Abstract
The function of the apoplastic enzyme ascorbate oxidase (AO) was investigated in tobacco (Nicotiana tabacum). The abundance of AO mRNA was up-regulated by light. Cytosolic ascorbate peroxidase (APX1) transcripts were also highest in the light. In contrast, l-galactono-γ-lactone dehydrogenase, stromal APX, and thylakoid APX transcripts remained constant over the day/night cycle. Salicylic acid inhibited growth, increased expression of the pathogenesis-related protein (PR) 1a, and decreased AO transcript abundance. In contrast, the application of auxin enhanced growth and increased AO and PR 1a gene expression. Therefore, AO transcript abundance varied in a manner similar to hormone-mediated changes in plant growth. To study the effects of modified AO expression on growth, transformed tobacco plants expressing AO in the sense and antisense orientations were generated. The resultant large changes in apoplastic AO activity in the transformed tobacco plants had little effect on whole leaf ascorbate (AA) content, but they had dramatic effects on apoplastic AA levels. Enhanced AO activity oxidized the apoplastic AA pool, whereas decreased AO activity increased the amount of AA compared with dehydroascorbate. A relationship was observed between AO activity and plant height and biomass. Native AO transcript levels were no longer subject to light/dark regulation in AO sense and antisense plants. Taken together, these data show that there is an interaction between hormone, redox, and light signals at the level of the apoplast via modulation of ion of AA content.
Ascorbate (AA) plays a key role in defense against oxidative stress and is particularly abundant in photosynthetic tissues (Foyer et al., 1983; Smirnoff, 2000). Most (over 90%) of the AA is localized in the cytoplasm, but unlike other soluble antioxidants, a substantial proportion is exported to the apoplast, where it is present at millimolar concentrations. Apoplastic AA is believed to represent the first line of defense against potentially damaging external oxidants such as ozone, SO2, and NO2 (Plöchl et al., 2000; Barnes et al., 2002). In the apoplast, AA is oxidized to monodehydroascorbate (MDHA) by the enzyme ascorbate oxidase (AO). MDHA is an unstable radical and rapidly disproportionates to yield DHA and AA. DHA is then transported into the cytosol through the plasma membrane by a specific carrier that preferentially translocates the oxidized form in exchange for the reduced form, ensuring a continuous flux of reducing power to the cell wall (Horemans et al., 2000). Perhaps the most intriguing and poorly understood of the enzymes involved in AA metabolism in plants is the apoplastic AO. No clear biological functions for AO have been described to date. However, it is widely believed that AO plays a role in cell elongation because of its extracellular localization and its high activity in rapidly expanding tissues (Esaka et al., 1992; Moser and Kanellis, 1994; Ohkawa et al., 1994; Kato and Esaka, 1999). Recent work has shown that tobacco (Nicotiana tabacum) Bright Yellow-2 protoplasts overexpressing a pumpkin (Cucurbita pepo) AO cDNA elongate more rapidly than untransformed controls (Kato and Esaka, 2000). Several mechanisms whereby AO controls cell growth have been proposed (Esaka, 1998; Smirnoff, 2000). MDHA generated from AA by AO in the apoplast, stimulates cell growth through enhanced vacuolization (Hidalgo et al., 1989) and ion uptake caused by depolarization of the plasma membrane. Moreover, DHA is considered to be responsible for cell enlargement by promoting cell wall loosening (Lin and Varner, 1991). Local auxin-mediated production of free radicals has also been shown to induce cell wall extension (Joo et al., 2001). Because AA is believed to be the most important antioxidant in the apoplast of leaves and stems, its destruction via AO may be important in facilitating cell expansion.
Plant growth is triggered by auxin. Exogenous applications of auxin can trigger a variety of cellular processes such as cell elongation and lateral root growth (Laskowski et al., 1995). Auxin promotes cell enlargement via the enhancement of proton excretion into the cell wall (Cleland, 1987). The transduction process leading to activation of the associated ATPase proton pump localized in the plasma membrane may involve oxidant/antioxidant signals (Mills et al., 1980). The similarities and relationship between auxin and AO action in inducing cell growth are intriguing and merit further study.
Relatively little information is available on other enzymes involved in AA synthesis and metabolism. The last step of the pathway of AA synthesis in plants is relatively well characterized and involves the cytochrome c-dependent oxidation of l-galactono-1, 4-lactone by the mitochondrial enzyme l-galactono-γ-lactone dehydrogenase (GLDH; Ostergaard et al., 1997; Bartoli et al., 2000). Similarly, the AA peroxidase (APX) family that is responsible for the scavenging of H2O2 is well characterized. There are at least seven APX isoforms in plants, some of which are encoded by discrete genes with expression localized in different subcellular compartments: cytosol, mitochondria, peroxisomes, apoplast, and chloroplasts (Asada, 1997). Two isoforms are located in the cytosol (cAPX1 and cAPX2), with cAPX2 expression restricted to high photosynthetic photon flux densities in Arabidopsis leaves (Karpinski et al., 1999). In chloroplasts, APX is located in the stroma (sAPX) and is bound to the thylakoids (tAPX).
The present study was undertaken to elucidate the function of AO in tobacco leaves. We show that expression of AO is regulated by light and by plant hormones that modulate growth. Moreover, we demonstrate that enhanced apoplastic AO activities in transformed tobacco plants favor increased growth. Taken together, these observations provide evidence that AO has a role in the control of growth.
RESULTS
Diurnal Changes in Transcript Abundance of Enzymes Involved in the AA Metabolism
The expression of GLDH, sAPX, and tAPX were similar in the dark and in the light (Fig. 1), giving similar values after 16 h of dark or 24 h of light. In contrast, AO transcripts fell to very low levels in the dark and increased only slowly in the light (Fig. 1). The abundance of cAPX1 mRNA was also influenced by light, being minimum in the dark and attaining maximum values in the light (>6 h; Fig. 1). Actin was used as the internal control in these and all of the following experiments after confirmation that the abundance of actin transcripts was unaffected by any of the treatments used. This was achieved by performing actin-specific RT-PCR on RNA samples extracted from leaves subjected to a wide range of light-dark transitions and hormone treatments.
Figure 1.

Light-dependent changes in transcripts of genes of AA metabolism. Leaf discs were harvested in plants exposed to 16 h of light or 16 h of dark and then, after 16 h dark, after 1, 6, or 24 h of light. Semiquantitative reverse transcriptase (RT)-PCR was performed on 3-week-old tobacco plants 1, 6, 16, and 24 h after transfer to light (250 μmol m–2 s–1) or 16 h of dark. Actin was used as the internal control. The relative intensity of each band is expressed as a density number, which can be used as a indication of the relative abundance of transcripts. Values are normalized to the 16-h value. This experiment was repeated three times and similar results were obtained.
