Abstract
Gentian (Gentiana triflora) blue petals predominantly contain an unusually blue and stable anthocyanin, delphinidin 3-O-glucosyl-5-O-(6-O-caffeoyl-glucosyl)-3′-O-(6-O-caffeoyl-glucoside) (gentiodelphin). Glucosylation and the subsequent acylation of the 3′-hydroxy group of the B-ring of anthocyanins are important to the stabilization of and the imparting of bluer color to these anthocyanins. The enzymes and their genes involved in these modifications of the B-ring, however, have not been characterized, purified, or isolated to date. In this study, we purified a UDP-glucose (Glc):anthocyanin 3′-O-glucosyltransferase (3′GT) enzyme to homogeneity from gentian blue petals and isolated a cDNA encoding a 3′GT based on the internal amino acid sequences of the purified 3′GT. The deduced amino acid sequence indicates that 3′GT belongs to the same subfamily as a flavonoid 7-O-glucosyltransferase from Schutellaria baicalensis in the plant glucosyltransferase superfamily. Characterization of the enzymatic properties using the recombinant 3′GT protein revealed that, in contrast to most of flavonoid glucosyltransferases, it has strict substrate specificity: 3′GT specifically glucosylates the 3′-hydroxy group of delphinidin-type anthocyanins containing Glc groups at 3 and 5 positions. The enzyme specifically uses UDP-Glc as the sugar donor. The specificity was confirmed by expression of the 3′GT cDNA in transgenic petunia (Petunia hybrida). This is the first report of the gene isolation of a B-ring-specific glucosyltransferase of anthocyanins, which paves the way to modification of flower color by production of blue anthocyanins.
Anthocyanins, a colored class of flavonoids, are major constituents of flower color. Hundreds of different forms of anthocyanins have been isolated from plants, and their structures have been determined (Harborne and Williams, 2000). Such variation partly explains the extensive variety of observed flower colors. There are only six chromophore forms (aglycones) in anthocyanins despite the structural varieties. The biosynthetic pathway leading to anthocyanidin 3-glycosides (usually glucosides) is well conserved in plants and is well characterized. All structural genes that encode enzymes involved in the pathway and many regulatory genes for transcriptional regulation of the structural genes have been cloned from a wide variety of plants, and this field has been reviewed (Forkmann and Heller, 1999; Mol et al., 1999; Winkel-Shirley, 2001). Anthocyanidin 3-glycosides are further modified with glycosyl and acyl groups in species-specific manners. These modifications are less understood, although the enzymes involved in the modification of anthocyanins are commercially important for metabolic engineering of the flavonoid biosynthetic pathway (Tanaka et al., 1998; Forkmann and Martens, 2001).
Glycosylation and subsequent acylation play important roles not only in modifying flower color but also in increasing solubility and stability of hydrophobic flavonoids by means of external hydrogen bonding of sugar residues with surrounding water molecules (Goto et al., 1982; Yoshida et al., 2000). Glycosylation of flavonoids is also regarded as an indispensable signal for vacuolar transport (Marrs et al., 1995). Thus most of the naturally occurring flavonoids are glycosylated with one or more sugar groups. The best-studied flavonoid glucosyltransferase is UDP-Glc:flavonoid 3-O-glucosyltransferases (3GT). The 3GT enzymes have been characterized biochemically and molecular biologically in maize (Zea mays; Futtek et al., 1988), snapdragon (Antirrhinum majos; Martin et al., 1991), barley (Hordeum vulgare; Wise et al., 1990), grape (Vitis vinifera; Ford et al., 1998) and gentian (Gentiana triflora; Tanaka et al., 1996) to name a few. Interestingly, UDP-Gal:anthocyanidin 3-O-galactosyltransferases from red bean (Mato et al., 1998) and petunia (Petunia hybrida; Miller et al., 1999) are structurally closely related to 3GT. The cDNA encoding UDP-rhamnose:anthocyanidin 3-O-glucoide rhamnosyltransferase has been isolated from petunia (Brugliera et al., 1994; Kroon et al., 1994). We have isolated UDP-Glc:anthocyanin 5-O-glucosyltransferase (5GT) from perilla (Yamazaki et al., 1999), verbena (Yamazaki et al., 1999), petunia (Yamazaki et al., 2002), and torenia (DNA data bank of Japan [DDBJ] accession no. AB076698). The cDNA encoding UDP-Glc: flavonoid 7-O-glucosyltransferase (7GT), with broad substrate specificity, was cloned from hairy root culture of Scuttellaria baicalensis (Hirotani et al., 2000). It has been proposed that 3GTs, 5GTs, and 7GTs involved in the flavonoid biosynthesis pathway form distinct groups within the glycosyltransferase superfamily (Lie et al., 2001). The group of 7GT also contains betanidin 5-O-glucosyltransferase (betanidin 5GT) from Dorotheanthus bellidiformis (Vogt et al., 1997, 1999) and a few jasmonic acid-inducible (EMBL accession no. T07404) or salicylic acid-inducible glucosyltransferases (Horvath and Chua, 1996). The betanidin 5GT was shown to glucosylate the 4′-hydroxy group of flavonols and flavones as well as its natural substrate betanidin when the reaction was carried out in vitro (Vogt et al., 1997). Recently, it was revealed that Arabidopsis contains 99 glycosyltransferase homologs, and their molecular phylogenetic relationship has been analyzed (Lie et al., 2001). However, there has been no report on the isolation of a glycosyltransferase that transfers a glycosyl group to the B-ring of flavonoids both in vitro and in vivo, although such glycosyl transfer is important in the natural biosynthetic pathway.
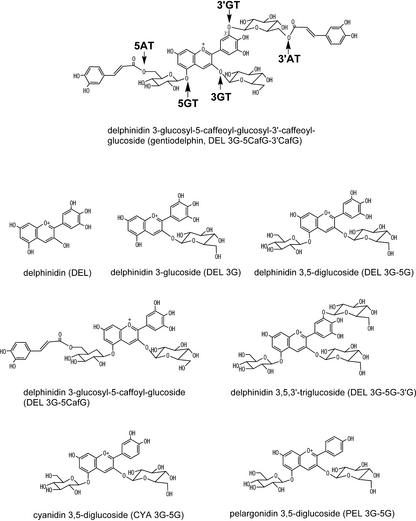
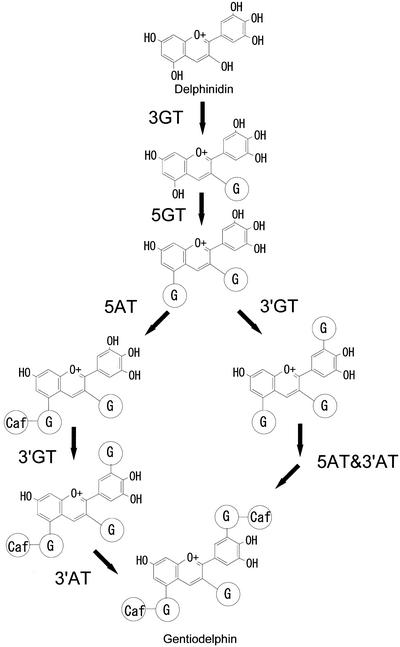
Gentian produces blue flowers predominantly containing an anthocyanin modified with two aromatic acyl groups, delphinidin 3-O-glucosyl-5-O-(6-O-caffeoyl-glucosyl)-3′-O-(6-O-caffeoyl-glucoside) (gentiodelphin), which is blue at a wide range of pH values and unusually stable not only in acidic conditions but also at neutral pH (Goto et al., 1982). Yoshida et al. (2000) suggested that the 3′-O-caffeoyl-glucosyl moiety, not the 5-O-caffeoyl-glucosyl moiety, plays a critical role in both blue color development and stabilization of gentiodelphin by intramolecular stacking of the caffeoyl moiety with the B-ring (Yoshida et al., 2000). Thus, the glucosylation at the 3′-hydroxy group of delphinidin (DEL), as a prerequisite for acylation, is essential to the stabilization and the imparting of a bluer color to the anthocyanin. The gene encoding 3′GT is therefore a useful molecular tool for modification of flower color using genetic engineering, and the cloning of a 3′GT gene will contribute to further understanding of structurefunction relationships of glucosyltransferases, especially glucosyltransferases involved in flavonoid biosynthesis.
