Abstract
β-alanine (Ala) betaine, an osmoprotectant suitable under saline and hypoxic environments, is found in most members of the halophytic plant family Plumbaginaceae. In Limonium latifolium (Plumbaginaceae), it is synthesized via methylation of β-Ala by the action of a trifunctional S-adenosyl l-methionine (Ado-Met): β-Ala N-methyltransferase (NMTase). Peptide sequences from purified β-Ala NMTase were used to design primers for reverse transcriptase-PCR, and several cDNA clones were isolated. The 5′ end of the cDNA was cloned using a 5′-rapid amplification of cDNA ends protocol. A 500-bp cDNA was used as a probe to screen a λ-gt10 L. latifolium leaf cDNA library. Partial cDNA clones represented two groups, NMTase A and NMTase B, differing only in their 3′-untranslated regions. The full-length NMTase A cDNA was 1,414 bp and included a 1128-bp open reading frame and a 119-bp 5′-untranslated region. The deduced amino acid sequence of 375 residues had motifs known to be involved in the binding of Ado-Met. The NMTase mRNA was expressed in L. latifolium leaves but was absent in Limonium sinuatum, a member of the genus that lacks the synthetic pathway for β-Ala betaine. NMTase mRNA expression was high in young and mature leaves and was enhanced by light. NMTase cDNA was expressed in yeast (Saccharomyces cerevisiae) under the control of a galactose-inducible promoter. Protein extracts of galactose-induced recombinant yeast had Ado-Met-specific NMTase activities that were highly specific to β-Ala, N-methyl β-Ala, and N,N-dimethyl β-Ala as methyl acceptors. NMTase activities were not detectable in comparable protein extracts of yeast, transformed with vector control. The NMTase protein sequence shared homology with plant caffeic acid O-methyltransferases and related enzymes. Phylogenetic analyses suggested that β-Ala NMTase represents a novel family of N-methyltransferases that are evolutionarily related to O-methyltransferases.
Drought, salinity, flooding, and freezing adversely affect agricultural productivity (Boyer, 1982). However, many plants have evolved metabolic adaptations to these abiotic stress factors. Accumulation of osmoprotectants is a common adaptation found in stress-tolerant plants, bacteria, and marine algae (Yancey et al., 1982; Bohnert and Sheveleva, 1998). Quaternary ammonium compounds (QACs) represent some of the most effective osmoprotectants known in biology (Anthoni et al., 1991; Rhodes and Hanson, 1993).
Because only certain stress-tolerant plants accumulate the common QAC Gly betaine and many crops do not, it was suggested that engineering crops for Gly betaine overproduction could be a way to improve their stress tolerance (McCue and Hanson, 1990). Transgenic plants, overexpressing bacterial and plant pathways for Gly betaine synthesis, accumulated relatively small quantities of the QAC (Rontein et al., 2002), but nonetheless exhibited stress-tolerant phenotypes (Sakamoto and Murata, 2000, 2001; Hibino et al., 2002). However, reiterative metabolic engineering experiments indicated that availability of the substrate choline limited Gly betaine synthesis in transgenic plants (McNeil et al., 2000).
In the evolution of the stress-tolerant plant family Plumbaginaceae, β-Ala betaine replaced Gly betaine (Hanson et al., 1991, 1994). β-Ala betaine synthesis is not constrained by choline availability, because it is derived by the methylation of the non-protein amino acid, β-Ala. Unlike Gly betaine synthesis, β-Ala betaine synthesis does not require oxygen, and hence it was suggested to be suitable for osmoprotection under saline and hypoxic conditions (Hanson et al., 1994; Rathinasabapathi, 2000). Accordingly, β-Ala betaine was distributed among species of the Plumbaginaceae, adapted to a wide range of adverse stress environments including saline and hypoxic conditions (Hanson et al., 1994). Although β-Ala betaine accumulation has been intensely studied for its role in osmoprotection (Hanson et al., 1994; Rathinasabapathi et al., 2000, 2001), early work on this com-pound in marine algae also suggested it to have a cholesterol-reducing effect in animal feeding experiments (Abe and Kaneda, 1973).
We have characterized the β-Ala betaine synthetic pathway in Limonium latifolium, a member of the Plumbaginaceae. β-Ala betaine is synthesized by N-methylation of β-Ala via N-methyl and N,N-dimethyl β-Alas (Rathinasabapathi et al., 2000). β-Ala methylation, catalyzed by a S-adenosyl l-Met (Ado-Met)-dependent N-methyltransferase (β-Ala N-methyltransferase [NMTase]) was specific to β-Ala betaine-accumulating members of the Plumbaginaceae and was absent in species lacking this pathway (Rathinasabapathi et al., 2000). An 86-kD leaf protein, a dimer of 43-kD subunits, purified from L. latifolium was trifunctional, catalyzing all three methylations of β-Ala betaine synthesis (Rathinasabapathi et al., 2001). This suggested that β-Ala betaine synthesis could be engineered in transgenic plants by simply expressing a single gene. We report here the isolation and functional expression of a full-length cDNA for β-Ala NMTase as a step toward a metabolic engineering approach to understanding the biological roles of β-Ala betaine.
RESULTS
cDNA Cloning Using a PCR Strategy
Active β-Ala NMTase protein was purified from L. latifolium leaves using a previously described protocol (Rathinasabapathi et al., 2001). The 43-kD band was extracted from a polyacrylamide gel and subjected to degradation by lysylendo-peptidase (lysC), and several peptides were sequenced (Table I). Degenerate primers were designed based on the peptides 1, 2, and 3 in both sense (F) and antisense (R) orientations (Table II). Under optimum conditions, primers 2F and 3R amplified a 500-bp cDNA. Primer combinations 2F and 1R and 1F and 3R produced shorter products of 400 and 100 bp, respectively (data not shown). The 500-bp PCR product was named clone 23 and was sequenced (accession no. AY216896).
Table I.