Further controls were performed to rule out the possibility that the observed light-dark changes in AO and cAPX1 transcript levels were due to circadian rhythmicity. No circadian control was found in AO or cAPX1 (data not shown).
Effects of SA and α-Naphthaleneacetic Acid (NAA) on AO Transcript Abundance and Plant Growth
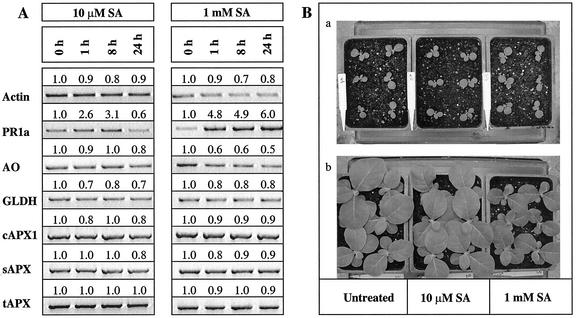
Pathogenesis-related protein (PR) 1a was used as a positive control for the efficacy of the following treatments as its expression has previously been shown to be triggered by SA (Durner et al., 1998) and by auxin (Ohashi and Matsuoka, 1987) in tobacco leaves. Application of 10 μm SA led to transient expression of PR1a, with maximum induction occurring between 1 and 8 h after spraying. The abundance of GLDH, AO, sAPX, tAPX, or cAPX1 transcripts was unaffected by the application of 10 μm SA (Fig. 2A). On the other hand, application of 1 mm SA resulted in a sustained increase in PR1a expression. Although AO transcripts rapidly decreased after application of 1 mm SA, the abundance of GLDH, sAPX, tAPX, or cAPX1 mRNA was unaffected (Fig. 2A).
Figure 2.
Effects of SA on transcripts abundance and plant growth. A, Transcript abundance; whole shoot samples were harvested 1, 8, and 24 h after spraying, which was performed halfway through the light period. Semiquantitative RT-PCR was performed on 3-week-old tobacco plants sprayed with 10 μm or 1 mm SA and then exposed to an extended photoperiod (24 h at 250 μmol m–2 s–1). Actin was used as the internal control. B, Plant growth; phenotype of SA-treated and -untreated plants showing seedlings before treatment (a) and 1 week after the treatment (b). This experiment was repeated three times and similar results were obtained.
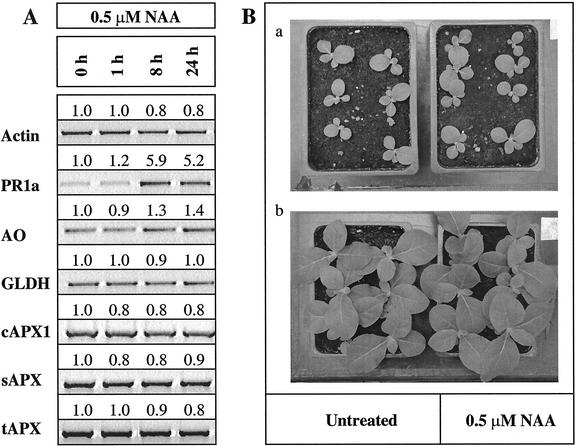
A marked increase in AO and PR1a expression was observed 8 h after spraying with the auxin, NAA. In contrast, the expression of GLDH, sAPX, tAPX, or cAPX1 mRNA was unchanged by this treatment (Fig. 3A). SA and NAA had inverse effects on plant growth (Figs. 2B and 3B). The growth of plants sprayed with 1 mm SA was much slower than that of control plants sprayed with low SA concentrations (10 μm; Fig. 2B). In contrast, low concentrations (0.5 μm) of NAA stimulated growth (Fig. 3B).
Figure 3.
Effects of auxin on transcript abundance and plant growth. A, Transcript abundance; whole shoot samples were harvested 1, 8, and 24 h after spraying, which was performed halfway through the light period. Semiquantitative RT-PCR was performed on 3-week-old tobacco plants sprayed with 0.5 μm NAA and then exposed to an extended photoperiod (24 h at 250 μmol m–2 s–1). B, Plant growth; phenotype of NAA-treated and -untreated plants showing seedlings before treatment (a) and 1 week after the treatment (b). This experiment was repeated three times and similar results were obtained.
Transgenic Tobacco Overexpressing AO in Sense and Antisense Orientations
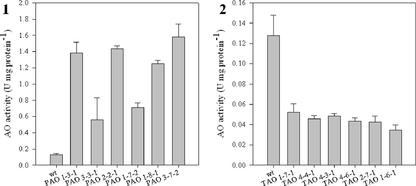
To further explore the function of AO in tobacco leaves, two constructs for the expression of AO in the sense and antisense orientations were produced. Six independent primary transformants (T0 generation) were selected for each construct on the basis of their ability to survive growth on kanamycin, the presence of the transgene, and their leaf AO activity. Leaf AO activity was markedly different in all the transformants compared with the wild type. AO activity was five to 16 times that of PAO leaves and 0.3 to 0.5 times that of the TAO leaves than the wild type (Fig. 4). Transgene-dependent differences in leaf AO were similar whether plants were grown in tissue culture or in soil (data not shown). Genomic DNA from each of the lines was probed with 2X35S-PAO or 2X35S-TAO DNA fragments to confirm the presence of the transgene and check the copy number of the insertion. All lines tested positive for the transgene (data not shown). Three sense (PAO2-2-1, PAO1-7-2, PAO3-7-2) and two antisense (TAO2-7-1, TAO1-6-1) lines containing only one copy of the introduced sequence were selected for further analysis. These lines were grown for two additional generations (T1 and T2) and the relationship between AO and plant development was followed in each generation.
Figure 4.
Characterization of primary transformants. AO activity in leaf samples of PAO sense (1) and TAO antisense (2) primary transgenic lines. Each value represents the mean ± sd of three independent measurements.