In the present study, we have purified to homogeneity 3′GT protein from gentian petals. Partial sequence of the purified 3′GT protein allowed us to isolate a cDNA encoding the gentian 3′GT. The phylogenic analysis of the deduced amino acid sequence indicates that it belongs to the same glucosyltransferase subfamily as 7GT. The 3′GT was biochemically characterized using Escherichia coli and petunia as hosts. The biosynthetic pathway of gentiodelphin is also discussed on the basis of the enzymatic properties of the recombinant 3′GT.
RESULTS
Purification of 3′GT from Gentian Petals
Incubation of the crude protein extract from gentian petals with DEL 3,5-diglucoside (DEL 3G-5G) and UDP-Glc yielded a new anthocyanin product, which eluted earlier than DEL 3G-5G on HPLC and whose λmax of 515 nm, showed hypsochromic shift compared with that for DEL 3G-5G, at 524 nm. Considering that the major anthocyanin in gentian petals is gentiodelphin, which is triglucosylated at the 3, 5, and 3′ positions (Fig. 1), the substrate DEL 3G-5G was most likely to be glucosylated at 3′-hydroxy group by 3′GT activity present in gentian petals.
Figure 1.
Structures of gentiodelphin and anthocyanins used in this study. The abbreviation for each compound is in parentheses. The enzymes involved in biosynthesis of gentiodelphin from DEL were indicated on the structure of gentiodelphin (top of figure). 3′AT, Hydroxycinnamoyl-CoA:anthocyanin 3′-O-glucoside-6′′′-O-hydroxycinnamoyltransferase.
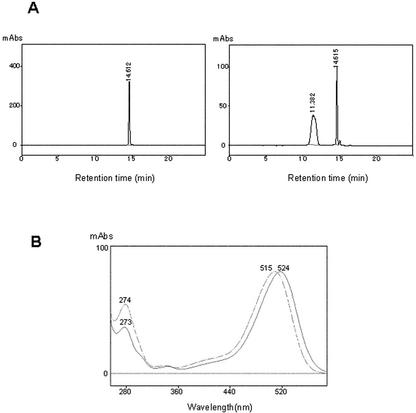
The putative 3′GT protein was purified from gentian petals following glucosylating activity of DEL 3G-5G. After five steps of column chromatography (Sephadex G-25, Q-Sepharose, Blue A, DEAE-Sepharose, and Superose 12), the protein was purified to near homogeneity on SDS-PAGE (Fig. 2). The molecular mass of gentian 3′GT was estimated to be 55 kD by reference to molecular markers in SDS-PAGE. The enzyme assay using purified enzyme with DEL 3G-5G and UDP-Glc as substrates yielded a new peak (compared with controls) on HPLC analysis whose retention time was approximately 11.4 min; 3.3 min earlier than that for DEL 3G-5G (Fig. 3A). The λmax of absorption spectra of this product was 515 nm, which showed hypsochromic shift by 9 nm from the λmax for DEL 3G-5G, at 524 nm (Fig. 3B). In addition, liquid chromatography-mass spectrometry (MS) analysis of this product detected a molecular ion peak m/z 789 [M]+, which corresponds to the Mr of DEL triglucoside. These results and the structure of gentiodelphin strongly suggested that the substrate DEL 3G-5G was glucosylated at the 3′-hydroxy residues in this reaction and that the purified protein from gentian petals was most likely 3′GT. This was subsequently verified by NMR analysis of a large amount of the product produced by the recombinant protein (see below).
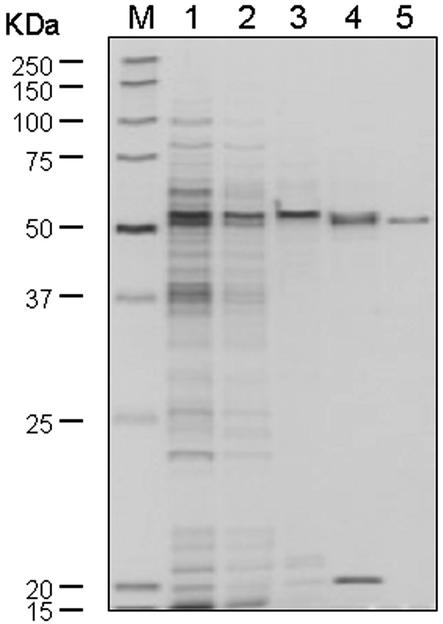
Figure 2.
SDS-PAGE analysis of the 3′GT protein purified from gentian petals. The active fractions after each purification step were analyzed through a 10% to 20% (m/v) polyacrylamide gel and detected by silver-staining. Lane M, Mr marker; lane 1, fraction of 40% to 70% saturation with ammonium sulfate; lane 2, Sephadex G-25 elute; lane 3, Q-Sepharose elute; lane 4, Blue A elute; lane 5, Superose 12 elute. Mr for each band of the marker is indicated to the left of the panel.
Figure 3.
HPLC analysis of the reaction products of the purified gentian 3′GT. A, HPLC profiles of the substrate DEL 3G-5G (left) and the reaction product by the purified gentian 3′GT using DEL 3G-5G and UDP-Glc as substrates (right). A520 was monitored. The retention times corresponding to DEL 3G-5G and the major product were approximately 14.6 and 11.4 min, respectively. B, UV-visible spectra of the DEL 3G-5G (solid line) and the major product detected at 11.4 min in HPLC analysis (dashed line). The absorbance maximum for DEL 3G-5G and the major product were 524 and 515 nm, respectively (as indicated).
The purified protein did not use UDP-Gal, UDP-GlcUA, or UDP-N-acetylgalactosamine as sugar donors and showed significantly reduced Glc transfer when DEL or DEL 3-glucoside (DEL 3G) was used as a substrate (data not shown).
Cloning of a Gentian 3′GT cDNA
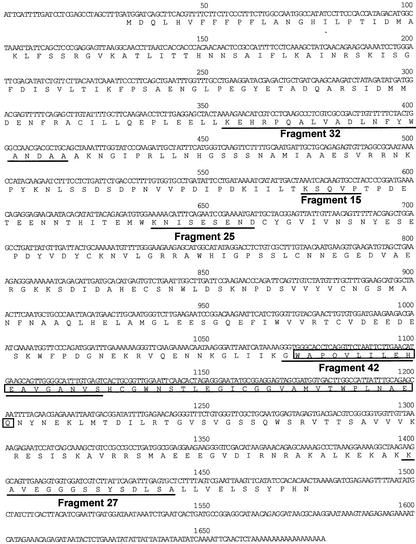
Approximately 15 μg of purified enzyme was obtained and was subject to peptide sequencing. As the N terminus of the protein was blocked, the purified protein was digested with a lysylendopeptidase to generate peptides that were then purified by HPLC before determination of their amino acid sequences. The amino acid sequence of five peptide fragments were obtained (Fig. 4). The sequence of fragment 32 was used to design a degenerate oligonucleotide primer, GT-2. PCR using the synthetic GT-2 and XhoI-T primers with the gentian petal cDNA library as a template successfully amplified a DNA fragment, which was then labeled with digoxigenin to screen the gentian petal cDNA library. Approximately 100 positives were detected in the 180,000 phages of the library. Of these, 20 were randomly selected and sequenced. The sequences from both 5′ and 3′ ends of the cDNAs revealed that they appeared to originate from the same genetic allele. The longest clone (pSPB479) containing a putative initiation Met codon was subjected to further analysis.