Peptide sequences obtained from purified L. latifolium β-Ala NMTase
Amino acid residues in parentheses are alternate possibilities indicating ambiguity in sequencing.
| Peptide | Identification Code | Sequence |
|---|---|---|
| 1 | A-1019-b | H(S/A/Q)RTE(Q)EE(L)YRQLGLLAG |
| 2 | A-1018-a | ALLGSGYDGFEGVK |
| 3 | A-1017-a | FRVIHVDYFFPVVEF |
| 4 | A-1017-a-b | AVI (P)ELXTAILDA(K)S |
| 5 | D-542-a | S(Q/A)LDG(A)SG(Y/E)DGFEG |
| 6 | D-541-a | H (S/Q/A)RTE(Q)EE(L)YRQLGLLAG |
| 7 | A-1019-b-c | R(H/A/Q)INFDLPXVVA |
Table II.
Degenerate primer sequences used for RT-PCR
Primers were named for the peptide from which they were derived. F stands for forward and R for reverse direction of the primers. I is inosine.
| Primer Name | Sequence (5′ to 3′) |
|---|---|
| 1F | CAYMGIACYGARGARGARTAYMG |
| 1R | TGICKRTAYTCYTCYTCIGT |
| 2F | GGITAYGAYGGITTYGARGGIGT |
| 2R | TTIACICCYTCRAANCCRTC |
| 3F | ATHCAYGTIGAYTAYTTYTTYCC |
| 3R | ACIGGRAARAARTARTCIACRTG |
| 23R | CCACCAACATCTACCAATG |
| 4F | ACIGCNATHYTIGAYGCIWS |
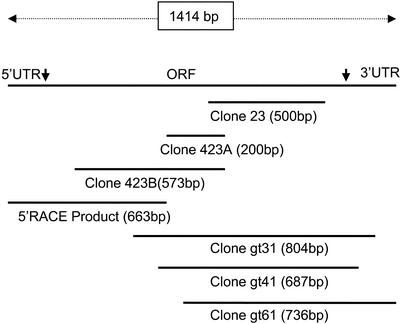
PCR primers were designed to amplify cDNA, 5′ to clone 23. This was done by using a degenerate sense primer based on peptide 4 and an antisense primer based on the 5′ end of clone 23. This resulted in two overlapping products, 423a (accession no. AY216897) and 423b (accession no. AY216898). The longer product, 423b (Fig. 1) was obtained by chance because the 5′ end primer had shared a small region of homology in the target sequence.
Figure 1.
cDNA cloning using RT-PCR, 5′-RACE, and cDNA library screening.
A cDNA partially overlapping clone 423b was further amplified employing a 5′-RACE protocol (Fig. 1) using the gene-specific primer 423R 5′ to 3′-TCAACTCGTCATTCTTCCCGTAGTAC. We used a high-fidelity enzyme system to reduce potential errors during 5′-RACE PCR. However, to detect possible mutations during PCR amplification, this reaction was done in duplicate in independent reactions, and the products were sequenced. Both 5′-RACE sequences matched exactly with each other (accession no. AY216899).
cDNA Library Screening
To validate the PCR-based cloning strategy, we also screened a λ-gt10 L. latifolium cDNA library using clone 23 cDNA as a probe. Among the many positive clones identified, three clones, gt31, gt41, and gt61 (accession nos. AY216900, AY216901, and AY216902, respectively), were sequenced in both strands (Fig. 1). All three were partial cDNAs. The cDNA sequences of these clones verified the PCR-based cDNA cloning in that the open reading frame obtained via both methods was the same. The 3′-untranslated region, however, differed among clones. Clones gt41 and gt61 represented NMTase A. The 3′-untranslated region of clone gt31 differed from the other clones in several nucleotides (data not shown), representing NMTase B.
Characterization of the Full-Length NMTase cDNA
The full-length (1,414-bp) NMTase A cDNA (accession no. AY216903), reconstructed by splicing clone gt61 to the 5′-RACE product, had 119 bp of 5′-untranslated region, an open reading frame of 1,128 bp, and a 3′-untranslated region of 167 bp. The open reading frame had two ATGs within the first 30 bp. The sequence context near the first Met had matched well with the sequence context conserved around the initiation codon for plant genes (Joshi, 1987; Cavener and Ray, 1991). A putative polyadenylation signal, AAATAAT (Heidecker and Messing, 1986), preceded poly(A+) by 17 bp.
The deduced amino acid sequence had 375 amino acid residues (Fig. 2). It had all of the peptides that were sequenced (Table I) from purified β-Ala NMTase (Fig. 2). All three motifs implicated in Ado-Met binding (Joshi and Chiang, 1998) were conserved in β-Ala NMTase (Fig. 2). There is no recognizable signal sequence (Emanuelsson et al., 2000) based upon sequence analyses.
Figure 2.
L. latifolium NMTase deduced amino acid sequence. Underlined sequences match to peptides sequenced from purified NMTase. Ado-Met-binding motifs, A, B, and C are shown in bold in that order.
NMTase Expression Is Unique to β-Ala Betaine-Accumulating Species
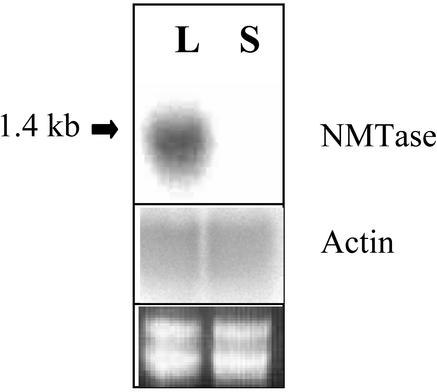
RNA blots probed with β-Ala NMTase cDNA indicated that a 1.4-kb NMTase mRNA is expressed in L. latifolium leaves but not in the leaves of Limonium sinuatum (Fig. 3), a species that does not methylate β-Ala (Hanson et al., 1991). RNA blots from both of these species had nearly equal signal intensities when the filter was probed with a L. latifolium actin probe (Fig. 3), and equal loading was verified by an ethidium bromide-stained gel (Fig. 3).