The AO Over- and Underexpression Phenotypes
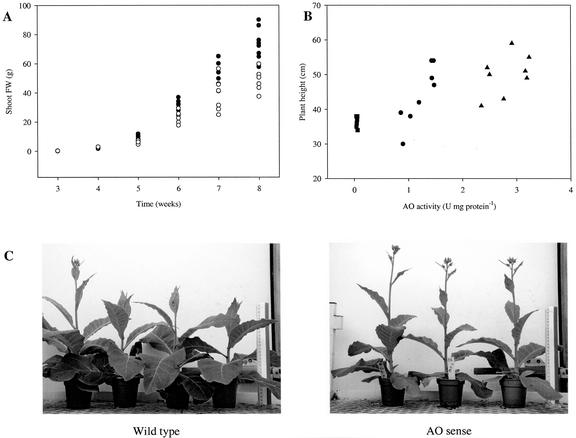
Plant growth, measured as biomass accumulation over time, was monitored in T1 and T2 populations derived from each selected transgenic line. Each population was composed of 25 individuals germinated on nonselective media, allowing simultaneous measurements of individuals that had passed through the transformation procedure but did not express the transgene and AO-expressing plants. All plants were grown under controlled environment conditions at the same time and shoot fresh weights were recorded weekly from three plants per population. Additional wild-type populations were not necessary in these experiments, given the presence of internal controls within each population. No statistically significant differences in shoot fresh weight were observed in 3-, 4-, or 5-week-old sense (PAO) or antisense (TAO) plants (Fig. 5A). However, between weeks 6 and 8, plants expressing the AO transgene in the sense orientation (PAO) accumulated greater biomass than plants expressing the AO transgene in the antisense orientation (TAO; Fig. 5A). By the end of the experiment (8 weeks), AO sense-expressing plants (PAO) showed a statistically significant increase in fresh weight (P < 0.05 at 7 weeks, and P < 0.01 at 8 weeks) compared with the AO antisense plants (TAO; Fig. 5A). The relative growth (measured as weekly biomass accumulation) rate was linear (r2 > 0.95) between weeks 6 and 8. At this point, the growth rate of AO sense plants (PAO) was 30% higher than that of AO antisense (TAO) plants. Values of 20.8 g fresh weight gain per week were measured in AO sense plants (PAO) compared with 14.1 g fresh weight gain per week in antisense (TAO) plants (Fig. 5A).
Figure 5.
Phenotypes of AO sense and antisense transgenic plants. A, Effect of AO over- and underexpression on growth rates of T1 populations: three AO sense and two AO antisense. Growth rate of sense (•) and antisense (○) AO transgenic T1 populations measured as the increase in shoot fresh weight per week. All plants were grown under controlled environment conditions. B, Correlation between AO activity and height in 7-week-old AO sense (PAO) and wild-type plants. Squares, wild type; circles, T1 generation; triangles, T2 generation. Each activity value represents the mean of three independent measurements. Mean values for AO activities are 0.05 ± 0.00 for wild type; 1.4 ± 0.5 for T1 generation; 3.0 ± 0.4 for T2 generation. Mean values for plant heights are 33 ± 5.0 for wild type; 42 ± 9.8 for T1 generation; 50 ± 8.0 for T2 generation. This experiment was repeated three times and similar results were obtained. C, Phenotype of wild-type and 7-week-old AO sense tobacco plants (PAO).
Once established that AO sense plants (PAO) showed an increased growth rate when compared with antisense (TAO) plants, growth of sense plants (PAO), measured this time as plant height, was compared with that of wild-type plants. This experiment was conducted to establish whether a correlation between AO activity and plant height could be drawn. T1 and T2 seeds from two AO sense lines (PAO2-2-1 and PAO3-7-2) that had been selected on the basis of high AO activity, were germinated on media containing kanamycin to select for plants containing the transgene. Once this was confirmed by PCR (data not shown), PAO plants were transferred to compost and were grown in controlled environment chambers together with wild-type populations. After 6 weeks, AO activities were measured in young fully developed leaves of four to five plants per line, and the height of the corresponding plants was recorded. Overall, T2 plants exhibited higher AO activity and a taller phenotype than their respective T1 progenitors (Fig. 5B). A positive correlation was demonstrated between AO activity and plant height (Fig. 5B; r2 = 0.85). Analysis of variance of the data confirmed that T2 PAO lines grew significantly taller than T1 PAO lines and that T1 and T2 PAO lines grew taller than the wild type (P < 0.01; Fig. 5, B and C).
Relationships between AO Activity and AA in the Leaf Apoplast
Whole leaf AA, DHA contents, and AO activities were measured in extracts from discs excised from young fully expanded leaves of 6-week-old T1 sense (PAO lines PAO2-2-1 and PAO3-7-2) and antisense (TAO lines TAO2-7-1 and TAO1-6-1) plants. PAO plants exhibited about a 40-fold higher leaf AO activity than the wild type (Table I), whereas TAO lines exhibited 0.4-fold less leaf AO activity. Apoplastic AO activity was measured in whole leaf extracts, soluble, and ionically bound fractions (Table I). Total AO activity measured in whole leaf extracts was found to be entirely associated with the cell wall (ionically bound) and localized in the apoplast (Table I). AO activity was undetectable in soluble fractions of leaf extracts. TAO leaves contained similar amounts of AA and DHA to the wild type, but PAO leaves contained less AA and total (AA plus DHA) AA (Table I). DHA contents were higher in PAO lines, but the leaf ascorbate pool was always largely reduced, values being slightly higher for wild-type (92%) than PAO plants (85%). In contrast to whole leaf AA, modified AO expression resulted in dramatic changes in the apoplastic AA pool. The total (AA plus DHA) AA content of the apoplast was at least double that of the wild type in sense and antisense plants (Table I). However, the greatest effect was on the AA redox state (Table I). In the wild type, the AA pool in the apoplast was about 40% reduced, whereas it was 66% reduced in the antisense plants and only 3% reduced in the sense plants. The DHA content in the apoplast of sense plants was 3.5-fold greater than in wild-type or antisense plants. For apoplastic AA and DHA measurements, cytosolic contamination of the apoplast (monitored by measuring Glc-6-P in intercellular washing fluid (IWF) was maintained below 0.1% (data not shown). However, it should be noted that the presence of apoplastic acid phosphatases may lead to underestimation of the contamination of the apoplastic fraction as well.
Table I.
AO activity and ascorbate content in whole leaves and apoplast of T1 and T2 PAO sense and TAO antisense transgenic lines and in wild-type plantsa
| Whole-Leaf Parameters | PAO Senseb | TAO Antisensec | Wild Type |
| AO activityd | 1.92 ± 0.1 | 0.02 ± 0.0 | 0.05 ± 0.0 |
| Total ascorbatee | 218.7 ± 7.8 | 282.1 ± 8.6 | 270.3 ± 34.4 |
| AAe | 186.6 ± 6.5 | 258.8 ± 8.1 | 250.5 ± 30.2 |
| DHAe | 33.9 ± 3.4 | 24.1 ± 2.3 | 20.9 ± 5.8 |
| Apoplastic parameters | |||
| AO activityd (ionically bound) | 1.92 ± 0.1 | 0.02 ± 0.0 | 0.05 ± 0.0 |
| AO activityd (soluble) | 0.00 | 0.00 | 0.00 |
| Total ascorbatee | 32.6 ± 2.9 | 26.0 ± 6.2 | 13.7 ± 3.7 |
| AAe | 1.1 ± 0.2 | 17.3 ± 5.4 | 5.2 ± 0.9 |
| DHAe | 31.5 ± 2.8 | 8.6 ± 1.1 | 8.1 ± 2.9 |
Leaf samples were taken from 6-week-old-plants grown in controlled environment. Transgenic plants were generated from T1 seeds germinated in selective medium. Values are the mean of three samples per leaf and 20 leaves per line. b Lines PAO 2-2-1 and PAO 3-7-2. c Lines TAO 2-7-1 and TAO 1-6-1. d Given as units per milligram of total protein content. e Given as nanomoles per milligram of total protein content.