Figure 4.
The nucleotide and deduced amino acid sequences of a gentian 3′GT cDNA. The sequences of five peptide fragments generated by lysylendopeptidase digestion of the purified gentian 3′Gt are underlined.
The entire sequence of the pSPB479 cDNA was determined and has been submitted to the GenBank/EMBL/DDBJ nucleotide databases with accession number AB076697. The pSPB479 cDNA appeared to encode a protein consisting of 482 amino acids (Fig. 4). The molecular mass of the protein was calculated to be 54,037 D, which is consistent with the estimate for the purified protein from SDS-PAGE (Fig. 2). The deduced amino acid sequence contained all of the peptide sequences determined for the digestion products of purified protein (Fig. 4, underlined).
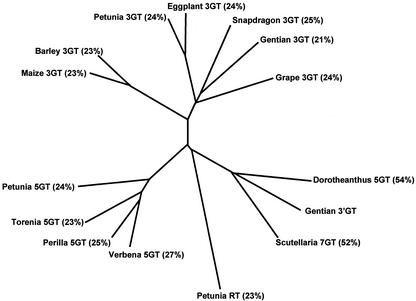
The amino acid sequence encoded by the pSPB479 cDNA showed significant homologies with glucosyltransferases involved in plant secondary metabolism. Among the enzymatically characterized glucosyltransferases, the deduced amino acid sequence encoded in the pSPB479 cDNA was closest to a betanidin 5GT from D. bellidiformis (55% identity), followed by a 7GT from S. baicalensis (52%), whereas the amino acid identity with flavonoid 3GTs and 5GTs was 21% to 25% and 23% to 27%, respectively. The phylogenetic tree based on ClustalW analysis also shows that the gentian 3′GT belongs to the same subfamily as the 7GT and betanidin 5GT (Fig. 5) and that the gentian 3′GT is distant from the 3GT and 5GT subfamilies.
Figure 5.
Molecular phylogenetic tree of the amino acid sequences of the plant glycosyltransferase superfamily. The tree was obtained by ClustalW analysis program (Thompson et al., 1994). GenBank accession numbers of the GTs: Gentian 3′GT (AB076697); D. bellidiformis 5GT (Y18871); scuttellaria 7GT (BAA83484); verbena 5GT (BAA36423); perilla 5GT (AB013596); torenia 5GT (AB076698); petunia 5GT (AB027455); petunia RT (Z25802); maize 3GT (X13501); barley GT (X15694); petunia 3GT (AB027454); egg-plant 3GT (X77369); gentian 3GT (D85186); and grape 3GT (AF000371). The sequence of snapdragon 3GT appeared in Martin et al. (1991). The percent identity of amino acid residues between each glucosyltransferase and the gentian 3′GT is indicated in parentheses.
Enzymatic Characterization of the Recombinant Gentian 3′GT
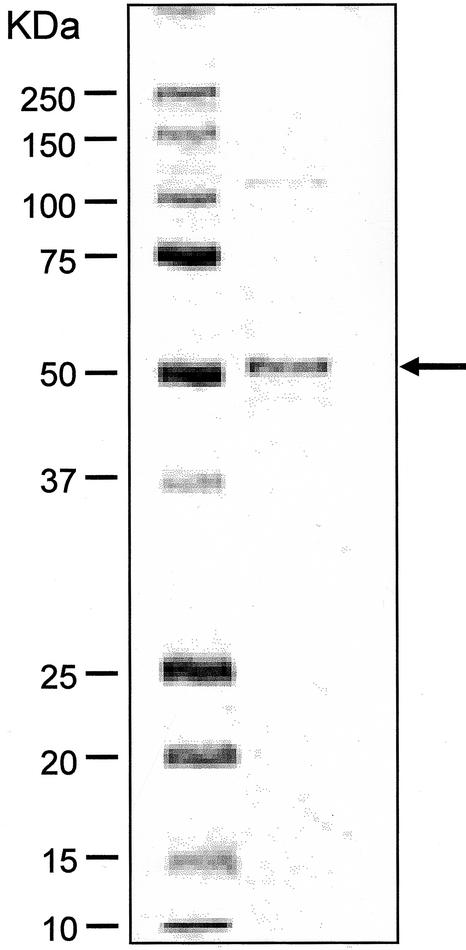
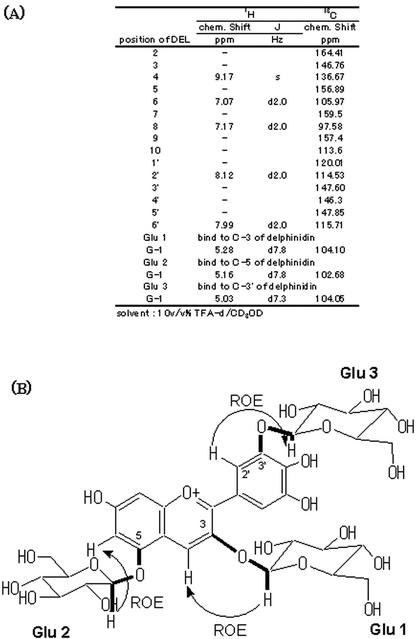
The recombinant 3′GT cDNA was expressed in E. coli. The expressed protein accumulated in the soluble fraction at high levels as observed on an SDS-PAGE gel. The recombinant 3′GT was purified to near homogeneity on SDS-PAGE using a Blue A column and followed by a Phenyl Sepharose column (Fig. 6) and was subjected to enzymatic characterization. Incubation of the recombinant protein with DEL 3G-5G and UDP-Glc generated a product whose retention times on HPLC and absorption spectra were identical with those of the product generated by the purified native 3′GT (Fig. 3). A large-scale reaction with the purified recombinant protein and DEL 3G-5G and UDP-Glc as substrates yielded approximately 2.5 mg of the product. This product was subjected to NMR analysis to determine its structure. Rotating frame overhauser enhancement spectroscopy (ROESY) detects a cross peak if two protons are located spatially in close to each other. A cross peak of ROESY was observed between the aromeric proton of a Glc (H-1 in Fig. 7; δ 5.03) and the proton at 2′ position of B-ring (H-2′ in Fig. 7; δ 8.12), in addition to the ROESY observed for the aromeric protons of 3- and 5-Glc (Glu 1 and Glu 2 in Fig. 7, respectively). In addition, on 1H {13C}-HMBC analysis, which detects the coupling of a proton to a 13C located far from the proton by two or three bonds, a cross peak was observed between the aromeric proton of a Glc (H-1 in Fig. 7) and carbon at 3′ position (C-3′ in Fig. 7; δ 147.60). Thus, these results revealed that the product is DEL3G-5G-3′G (Fig. 7). Therefore, the protein encoded in pSPB479 cDNA proved to encode gentian 3′GT, which catalyzes the transfer of a glucosyl moiety from UDP-Glc to the 3′-hydroxy group of DEL 3G-5G to produce DEL 3G-5G-3′G. The Km value for DEL 3G-5G was determined to be 120 μm, and Vmax was 1,370 nmol min–1 mg–1 protein when UDP-Glc (1 mm) was used as a Glc donor.
Figure 6.