Figure 3.
NMTase mRNA expression in Limonium sp. Total RNA (10 μg) per lane was used and probed with clone 23 DNA probe (top) or a L. latifolium actin probe (middle). The bottom panel shows an ethidium bromide-stained gel as a loading control. Mature leaf RNA was from L. latifolium (L) and L. sinuatum (S). The arrow points to 1.4-kb size.
Tissue Specificity and Regulation of NMTase Expression
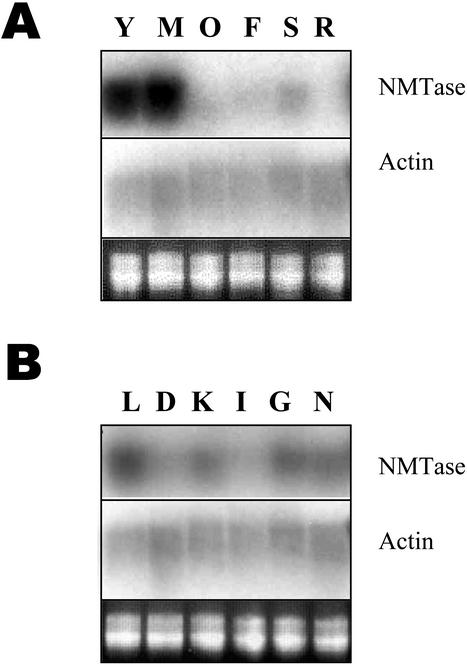
Young and mature leaves had high levels of β-Ala NMTase steady-state mRNA expression, whereas old leaves, floral stems, flowers, and roots had relatively lower levels of expression (Fig. 4A). Corroborating expression at the RNA level, crude extracts of young and mature leaves had detectable NMTase activities (150–200 pmol h–1 mg–1 protein), and floral stems, flowers, and roots had no detectable activity. We used excised mature leaves to test what factors may regulate β-Ala NMTase mRNA levels. Leaves incubated in a nutrient medium were exposed to light or dark conditions for 24 h. Some of the leaves incubated under light were also exposed to sodium chloride, indole butyric acid, GA3, and kinetin in the nutrient medium for 24 h. β-Ala NMTase expression was about 10-fold higher under light than dark (Fig. 4B), which was also verified by incubating whole plants under dark and light conditions (data not shown). Under light, exogenous auxin to excised leaves directly or indirectly down-regulated NMTase, and other treatments including sodium chloride did not significantly affect NMTase RNA levels (Fig. 4B).
Figure 4.
A, β-Ala NMTase mRNA expression in various tissues of L. latifolium. Total RNA from young leaves (Y), mature leaves (M), old leaves (O), flower (F), floral stem (S), and root (R) was separated and probed with either NMTase cDNA (clone 23) probe (top) or actin cDNA probe (middle). The bottom panel shows an ethidium bromide-stained RNA as a loading control. B, β-Ala NMTase mRNA expression in excised leaves of L. latifolium as modulated by external treatments. L, Exposure to light; D, dark; K, kinetin; I, IBA; G, GA3; and N, NaCl. The middle panel shows corresponding RNA blot probed with L. latifolium actin cDNA. The bottom panel shows an ethidium bromide-stained RNA as a loading control.
β-Ala NMTase Is Coded by a Single or Low Copy Number of Genes
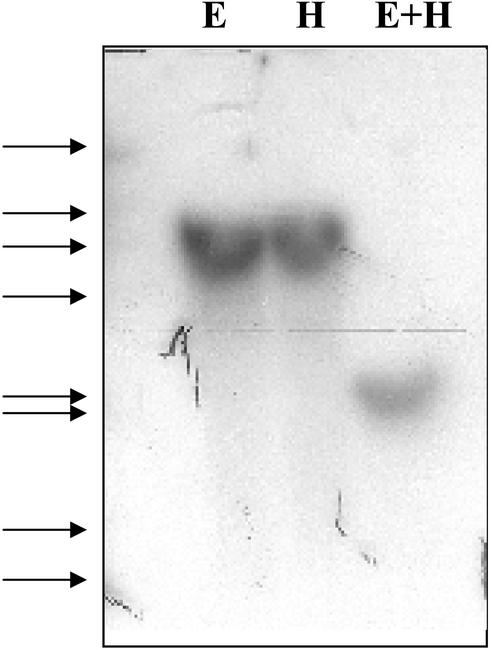
Analysis of L. latifolium genomic DNA by Southern-blot hybridization showed a 5,000-bp band for an EcoRI digest, a 5,000-bp band for a HindIII digest, and a 2,000-bp band for a double digest (Fig. 5). The simple patterns obtained were consistent with a single or a few copies of the gene in L. latifolium genome.
Figure 5.
Southern-blot hybridization of L. latifolium genomic DNA probed with NMTase clone 23 probe. Lanes contained 20 μg of genomic DNA digested with EcoRI (E), HindIII (H), and both EcoRI and HindIII (E+H). Arrows at the left indicate the positions of λ-HindIII markers.