Light/Dark Effects on AO Transcript Abundance in AO Transgenic Plants
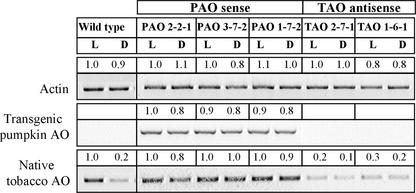
AO transcript abundance was highest in the light and showed a marked light/dark expression pattern in wild-type plants (Fig. 1). The following experiments were performed to investigate whether such marked diurnal patterns of expression were maintained in the native wild-type AO of PAO and TAO transgenic plants. Due to the sequence similarity between the tobacco and the pumpkin AO (used for the generation of the AO sense plants), careful design of species-specific primers was necessary to allow expression patterns of native and transgenic AO to be discriminated. No expression patterns in the expression of the transgenic AO (PAO) were expected given the presence of the strong constitutive promoter 35S cauliflower mosaic virus (CaMV). The specificity of TAO (specific for tobacco AO) and PAO (specific for pumpkin AO) primers was confirmed by sequencing of the relative PCR products. They were then used to analyze by RT-PCR the expression of the native tobacco AO and the introduced pumpkin AO (PAO) in the leaves of T1 and T2 generations of transformants grown in soil under controlled environment conditions. Samples from wild type, three sense (PAO), and two antisense (TAO) lines were harvested after 16 h of light or 16 h of dark (Fig. 6). PAO was highly and equally expressed in all the sense PAO leaves in the light and in the dark. This is consistent with the constitutive expression of PAO under the control of the CaMV35S promoter. Moreover, PAO plants showed no cosuppression of the native wild-type AO because the transcript abundance of the native wild-type tobacco AO transcripts was similar in the sense PAO lines and in the wild type. In contrast to the strong dark/light pattern of AO expression observed in wild-type plants, no dark/light effects on native wild-type tobacco AO transcript abundance was detected in the PAO sense lines. In this case, the amounts of native AO transcripts were similar in the light and dark (Fig. 6). There was a large overall decrease in native AO transcripts in TAO leaves compared with the wild type (Fig. 6), demonstrating the efficacy of the antisense suppression strategy adopted. However, in the same TAO plants, there was also a decrease in the observed stimulation of native wild-type AO expression by light (Fig. 6).
Figure 6.
Effects of light and dark on native wild-type AO transcript abundance and transgenic pumpkin AO in wild-type and transgenic plants by semiquantitative RT-PCR. Expression analysis of native wild-type tobacco in sense (PAO) and antisense (TAO) primary transgenic lines. L, after 16 h of light; D, after 16 h of dark. Sense lines: PAO 2-2-1, PAO 3-7-2, PAO 1-7-2; antisense lines: TAO 2-7-1, TAO 1-6-1. The density numbers above each band represent the relative intensity of each band and can be used as an indication of the relative abundance of transcripts. Actin was used as the internal control. Similar results were obtained in three other independent experiments.
DISCUSSION
Although AO activity was first described in plants many years ago, its biological function has remained elusive. Much circumstantial evidence has linked tissue AA contents to growth (Chinoy, 1984), but to date, the mechanism has remained unknown and largely unexplored. Recent evidence has provided an indication of the mechanism whereby AA is involved in the regulation of growth (Pastori et al., 2003). Several lines of evidence also suggest AO may be involved in this process (for example, see Esaka, 1998). The results presented here show that the expression of native AO is regulated by light but that this control is lost when overall AO transcript abundance is manipulated in transgenic plants; AO transcripts are modified by auxin and SA in a manner consistent with effects on growth, and enhanced AO activity oxidizes the apoplastic AA pool and this can stimulate growth in certain conditions. Taken together, these observations demonstrate that AO activity can influence plant growth.
Light Regulation of Gene Expression in Enzymes of the AA Metabolism
We report here the effects of light and dark over a 24-h period on the expression patterns of genes involved in AA metabolism. The expression of cAPX1 is regulated by light in tobacco (Fig. 1), consistent with earlier observations of APX1 mRNA abundance in tobacco (Tabata et al., 2002), Arabidopsis (Kubo et al., 1995), and APX activity in mustard (Sinapis alba; Thomsen et al., 1992). The expression of GLDH, the last enzyme of AA biosynthesis, was not induced by light over a 24-h period in tobacco leaves in the growth conditions used here (Fig. 1). Therefore, we conclude that the light-dependent AA accumulation that we observe under these conditions, increasing from 800 ± 200 nmol g–1 fresh weight at the end of a 16-h dark period to 3000 ± 400 nmol g–1 fresh weight after 16 h in the light is not the result of modulation of GLDH expression. Therefore, the light dependency of AA accumulation observed over the day/night cycle in leaves must result from other factors such as the requirement for carbon skeletons produced by photosynthesis (Smirnoff and Pallanca, 1996). Several of the genes encoding enzymes of the AA biosynthetic pathway increase in expression after transfer of plants to high light intensity (Tabata et al., 2002). GLDH transcripts were found to decrease after a prolonged (5 d) dark treatment (Tabata et al., 2002). Although a recent study in Arabidopsis leaves has reported that GLDH transcripts are lowest in the morning, increasing during the day, and decreasing in the subsequent night (Tamaoki et al., 2003), this is clearly not the case in tobacco.
Complex transcriptional and translational controls modulate AO expression (Esaka et al., 1992; Kato and Esaka, 1999). Phytochrome-dependent modulation of AO activity by light has been reported (Leaper and Newbury, 1989; Hayashi and Morohashi, 1993). Although our data are not exhaustive in exploring the regulation of AO in response to light, they provide corroborative evidence that AO expression is modulated by light and is repressed in the dark (Fig. 1). Controls showed that this diurnal pattern of regulation was not due to circadian rhythmicity (data not shown). It is surprising that sense and antisense manipulation of AO expression led to the elimination of light-induced changes in abundance of native wild-type AO transcripts (Fig. 6). None of the PAO lines showed a decrease in native wild-type AO transcript abundance after a 16-h dark period (Fig. 6). It is our hypothesis that the observed perturbation of the redox state of the apoplast, as discussed below, is responsible for the loss the endogenous diurnal regulation of AO.
AO Transcript Abundance Is Regulated by Plant Hormones in Wild-Type Tobacco
An induction of AO by auxin, similar to that observed here, was reported in pumpkin (Esaka et al., 1992). Moreover, the existence of a cis-acting region in the AO promoter responsible for auxin regulation has been suggested (Kisu et al., 1997). We demonstrate that AO expression is also induced by auxin in tobacco and that the effect is associated with the promotion of plant growth. AO has been shown to catalyze the oxidative decarboxylation of auxin (Kerk et al., 2000), suggesting a role for AO in the regulation of auxin levels within the quiescent centers of maize roots.