SDS-PAGE analysis of the 3′GT protein expressed in E. coli. The active fraction after a Phenyl Sepharose column was analyzed through a 10% to 20% (m/v) polyacrylamide gel and stained by Coomassie Blue. Left lane, Mr marker; right lane, the active fraction after a Phenyl Sepharose. Mr for each band of the marker is indicated to the left of the panel. The arrow indicates the size for 3′GT.
Figure 7.
1H- and 13C-NMR assignment and structure of the reaction produced by 3′GT. A, Assignment of the NMR spectra of the reaction product by the recombinant 3′GT protein using DEL 3G-5G and UDP-Glc as substrates. B, Structure of the reaction product confirmed by ROESY and heteronuclear multiple bond correlation (HMBC) analyses. Rotating frame overhauser enhancement (ROE) detected on ROESY are indicated by arrow curves and HMBC cross peaks are indicated by thick lines.
The effect of several different metal cations and chealating reagents on 3′GT activity was analyzed. 3′GT activity was significantly inhibited by Al3+ (20% compared with the reaction without inhibitors) and Co2+ (31%) as reported for perilla 5GT. Zn2+ moderately reduced gentian 3′GT activity (74%). The metal cations Mg2+, Ca2+, and Mn2+ and chealating reagents EDTA and EGTA had negligible effects on 3′GT activity. This results contrasts to a soybean (Glycine max) UDP-GlcA:soyasapogenol glucosyltransferase, which requires divalent cations for its activity (Kurosawa et al., 2002).
Similar to the native protein, the recombinant 3′GT showed strict specificity for its sugar donor; neither UDP-acetylglucosamine, UDP-Gal, nor UDP-GlcUA was used by the 3′GT (Table I). This result contrasts with the UDP-Gal:flavonoid 3-O-galactosyltransferase from Vigna mungo, which also uses UDP-Glc as a sugar donor (Mato et al., 1998), and tobacco (Nicotiana tabacum) glucosyltransferases, which can use UDP-Xyl as a weak donor (Taguchi et al., 2001). We analyzed the inhibition of 3′GT activity with UDP. The 3′GT activity was competitively inhibited with UDP when DEL 3G-5G and UDP-Glc were used as substrates and the Ki of UDP was calculated to be 760 μm.
Table I.
Activity of the recombinant 3′GT forvarbus flavonoid and sugar donors
The recombinant 3′GT was expressed in E. coli and partially purified. The 3′GT was incubated with 250 μm of each flavonoid and 500 μm of a sugar donor as described in “Materials and Methods.” The relative activity was calculated by the activity toward DEL 3G-5G as 100%.
| Substrate
|
||
|---|---|---|
| Anthocyanin | Sugar Donor | Relative Activity |
| % | ||
| DEL 3G-5G | UDP-Glc | 100 |
| CYA 3G-5G | UDP-Glc | 0 |
| PEL 3G-5G | UDP-Glc | 0 |
| DEL | UDP-Glc | 3.3 |
| DEL 3G | UDP-Glc | 2.9 |
| DEL 3G-5CafG | UDP-Glc | 50.9 |
| DEL 3G-5G | UDP-Gal | 0 |
| DEL 3G-5G | UDP-GlcUA | 0 |
| DEL 3G-5G | UDP-N-acetylgalactosamine | 0 |
| Dihydrokaempferol | UDP-Glc | 0 |
| Dihydroquercetin | UDP-Glc | 0 |
| Dihydromyricetin | UDP-Glc | 0 |
| Tricetin | UDP-Glc | 0 |
The recombinant 3′GT did not transfer a glucosyl moiety to pelargonidin 3,5-diglucoside or to cyanidin 3,5-diglucoside (CYA 3G-5G) that carry one or two hydroxy groups on the B-ring, respectively (Table I; Fig. 1), which suggests that the 3′GT strictly requires three hydroxy groups on the B-ring for its substrates.
None of the dihydroflavonols, dihydrokaempferol, dihydroquercetin, or dihydromyricetin, nor a flavone, tricetin, was used as a substrate for the recombinant 3′GT, even though dihydromyricetin and tricetin have three hydroxy groups on the B-ring (Table I).
Despite the efficient 3′-glucosylation of DEL 3G-5G, the recombinant 3′GT shows much lower glucosylation activity on DEL or DEL 3G (Table I; Fig. 1). These results are consistent with those for the purified native protein.
The 5- and 3′-glucosyl moieties are further acylated with caffeoyl groups in gentiodelphin (Goto et al., 1982; Fig. 1). In view of the substrate specificity of gentian 3′GT described above, the glucosylations at both the 3 and 5 positions must precede the 3′-glucosylation. There then appear to be two possible paths for the biosynthesis of gentiodelphin. Either the 5-acylation precedes the 3′-glucosylation or both 5- and 3′-acylations occur after glucosylations at the 3, 5, and 3′ positions are complete. Both pathways probably occur in gentian petals because the recombinant 3′GT glucosylates both DEL 3G-5G and DEL 3-O-glucosyl-5-O-(6-O-caffeoyl-glucoside) (DEL 3G-5CafG) in vitro with an approximate preference for DEL 3G-5G (Table I) twice that for DEL 3G-5CafG (Table I).
Analysis of Expression of the 3′GT Gene in Gentian
Expression of 3′GT in gentian leaves and petals at each developmental stage was analyzed (Fig. 8). The 3′GT transcripts were abundant in petals at three developmental stages but dramatically decreased in petals at stage 4 (fully opened flowers) and were barely detected in leaves. This result is consistent with the expression pattern of the gentian gentian hydroxycinnamoyl-CoA:anthocyanin 3, 5-O-diglucoside 5-O-glucoside-6″-O-hydroxycinnamoyltransferase (5AT) gene (Fujiwara et al., 1998). These results and the expression of other genes involved in anthocyanin biosynthesis in gentian petals such as genes of flavonoid 3′,5′-hydroxylase, dihydroflavonol 4-reductase, and 3GT (Fujiwara et al., 1998) suggest a finely tuned and concerted expression of structural genes involved in anthocyanin biosynthesis in gentian petals.
Figure 8.
Northern-blot analysis of the expression of the gentian 3′GT gene. Ten micrograms of total RNA from gentian petals at each developmental stage and from leaves was loaded on each lane and probed with DIG-labeled gentian 3′GT cDNA (middle panel) or gentian 5AT cDNA (bottom panel; Fujiwara et al., 1998) after electrophoresis and transfer to nylon membrane. The top panel shows RNA samples on an agarose gel before transfer to a nylon membrane. RNA was from petals of unpigmented buds tightly closed (lane 1), petals of pigmenting buds (lane 2), petals of fully pigmented buds before opening (lane 3), petals of fully opened flowers (lane 4), and leaves (lane 5).
