Abstract
Maize (Zea mays) Viviparous1 (VP1) and Arabidopsis ABI3 are orthologous transcription factors that regulate key aspects of plant seed development and ABA signaling. To understand VP1-regulated gene expression on a global scale, we have performed oligomicroarray analysis of transgenic Arabidopsis carrying 35S::VP1 in an abi3 null mutant background. We have identified 353 VP1/ABA-regulated genes by GeneChip analysis. Seventy-three percent of the genes were affected by both VP1 and ABA in vegetative tissues, indicating a tight coupling between ABA signaling and VP1 function. A large number of seed-specific genes were ectopically expressed in vegetative tissue of 35S::VP1 plants consistent with evidence that VP1 and ABI3 are key determinants of seed-specific expression. ABI5, a positive regulator of ABA signaling, was activated by VP1, indicating conservation of the feed-forward pathway mediated by ABI3. ABA induction of ABI1 and ABI2, negative regulators of ABA signaling, was strongly inhibited by VP1, revealing a second pathway of feed-forward regulation. These results indicate that VP1 strongly modifies ABA signaling through feed-forward regulation of ABI1/ABI5-related genes. Of the 32 bZIP transcription factors represented on the GeneChip, genes in the ABI5 clade were specifically coregulated by ABA and VP1. Statistical analysis of 5′ upstream sequences of the VP1/ABA-regulated genes identified consensus abscisic responsive elements as an enriched element, indicating that many of the genes could be direct targets of the ABI5-related bZIPs. The Sph element is an enriched sequence motif in promoters of genes co-activated by ABA and VP1 but not in promoters of genes activated by ABA alone. This analysis reveals that distinct combinatorial patterns of promoter elements distinguish subclasses of VP1/ABA coregulated genes.
Abscisic acid (ABA) has a central role in regulation of seed development as well as plant responses to stresses such as cold and drought. Identification of abscisic acid response mutants in Arabidopsis and maize (Zea mays) has provided insight into the molecular components of ABA signaling in developing seeds. There are two classes of the ABA response mutants in seeds, insensitive and hypersensitive (for review, see Finkelstein et al., 2002). The ABA-insensitive mutants include abi1, abi2, abi3 (Koornneef et al., 1984; Finkelstein and Somerville, 1990), abi4, abi5 (Finkelstein, 1994), cho1, cho2 (Nambara et al., 2002), rcn1 (Kwak et al., 2002) of Arabidopsis and viviparous1 (vp1; McCarty et al., 1989) of maize. The ABA-hypersensitive mutants include era1 (Cutler et al., 1996), ein2 (allelic to era3; Beaudoin et al., 2000; Ghassemian et al., 2000), hyl1 (Lu and Fedoroff, 2000), sad1 (Xiong et al., 2001a), abh1 (Hugouvieux et al., 2001), rop10 (Zheng et al., 2002), and fiery1 (Xiong et al., 2001b). The respective genes for these mutants have been cloned and shown to encode variety of proteins. The ABI1, ABI2, and RCN1 genes encode protein phosphatases (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998a; Kwak et al., 2002). The ABI3 gene encodes a transcription factor homologous to maize VP1 (McCarty et al., 1991; Giraudat et al., 1992). The ABI4 and ABI5 genes encode an AP2 domain transcription factor (Finkelstein et al., 1998) and a bZIP transcription factor (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), respectively. The ERA1 gene encodes a protein farnesyl transferase (Cutler et al., 1996). ERA3, an EIN2 allele of ethylene-signaling pathway, encodes a membrane metal transporter (Alonso et al., 1999). HYL1, SAD1, and ABH1 genes encode various types of RNA-binding proteins (Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001a). ROP10 and FIERY1 encode a small G protein and an inositol polyphosphate 1-phosphatase, respectively (Xiong et al., 2001b; Zheng et al., 2002). In addition to these mutants, silencing of a calcium sensor, SCaBP5, and protein kinases, PKS3 and PKS18, causes ABA hypersensitivity (Gong et al., 2002; Guo et al., 2002).