High concentrations of SA (1 mm) caused a pronounced inhibition of the growth of tobacco seedlings (Fig. 2B). SA inhibition of plant growth and development is a well-known phenomenon (Petersen et al., 2000). SA can affect cell growth, positively or negatively, depending on the context in which signaling occurs, as studies on Arabidopsis mutants have illustrated (Vanacker et al., 2001). Of the AA-related enzymes analyzed, only the expression of AO was changed by the application of SA and NAA (Figs. 2A and 3A).
Sense and Antisense Manipulation of AO
The modification of AO expression in tobacco by sense (PAO) and antisense (TAO) technologies has allowed us to explore the relationship between AO and growth in planta. To minimize the risk of gene silencing by cosuppression, we used a pumpkin AO cDNA for the sense construct, which shares approximately 68% homology with the tobacco sequence. This choice proved to be successful, as no silencing of the native AO was found in the sense (PAO) lines. The native AO gene was expressed at wild-type levels in PAO lines (Fig. 6). For the antisense suppression of AO, we used one-third of the 5′ sequence of the tobacco cDNA, as in recent years, the use of only partial cDNA sequences proved to be very effective in antisense suppression (Bourque, 1995). In antisense (TAO) lines, we obtained a dramatic reduction in AO transcript abundance (Fig. 6). However, none of the TAO lines exhibited complete suppression of AO transcripts, perhaps because of the limited efficiency of the chosen antisense fragment or because of the genomic position of the transgene.
The data presented here demonstrate that our genetic approach was successful in significantly modifying apoplastic AO activity in leaves, as increases of up to 40-fold were measured in PAO sense transformed lines, whereas activities were halved in antisense TAO lines. We were unable to measure AO protein abundance in this study, as the AO antibodies that we prepared did not have sufficient specificity. Nevertheless, the parallel changes in transcripts and AO activities reported here allow us to assume that our manipulation also modified amounts of AO protein.
AA Content of the Apoplast Was Modulated by AO Activity
PAO sense lines contained less (22%–27%) total AA and AA and more (50%) DHA than TAO antisense lines or wild-type plants (Table I). No differences in leaf AA content were detected between TAO antisense and wild-type plants. The effect of high AO activity on the whole leaf AA pool is minimal, taking into account the fact that AO activities in sense lines were increased by up to 40-fold. The observation that such large increases in AO activity did not lead to major changes in leaf AA may be explained by differential localization of the two components (Lin and Varner, 1991). AO is located in the apoplast, whereas most of the AA is in the cytoplasm (Table I). MDHA and DHA generated in the apoplast have to be translocated to the cytoplasm where they are efficiently regenerated. The observation that the DHA content was doubled in sense lines suggests that the rate of DHA transport or the rate of AA regeneration (or both) were too slow to compensate for the enhanced AO activity in sense lines. We measured the AA content and redox state in the leaf apoplast in the same plants as those used for total leaf AA determinations. The AA pool was virtually all oxidized (97%) in the PAO sense lines and was more reduced than the wild type in TAO antisense lines (66% as opposed to 40%; Table I). These data, together with the observation that the sense lines accumulated almost four times more DHA than the wild type in the apoplast, confirmed two points. First, our sense construct successfully targeted AO to the apoplast. Second, manipulation of AO produced a marked localized effect on (apoplastic) AA. It is interesting to note that in PAO sense and TAO antisense lines, the total AA content of the apoplast was twice that of the wild type. This may suggest that AO-mediated perturbation of the AA and DHA contents of the apoplast influences the transport of reduced and oxidized forms, such that when the apoplastic pool is very reduced or very oxidized, transport processes are increased in an attempt to redress the balance.
AO and Plant Development
In the present study, we demonstrate that increasing AO activity by 40-fold leads to enhanced biomass accumulation and elongation in tobacco plants (Fig. 5). In contrast, reduction of AO activity by antisense technology results in a reduction in growth rate (Fig. 5A). Taken together, the data presented here suggest that enhanced AO activity can have a positive effect on growth (Fig. 5B). Increased biomass production in PAO sense plants appears to be mainly due to enhanced internode elongation. It is possible that AO action alone stimulates growth through the generation of MDHA radicals in the apoplast. This enhanced growth could occur by a chemorheological wall-loosing reaction as described for superoxide- and hydroxyl radical-mediated extension growth (Schopfer et al., 2002), or by enhancing cell enlargement via the polarization of the plasmalemma, leading to expansion by vacuolization as suggested previously (Esaka, 1998; Smirnoff, 2000), or both.
Plant growth is the outcome of various mechanisms of regulation, signaling, and crosstalk. We have provided in planta evidence that AO is one of these components and therefore participates to this broad dialogue between signaling molecules and environmental cues. It should be noted that in another study on modulation of AO in tobacco, no effects on growth were reported (Sanmartin at al., 2003). In the experimental conditions described here, changes in AO activity alone produced marked effects on plant growth, measured as shoot biomass accumulation and stem height. However, the growth effect was clearly environment dependent as observed in the AA-deficient Arabidopsis vtc 1 mutant (Conklin et al., 1997; Veljovic-Jovanovic et al., 2001). Vtc1 shows a marked conditional phenotype, such that when grown on short days where vegetative growth predominates, vtc1 is much slower growing than the wild type (Col0). Under long days where reproductive growth predominates, there is no significant difference in growth. In this case, AA has been shown to regulate genes controlling plant development through hormone signaling (Pastori et al., 2003). We conducted a large-scale greenhouse experiment in the summer months, with growth conditions markedly different from those described here. In these conditions, where light intensities were higher, the photoperiod was longer, and temperatures were higher, the effects of modified AO activity were much less marked. However, it is interesting to note that although no significant differences in plant height and biomass were recorded between wild-type and transgenic plants, other effects were observed under these conditions. In particular, AO sense plants showed a significantly higher number of smaller flowers compared with wild-type and antisense plants (data not shown). This effect on the flowers of the AO sense lines caused a reduction of 6% to 14% in the weight of seeds. Such differences were not observed when plants were grown in controlled environment chambers, as described here. We conclude that the phenotype associated with increased AO activity in tobacco is conditional and is modulated by environmental cues.
In conclusion, the data presented here suggest that factors such as apoplastic redox state that is dominated by AA and regulated by AO modulate receptor function and signal transduction and that there is scope for modulation and interaction between different signals (hormone, redox, and light) in the apoplast.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Tobacco (Nicotiana tabacum cv Petit Havana, mutant SRI) seeds were germinated in petri dishes on moistened filter paper. After 10 d, seedlings were transferred to compost (Petersfield Products, Leicester, UK) in pots. Plants were grown at 22°C day/night in controlled environment chambers supplying a photosynthetic photon flux density of 250 μmol m–2 s–1 at plant height as a 16-h photoperiod. Young fully expanded leaves from 6-week-old plants were used for AA measurements in the light and dark. Three-week-old plants were used for all treatments and RT-PCR analyses.