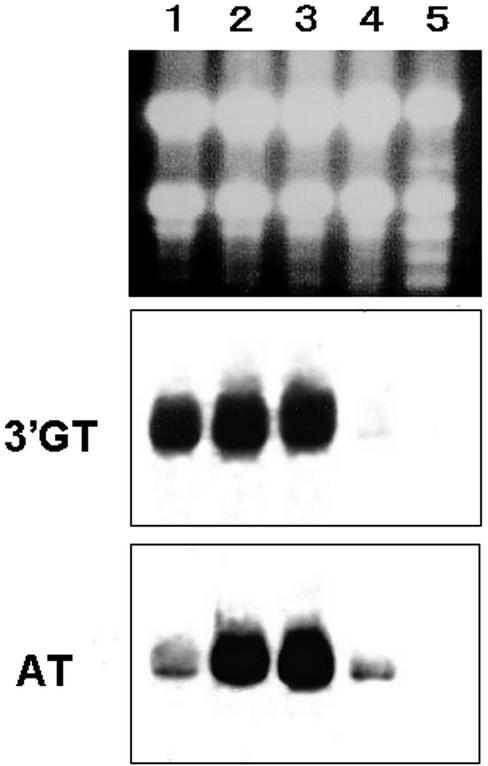
Expression of Gentian 3′GT in Petunia Flowers
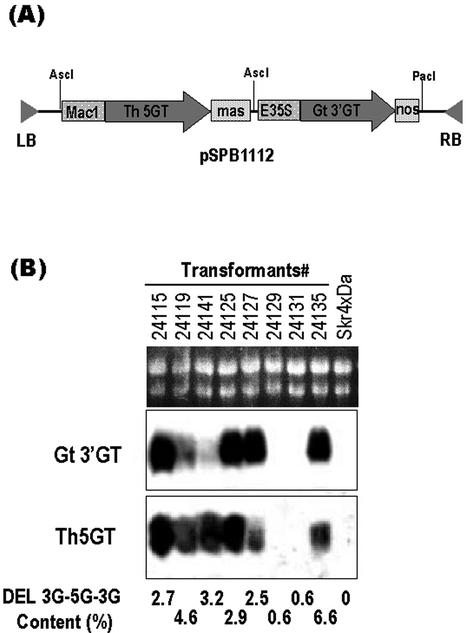
Petunia cv Skr4 × Da, which produces DEL 3G as its major anthocyanin due to deficiency of the UDP-rhamnose:anthocyanidin 3-O-glucoide rhamnosyltransferase gene (Brugliera et al., 1994; Kroon et al., 1994), was transformed with a binary vector pSPB1112 containing both gentian 3′GT and torenia 5GT (GenBank/EMBL/DDBJ accession no. AB076698) expression cassettes. More than 50 transgenic plants were obtained and subjected to northern blot analysis probed with the32P-labeled 3′GT and 5GT cDNA fragments. It was revealed that most plants expressed both of the 3′GT and 5GT transgenes, and typical results are shown in Figure 9. HPLC analysis of the anthocyanins from the petals of the transgenic petunia detected an anthocyanin compound that was not present in petals of the host plants. Absorbance spectra of the product and cochromatography with the authentic DEL 3G-5G-3′G confirmed that the compound produced in petunia transformants was DEL 3G-5G-3′G. The relative amount of DEL 3G-5G-3′G was 2% to 6% (mol/mol) of the total anthocyanins depending on the transformant (Fig. 9). DEL 3G-5G, DEL 3′-glucoside, or DEL 3,3′-diglucoside was not detected in the petals of the transformants. The petunia transformants producing DEL 3G-5G-3′G did not show a significant change in flower color compared with their host plants, probably due to the low levels of DEL 3G-5G-3′G produced.
Figure 9.
Northern-blot analysis of the expression of gentian 3′GT and torenia 5GT transgenes in petunia transformants. A, Schematic view of the binary plasmid pSPB1112 expression cassettes for both gentian 3′GT and torenia 5GT cDNAs. B, Ten micrograms of total RNA from petals of each transformant and host petunia cv Skr4 × Da was loaded on each lane, and the membrane was probed with 32P-labeled gentian 3′GT cDNA (middle panel) and torenia 5GT cDNA (bottom panel). The top panel shows RNA samples on an agarose gel before transfer to a nylon membrane. The numbers above the panel identify each transformant. The content of DEL 3G-5G-3′G in fully opened flowers of each transformant (as a percentage of total anthocyanins analyzed by HPLC) are described below the bottom panel.
DISCUSSION
The enzyme that catalyzes the transfer of the glucosyl moiety from UDP-Glc to the 3′-hydroxy group of anthocyanin was successfully purified to near homogeneity. Determination of partial amino acid sequence of the purified protein allowed us to isolate a cDNA encoding the enzyme. Expression of this cDNA in E. coli and in petunia plants confirmed that it encodes an active 3′GT.
A molecular phylogenetic tree consisting of gentian 3′GT and various plant glycosyltransferases indicates that the gentian 3′GT belongs to a subgroup consisting of betanidin 5GT from D. bellidiformis and flavonoid 7GT from S. baicalensis (Fig. 5). The betanidin 5GT glycosylates not only the 5-hydroxy group of betanidin but also the 7-hydroxy and 4′-hydroxy groups of quercetin and myricetin (flavonols) and cyanidin (an anthocyanidin) in vitro (Vogt et al., 1997, 1999). The 7GT from S. baicalensis was reported to glucosylate a broad range of substrates in vitro including the 7- and 4′-hydroxy groups of kaempferol (a flavonol) in addition to its prospective substrate baicalein (Hirotani et al., 2000). In the case of betanidin 5GT, its reactivity on the 7- and the 4′-hydroxy groups of myricetin/cyanidin was explained as due to an equivalent superimposed structure in which the 7- and the 4′-hydroxy groups of myricetin/cyanidin are equivalent to the 5-hydroxy group of betanidin (Vogt et al., 1999). This interpretation can be also applied in the case of S. baicalensis 7GT. Thus, some structural similarity between gentian 3′GT, betainidin 5GT, and schuterallia 7GT may not be surprising.
Enzymatic characterization using the recombinant 3′GT protein revealed some unique properties of gentian 3′GT with respect to its substrate. It has been suggested that most glucosyltransferases in the flavonoid biosynthesis pathway exhibit high regiospecificity for a specific position of a hydroxy group to be glucosylated but are less specific to the overall flavonoid structure (Heller and Forkmann, 1993). For example, 3GTs from gentian (Tanaka et al., 1996), grape (Ford et al., 1998), and petunia (Yamazaki et al., 2002) can glucosylate the 3-hydroxy group of PEL, CYA, and DEL. The perilla and verbena 5GTs glucosylate the 5-hydroxy group of all of three anthocyanins, DEL 3G, CYA 3G, and PEL 3G (Yamazaki et al., 1999). In contrast, the gentian 3′GT enzyme has an absolute requirement for three hydroxy groups at the 3′, 4′, and 5′ positions of the B-ring as its substrate. Furthermore, it glucosylates the 3′-hydroxy group of DEL 3G-5G but shows no activity toward CYA 3G-5G, which has a 3′-hydroxy residue, and, needless to say, it has no activity on pelargonidin 3,5-diglucoside, which does not have 3′-hydroxy group. These results are consistent with the fact that blue gentian petals contain gentiodelphin as a major anthocyanin with some other DEL derivatives as minor components (Fujiwara et al., 1997). The flowers of a pink Gentiana sp., on the other hand, were reported to contain cyanidin derivatives, one of which is triglucosylated at the 3, 5, and 3′ positions (Hosokawa et al., 1995). This may imply that the pink gentian has another type of 3′GT recognizing CYA 3G-5G or that the 3′GT studied here transfers a Glc group to CYA 3G-5G in vivo.
Besides the requirement for DEL-type anthocyanin as its substrate, the gentian 3′GT showed strict specificity for the overall structure of its substrate. DEL and DEL 3G are poor substrates compared with a high preference for DEL 3G-5G (Table I). This result is also supported by the in vivo expression of the 3′GT. The transgenic petunia plants expressing the gentian 3′GT and torenia 5GT produced DEL 3G-5G-3′G using DEL 3G as a substrate, which accumulates in petals of the host plants, but neither DEL 3′-glucoside nor DEL 3,3′-diglucosides was detected in the transgenic petunias. These results, together with the fact that partially purified gentian 5GT can glucosylate DEL 3G (M. Fukuchi-Mizutani, unpublished data), suggest a strict order for three glucosylatioin steps in the biosynthesis of gentiodelphin: first glucosylation of the 3-hydroxy group, then the 5-hydroxygroup, and finally glucosylation of the 3′-hydroxy group (Fig. 10). This definitive order of glucosylation and the substrate specificity of gentian 3′GT contribute to the efficient accumulation of gentiodelphin in the gentian petals.