Among these mutants, the Arabidopsis abi3 and maize vp1 mutants have the most profound effect on seed development. Null alleles of ABI3 and VP1 result in loss of ABA sensitivity, leading to non-dormancy or vivipary in Arabidopsis and maize, respectively (McCarty et al., 1989; Nambara et al., 1992, 1994). In Arabidopsis, three other mutants, lec1, lec2, and fus3, have a precocious germination phenotype, despite displaying nearly normal ABA sensitivity (Meinke, 1992; Keith et al., 1994; Meinke et al., 1994; West et al., 1994). LEC2 and FUS3 genes encode transcription factors that are structurally related to ABI3/VP1 (Luerssen et al., 1998; Stone et al., 2001). LEC1 encodes a transcription factor homologous to HAP3 (Lotan et al., 1998). Although these genes genetically interact with each other (Parcy et al., 1997; Nambara et al., 2000; Vicient et al., 2000; Raz et al., 2001) and other abi loci (Brocard-Gifford et al., 2003), only the ABI3/VP1 factor is directly implicated in ABA signaling.
ABI3/VP1 is a multidomain transcription factor that functions as both an activator and a repressor depending on the promoter context (McCarty et al., 1991; Hattori et al., 1992; Hoecker et al., 1995; Nambara et al., 1995). Three basic protein domains, B1, B2, and B3, are highly conserved among ABI3/VP1 factors from various plant species (McCarty et al., 1991; Giraudat et al., 1992; Hattori et al., 1994; Bobb et al., 1995; Chandler and Bartels, 1997; Rohde et al., 1998; Shiota et al., 1998). The C-terminal B3 domain of VP1 binds specifically to the Sph DNA element in the maize C1 promoter (Suzuki et al., 1997), whereas the N-terminal B1 and B2 domains are implicated in nuclear localization and interactions with other proteins (Giraudat et al., 1992; Ezcurra et al., 2000). The N-terminal co-activation repression domain is necessary and sufficient for ABA-dependent co-activation functions and repressor activities (Hoecker et al., 1995; Carson et al., 1997) of VP1/ABI3, whereas the C-terminal B3 is required for a discrete subset of gene activation functions (Carson et al., 1997). Recent genetic analysis of abi3 alleles has revealed further complexity of the role of ABI3 in ABA signaling, suggesting that multiple ABA-signaling pathways are perceived through ABI3 (Nambara et al., 2002). Moreover, modification of chromatin structure by PvALF, the Phaseolus sp. ABI3 ortholog, has been shown, suggesting that VP1/ABI3 has a potential to recruit additional DNA-binding proteins to promoters (Li et al., 1999, 2001).
A number of ABA response elements (ABREs) have been identified in promoters of ABA-induced genes. In most cases, the ABREs contain a core ACGT motif, the most common of those is designated the G-box (Giuliano et al., 1988; Marcotte et al., 1989; Mundy et al., 1990; Marcotte and Quatrano, 1993). A number of basic-Leu zipper (bZIP) proteins have been shown to bind ABREs identified in promoters of ABA-induced genes (Guiltinan et al., 1990; for review, see Jakoby et al., 2002). VP1 activates expression of ABA-inducible genes through the G-box. However, VP1 does not appear to bind to the element directly (Suzuki et al., 1997). Rather, VP1 regulation is likely mediated by protein-protein interactions with G-box-binding factors (McCarty et al., 1991; Hattori et al., 1995; Vasil et al., 1995; Shen et al., 1996; Carson et al., 1997). In support of that model, the rice (Oryza sativa) TRAB1 bZIP protein was shown to interact with OsVP1 by yeast two-hybrid system (Hobo et al., 1999b). The Arabidopsis ABI5 is homologous to TRAB1 and similarly interacts with ABI3 in the two-hybrid system (Nakamura et al., 2001). In the abi5 mutant, expression of ABI3-regulated genes is reduced during seed maturation (Finkelstein and Lynch, 2000). A clade of 13 ABI5-related genes exists in the Arabidopsis genome (Arabidopsis Genome Initiative, 2000; Bensmihen et al., 2002), and these genes are expressed differentially in plant tissues (Choi et al., 2000; Uno et al., 2000; Brocard et al., 2002; Bensmihen et al., 2002; Kang et al., 2002; Kim et al., 2002). In addition, phosphorylation of ABI5 and related proteins also likely regulates the protein activity and stability (Uno et al., 2000; Lopez-Molina et al., 2001, 2003; Johnson et al., 2002; Kagaya et al., 2002; Lu et al., 2002). These findings have indicated complexity of ABA signaling mediated by the ABI5-related proteins.