Heterologous Expression of β-Ala NMTase cDNA
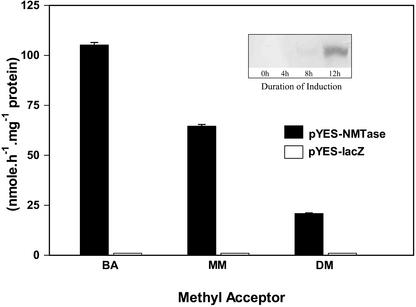
β-Ala NMTase cDNA was cloned into yeast expression vector pYES-NT/B to derive pYES-NMTase construct and was introduced into Brewer's yeast (Saccharomyces cerevisiae) strain InVSc1 (Invitrogen, Carlsbad, CA). This system allowed Gal-induced expression of the recombinant fusion protein with a hexa-His tag at the N terminus. After Gal induction for 12 h and analysis of the protein extracts in immunoblots, a single protein band was stained with hexa-His-specific antibodies (Fig. 6, inset). The molecular mass of the immunostained protein band, 47.5 kD, was comparable with the expected mass of the recombinant protein, 45.3 kD. Protein extracts of yeast containing pYES-NMTase construct had detectable NMTase activities with β-Ala, N-methyl β-Ala, and N,N-dimethyl β-Ala as methyl acceptors (Fig. 6). Protein extracts of yeast containing a control pYES vector with β-galactosidase gene cloned under the Gal promoter (pYES-lacZ) had the expected immunoreactive β-galactosidase recombinant protein when probed with hexa-His-specific antibodies (data not shown) but had no detectable NMTase activities (Fig. 6).
Figure 6.
Expression of functional NMTase in yeast. NMTase activities measured in protein extracts of yeast expressing pYES-NMTase (black bar) and control yeast containing pYES-lacZ (white bar). For the control yeast, no NMTase activities were detected and the minimum detectable activity (×100) is shown. The values are means and se for three determinations. The methyl acceptors, at 10 mm concentrations, were β-Ala (BA), N-methyl β-Ala (MM), or N,N-dimethyl β-Ala (DM). The protein extracts were subjected to enterokinase, before assay. Inset, Immunoblot of protein extract of recombinant yeast containing pYES-NMTase (His6), 0, 4, 8, and 12 h after Gal induction, showing an immunopositive reaction corresponding to 47.5-kD band at 12 h.
β-Ala NMTase Has High Substrate Specificity
Protein extracts of yeast containing pYES-NMTase or pYES-lacZ were assayed for Ado-Met-specific methyltransferase activities with the following potential substrates as methyl acceptors: β-alanyl Gly, dl-β-aminoisobutryic acid, l-Ala, l-Pro, trans-4-hydroxy l-Pro, Gly, putrescine, γ-amino-n-butyric acid, N-methyl dl-Ala, and N,N-dimethyl Gly (Table III). The compounds tested, when used as methyl acceptors, could support only less than 2% of the activity found with β-Ala (Table III). This suggested that β-Ala NMTase has high substrate specificity. Little (<1.2%) methyltransferase activity was found in protein extracts of yeast containing pYES-lacZ (Table III), used as a negative control.
Table III.
Substrate specificity of β-Ala NMTase expressed in yeast (pYES-NMTase)
Relative activities are shown as percentage of that found with β-Ala. Specific activity with β-Ala (100%) was 105.2 nmol h–1 mg–1 protein. A protein extract from yeast (pYES-lacZ) was assayed as a negative control. ND, Not detectable. The minimum detectable activity was 10 pmol h–1 mg–1 protein. Values are means and se from triplicate assays.
| Methyl Acceptor Substrate (10 mM) | pYES-NMTase | pYES-lacZ |
|---|---|---|
| β-Ala | 100 | ND |
| N-Methyl β-Ala | 61.4 ± 0.34 | ND |
| N,N-Dimethyl β-Ala | 19.8 ± 0.22 | ND |
| β-Alanyl Gly | 0.39 ± 0.03 | 0.31 ± 0.01 |
| DL-β-Aminoisobutryic acid | 2.01 ± 0.13 | 0.31 ± 0.01 |
| L-Ala | 0.47 ± 0.12 | 0.21 ± 0.08 |
| L-Pro | ND | 0.17 ± 0.04 |
| trans-4-Hydroxy L-Pro | ND | ND |
| Gly | 1.77 ± 0.04 | 0.9 ± 0.1 |
| Putrescine | 1.36 ± 0.11 | 1.61 ± 0.12 |
| γ-Amino-n-butyric acid | ND | 1.15 ± 0.04 |
| N-Methyl DL-Ala | 0.65 ± 0.09 | 0.41 ± 0.02 |
| N,N-Dimethyl Gly | ND | ND |
β-Ala NMTase Is Related to O-Methyltransferases
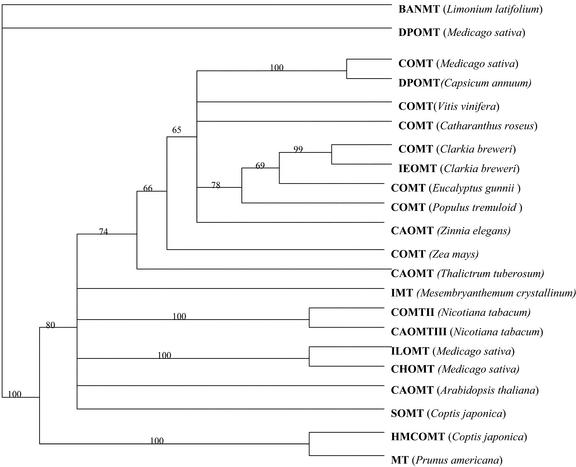
The deduced amino acid sequence of β-Ala NMTase showed sequence homology to caffeic acid O-methyltransferases and related O-methyltransferases (Fig. 7). The homology was highest at the carboxyl end of the protein including the conserved motifs described for Ado-Met-binding proteins. A phylogenetic analysis indicated that β-Ala NMTase was monophyletic, whereas nodes with caffeic acid O-methyltransferases and related enzymes were either bifurcating or polytomous in nature (Fig. 8).
Figure 7.
Alignment of β-Ala NMTase deduced amino acid sequence with sequences of other well-characterized O-methyltransferases from alfalfa (Medicago sativa). The sequences are identified by their GenBank identification numbers. COMT, Caffeic acid 3-O-methyltransferase; CHOMT, chalcone O-methyltransferase; IFOMT, isoflavone O-methyltransferase; and BAMT, β-Ala NMTase. Conserved residues are marked by an asterisk, and conserved substitutions are marked by one or two dots.
Figure 8.