Hormone Treatments
Three-week-old seedlings were sprayed to run-off (using an aerosol spray bottle supplied by Nalgene, Rochester, NY) with 10 μm and 1 mm SA and 0.5 μm NAA. After treatment, plants were kept in continuous light for 24 h and whole shoots were harvested for RT-PCR analysis. For the evaluation of the effects of hormones on growth, sprayed plants were subsequently returned to “growth conditions” until further analysis 1 week after the initial harvest.
PCR Amplification of AO cDNAs for Cloning
Cloning of the genes encoding AO from pumpkin (Cucurbita pepo; PAO; EMBL GenBank accession no. X55779) and tobacco (TAO; accession no. D43624) was achieved from PCR amplification of full-length cDNAs using primers designed to add additional restriction sites to facilitate the cloning. For PAO the primers were 5′-ACCACTCGAGATGCTTCAGATG -3′ (nucleotide position 18–29 of cDNA; added XhoI site is underlined) and 5′-ACCAGAGCTCTTAGGGGTTATTT-3′ (nucleotide position 1,757–1,745; added SacI site is underlined). For TAO the primers were 5′-ACCAGAGCTCATGGCTTCCTTA-3′ (nucleotide position 88–100 of cDNA; added SacI site is underlined) and 5′-ACCACTCGAGTTTGTGCCACC-3′ (nucleotide position 609–559; added XhoI site is underlined). The PCR conditions were 10 cycles of 1 min at 94°C, 1 min at 40°C, and 1.5 min at 72°C, and 15 cycles of 1 min at 94°C, 1 min at 61°C, and 1.5 min at 72°C, using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), 1 ng of cDNA, and 0.2 μm each primer in a 50-μL reaction. At the end of the cycles, the reactions were incubated at 72°C for 10 min. The identity of the 1.7-kb (PAO) and 520-bp (TAO) PCR products was confirmed by single-strand sequencing (ABI PRISM, 310 genetic analyzer; Perkin-Elmer, Warrington, Cheshire, UK).
Construction of Sense and Antisense AO in cj102 and Transformation into Agrobacterium tumefaciens
PAO and TAO PCR products were subcloned as XhoI-SacI fragments into the corresponding sites of pp5ln (derived from pUC19; Frenken et al., 1999) by standard in vitro recombination techniques (Sambrook et al., 1989), generating the fusion constructs PAO/pp5ln and TAO/pp5ln. In PAO/pp5ln, the AO cDNA is placed in sense orientation under the control of the CaMV35S promoter with duplicated enhancer region (2x35S), originating the 2x35S-PAO expression cassette. In TAO/pp5ln, the AO cDNA is placed in antisense orientation under the control of 2x35S, originating the 2x35S-TAO expression cassette. Sense and antisense expression cassettes were ligated into cj102, a derivative plasmid of pGPTV (Becker et al., 1992), as HindIII-SacI fragments into the corresponding sites of cj102 in replacement of the 2x35S-β-glucuronidase cassette. The cj102-PAO and cj102-TAO constructs obtained were transformed into A. tumefaciens LBA4404 by electroporation (Parry et al., 1998).
Plant Material and Transformation
Sterile cultures of tobacco were transformed with cj102-PAO and cj102-TAO constructs by A. tumefaciens leaf disc infection (Gallois and Marinho, 1995). T1 seeds from primary transformants were germinated in petri dishes containing 1.5% (w/v) agar in distilled water (nonselective medium), supplied with 100 mg L–1 kanamycin (selective medium). T2 seeds were obtained from individual T1 plants by self-pollination and were used to generate T2 transgenic progeny.
Southern-Blotting Analysis
Genomic DNA (5 μg) from PAO sense and TAO antisense lines was digested with HindIII, which cuts once within the transgenic sequence, run on an 0.8% (w/v) agarose gel and blotted onto a nylon membrane (Hybond-NX; Amersham Life Science, Buckinghamshire, UK). The membranes were prehybridized at 55°C for 2 h in hybridization buffer (Amersham Life Science) and were hybridized overnight in the same buffer. The HindIII/SacI fragments 2x35S-PAO and 2x35S-TAO were alkaline phosphatase labeled and were used as probes (AlkPhos Direct; Amersham Life Science). Hybridization temperatures were 65°C for 2x35S-PAO and 68°C for 2x35S-TAO. The membranes were washed according to Amersham's recommendations and detection was performed with CDP-Star (Amersham Life Science) by exposing Hyperfilm enhanced chemiluminescence film (Amersham Life Science) to the membrane for 1.5 h.
Total RNA Extraction and Gene Expression Analysis by RT-PCR
Total RNA was extracted from using RNeasy plant mini kit (Qiagen, West Sussex, UK) according to the supplier's recommendation. Residual DNA was removed with DNase I, Amp Grade (Invitrogen, Strathclyde, UK). The absence of DNA contamination in the samples was confirmed by a saturating PCR of 40 cycles using actin- (X63603) specific primers (5′-CGCGAAAAGATGACTCAAATC-3′ and 5′-AGATCCTTTCTGATATCCACG-3′), which give a 687-bp product with genomic DNA and a 533-bp product with cDNA. One microgram total RNA was reverse transcribed using 0.5 μg of Oligo (dT)12–18 (Invitrogen) and 200 units of Superscript II (Invitrogen) following the supplier's recommendation. cDNA samples were standardized by PCR for actin content using the gene-specific primers. On the basis of the published sequences, the following gene-specific primers were designed and used for amplification: PR-1a (X12737), 5′-GCCTTCATTTCTTCTTGTCTC-3′ and 5′-TTAGTATGGACTTTCGCCTC-3′; GLDH (AB048530), 5′-TTTTAGGCTTTGACTGTGGTG-3′ and 5′-TCAGATGAAGAGCTTCTCAAG-3′; cAPX1 (X59600), 5′-CTCAAGCTGTTGACAAATG-3′ and 5′-AGCTTCAGCAACCAATTC-3′; sAPX (AB02 2274), 5′-TTGT TTCAGTTGGCCAGTGC-3′ and 5′-CGCTGCCTTGTGTAGG-3′; tAPX (AB022273), 5′-TGTTTTCTACAGAATGGGC-3′ and 5′-GTTGAGTATTTTG CTGCCAC-3′; PAO (X55779), 5′-TTGACCGGAGCAAAAACTTC-3′ and 5′-AATTCAATGACGACTCCTCC-3′; and TAO (D43624), 5′-AACCAAAAACACCTCAAGGC-3′ and 5′-GGTGCTTGTTTTAGGACATC-3′.