Recently, transcriptome analyses of ABA-regulated genes have been reported (Hoth et al., 2002; Seki et al., 2002). These analyses reveal global patterns of ABA-regulated gene expression in various tissues. In addition, genetic and biochemical studies of ABI5-related proteins mentioned above have clearly shown that these proteins play key roles for various ABA-signaling events including seed maturation. However, the precise role of ABI3/VP1 in mediating ABA signaling remains elusive. In this study, we analyze the global effects of VP1/ABI3 expression on ABA-regulated gene expression. We have used an oligomicroarray analysis of 35S::VP1 transgenic Arabidopsis to dissect and identify a large number of genes regulated by VP1/ABI3 and ABA. The results reveal complex combinatorial interactions between ABA signaling and VP1. We show that VP1 has a capacity to modify ABA signaling through feed-forward and feedback interactions mediated by members of the ABI5- and ABI1-related gene families, respectively. We furthermore show that different classes of VP1- and ABA-regulated genes have distinct patterns of enriched cis-elements in their promoters.
RESULTS
VP1 Alters the Global Pattern of ABA-Regulated Gene Expression
Our previous study showed that maize VP1 complements the Arabidopsis abi3 mutant (Suzuki et al., 2001). Moreover, like ABI3 (Parcy et al., 1994), expression of VP1 in vegetative tissue causes ectopic expression of seed-specific genes in an ABA-dependent manner. The strong functional conservation indicates that maize VP1 and Arabidopsis ABI3 share a capacity to regulate a significant number of genes in Arabidopsis. To understand the global patterns of VP1/ABI3- and ABA-mediated gene expression in Arabidopsis, we performed oligomicroarray analysis of 35S::VP1/abi3-6 plants (Suzuki et al., 2001) and abi3-6 control plants. Two-week-old seedlings of 35S::VP1/abi3-6 and abi3-6 were treated with or without 5 μm ABA for 12 h and then harvested for total RNA preparation. We expected that the relatively short duration treatment with the relatively low concentration of ABA would allow us to identify a set of genes that are directly regulated by ABA and VP1. The comparisons were made independently in the two replicate experiments, and genes were identified with an average response of 3-fold or greater with at least 2.5-fold effects present in both replicates.
We identified 353 genes that were 3-fold or greater activated or repressed by 35S::VP1 and/or ABA treatments relative to untreated abi3 mutant control plants (Supplemental Data 1; supplemental material can be viewed at http://www.plantphysiol.org). Consistent with our previous report (Suzuki et al., 2001), ectopic expression of VP1 dramatically alters ABA-regulated gene expression in vegetative tissues. Activations of as high as 800-fold were detected (Supplemental Data 1). Fifty-four genes were activated 30-fold or greater by VP1 and/or ABA treatments. The affected genes made up nearly 4.8% of 7,402 annotated nuclear genes represented on the GeneChip (Affymetrix, Santa Clara, CA). Extrapolation to the whole Arabidopsis genome suggests that nearly 1,200 genes may be similarly regulated by VP1/ABI3 (Arabidopsis Genome Initiative, 2000).
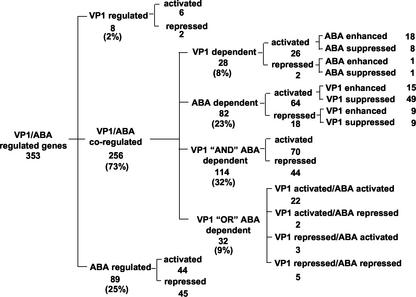
The 353 affected genes fell into at least 22 distinct response classes using a 2.5-fold cut-off for classification (Fig. 1). Two hundred and sixty-four of the affected genes (75%) were regulated by VP1. All but eight of these genes (97% of the 264 VP1-regulated genes) were regulated by both VP1 and ABA, indicating a tight coupling between ABA signaling and VP1 action. In contrast, a quarter of the 353 genes (89 genes) were affected by ABA treatment alone but not by VP1, indicating that a subset of ABA signaling is independent of regulation by VP1 in vegetative tissues. This class included a number of known ABA-regulated genes (Hoth et al., 2002) including RD29a (At5g52310), RD22 (At5g25610; Yamaguchi-Shinozaki and Shinozaki, 1993a, 1993b), KIN1 (At5g15960; Kurkela and Franck, 1990), and GBF3 (At2g46270; Lu et al., 1996). Detection of these genes verified the efficacy of our ABA treatment (12 h, 5 μm) conditions.
Figure 1.
Classification of VP1/ABA-regulated genes. The classification was made based on comparisons between treatments by using a 2.5-fold cut-off (see Supplemental Data 1). The numbers of genes in each class are indicated. The distribution of six major classes (VP1 regulated, four classes of VP1/ABA regulated, and ABA regulated) is shown in parentheses as a percentage of 353 total genes.