A rooted phylogenetic tree showing the relationships between β-Ala NMTase and other methyltransferases. The enzymes are BANMT, β-Ala N-methyltransferase; DPOMT, O-diphenol-O-methyltransferase; COMT, caffeic acid O-methyltransferase; IEOMT, isoeugenol O-methyltransferase; CAOMT, catechol O-methyltransferase; IMT, myo-inositol O-methyltransferase; ILOMT, isoliquirifigenin O-methyltransferase; CHOMT, chalcone O-methyltransferase; SOMT, scoulerine-9-O-methyltransferase; MT; methyltransferase; and HMCOMT, hydroxy N-methyl S-coclaurine 4-O-methyltransferase. The GenBank identification numbers are given in Materials and Methods, and the species name is shown in brackets. The bootstrap values above 60 are shown.
DISCUSSION
We have cloned, for the first time to our knowledge, a full-length cDNA and several partial cDNA clones for L. latifolium β-Ala NMTase (Fig. 1) using peptide sequence data for the purified β-Ala NMTase. The deduced amino acid sequence (Fig. 2) resulted in a theoretical mass of 41,286 D and a pI of 5.84, closely resembling experimental determinations of 43,000 D and 5.15 for the NMTase (Rathinasabapathi et al., 2001). β-Ala NMTase had all three motifs conserved for Ado-Met binding (Joshi and Chiang, 1998).
Our results suggest that β-Ala NMTase is probably a cytoplasmic enzyme because no signal sequences can be identified in the deduced amino acid sequence (Fig. 2). It is interesting to note that pantothenate synthetase, involved in ligating d-pantoate and β-Ala to make pantothenate, is also a cytoplasmic enzyme (Genschel et al., 1999). There are perhaps regulatory features that allow the β-Ala pool to be shared by both pantothenate and β-Ala betaine synthetic pathways in the same compartment.
Our previous radiotracer and enzyme measurements showed that β-Ala betaine synthesis was specific to β-Ala betaine-accumulating members of the Plumbaginaceae (Hanson et al., 1991; Rathinasabapathi et al., 2000). Consistent with this, β-Ala NMTase mRNA expression was present in L. latifolium and absent in L. sinuatum (Fig. 3).
In L. latifolium, β-Ala NMTase mRNA was expressed at relatively high levels in young and mature leaves but at lower levels in floral stem, flowers, and old leaves (Fig. 4A). NMTase activity measurements in protein extracts from these tissues also matched these mRNA expression patterns, suggesting that β-Ala betaine synthesis in L. latifolium is developmentally regulated at the transcriptional level. Previous radiotracer studies showed that root tissue of L. latifolium had a capacity to synthesize β-Ala betaine (Hanson et al., 1991). In contrast, RNA blots indicated poor expression of β-Ala NMTase in roots (Fig. 4A), and no detectable NMTase activities could be measured in crude protein extracts of roots, using the assay routinely employed to investigate NMTase in leaves. This contradiction suggests the possibility of another root-specific NMTase isoform diverged both in its sequence and properties from the leaf isoform.
β-Ala NMTase expression was not significantly affected by salinity stress treatment imposed on excised leaves (Fig. 4B). This was consistent with our previous observations that β-Ala NMTase activity was not induced by salinity stress treatment (Rathinasabapathi et al., 2000) and that β-Ala betaine levels are somewhat constitutive in L. latifolium leaves (Hanson et al., 1991).
Light promoted the expression of β-Ala NMTase (Fig. 4B), similar to what was observed for phosphobase N-methyltransferase in the synthesis of choline (Weretilnyk et al., 1995). Exogenous auxin directly or indirectly down-regulated the NMTase (Fig. 4B). Genes that are up-regulated by light and down-regulated by auxin are known in other plants (Datta et al., 1993). Further studies on the physiological significance of this regulation are in progress.
When the full-length NMTase cDNA was expressed in yeast, a protein of expected molecular mass was made upon induction with Gal (Fig. 6, inset). The difference between the mass of the band labeled by the immunoblot and the expected mass is likely due to imprecision of the SDS-PAGE technique, but modifications of the protein in yeast cannot be ruled out. Protein extracts of Gal-induced yeast had NMTase activities with all three methyl acceptor substrates β-Ala, N-methyl β-Ala, and N,N dimethyl β-Alas (Fig. 6). Such activities were not observed in cultures of vector control yeast, which were grown and extracted similarly (Fig. 6). This indicated that the NMTase cDNA cloned was stably expressed in yeast and coded for the trifunctional NMTase. The specific activity toward N,N-dimethyl β-Ala was less than that found with β-Ala or N-methyl β-Ala as the methyl acceptors (Fig. 6), like what was found with the enzyme purified from L. latifolium leaves (Rathinasabapathi et al., 2001).
When a range of amino acids and methyl amines were tested as potential methyl acceptors, none of them acted as a substrate. This confirmed that the recombinant β-Ala NMTase expressed in yeast had high substrate specificity, as observed previously in partially purified L. latifolium leaf protein (Rathinasabapathi et al., 2000). This suggests that metabolic engineering of β-Ala methylation using this enzyme will result in specific and predictable synthesis of β-Ala betaine.
β-Ala NMTase protein sequence shared homology to O-methyltransferases (Fig. 7) but did not have significant homology to N-methyltransferases. Several O-methyltransferases involved in important secondary product synthesis pathways have recently been structurally characterized (Parvathi et al., 2001; Zubieta et al., 2001, 2002). Our analysis comparing β-Ala NMTase with those sequences (Fig. 7) revealed the structural elements common and unique among them. Like the O-methyltransferases, β-Ala NMTase had a large C-terminal catalytic domain responsible for Ado-Met binding. The N terminus of β-Ala NMTase had several unique regions, and future work will examine the biochemical significance of these regions in determining substrate specificity.