For semiquantitative RT-PCR, the cycle number was kept within the linear range (30 cycles) and the conditions were 3 min at 94°C, cycle of 45 s at 94°C, 30 s at 52°C, and 45 s at 72°C, followed by 10 min at 72°C, using 0.5 μL of the RT reaction and 0.2 μm each oligonucleotide primer in a total volume of 25 μL. The identity of the PCR products was verified by single-strand sequencing (ABI PRISM, 310 genetic analyzer; Perkin-Elmer). RT-PCR products were loaded on 2% (w/v) agarose gel containing 0.5 μg mL–1 ethidium bromide and the band intensities were quantified with the Eagle Eye II (Stratagene).
Determination of AA and Glc-6-P Content
Leaf samples were powdered in liquid nitrogen, then 1 mL of ice-cold 1 n HClO4 was added per 0.1-g sample (fresh weight). Extracts were then centrifuged at 14,000 rpm for 5 min at 4°C. HEPES buffer (0.1 m, pH 7.0) was added at a buffer:extract ratio of 1:5 (v/v). K2CO3 (5 m) was added until the extract reached pH 5.6. The extracts were again centrifuged at 14,000 rpm, this time for 2 min, to allow the removal of precipitated K2ClO4, and were then assayed for AA and DHA as described by Foyer and coworkers (1983). Glc-6-P was measured according to Latzko and Gibbs (1972).
Preparation of IWF
IWF was prepared using a method similar to that adopted by Turcsànyi et al. (2000). Young fully expanded leaves were vacuum infiltrated at –70 kPa with chilled 10 mm citrate buffer (pH 3.0) for 5 min. Leaves were then blotted dry, carefully rolled and inserted into a prechilled syringe, and centrifuged at 2,000 rpm for 10 min at 4°C. For ascorbate and Glc-6-P analyses, IWF was collected in empty preweighed Eppendorf tubes. Controls centrifuged into tubes containing cold metaphosphoric acid (2%, w/v) gave similar results.
AO Assay
For total AO activity, leaf tissue was powdered in liquid nitrogen and then homogenized with 0.1 m sodium phosphate, pH 6.5 (1 mL 0.1 g–1 fresh weight). The extract was then diluted 10-fold in the same buffer and 50 μL was used in the assay. Measurements of soluble and ionically bound AO activity were performed according to Sanmartin et al. (2003). Leaf extracts were centrifuged at 15,000g for 10 min at 4°C and soluble AO activity was measured on the supernatant. The pellet (ionically bound fraction) was resuspended in 0.1 m sodium phosphate, pH 6.5, with the addition of 1 m NaCl, vortexed for 10 min at 4°C, and then centrifuged at 15,000g for 10 min at 4°C. AO activity was measured on the supernatant.
AO activity was determined from the decrease in A265 at 25°C in a reaction mixture containing 0.1 m sodium phosphate, pH 5.6, 0.5 mm EDTA, and 100 μm AA. One unit of AO activity was defined as the oxidation of 1 μmol AA min–1 at 25°C. An extinction coefficient for AA of 14 mm–1 cm–1 at 265 nm was used in calculations (Nakano and Asada, 1981). Recovery experiments with purified AO were performed to ensure that maximal activity was extracted and maintained during isolation procedures.
Growth Rates of T1 Populations and Phenotypic Analysis
T1 populations were obtained by germinating T1 seeds on nonselective medium. Twenty-five plants per each transgenic line (three AO sense and two AO antisense) were used for growth experiments. Growth curves were drawn by measuring every week shoot fresh weight of three plants per line growing in compost in controlled environment. Shoots were excised at the base of the stem and were weighed for the fresh weight determination. Plant height was measured on 7-week-old plants from the basis of the stem to inflorescence.
Statistical Analyses
Statistical analyses were performed using GENSTAT 5 (Peyne et al., 1993). Data were subjected to analysis of variance and t test to investigate the statistical significance of the effects of modified AO activity on growth. Correlations were performed using Sigmaplot 2001 (SPSS, Chicago) using least square linear regression methods to test the goodness of fit.
Acknowledgments
We thank Prof. Muneharu Esaka (Hiroshima University, Japan) for the kind donation of the pumpkin and tobacco AO cDNAs and Sue Robinson and Dr. Karl Hunter (Unilever Research) for performing the large-scale greenhouse study of transgenic tobacco plants.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022798.
This work was supported by Unilever Foods.
References
- Asada K (1997) The role of ascorbate peroxidase and monodehydroascorbate reductase in H2O2 scavenging in plants. In JG Scandalios, ed, Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 715–735
- Barnes JD, Zheng Y, Lyons TM (2002) Plant resistance to ozone: the role of ascorbate. In K Omasa, H Saji, S Youssefian, N Kondo, eds, Air Pollution and Plant Biotechnology. Springer-Verlag, Tokyo, pp 235–254
- Bartoli CG, Pastori GM, Foyer CH (2000) Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol 123: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Bourque JE (1995) Antisense strategies for genetic manipulations in plants. Plant Sci 105: 125–149 [Google Scholar]
- Chinoy NJ (1984) Ascorbic acid in plant growth and development. In M Nijhoff, W Junk, eds, The Role of Ascorbic Acid in Growth, Differentiation, and Metabolism of Plants. Martinus Nijhoff, The Netherlands, The Hague, Dr Junk, pp 68–195
- Cleland RE (1987) How hormones work: auxin and cell elongation. In PJ Davies, ed, Plant Hormones and Their Role in Plant Growth and Development. Kluwer Academic Publishers, Lancaster, UK, pp 132–148
- Conklin PL, Pallanca JE, Last RL, Smirnoff N (1997) l-Ascorbic acid metabolism in the ascorbate-deficient Arabidopsis mutant vtc1. Plant Physiol 115: 1277–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaka M (1998) Gene expression and function of ascorbate oxidase in higher plants. Recent Res Dev Phytochem 2: 315–326 [Google Scholar]
- Esaka M, Fujisawa K, Goto M, Kisu Y (1992) Regulation of ascorbate oxidase expression in pumpkin by auxin and copper. Plant Physiol 100: 231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Rowell J, Walker D (1983) Measurement of the ascorbate content of spinach leaf protoplasts and chloroplasts during illumination. Planta 157: 239–244 [DOI] [PubMed] [Google Scholar]
- Frenken LGJ, Jobling SA, The Y, Verhoeyen ME, Wilkinson JE, Van der Logt CPE (1999) Modifying a plant to produce an antibody useful for increasing pathogen resistance or to modulate metabolism comprises introducing a DNA sequence encoding a heavy chain immunoglobulin linked to a peptide that targets a cellular compartment. Patent number EP1118669 European Patent Office, Munich, Germany 8th December 2000