Figure 10.
Proposed modification pathway of DEL to gentiodelphin in gentian petals. G in an oval represents O-glucoside and Caf represent O-caffeic acid. 3′AT, Hydroxycinnamoyl-CoA:anthocyanin 3′-O-glucoside-6″-O-hydroxycinnamoyltransferase.
The recombinant 3′GT protein can glucosylate both 5-acylated and nonacylated substrates in vitro. It was shown to transfer a glucosyl moiety to both DEL 3G-5CafG and DEL 3G-5G. The results indicate that the 3′-glucosylation and both acylations may form a metabolic grid in the biosynthetic pathway of gentiodelphin (Fig. 10).
Production of DEL by expressing a flavonoid 3′,5′-hydroxylase gene in non-DEL-producing plants such as carnation (Dianthus caryophyllus) successfully produced a violet variety of carnation (Mol et al., 1999). However, in most blue flowers such as gentian, butterfly pea, and lobelia, DEL derivatives are highly glucosylated and subsequently aromatically acylated. Yoshida et al. (2000) reported that a caffeoylglucosyl moiety at the 3′ position of B-ring of gentiodelphin contributes to both bluer color and stabilization of anthocyanins via intramolecular stacking. Cloning of the 3′GT cDNA in this study therefore paves the way to engineer blue flowers with metabolic engineering. This study also sheds light on the phylogenetic relationship of the glucosyltransferase superfamily which is involved in a wide range of plant metabolic pathways.
MATERIALS AND METHODS
Plant Materials
Fresh and pigmenting petals were collected from commercially available gentian (Gentiana triflora var japonica) cut flowers at a pre-anthesis stage, stored at –80°C, and used for purification of 3′GT protein. For extraction of total RNA, petals were collected from flowers at each developmental stage as follows; stage 1, unpigmented buds tightly closed; stage 2, pigmenting buds; stage 3, fully pigmented buds before opening; and stage 4, fully opened flowers.
Substrates
All anthocyanins used in this study were purified from appropriate plant materials as described by Fujiwara et al. (1997) unless otherwise mentioned.
Enzyme Assay
3′GT activity was measured using DEL 3G-5G and UDP-Glc as substrates. A standard reaction mixture consisted of 250 μm DEL 3G-5G, 500 μm UDP-Glc, and 20 μL of enzyme solution in 50 μL of 0.1 m potassium phosphate buffer (pH 8.5). The mixture was incubated for 15 min at 30°C, and the reaction was terminated by adding an equal volume of 90% (v/v) CH3CN with 0.1% (v/v) trifluoroacetic acid (TFA). After centrifugation, the supernatant was analyzed by thin-layer chromatography on a cellulose plate (HPTLC-Fertigplatten cellulose F254s, Merck, Darmstadt, Germany) using a solvent system of CH3COOH:HCl:H2O (1:1:2) and/or by HPLC equipped with a reverse phase column RS pack DE-413L (4.6 × 250 mm; Shodex, Tokyo, Japan). The anthocyanins were eluted with a linear gradient of 20% to 50% (v/v) CH3CN containing 0.5% (v/v) TFA at a flow rate of 0.6 mL min–1 for 15 min and 50% isoclatic elution for 10 min and were detected by a photodiode array detector SPD-M10A (250–600 nm; Shimadzu, Kyoto).
Purification of 3′ GT Protein from Gentian Petals
The gentian 3′GT protein was purified by monitoring 3′GT activity. Frozen petals (650 g fresh mass) were ground in liquid nitrogen with an Excel-auto homogenizer (Nihonseiki Kansai Ltd., Osaka), and the powdered tissues were suspended in 3 L of buffer A (100 mm Tris-HCl, pH 7.5, 10 mm sodium ascorbate, 5 mm dithiothreitol, 0.1% [v/v] β-mercaptoethanol, and 10 μm (p-aminophenyl) methanesulfonyl fluoride [p-APMSF]). After the addition of Polyclar SB-100 (Gokyosangyo Ltd., Osaka) to a final concentration of 5% (w/v), the suspension was filtered through four layers of gauze. After centrifugation at 8,000g for 30 min, the supernatant was fractionated with ammonium sulfate to between 40% and 70% saturation. The precipitated protein was dissolved in a minimum volume of buffer B (20 mm Tris-HCl, pH 7.5, 0.1% [v/v] β-mercaptoethanol, 10% [v/v] glycerol, and 10 μm p-APMSF). All of the procedures described above were performed at 0°C to 4°C, and the following purification proceeded using an FPLC system (Amersham Biosciences, Buckinghamshire, UK) at room temperature. The crude enzyme solution was desalted on a Sephadex G-25 column (5.0 × 40 cm, Amersham Biosciences) equilibrated with buffer B and then applied to a Q-Sepharose fast flow column (2.6 × 10 cm, Amersham Biosciences) equilibrated with buffer B. The column was washed extensively with buffer B, and the proteins were eluted with a linear gradient of 0 to 0.5 m NaCl in buffer B at a flow rate of 8 mL min–1. The active fractions eluted at 0.3 to 0.4 m NaCl were loaded on a Blue A Dyematrix column (0.5 × 5 cm, Amicon, Toronto). After extensive washing, proteins were eluted by a linear gradient of 0.3 to 1.5 m KCl in buffer B. The fractions with 3′GT activity eluted at 1.5 m KCl were concentrated and desalted using a Centricon 30 (exclusion limit 30 kD, Amicon) followed by buffer replacement in buffer C (20 mm sodium phosphate buffer, pH 7.0, 10% [v/v] glycerol, 0.1% [v/v] β-mercaptoethanol, and 10 μm p-APMSF). The protein was loaded on a DEAE-Sepharose column (0.5 × 10 cm, Amersham Biosciences) equilibrated with buffer C and eluted by a linear gradient of 0 to 0.5 m NaCl in buffer C at a flow rate of 0.5 mL min–1. The active fractions eluted at 0.33 to 0.5 m NaCl were desalted and concentrated by a Centricon 30 (Amicon), then loaded on a Superose 12 column (Amersham Biosciences) equilibrated with buffer C, and eluted by buffer C at a flow rate of 0.5 mL min–1. The active fractions at each purification step were subjected to SDS-PAGE analysis and were detected by silver-staining with a silver-staining kit (Daiichikagaku Chemical, Osaka).
Determination of Amino Acid Sequences
The amino acid sequence of the purified peptides were determined essentially as described previously (Fujiwara et al., 1998). Approximately 100 pmol of purified protein was used for determining the amino terminal sequence. To determine internal sequences, the purified protein (100 pmol) was digested with lysyl-endopeptidase from Achromobacter lyticus (Wako Pure Chemicals, Osaka), and the resultant peptides were separated by reverse-phase HPLC. Amino acid sequences were determined using a liquid-phase sequencer (PSQ-1, Shimadzu).
Standard Molecular Procedures
Standard molecular procedures were as described previously unless specified (Fujiwara et al., 1997). The amino acid sequences were aligned using the multiple alignment program, ClustalW (Thompson et al., 1994). On the basis of this alignment, a phylogenetic analysis was carried out by the neighbor-joining method (Sato and Nei, 1987), and a phylogenetic tree was drawn using the software TREEVIEW (Page, 1996).