Our phylogenetic analyses (Fig. 8) are consistent with the hypothesis that β-Ala NMTase had evolved from an O-methyltransferase ancestral to caffeic acid O-methyltransferases and related enzymes. This is similar to what was known for putrescine N-methyltransferase in tobacco (Nicotiana tabacum), where some homology to O-methyltransferases was observed (Hibi et al., 1994). We suggest that diverse O-methyltransferases can be progenitors to N-methyltransferases and some sequences annotated as O-methyltransferases in the genome databases based on sequence homology may actually be N-methyltransferases.
Although a variety of QAC osmoprotectants are known in plants (Rhodes and Hanson, 1993), only the biosynthesis of the most common QAC, Gly betaine, has thoroughly been investigated. Engineering of Gly betaine synthesis requires installing two genes for choline oxidation and additional gene(s) to increase the availability of choline (McNeil et al., 2001; Rontein et al., 2002).
It may be possible to avoid the problem of poor availability of choline by engineering β-Ala betaine synthesis instead of Gly betaine synthesis in transgenic crops. Recent research in our laboratory indicates that d-pantoate is what limits pantothenate synthesis in plants and not β-Ala (B. Rathinasabapathi, C. Sigua, and S.B. Raman, unpublished data). This suggests that it should be possible to engineer transgenic plants synthesizing β-Ala betaine without adversely reducing pantothenate levels. Additionally, because β-Ala betaine synthesis will not require oxygen for its activity, it would be suitable under hypoxic conditions.
Current research in our laboratory therefore focuses on the expression of β-Ala NMTase in model plants and testing whether such manipulation would result in the synthesis and accumulation of β-Ala betaine. Cloning, characterization, and functional validation of the novel β-Ala NMTase described here has opened up this important opportunity.
MATERIALS AND METHODS
Chemicals
If not otherwise indicated, chemicals used were from Sigma-Aldrich (St. Louis) and were of the highest purity available. Magna Lift nylon (0.45 μm, 137 mm) circles were from Osmonics Inc. (Minnetonka, MN). dGTP (α-32P, 800 Ci mmol–1) was purchased from Amersham Biosciences (Piscataway, NJ). Plasmid purification and gel extraction kits were from Qiagen (Valencia, CA). RACE protocol kit was from BD Biosciences Clontech (Palo Alto, CA). Mr markers, Taq polymerase, dNTPs, and restriction enzymes were from Promega (Madison, WI). Oligonucleotide primers were synthesized from the custom primer synthesis unit of Invitrogen.
Plant Material
Limonium latifolium and Limonium sinuatum plants were grown under controlled conditions in a greenhouse as described previously (Rathinasabapathi et al., 2001). For salinity treatment, plants were grown in vermiculite and irrigated daily with one-half-strength Hoagland medium (Hoagland and Arnon, 1950). Sodium chloride was added to the nutrient solution at 50 mm per every 3 d until reaching 200 mm. The plants were kept in this salinity for another week. Fully expanded leaves were harvested for RNA extraction to prepare a cDNA library.
RNA Extraction
Young, mature (i.e. fully expanded), and old (i.e. senescing) leaves, roots, floral stems, and flowers were harvested from 2-year-old plants grown in a greenhouse. Total RNA was extracted using a modified hot borate method (Wan and Wilkins, 1994). In brief, tissue was ground in a mortar in liquid nitrogen and dithiothreitol powder (2 mm final concentration) and was further extracted in a boiling medium containing 0.2 m sodium borate, 30 mm EDTA, and 1% (w/v) SDS, pH 9. Proteins were digested using proteinase K (5 mg g–1 fresh weight tissue) at 42°C for 2 h. SDS was removed by precipitation by the addition of potassium chloride solution to a final concentration of 145 mm. The extract was then filtered through four layers of cheesecloth and centrifuged 20,000 g. Barium chloride at 75 mm was added to the supernatant to remove carbohydrates. Following this step, lithium chloride at 2 m was used to precipitate the total RNA. Lithium chloride step was repeated two more times to achieve high-purity total RNA. The RNA was concentrated by precipitation in ethanol, redissolved in RNase-free water, quantified using a UV-visible spectrophotometer, and analyzed by gel electrophoresis.
Poly(A+) RNA was isolated from leaf total RNA using Oligo(dT)-cellulose (GenElute-mRNA miniprep kit, Sigma-Aldrich). First-strand cDNA was synthesized using oligo(dT)20 primers and reverse transcriptase (RT; Thermoscript RT-PCR System, Invitrogen).
Degenerate Primers
Active NMTase protein was purified from leaves of Limonium latifolium using the procedure described previously (Rathinasabapathi et al., 2001). The purified protein (20 μg) was separated on an SDS-PAGE gel and stained with Coomassie Blue. The 43-kD band was eluted from the gel and was digested with lysyl endopeptidase C (LysC). Peptide sequencing was done by Edman degradation (Tempst et al., 1990). On the basis of the peptide sequences, degenerate primers were designed for RT-PCR. Each primer included one or two inosines.
RT-PCR
RT-PCR was performed using a RT-PCR kit (Thermoscript RT-PCR system, Invitrogen) according to manufacturer's instructions. RT-PCR reactions were in a volume of 50 μL in thin-walled amplification tubes. The reactions contained 10 μL of first-strand reaction, 200 μm of each of the four dNTPs, 2 mm magnesium chloride, 4 μm each of the sense and antisense primers, and 5 units of Taq DNA polymerase in 10 mm Tris-HCl, pH 9, 50 mm KCl, and 0.1% (v/v) Triton X-100. Forty cycles each with 93°C for 30 s for denaturation, 60°C for 30 s for annealing, and 72°C for 1.5 min for extension were performed in a thermal cycler (MiniCycler, MJ Research, Watertown, MA). The products were analyzed in an agarose (1%, w/v) gel and stained with ethidium bromide.
cDNA Library Construction
Poly(A+) RNA was isolated from L. latifolium leaves from plants salinized with 200 mm NaCl. First- and second-strand cDNAs were made using Moloney murine leukemia virus RT (Sambrook et al., 1989). cDNAs size-selected for 1,000 bp were cloned into the EcoRI site of λ-vector gt10 (BD Biosciences Clontech). The primary library had a titer of 1.5 × 106 plaque forming units mL–1.