- Gallois P, Marinho P (1995) Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. In H Jones, ed, Methods in Molecular Biology, Vol 49: Plant Gene Transfer and Expression Protocols. Humana Press, Totowa, NJ, pp 39–48 [DOI] [PubMed] [Google Scholar]
- Hayashi R, Morohashi Y (1993) Phytochrome control of the development of ascorbate oxidase activity in mustard (Sinapis alba) cotyledons. Plant Physiol 102: 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo A, Gonzalez-Reyes JA, Navas P (1989) Ascorbate free radical enhances vacuolisation in onion root meristems. Plant Cell Environ 12: 455–460 [Google Scholar]
- Horemans N, Foyer CH, Asard H (2000) Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci 5: 263–267 [DOI] [PubMed] [Google Scholar]
- Joo JH, Bae YS, Lee JS (2001) Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol 126: 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P (1999) Systemic signalling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Kato N, Esaka M (1999) Changes in ascorbate oxidase gene expression and ascorbate levels in cell division and cell elongation in tobacco cells. Physiol Plant 105: 321–329 [Google Scholar]
- Kato N, Esaka M (2000) Expansion of transgenic tobacco protoplasts expressing pumpkin ascorbate oxidase is more rapid that that of wild-type protoplasts. Planta 210: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Kerk NM, Jiang K, Feldman LJ (2000) Auxin metabolism in the root apical meristem. Plant Physiol 122: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisu Y, Harada Y, Goto M, Esaka M (1997) Cloning of the pumpkin ascorbate oxidase gene and analysis of a cis-actin region involved in induction by auxin. Plant Cell Physiol 38: 631–637 [DOI] [PubMed] [Google Scholar]
- Kubo A, Saji H, Tanaka K, Kondo N (1995) Expression of Arabidopsis cytosolic ascorbate peroxidase gene in response to ozone or sulphur dioxide. Plant Mol Biol 29: 479–486 [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root-meristems is a 2-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Latzko E, Gibbs M (1972) Measurement of the intermediate of the photosynthetic carbon reduction cycle, using enzymatic methods. Methods Enzymol 24: 261–293 [DOI] [PubMed] [Google Scholar]
- Leaper L, Newbury HJ (1989) Phytochrome control of the accumulation and rate of synthesis of ascorbate oxidase in mustard cotyledons. Plant Sci 64: 79–90 [Google Scholar]
- Lin LS, Varner JE (1991) Expression of ascorbate oxidase in zucchini squash (Cucurbita pepo). Plant Physiol 96: 159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JD, Mitchell P, Schürmann P (1980) Modulation of coupling factor ATPase activity in intact chloroplasts: the role of the thioredoxin system. FEBS Lett 112: 173–177 [Google Scholar]
- Moser O, Kanellis AK (1994) Ascorbate oxidase of Cucumis melo L. var. reticulatus: purification, characterisation and antibody production. J Exp Bot 45: 717–724 [Google Scholar]
- Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplast. Plant Cell Physiol 22: 867–880 [Google Scholar]
- Ohashi Y, Matsuoka M (1987) Induction and secretion of pathogenesis-related proteins by salicylate or plant hormones in tobacco suspension cultures. Plant Cell Physiol 28: 573–580 [Google Scholar]
- Ohkawa J, Ohya T, Ito T, Nozawa H, Nishi Y, Okada N, Yoshida K, Takano M, Shinmyo A (1994) Structure of the genomic DNA encoding cucumber ascorbate oxidase and its expression in transgenic plants. Plant Cell Rep 13: 481–488 [DOI] [PubMed] [Google Scholar]
- Ostergaard J, Persiau G, Davey MW, Bauw G, Van Montagu M (1997) Isolation of a cDNA coding for l-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J Biol Chem 272: 30009–30016 [DOI] [PubMed] [Google Scholar]
- Parry MA, Colliver SP, Madgwick PJ, Paul MJ (1998) Manipulation of photosynthetic metabolism. In C Cunningham, AJR Porter, eds, Methods in Biotechnology, Vol 3: Recombinant Proteins from Plants: Production and Isolation of Clinically Useful Compounds. Humana Press, Totowa, NJ, pp 229–249 [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH (2003) Leaf vitamin C contents modulate plant defense transcripts and regulate genes controlling development through hormone signaling. Plant Cell 15: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Peyne RW, Lane PW, Digby PGN, Harding SA, Leech PK, Morgan GW, Todd AD, Thompson R, Tunnicliffe WG, Welham SJ et al. (1993) Genstat 5, Release 3, Reference Manual. Clarendon Press, Oxford, UK
- Plöchl M, Lyons T, Ollerenshaw J, Barnes J (2000) Simulating ozone detoxification in the leaf apoplast through the direct reaction with ascorbate. Planta 210: 454–467 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis TA (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sanmartin M, Drogouti PD, Lyons T, Barnes J, Kanellis AK (2003) Overexpression of ascorbate oxidase in the apoplast of transgenic tobacco results in altered ascorbate and glutathione redox states and increased sensitivity to ozone. Planta 216: 918–928 [DOI] [PubMed] [Google Scholar]
- Schopfer P, Liszkay A, Bechtold M, Frahry G, Wagner A (2002) Evidence that hydroxyl radicals mediate auxin-induced extension growth. Planta 214: 821–828 [DOI] [PubMed] [Google Scholar]
- Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multifacetted molecule. Curr Opin Plant Biol 3: 229–235 [PubMed] [Google Scholar]
- Smirnoff N, Pallanca JE (1996) Ascorbate metabolism in relation to oxidative stress. Biochem Soc Trans 24: 472–478 [DOI] [PubMed] [Google Scholar]
- Tabata K, Takaoka T, Esaka M (2002) Gene expression of ascorbic acid-related enzymes in tobacco. Phytochemistry 61: 631–635 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Mukai F, Asai N, Nakajimi N, Kubo A, Aono M, Saji H (2003) Light-controlled expression of a gene encoding l-galactono-γ-lactone dehydrogenase which affects ascorbate pool size in Arabidopsis thaliana. Plant Sci (in press)
- Thomsen B, Drumm-Herrel H, Mohr H (1992) Control of the appearance of ascorbate peroxidase (EC 1.11.1.11) in mustard seedling cotyledons by phytochrome and photo-oxidative treatments. Planta 186: 600–608 [DOI] [PubMed] [Google Scholar]
- Turcsànyi E, Plöchl M, Lyons TM, Barnes JD (2000) Does ascorbate in the mesophyll cell walls form the first line of defence against ozone? Testing the concept using broad bean (Vicia faba). J Exp Bot 51: 901–910 [PubMed] [Google Scholar]
- Vanacker H, Lu H, Rate DN, Greenberg JT (2001) A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S, Pignocchi C, Noctor G, Foyer CH (2001) Low vitamin C in the vtc 1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol 127: 426–435 [PMC free article] [PubMed] [Google Scholar]