Forty-five percent of the 256 VP1/ABA coregulated genes (114 genes) showed a response that strictly required both factors. These were designated the VP1 AND ABA-dependent class. This group included 70 activated and 44 repressed genes. The responses of 28 genes in the VP1-dependent set (11% of the 256 genes) showed a strict requirement for presence of VP1 and with an additional modifying effect in ABA-treated 35S::VP1 plants. Conversely, responses of 82 ABA-dependent genes, consisting of 64 activated genes and 18 repressed genes, showed a strict requirement for ABA signaling that was further modified by the presence of VP1. Sixty percent of the ABA-dependent class (49 genes) was ABA-inducible but the induction was inhibited by VP1. Finally, 32 genes in the VP1 OR ABA class were activated independently by either presence of VP1 or ABA treatments with little or no combined effect. In total, 68 genes (eight genes from the VP1 only class, 28 genes from VP1-dependent class, and 32 genes from VP1 OR ABA class) were activated or repressed by VP1 overexpression in the absence of exogenous ABA, whereas 196 genes required ABA for VP1 effects. Hence, in a vegetative context at least, the ABA-dependent effects of VP1 on gene expression out-numbered the hormone-independent effects by factor of three.
Functional Classification of VP1 and ABA-Regulated Genes
Seed storage protein genes were prominent among the VP1-activated genes, whereas this functional classes was absent from the repressed category (Table I). In contrast, there were a greater number of signaling protein genes in the VP1-repressed class (12 genes) than in the VP1-activated class (three genes). This difference in distribution indicates that activation and repression by VP1 have distinct functional classes of genes as targets. Other functional categories including metabolism genes and transcription factor genes were more evenly represented in the repressed and activated gene categories. These results indicate VP1 and ABA together have the potential to cause a substantial reprogramming of metabolism and transcription in vegetative cells.
Table I.
Functional groups of the VP1/ABA-regulated genes in vegetative tissue
| Positively regulated by VP1 | Negatively regulated by VP1 | Activated by ABA only | Repressed by ABA only | |
|---|---|---|---|---|
| Seed storage proteins | 27 | 0 | 0 | 0 |
| Metabolism/homeostasis | 61 | 45 | 12 | 21 |
| Signaling | 3 | 12 | 4 | 3 |
| Transporters | 5 | 8 | 4 | 2 |
| Stress/Defense | 10 | 6 | 5 | 5 |
| Transcription | 3 | 8 | 4 | 1 |
| Unknown | 40 | 33 | 13 | 12 |
| Nucleic acid | 1
|
2
|
2
|
1
|
| 150
|
114
|
44
|
45
|
|
| 264 (3.6%)
|
89 (1.2%)
|
|||
| 353 (4.8%) | ||||
35S::VP1 and ABA Cause Ectopic Induction of a Seed-Specific Developmental Program
Genes encoding known seed protein genes or putative seed protein genes made up 10% (27 genes) of all of the VP1-regulated genes. All but two of these genes showed a strict requirement for VP1 in vegetative tissue, indicating that VP1 is sufficient to confer ABA induction to a broad range of seed protein genes. This is consistent with the previous studies showing ectopic activation of specific seed expressed genes in vegetative tissues of ABI3- and VP1-expressing plants (Parcy et al., 1994; Suzuki et al., 2001, respectively).
As predicted by our earlier study (Suzuki et al., 2001), the ectopically expressed seed genes detected in this experiment include a number of known ABI3-regulated genes. These include AtEm1 (At3g51810), cruciferin A (At5g44120), cruciferin C (At4g28520), AT2S1 (At4g27140), M17 (At2g41260), and oleosins genes that show reduced expression in developing seeds of the abi3 mutant (Parcy et al., 1994; Nambara et al., 1995; Crowe et al., 2000). For most of these genes, ABI3 may be sufficient in the vegetative context, although not strictly necessary in the seed context. This is presumably a reflection of the partial functional redundancy of ABI3 with the related FUS3 and LEC2 factors and other factors such as ABI4 in seeds (Keith et al., 1994; Parcy et al., 1997; Soderman et al., 2000; Stone et al., 2001).