Library Screening
Clone 23 was labeled with [32P]dGTP (800 Ci mmol–1, Amersham BioSciences, Piscataway, NJ) using a random primer labeling kit (Invitrogen) according to manufacturer's instructions. Plaque lifts of the library on nylon membranes were screened using the radiolabeled probe by following the formamide procedure (Sambrook et al., 1989). Positive clones were identified after autoradiography and were purified using standard protocols (Sambrook et al., 1989). λ-DNA was extracted using Wizard Lambda prep system (Promega) according to manufacturer's protocol.
DNA Sequencing and Analysis
DNA sequencing was in both strands using the fluorescent chain-terminating dideoxynucleotides method. DNA sequences were analyzed by several software packages including BLAST (Patnaik and Blumenfeld, 2001).
RNA Blots
Total RNA from L. latifolium and L. sinuatum was loaded onto a formaldehyde agarose (1.2%, w/v) gel, 10 μg lane–1, and blotted onto nylon membranes (Sambrook et al., 1989). Equal loading of RNA in the gels was verified by ethidium bromide staining of the gel. Completion of RNA transfer was verified by methylene blue staining of the blots and ethidium bromide staining of the gels. The blots were probed using clone 23 cDNA probe labeled with [32P]dGTP. Band intensities in autoradiographs were analyzed using densitometry. The RNA blots were also probed with a 395-bp L. latifolium actin cDNA (S.B. Raman and B. Rathinasabapathi, unpublished data).
Treatment of Excised Leaves
For the experiments on β-Ala NMTase mRNA expression, mature leaves were excised and incubated in 50 mL of Hoagland nutrient medium (Hoagland and Arnon, 1950) for 24 h under light (photosynthetic photon flux density, 46 μmol m–2 s–1, cool-white fluorescent lamps) or complete darkness. Some of the leaves under light were treated with kinetin (1 mg L–1), indole-3-butyric acid (1 mg L–1), GA3 (1 mg L–1), or 200 mm NaCl in Hoagland nutrient medium.
Heterologous Expression
Coding sequences of NMTase cDNA were amplified with primers 5′ to 3′-GCGGATCCAATGGCGAACCACTCCTCAGCTG and 5′ to 3′-CTCGAGTCACTTCTGGAACTCTACCACGG with BamHI and XhoI restriction enzyme recognition sites at the primer ends, respectively. The amplified product was digested with the appropriate restriction enzymes and subcloned into BamHI and XhoI restriction sites of yeast expression vector pYES-NTB. The resulting plasmid was verified by sequencing and was introduced into INVSc1 strain using the lithium chloride method (Ausubel et al., 1995). Recombinants were selected and maintained on minimal medium lacking uracil. Preliminary experiments were carried out to determine the optimum duration of growth in Glc-free Gal-containing medium for optimal induction of recombinant protein (data not shown). Total protein was extracted from cells following breakage of the cells using glass beads (Ausubel et al., 1995) in breaking buffer containing 50 mm sodium phosphate buffer (pH 7.4) with 1 mm EDTA, 5% (v/v) glycerol, and 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride. Protein extracts were digested with enterokinase for 1 h at 30°C according to manufacturer's protocol (Invitrogen) and were assayed for NMTase activity using the radiometric method described previously (Rathinasabapathi et al., 2001). Heat-killed controls and cultures without Gal induction had no detectable NMTase activities. Substrate specificity assays were done with 10 mm potential methyl acceptors, following the radiometric method described previously (Rathinasabapathi et al., 2000). Total protein was estimated by the method of Peterson (1977), and bovine serum albumin was the standard.
Immunoblots
SDS-PAGE was performed according to the method of Laemmli (1970) in 12% (w/v) separation gel and 5% (w/v) stacking gel. Proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA) using an electroblotting device. The protein blots were probed according to manufacturer's protocol, with monoclonal antibodies against (His)6Gly conjugated to alkaline phosphatase (Invitrogen).
DNA Extraction and Southern-Blot Hybridization
Genomic DNA was isolated from leaves of L. latifolium using a cetyltrimethyl-ammonium bromide method (Rogers and Bendich, 1994). DNA was digested with restriction enzymes and separated on an agarose gel. DNA was blotted onto a nylon membrane following the procedure described by Sambrook et al. (1989). Southern-blot hybridization was performed following the formamide procedure as described (Sambrook et al., 1989).
Multiple Sequence Comparisons and Phylogeny
BLAST analysis indicated sequences highly homologous to β-Ala NMTase. Multiple sequence comparisons were done using ClustalX (Thompson et al., 1997). A phylogenetic tree was drawn using the neighbor-joining method in PAUP (Phylogenetic Analysis Using Parsimony, v4.0b10, Sinaur Associates, Sunderland, MA). Bootstrap values were obtained by running the PAUP program, and the tree was rooted using midpoint rooting. GenBank identification numbers of sequences used for these analyses are as follows, listed top-down in the phylogenetic tree: GI:6688808, GI: 3421382, GI:7488967, GI:7271883, GI:18025321, GI:3913289, GI:2832224, GI: 1169009, GI:231757, GI:642952, GI:729135, GI:4808524, GI:1170555, GI:19550749, GI:542050, GI:7447884, GI:13399464, GI:15223364, GI:758580, GI:17366954, and GI:2282586.
Acknowledgments
We thank Celia Sigua for excellent technical assistance, Dr. Mohammed Aly for useful discussions, Dr. Andrew Hanson for supplying a Limonium sp. cDNA library, Dr. Douglas Soltis for advice on phylogenetic analyses, and Dr. Harry Klee for useful comments on the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.020453.