Cloning of 3′GT cDNA
The cDNA library derived from gentian petals has been described previously (Fujiwara et al., 1998). The oligonucleotide primer, GT-2, contained the sequence 5′-TGGGCUAA(T/C) GA(T/C) GCIGC-3′, which corresponded to the amino acid sequence, WANDAA, in peptide fragment 32. The 3′ primer, XhoI-T, corresponded to the 3′ end of cDNAs in the gentian cDNA library and contained the sequence 5′-CTCGAGTTTTTTTTTTTTTTTTT-3′. A PCR using GT-2 and XhoI-T oligonucleotide primers with the gentian cDNA library as a template amplified DNA fragments, one of which encoded an amino acid sequence homologous to those of flavonoid GTs, and was used as a probe to screen the cDNA library.
Expression of a 3′GT cDNA in Escherichia coli and Purification of the Recombinant Protein
An NcoI site was introduced by PCR to overlap the putative
initiation Met codon in the gentian 3′GT cDNA using an oligonucleotide
(5′-ATTCC GATCAGCTTCACGTTTTC-3′, the Met codon is
double-underlined and the NcoI site is underlined). The amplified
fragment was digested with NcoI and BglII and ligated with
the 3′-half of the gentian 3′GT cDNA between BglII and
KpnI sites, into NcoI and KpnI sites of pQE61
vector (Qiagen, Chatsworth, CA) to generate an expression plasmid
pQE3′GT. The E. coli strain JM105 was transformed with
pQE3′GT, and cells containing pQE3′GT were precultured at 37°C
overnight and then inoculated into 8 L of Luria-Bertani media containing 50
μg mL–1 ampicillin and incubated at 27°C
until the A600 reached 0.6. After addition of
isopropylthio-β-galactoside to a final concentration to 0.4
mm, the cells were further cultured at 27°C overnight, then
collected and suspended in sonication buffer (25 mm Tris-HCl, pH
7.5, 250 mm NaCl, 1 mm EDTA, and 0.5% [v/v]
β-mercaptoethanol). Following disruption of the cells by sonication, the
soluble fraction was fractionated with 40% to 70% (w/v) saturated ammonium
sulfate. The fraction was dialyzed and suspended in buffer D (25 mm Tris-HCl [pH 7.5] and 0.5% [v/v] β-mercaptoethanol), then loaded on
DEAE-TOYOPEARL (TOSO, Tokyo) equilibrated with buffer D, and eluted by a
linear gradient of 0 to 0.5 m NaCl in buffer D. The active
fractions eluted at 0.25 to 0.35 m NaCl were collected and loaded
on a Blue A Dyematrix column (0.5 × 5 cm, Amicon) equilibrated with
buffer D. After thorough washing with buffer D, the active fraction was eluted
by 2 m KCl in buffer D. The fraction was desalted and suspended in
50 mm Tris-HCl (pH 7.5) containing 30% (w/v) ammonium sulfate, then
loaded on a Phenyl Sepharose column (1 mL), and eluted by linear gradient of
30% to 0% (w/v) ammonium sulfate in 50 mm Tris-HCl (pH 7.5). The
most active fraction eluted at 0% (w/v) ammonium sulfate was used to
characterize the enzymatic properties.
GATCAGCTTCACGTTTTC-3′, the Met codon is
double-underlined and the NcoI site is underlined). The amplified
fragment was digested with NcoI and BglII and ligated with
the 3′-half of the gentian 3′GT cDNA between BglII and
KpnI sites, into NcoI and KpnI sites of pQE61
vector (Qiagen, Chatsworth, CA) to generate an expression plasmid
pQE3′GT. The E. coli strain JM105 was transformed with
pQE3′GT, and cells containing pQE3′GT were precultured at 37°C
overnight and then inoculated into 8 L of Luria-Bertani media containing 50
μg mL–1 ampicillin and incubated at 27°C
until the A600 reached 0.6. After addition of
isopropylthio-β-galactoside to a final concentration to 0.4
mm, the cells were further cultured at 27°C overnight, then
collected and suspended in sonication buffer (25 mm Tris-HCl, pH
7.5, 250 mm NaCl, 1 mm EDTA, and 0.5% [v/v]
β-mercaptoethanol). Following disruption of the cells by sonication, the
soluble fraction was fractionated with 40% to 70% (w/v) saturated ammonium
sulfate. The fraction was dialyzed and suspended in buffer D (25 mm Tris-HCl [pH 7.5] and 0.5% [v/v] β-mercaptoethanol), then loaded on
DEAE-TOYOPEARL (TOSO, Tokyo) equilibrated with buffer D, and eluted by a
linear gradient of 0 to 0.5 m NaCl in buffer D. The active
fractions eluted at 0.25 to 0.35 m NaCl were collected and loaded
on a Blue A Dyematrix column (0.5 × 5 cm, Amicon) equilibrated with
buffer D. After thorough washing with buffer D, the active fraction was eluted
by 2 m KCl in buffer D. The fraction was desalted and suspended in
50 mm Tris-HCl (pH 7.5) containing 30% (w/v) ammonium sulfate, then
loaded on a Phenyl Sepharose column (1 mL), and eluted by linear gradient of
30% to 0% (w/v) ammonium sulfate in 50 mm Tris-HCl (pH 7.5). The
most active fraction eluted at 0% (w/v) ammonium sulfate was used to
characterize the enzymatic properties.
NMR and MS Analysis of Anthocyanins
1H- and 13C-NMR spectra were obtained on a DMX-500 spectrometer (Bruker Biospin, Rheinstetten, Germany). About 2.5 mg of the anthocyanin produced by the recombinant protein with DEL 3G-5G and UDP-Glc as substrates was dissolved in 10% (v/v) TFA-d/CD3OD mixture. The residual proton peak of CD3OD was used as the internal standard (3.3 ppm). Electrospray ionization-MS spectrum was measured on a LC-Q system with X-Calibur in positive mode (Thermo Finnigan, San Jose, CA).
Characterization of Enzymatic Properties
The purified native and the recombinant proteins were used for characterization of the enzymatic properties of 3′GT. A standard reaction mixture for the 3′GT protein consisted of 250 μm anthocyanin, 500 μm UDP-Glc, and 100 ng of protein in 50 μL of 0.1 m potassium phosphate buffer (pH 8.5). The reaction conditions and the subsequent HPLC analyses are as above. All reactions were at least duplicated. For determination of Km values, 3.2, 6.4, 16, 32, 64, 160, 320, and 640 μm DEL 3G-5G were used with 364 ng of the recombinant 3′GT protein and 1 mm UDP-Glc in 100 μL of 0.1 m potassium phosphate buffer (pH 8.5), and the reaction stopped at 2 min where the reaction velocity is linear. To determine the effect of metal cations, 1 mm of each metal cation or chealating reagent was added to the standard reaction. For analyzing specificity with respect to sugar donors, UDP-Gal, UDP-N-acetylglucosamine, or UDP-GlcUA was added instead of UDP-Glc. For the analysis of Ki of UDP, 3′GT activity was measured with 250 μm DEL 3G-5G and 1 mm UDP-Glc as substrates in the presence of 0, 0.25, 0.5, 1, 2.5, and 5 mm UDP.
Northern-Blot Analysis for the Expression of the 3′GT Gene in Gentian Petals
Total RNA was extracted from gentian leaves and petals at each developmental stage with a RNeasy RNA extraction kit (Qiagen). Ten micrograms of total RNA was electrophoresed and hybridized with DIG-labeled gentian 3′GT cDNA or 5AT cDNA (Fujiwara et al., 1998) according to the DIG northern Starter kit instructions (Roche Diagnostics, Mannheim, Germany).