Conversely, the GeneChip also includes several other known ABI3-regulated genes that were not detected at the 3-fold cutoff used in our analysis. These include the PAP10 (At2g16430) and M10 (At2g41280) genes. Ectopic activation of these genes may not have occurred for at least three reasons: (a) VP1/ABI3 may be necessary but not sufficient for seed-specific expression of some or all of these genes, possibly due to the functional redundancy with FUS3 and LEC2. (b) These genes may be regulated by a secondary cascade not detected on a 12-h time scale. (c) These genes may reveal non-conserved functional differences between VP1 and ABI3 (Suzuki et al., 2001). In any event, these markers define a distinct subclass of ABI3-dependent genes, revealing a new layer of complexity in VP1/ABI3-regulated gene expression.
VP1 Affects Key ABA-Signaling Intermediates
Our chip experiment revealed at least two classes of VP1/ABA-regulated genes that are directly implicated in ABA signaling. The first class includes positive regulators of ABA signaling, ABI5 (Finkelstein and Lynch, 2000) and related bZIP genes. The second class includes negative regulators of ABA signaling, ABI1 and ABI2 (Gosti et al., 1999; Merlot et al., 2001), and a related protein phosphatase 2C gene.
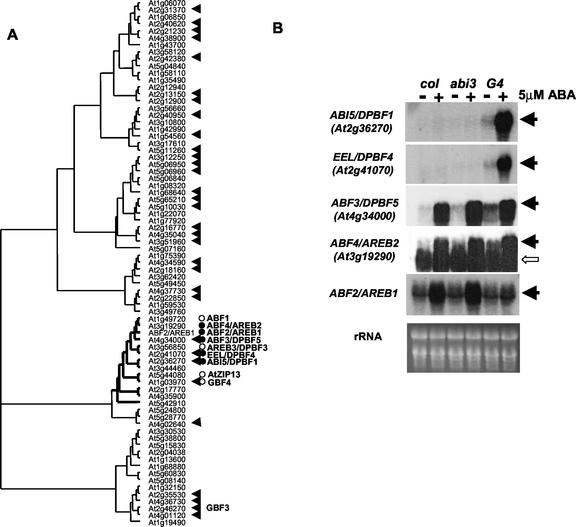
Four bZIP genes were responsive to VP1 and/or ABA in our chip experiment. To place the four bZIP genes, ABI5/DPBF1 (At2g36270), EEL/DPBF4 (At2g41070), ABF3/DPBF5 (At4g34000), and GBF3 (At2g46270), in the context of other bZIP genes in the Arabidopsis genome, we constructed a neighbor-joining tree of 75 bZIP domain proteins detected by psi-blast analysis of annotated Arabidopsis genes (Fig. 2A). The same number of Arabidopsis bZIPs were reported by Jakoby et al. (2002). At least 32 of the 75 bZIPs are represented on the GeneChip including representatives from each major clade. Three of the four VP1/ABA-regulated bZIPs (all except GBF3) belong to the ABI5 clade that includes a total of 13 genes. In addition to the bZIP domain, proteins in this subclass share a conserved N-terminal domain that is characteristic of this class (Hobo et al., 1999b; Bensmihen et al., 2002; Finkelstein and Lynch, 2000).
Figure 2.
Neighbor-joining tree of bZIP genes in Arabidopsis and expression of ABI5 and related genes. A, The tree was constructed by analysis of bZIP domains with ClustalW (Thompson et al., 1994). The bZIP genes represented on the GeneChip are marked by filled arrows. Bold lines denote 13 bZIP genes that form the ABI5 clade. Northern analysis was performed for all 13 genes. •, Genes responsive to VP1 and ABA. ○, Genes expressed in vegetative tissues but not responsive to either VP1 or ABA. B, Northern-blot analysis of ABI5-related genes in vegetative tissues of wild-type (Col), abi3-6, and 35S::VP1, abi3-6 transgenic Arabidopsis. Indicated probes were hybridized to 12.5 μg of total RNA prepared from aerial parts of 2-week-old seedlings treated with or without 5 μm ABA for 12 h. Black arrows, Positions of transcripts of each gene. White arrow, Signals of unknown identity.