This work was supported by the United States Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 2001–35318–10947 to B.R.) and by the Florida Agricultural Experiment Station. This is journal series no. R–09042.
References
- Abe S, Kaneda T (1973) Studies on the effect of marine products on cholesterol metabolism in rats: VIII. The isolation of hypocholesterolemic substance from green laver. Bull Jpn Soc Sci Fish 39: 383–389 [Google Scholar]
- Anthoni U, Christophersen C, Hougaard L, Nielsen PH (1991) Quaternary ammonium compounds in the biosphere: an example of a versatile adaptive strategy. Comp Biochem Biophys 99B: 1–18 [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1995) Short Protocols in Molecular Biology, Ed 3. John Wiley & Sons, New York
- Bohnert HJ, Sheveleva E (1998) Plant stress adaptations: making metabolism move. Curr Opin Plant Biol 1: 267–274 [DOI] [PubMed] [Google Scholar]
- Boyer JS (1982) Plant productivity and environment. Science 218: 443–448 [DOI] [PubMed] [Google Scholar]
- Cavener DR, Ray SC (1991) Eukaryotic start and stop translation sites. Nucleic Acids Res 19: 3185–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, LaFayette PR, Kroner PA, Nagao RT, Key JL (1993) Isolation and characterization of three families of auxin down-regulated cDNA clones. Plant Mol Biol 21: 859–869 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Genschel U, Powell CA, Abell C, Smith AG (1999) The final step of pantothenate biosynthesis in higher plants: cloning and characterization of pantothenate synthetase from Lotus japonicus and Oryza sativa (rice). Biochem J 341: 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rathinasabapathi B, Chamberlin B, Gage DA (1991) Comparative physiological evidence that β-alanine betaine and choline-o-sulfate act as a compatible osmolytes in halophytic Limonium species. Plant Physiol 97: 1199–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Rathinasabapathi B, Rivoal J, Burnet M, Dillon MO, Gage DA (1994) Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc Natl Acad Sci USA 91: 306–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G, Messing J (1986) Structural analysis of plant genes. Annu Rev Plant Physiol Plant Mol Biol 37: 439–466 [Google Scholar]
- Hibi N, Higashiguchi S, Hashimoto T, Yamada Y (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6: 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Waditee R, Araki E, Ishikawa H, Aoki K, Tanaka Y, Takabe T (2002) Functional characterization of choline monooxygenase, an enzyme for betaine synthesis in plants. J Biol Chem 277: 41352–41360 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Bull 347: 1–32 [Google Scholar]
- Joshi CP (1987) An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res 15: 6643–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-l-methionine-dependent methyltransferases. Plant Mol Biol 37: 663–674 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- McCue KF, Hanson AD (1990) Drought and salt tolerance: towards understanding and application. Trends Biotechnol 8: 358–362 [Google Scholar]
- McNeil SD, Nuccio ML, Ziemak MJ, Hanson AD (2000) Metabolic modeling identifies key constraints on an engineered glycine betaine synthesis pathway in tobacco. Plant Physiol 124: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil SD, Nuccio ML, Ziemak MJ, Hanson AD (2001) Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc Natl Acad Sci USA 98: 10001–10005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount JW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Patnaik SK, Blumenfeld OO (2001) Use of on-line tools and databases for routine sequence analyses. Anal Biochem 289: 1–9 [DOI] [PubMed] [Google Scholar]
- Peterson GL (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem 83: 346–356 [DOI] [PubMed] [Google Scholar]
- Rathinasabapathi B (2000) Metabolic engineering for stress tolerance: installing osmoprotectant synthesis pathways. Ann Bot 86: 709–716 [Google Scholar]
- Rathinasabapathi B, Fouad WM, Sigua CA (2001) β-Alanine betaine synthesis in the Plumbaginaceae: purification and characterization of a trifunctional, S-adenosyl-l-methionine-dependent N-methyltransferase from Limonium latifolium leaves. Plant Physiol 126: 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinasabapathi B, Sigua C, Ho J, Gage DA (2000) Osmoprotectant β-alanine betaine synthesis in the Plumbaginaceae: S-adenosyl-l-methionine dependent N-methylation of β-alanine to its betaine is via N-methyl and N,N-dimethyl β-alanines. Physiol Plant 109: 225–231 [Google Scholar]
- Rhodes D, Hanson AD (1993) Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu Rev Plant Physiol Plant Mol Biol 44: 357–384 [Google Scholar]
- Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. Plant Mol Biol Manual D1: 1–8 [Google Scholar]
- Rontein D, Basset G, Hanson AD (2002) Metabolic engineering of osmoprotectant accumulation in plants. Metab Eng 4: 49–56 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2000) Genetic engineering of glycinebetaine synthesis in plants: current status and implications for enhancement of stress tolerance. J Exp Bot 51: 81–88 [PubMed] [Google Scholar]
- Sakamoto A, Murata N (2001) The use of bacterial choline oxidase, a glycinebetaine-synthesizing enzyme, to create stress-resistant transgenic plants. Plant Physiol 125: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Tempst P, Link AJ, Riviere LR, Fleming M, Elicone C (1990) Internal sequence analysis of proteins separated on polyacrylamide gels at the picomole level: improved methods, applications and gene cloning strategies. Electrophoresis 11: 537–553 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223: 7–12 [DOI] [PubMed] [Google Scholar]
- Weretilnyk EA, Smith DD, Wilch GA, Summers PS (1995) Enzymes of choline synthesis in spinach: response of phosphobase N-methyltransferase activities to light and salinity. Plant Physiol 109: 1085–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN (1982) Living with water stress: evolution of osmolyte systems. Science 217: 1214–1222 [DOI] [PubMed] [Google Scholar]
- Zubieta C, He X, Dixon RA, Noel JP (2001) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol 8: 271–279 [DOI] [PubMed] [Google Scholar]
- Zubieta C, Kota P, Ferrer J, Dixon RA, Noel JP (2002) Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]