Expression of 3′GT cDNA in Petunia
A plant expression plasmid, pSPB1112, was constructed using a pBIN-PLUS binary vector (van Engelen et al., 1995) for co-expression of the gentian 3′GT cDNA and a torenia 5GT. In this construct, the gentian 3′GT cDNA is driven by an enhanced cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996), and a nopaline synthase terminator is used; whereas the torenia 5GT cDNA was downstream of a MAC promoter (Comai et al., 1990), and a mannopine synthase terminator was used. Thus, plants transformed with pSPB1112 are expected to constitutively co-express both the gentian 3′GT and the torenia 5GT. Petunia cv Skr4 × Da that accumulates DEL 3G was transformed using pSPB1112 and an Agrobacterium sp.-mediated method (Brugliera et al., 1994). Total RNA was extracted from petals of the petunia transformants and probed with each of the gentian 3′GT and torenia 5GT cDNAs. Procedure for northern-blot analysis of transformants has been described previously (Brugliera et al., 1994).
Acknowledgments
We are grateful to Florigene Ltd. for petunia transformation and to Dr. John Mason of Florigene Ltd. for critical review of the manuscript. We also thank Dr. Yuko Ohhashi for enhanced 35S promoter.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.018242.
This study was carried out in a joint research program of Suntory Ltd. and Florigene Ltd. (Australia).
References
- Brugliera F, Holton TA, Stevenson TW, Farcy E, Lu CY, Cornish E (1994) Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J 5: 81–92 [DOI] [PubMed] [Google Scholar]
- Comai L, Moran P, Maslyar D (1990) Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol Biol 15: 373–381 [DOI] [PubMed] [Google Scholar]
- Ford CM, Boss PK, Hoj PB (1998) Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem 273: 9224–9233 [DOI] [PubMed] [Google Scholar]
- Forkmann G, Heller W (1999) Biosynthesis of flavonoids. In U Sankawa, ed, Comprehensive Natural Products Chemistry. Elsevier, Amsterdam, pp 713–748
- Forkmann G, Martens S (2001) Metabolic engineering and application of flavonoid. Curr Opin Biotechnol 12: 155–160 [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Fukui Y, Nakao M, Ashikari T, Kusumi T (1997) Anthocyanin 5-aromatic acyltransferase from Gentiana triflora: purification, characterization and its role in anthocyanin biosynthesis. Eur J Biochem 249: 45–51 [DOI] [PubMed] [Google Scholar]
- Fujiwara H, Tanaka Y, Yonekura-Sakakibara K, Fukuchi-Mizutani M, Nakao M, Fukui Y, Yamaguchi M, Ashikari T, Kusumi T (1998) cDNA cloning, gene expression and subcellular localization of anthocyanin 5-aromatic acyltransferase from Gentiana triflora. Plant J 16: 421–431 [DOI] [PubMed] [Google Scholar]
- Futtek DB, Schiefelbein JW, Johnston F, Nelson JOE (1988) Sequence comparisons of three wild-type Bronze-1 alleles from Zea mays. Plant Mol Biol 11: 473–481 [DOI] [PubMed] [Google Scholar]
- Goto T, Kondo T, Tamura H, Imagawa H, Lino A, Takeda T (1982) Structure of gentiodelphin, an acylated anthocyanin isolated from Gentiana makinori, that is stable in dilute aqueous solution. Tetrahedron Lett 23: 3695–3698 [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Heller W, Forkmann G (1993) Biosynthesis of flavonoids. In JB Harborne, ed, The Flavonoids: Advances in Research since 1986. Chapman & Hall, London, pp 499–535
- Hirotani M, Kuroda R, Suzuki H, Yoshizawa T (2000) Cloning and expression of UDP-glucose:flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210: 1006–1013 [DOI] [PubMed] [Google Scholar]
- Hosokawa K, Fukushi R, Kawabata J, Fujii C, Ito T, Yamamura S (1995) Three acylated cyanidin glucosides in pink flowers of gentiana. Phytochemistry 40: 941–944 [Google Scholar]
- Horvath DM, Chua NH (1996) Identification of an immediate-early salicylic acid-inducible tobacco gene and characterization of induction by other compounds. Plant Mol Biol 31: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Kroon J, Souer E, de Graaff A, Xue Y, Mol J, Koes R (1994) Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J 5: 69–80 [DOI] [PubMed] [Google Scholar]
- Kurosawa Y, Takahara H, Shiraiwa M (2002) UDP-glucuronic acid:soyasapogenol glucuronosyltransferase involved in saponin biosynthesis in germinating soyabean seeds. Planta 215: 620–629 [DOI] [PubMed] [Google Scholar]
- Lie Y, Baldauf S, Lim E-K, Bowles DJ (2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Marrs KA, Alfenito MR, Lloyd AM, Walbot V (1995) A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375: 397–400 [DOI] [PubMed] [Google Scholar]
- Martin CR, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Anthirrhinum majus. Plant J 1: 37–49 [DOI] [PubMed] [Google Scholar]
- Mato M, Ozeki Y, Itoh Y, Higeta D, Yoshitama K, Teramoto S, Aida R, Ishikura N, Shibata M (1998) Isolation and characterization of a cDNA clone of UDP-galactose:flavonoid 3-O-galactosyltransferase (UF3GaT) expressed in Vigna mungo seedlings. Plant Cell Physiol 39: 1145–1155 [DOI] [PubMed] [Google Scholar]
- Miller KD, Guyon V, Evans JNS, Shuttleworth WA, Taylor LP (1999) Purification, cloning and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J Biol Chem 274: 34011–34019 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Mol J, Cornish E, Mason J, Koes R (1999) Novel coloured flowers. Curr Opin Biotechnol 10: 198–201 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Sato N, Nei N (1987) The neighbor-joining method: a new method for reconstructing phylogenic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Taguchi G, Yazawa T, Hayashi N, Okazaki M (2001) Molecular cloning and heterologous expression of a novel glucosyltransferases from tobacco cultured cells that have broad substrate specificity and are induced by salicylic acid and auxin. Eur J Biochem 268: 4086–4094 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Tsuda S, Kusumi T (1998) Metabolic engineering of modify flower color. Plant Cell Physiol 39: 1119–1126 [Google Scholar]
- Tanaka Y, Yonekura Y, Fukuchi-Mizutani M, Fukui Y, Fujiwara H, Ashikari T, Kusumi T (1996) Molecular and biochemical characterization of three anthocyanin synthetic enzymes from Gentian triflora. Plant Cell Physiol 37: 711–716 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap J-P, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- Vogt T, Grimm R, Strack D (1999) Cloning and expression of a cDNA encoding betanidin 5-O-glucosyltransferase, a betanidin- and flavonoid-specific enzyme with high homology to inducible glucosyltransferases from the Solanaceae. Plant J 19: 509–519 [DOI] [PubMed] [Google Scholar]
- Vogt T, Zimmermann E, Grimm R, Meyer M, Strack D (1997) Are the characteristics of betanidin glucosyltransferases from cell-suspension cultures of Dorotheanthus bellidiformis indicative of their phylogenetic relationship with flavonoid glucosyltransferases? Planta 203: 349–361 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2001) It takes a garden: How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 126: 485–493 [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Rohde W, Salamini F (1990) Nucleotide sequence of the Bronze-1 homologous gene from Hordeum vulgare. Plant Mol Biol 14: 277–279 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Saito K (1999) Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J Biol Chem 274: 7405–7411 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Yamagishi E, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K (2002) Two flavonoid glucosyltransferases from Petunia hybrida: molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48: 401–411 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Toyama Y, Kameda K, Kondo T (2000) Contribution of each cafferoyl residue of the pigment molecules of gentiodelphin to blue color development. Phytochemistry 54: 85–92 [DOI] [PubMed] [Google Scholar]