To determine whether other genes in the ABI5 clade were also ABA and/or VP1 regulated, we conducted a northern-blot analysis of all 13 genes in the clade. In total, nine genes were expressed in vegetative tissue. Five of the nine genes (ABI5/DPBF1, EEL/DPBF4, ABF3/DPBF5, ABF4/AREB2, and ABF2/AREB1) were affected by VP1 and ABA, whereas the remaining four (AREB3/DPBF3, ABF1, GBF4, and At5g44080) were not affected by either treatment. These data confirmed and extended the results of the chip analysis (Fig. 2B). Although we cannot rule out VP1/ABA regulation of other bZIP genes that are not represented on the chip, the results imply that members of the ABI5 clade respond specifically to VP1 and ABA. The fourth bZIP gene detected by the microarray experiment, GBF3, was ABA-inducible but not regulated by VP1.
The expression analysis further revealed distinctive interactions of VP1 and ABA in regulation of bZIP genes. ABF3/DPBF5, ABF4/AREB2, and ABF2/AREB1 genes were induced by ABA in wild-type and abi3 mutant, whereas ABA induction of ABI5 and EEL required the 35S::VP1 transgene. In addition, ABA induction of ABF3/DPBF5, ABF4/AREB2, and ABF2/AREB1 was affected differentially by expression of VP1. ABF3/DPBF5 and ABF4/AREB2 showed a positive response to VP1, whereas ABA regulation ABF2/AREB1 was inhibited by VP1. The expression patterns of the bZIPs are consistent with the previous studies of transgenic Arabidopsis ectopically expressing ABI3 (Brocard et al., 2002). The functional conservation between ABI3 and VP1 suggests that direct downstream targets of the genes in the ABI5 clade are regulated similarly by ABI3 and VP1 in Arabidopsis. VP1 activation of the positive regulators of ABA signaling is consistent with the ABI3-mediated feed-forward pathway in a combinatorial regulatory circuit proposed by Finkelstein and colleagues (Finkelstein and Lynch, 2000; Soderman et al., 2000; Finkelstein et al., 2002).
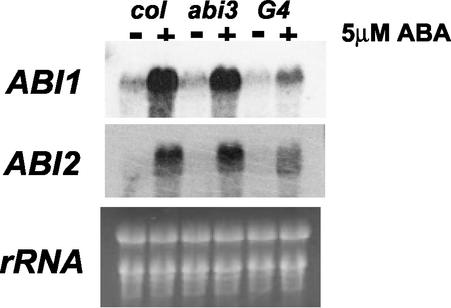
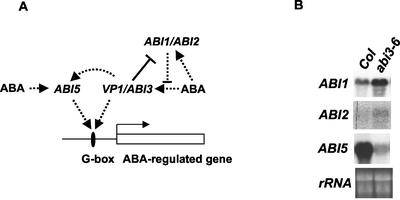
Five protein phosphatase 2C genes (ABI1, At4g26080; ABI2, At5g57050; AtP2C-HA, At1g72770, At4g31860, and At3g11410) were ABA inducible. Their induction was differentially affected by VP1 expression. Three of these genes (ABI1, ABI2, and AtP2C-HA) form a clade of closely related phosphatase 2C genes in Arabidopsis (Leung et al., 1994, 1997; Meyer et al., 1994; Rodriguez et al., 1998a, 1998b). ABI1 and ABI2 have genetically defined roles in ABA signaling as negative regulators (Finkelstein and Somerville, 1990; Gosti et al., 1999; Merlot et al., 2001). Northern analysis confirmed regulation of ABI1 and ABI2 genes (Fig. 3). ABA induction of ABI1 was significantly inhibited by VP1, consistent with expression pattern in our chip analysis. Although ABI2 regulation by VP1 was not detected by the microarray analysis (Supplemental Data 1), our northern analysis showed that VP1 expression significantly reduced ABA induction of ABI2 similar to ABI1. By inhibiting ABA induction of the negative regulators, ABI1 and ABI2, VP1 may potentiate a second independent positive feed-forward pathway (Fig. 4A). Although the present study focused on aerial portions of the plant, it is noteworthy that ectopic ABI3 and VP1 both confer hypersensitivity to ABA in roots (Parcy et al., 1994; Suzuki et al., 2001) Moreover, genetic studies have shown that the enhanced ABA sensitivity of roots is partially mediated by ABI1 (Parcy and Giraudat, 1997). Importantly, the regulation of ABI1 and ABI5 in vegetative tissues is consistent with effects of the abi3-6 mutant on expression of these genes in developing seeds (Fig. 4B), indicating that the implied positive feed-forward amplification of ABA signaling by VP1/ABI3 operates during seed development. Hence, VP1 regulation of ABI1 and ABI2 supports addition of a second branch to the proposed positive feed-forward regulation as a part of the combinatorial regulatory model (Soderman et al., 2000).
