in true potato seeds in relation to dormancy, gibberellin and abscisic acid. In JD Viemont, J Crabbé, eds, Dormancy in Plants. CAB International, Wallingford, UK, pp 347–364
- Anderberg RJ, Walker-Simmons MK (1992) Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc Natl Acad Sci USA 89: 10183–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A (1997) The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci 22: 12–13 [DOI] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Branier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry T, Bewley JD (1991) Seeds of tomato (Lycopersicon esculentum Mill.) which develop in a fully hydrated environment in the fruit switch from the developmental to germination mode without a requirement for desiccation. Planta 186: 27–34 [DOI] [PubMed] [Google Scholar]
- Berry T, Bewley JD (1992) A role for the surrounding fruit tissues in preventing germination of (Lycopersicon esculentum) seeds: a consideration of osmotic environment and abscisic acid. Plant Physiol 100: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bakó L, Ökrész L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer Mannheim Corporation (1995) Genius System User's Guide for Membrane Hybridization, Version 3.0. Boehringer Mannheim, Indianapolis
- Bouly J, Gissot L, Lessard P, Kreis M, Thomas M (1999) Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINα1, an SNF1-like protein kinase. Plant J 18: 541–550 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Chen F, Cooley MB, Dahal P, Downie B, Fukunaga KK, Gee OH, Gurusinghe S, Mella RA, Wu CT et al. (2000) Gene expression prior to radicle emergence in imbibed tomato seeds. In M Black, KJ Bradford, J Vázquez-Ramos, eds, Seed Biology: Advances and Applications. CABI Publishing, Wallingford, UK, pp 231–251
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Carlson M (1998) Regulation of Glc utilization in yeast. Curr Opin Genet Dev 8: 560–564 [DOI] [PubMed] [Google Scholar]
- Celenza JL, Carlson M (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol Cell Biol 9: 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, Eng FJ, Carlson M (1989) Molecular analysis of the SNF4 gene of Saccharomyces cerevisae: evidence for physical association of SNF4 protein with the SNF1 protein kinase. Mol Cell Biol 9: 5045–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Bradford KJ (2000) Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol 124: 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FQ, Foolad MR (1999) A molecular linkage map of tomato based on a cross between Lycopersicon esculentum and L. pimpinellifolium and its comparison with other molecular maps of tomato. Genome 42: 94–103 [Google Scholar]
- Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HI, Hong JH, Ha JO, Kang JY, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Cooley MB, Yang H, Dahal P, Mella RA, Downie B, Haigh AM, Bradford KJ (1999) Expression of vacuolar H+-ATPase in response to gibberellin during tomato seed germination. Plant Physiol 121: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RM, Halford NG, Hardie DG (2001) Cloning of DNA encoding a catalytic subunit of SNF1-related protein kinase-1 (SnRK1-α1), and immunological analysis of multiple forms of the kinase, in spinach leaf. Plant Mol Biol 45: 731–741 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Gurusinghe S, Dahal P, Thacker RR, Snyder JC, Nonogaki H, Yim K, Fukanaga K, Alvarado V, Bradford KJ (2003) Expression of a galactinol synthase gene in tomato is stress induced by an ABA-independent mechanism that is differentially responsive to cold in tomato seeds and leaves. Plant Physiol 131: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrás R, Ferrando A, Jásik J, Kleinow T, Ökréaz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20: 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Koncz-Kálmán Z, Farràs R, Tiburcio A, Schell J, Koncz C (2001) Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plants cells. Nucleic Acids Res 29: 3685–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2001) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5: 26–32 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000a) Abscisic acid inhibition of radicle emergence but not seedling growth is suppressed by sugars. Plant Physiol 122: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000b) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M (2002) Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham-Skelton AP, Chilley P, Lumbreras V, Reignoux S, Fenton TR, Dahm CC, Pages M, Gatehouse JA (2002) A novel higher plant protein tyrosine phosphatase interacts with SNF1-related protein kinases via a KIS (kinase interaction sequence) domain. Plant J 29: 705–715 [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D (2002) Proteomics of Arabidopsis seed germination: a comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol 129: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA (1997) Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203: 182–187 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387–391 [DOI] [PubMed] [Google Scholar]
- Gibson SI (2000) Plant sugar-response pathways: part of a complex regulatory web. Plant Physiol 124: 1532–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Lampen JO (1975) β-d-Fructofuranoside fructohydrolase from yeast. Methods Enzymol 42C: 504–511 [DOI] [PubMed] [Google Scholar]
- Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667–679 [PMC free article] [PubMed] [Google Scholar]
- Groot SPC, Karssen CM (1987) Gibberellins regulate seed germination in tomato by endosperm weakening: a study with gibberellin-deficient mutants. Planta 171: 525–531 [DOI] [PubMed] [Google Scholar]
- Halford NG, Bouly J-P, Thomas M (2000) SNF1-related protein kinases (SnRKs): regulators at the heart of the control of carbon metabolism and partitioning. Adv Bot Res 32: 405–434 [Google Scholar]
- Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon catabolism in plants? Plant Mol Biol 37: 735–748 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role for Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467–475 [DOI] [PubMed] [Google Scholar]
- Hannappel U, Vicente-Carbajosa J, Barker JHA, Shewry PR, Halford NG (1995) Differential expression of two barley SNF1-related protein kinase genes. Plant Mol Biol 27: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1-protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Hocher V, Sotta B, Maldiney R, Miginiac E (1991) Changes in abscisic acid and its β-d-glucopyranosyl ester levels during tomato (Lycopersicon esculentum Mill.) seed development. Plant Cell Rep 10: 444–447 [DOI] [PubMed] [Google Scholar]
- Holappa LD, Walker-Simmons MK (1995) The wheat abscisic acid-responsive protein kinase mRNA, PKABA1, is up-regulated by dehydration, cold temperature, and osmotic stress. Plant Physiol 108: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1996) Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev 10: 3105–3115 [DOI] [PubMed] [Google Scholar]
- Jiang R, Carlson M (1997) The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol 17: 2099–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Wagner R, Verhey SD, Walker-Simmons MK (2002) The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific ABA response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T (2002) Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen C (1995) Hormonal regulation of seed development, dormancy, and germination studied by genetic control. In J Kigel, G Galili, eds, Seed Development and Germination. Marcel Dekker, Inc., New York, NY, pp 333–350
- Kemp BE, Mitchelhill KI, Stapleton D, Michell BJ, Chen Z-P, Witters LA (1999) Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem Sci 24: 22–25 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchart K, Koncz C (2000) Functional identification of an Arabidopsis Snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J 23: 115–122 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33–38 [DOI] [PubMed] [Google Scholar]
- Lakatos L, Klein M, Höfgen R, Bánfalvi Z (1999) Potato StubSNF1 interacts with StubGAL83: a plant protein kinase complex with yeast and mammalian counterparts. Plant J 17: 569–574 [DOI] [PubMed] [Google Scholar]
- Laurie S, McKibbin RS, Halford NG (2003) Antisense SNF1-related (SnRK1) protein kinase gene represses transient activity of an α-amylase (α-Amy2) gene promoter in cultured wheat embryos. J Exp Bot 54: 739–747 [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967–971 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlan DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Lu C, Han M-H, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V, Mar Albà M, Kleinow T, Koncz C, Pageœ M (2001) Domain fusion between SNF1-related kinase subunits during plant evolution. EMBO Rep 21: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man AL, Purcell PC, Hannappel U, Halford NG (1997) Potato SNF1-related protein kinase: molecular cloning, expression analysis and peptide kinase activity measurements. Plant Mol Biol 34: 31–43 [DOI] [PubMed] [Google Scholar]
- Muranaka T, Banno H, Machida Y (1994) Characterization of the tobacco protein kinase, NPK5, a homologue of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose–repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol Cell Biol 14: 2958–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Mishimura M, Kawaide H, Kamiya Y, Naito S (2000) The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220: 412–423 [DOI] [PubMed] [Google Scholar]
- Niu X, Helentjaris T, Bate NJ (2002) Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell 14: 2565–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östling J, Carlberg M, Ronne H (1996) Functional domains in the Mig1 repressor. Mol Cell Biol 16: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Wyrzykowska J, Fleming AJ (2001) Novel marker genes for early leaf development indicate spatial regulation of carbohydrate metabolism within the apical meristem. Plant J 25: 663–674 [DOI] [PubMed] [Google Scholar]
- Pritchard SL, Charlton WL, Baker A, Graham IA (2002) Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J 31: 639–647 [DOI] [PubMed] [Google Scholar]
- Purcell PC, Smith AM, Halford NG (1998) Antisense expression of a sucrose non-fermenting-1-related protein kinase sequence in potato results in decreased expression of sucrose synthase in tubers and loss of sucrose-inducibility of sucrose synthase transcripts in leaves. Plant J 14: 195–202 [Google Scholar]
- Raz V, Bergervoet JHW, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14: S185–S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic signaling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Rose JD, Winston F, Hieter P (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmidt MC, McCartney RR (2000) β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J 19: 4936–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Laurie S, Bertini L, Beaudoin F, Dickinson JR, Halford NG (2002) Identification of SnIP1, a novel protein that interacts with SNF1-related protein kinase (SnRK1). Plant Mol Biol 49: 31–44 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG (1999) Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate HMG-CoA reductase, nitrate reductase and sucrose phosphate synthase in vitro. Plant Physiol 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Kajiya-Kanegae H, Funatsuki H, Kikuchi S (1998) Rice has two distinct classes of protein kinase genes related to SNF1 of Saccharomyces cerevisiae, which are differently regulated in early seed development. Mol Gen Genet 260: 388–394 [DOI] [PubMed] [Google Scholar]
- Thornton C, Snowden MA, Carling D (1998) Identification of a novel AMP-activated protein kinase β subunit isoform that is highly expressed in skeletal muscle. J Biol Chem 273: 12443–12450 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey SD, Walker-Simmons MK (1997) Abscisic acid-mediated responses in seeds involving protein kinases and phosphatases. In RH Ellis, M Black, AJ Murdoch, TD Hong, eds, Basic and Applied Aspects of Seed Biology. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 225–233
- Vincent O, Townley R, Kuchin S, Carlson M (2001) Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev 15: 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]