Figure 3.
Northern-blot analysis of ABI1 and ABI2 in vegetative tissues of wild-type (Col), abi3-6, and 35S::VP1, abi3-6 transgenic Arabidopsis. Indicated probes were hybridized to 12.5 μg of total RNA prepared from aerial parts of 2-week-old seedlings treated with or without 5 μm ABA for 12 h.
Figure 4.
Feed-forward regulation of ABA signaling mediated VP1 and ABI3. A, Feed-forward regulation of ABA signaling mediated by VP1 is mediated by the pathway proposed by Finkelstein and colleagues (Finkelstein and Lynch, 2000; Soderman et al., 2000). In addition, ABA induces two negative regulators of ABA signaling, ABI1 and ABI2, enabling feedback regulation of ABA signaling (Leung et al., 1997; Gosti et al., 1999; Merlot et al., 2001). These signaling pathways are shown with dotted lines. VP1 inhibits ABA induction of the negative regulators, potentiating further enhancement of ABA sensitivity (shown with bold line). B, Northern analysis of ABI1, ABI2, and ABI5 gene expression in maturing siliques of wild type (Col) and abi3-6. Total RNA samples were isolated from the late stage of developing siliques. Fifteen micrograms of the total RNA was hybridized with the indicated probes.
ABRE and Sph Elements Were Enriched in the Promoters of the VP1/ABA-Regulated Genes
A key question is how interactions of VP1 with ABA-signaling components give rise to the response classes we observe. We hypothesize that the classes are determined at least in part by combinatorial interactions of VP1 with an ensemble of ABA-regulated transcription factors, such as ABI5 and related bZIPs. In that case, it may be possible to discern patterns of conserved cis-elements in promoters of VP1/ABA-regulated genes that correlate with response classes. To test this idea, we performed a quantitative analysis of the promoter sequences (600 bp upstream of the annotated coding sequence) of the genes in two major classes that differ qualitatively in their dependence on VP1, the ABA-dependent-activated class (64 genes) and VP1 AND ABA-dependent-activated class (70 genes). Both classes respond to VP1 in the presence of ABA. We first constructed a database containing 600 nucleotide of 5′-flanking sequence of each annotated gene represented on the chip. Next, we constructed a motif dictionary comprising a complete nonredundant set of all possible 8-mer oligonucleotides that contain two degenerate bases (e.g. 43,168 sequences of form, acngtnct, excluding reverse complements as redundant). To identify the set of 8-mer motifs that are enriched in promoters of each response class, the frequency of each motif in the test set was compared with the frequency in a random set of 1,000 promoters selected from the promoter database. Statistical significance was evaluated by chi square with multiple copies in a promoter counting the same as a single copy. Using a cutoff P value of 10–4, we identified 197 motifs that are enriched in the VP1 AND ABA-dependent class and 41 motifs that are enriched in the ABA-dependent set. When mapped back onto the test promoters, the enriched motifs formed clusters over sequences that include consensus G-box-related ABREs as well as Sph-like elements that are potential binding sites for the B3 domain of VP1 (Supplemental Data 2). Detection of these known motifs confirmed the efficacy of this analysis. To simplify the analysis, we filtered the data to extract contiguous blocks of significant nucleotides that were eight bases or longer.
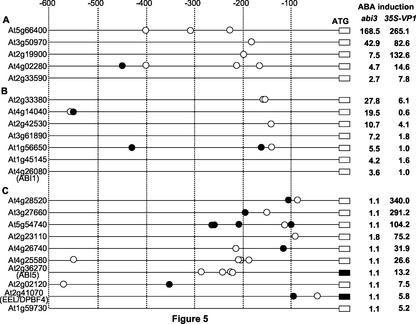
Table II shows that whereas ABRE and Sph elements are enriched in the promoters of both ABA-dependent and the VP1 AND ABA-dependent-activated classes, strong consensus Sph elements are significantly overrepresented in the VP1 AND ABA class (49 motifs/70 genes versus 16 motifs/62 genes). Forty percent of the promoters in this class possessed at least one copy of the consensus Sph element. The biased distribution of Sph motifs suggests that B3 DNA binding is a key determinant of the VP1 AND ABA-dependent class, although is not necessarily universally required within that class. Sph and ABRE motifis were identified and mapped on the promoters of the genes in the three classes. Locations of these motifs in the promoters of genes representing each class are shown in Figure 5, illustrating the enrichment of Sph motif in the VP1 AND ABA-dependent-activated class (Fig. 5, A and B versus C). With a few exceptions (e.g. At4g25580), the promoters lacking strong consensus Sph motifs (e.g. ABI5, At2g36270) were less strongly activated by ABA in 35S::VP1 plants. Overall, the density and proximity of the consensus elements to the putative transcription initiation sites were roughly correlated with the level of induction (Fig. 5). Because the transcription initiation sites are not annotated or known for the majority of genes analyzed, inaccurate mapping of the promoters may contribute to variation in this respect.
Table II.
The occurrence of Sph and ABRE sequences among the motifs enriched in the promoters of the ABA-dependent activated genes and VP1 AND ABA-dependent activated genes
| Sph
|
ABRE Total
|
ABREs
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| ACGTGTC | ACGTATC | ACGTTTC | ACGTGGC | ACGTGCC | ACGTGTA | ACGTGTT | |||
| ABA-dependent activated (62 genes)a | 16 (23%) | 47 (48%) | 10 (15%) | 3 (3%) | 0 (0%) | 10 (16%) | 1 (2%) | 3 (5%) | 20 (26%) |
| VP1 enhanced (15 genes) | 4 (27%) | 18* (67%) | 5 (33%) | 2 (7%) | 0 (0%) | 3 (20%) | 0 (0%) | 0 (0%) | 8 (40%) |
| VP1 suppressed (47 genes)a | 12 (21%) | 29 (43%) | 5 (9%) | 1 (2%) | 0 (0%) | 7 (15%) | 1 (2%) | 3 (6%) | 12 (21%) |
| VP1 AND ABA-dependent activated (70 genes) | 49*** (40%) | 93** (61%) | 33** (33%) | 7 (10%) | 0 (0%) | 23 (27%) | 1 (1%) | 11 (14%) | 18 (23%) |
Numbers of each element in indicated functional classes are shown in this table. Numbers in parentheses indicate frequency of promoters that possess indicated motif at least once. Asterisks indicate significant enrichment in indicated classes. Number of asterisks represents degree of probability by chi-square test. ***, P < 0.0003; **, P < 0.0019; *, P < 0.024.
Forty-seven of a total of 49 genes in the VP1 suppressed class were analyzed due to availability of the promoter sequences at the time of our analysis.
Figure 5.
Distribution of Sph and ABREs in the promoters of genes in the ABA-dependent-activated and VP1 AND ABA-dependent-activated classes. Twelve representative genes from the ABA-dependent-activated class (A, five from VP1 enhanced subclass; B, seven from VP1-suppressed subclass, respectively) and 10 representative genes from the VP1 AND ABA-dependent-activated class (C) were selected that span a range of ABA induction values. Location and distribution of Sph (CATGCA, •) and ABREs (Table II, ○) consensus motifs are shown in the 600-bp 5′ upstream region from annotated coding sequence of each gene. The 600-bp up-stream region of the known transcription start sites is shown for At2g36270 (ABI5) and At2g41070 (EEL/DPBF4).
ABRE sequences showed a similar enrichment in the VP1 AND ABA class relative to the ABA-dependent class (93 motifs/70 genes versus 47 motifs/62 genes). To discern potential patterns in the distribution of ABRE sequences, we analyzed the distributions of seven distinct ABRE variants functionally analyzed by Hattori et al. (2002). The variants contain an ACGT core motif and differ in flanking nucleotides. Six of the seven ABRE variants were detected as enriched motifs in ABA- and VP1-regulated promoters (Table II). Of these, three motifs (ACGTGTC, ACGTGGC, and ACGTGTT) were enriched in both the VP1 AND ABA class and the ABA-dependent class, implicating these motifs as potential targets of ABA signaling. Interestingly, one of the three motifs, ACGTGTC, was significantly enriched in the VP1 AND ABA class relative to the ABA-dependent class, whereas the others were evenly present between these classes. This result suggested that the ACGTGTC sequence might have a specific role in mediating interactions of VP1 and ABA signaling.
We identified 18 and 29 ABREs, respectively, in the VP1-enhanced subclass and VP1-suppressed subgroups of the ABA-dependent set, indicating significant enrichment of ABREs in the VP1 enhanced promoters. Hence, ABREs are positively correlated with positive regulation by VP1 within the set of ABA-activated genes.
DISCUSSION
Our oligomicroarray analysis reveals a complex interdependence between ABA signaling and VP1-regulated gene expression. Ectopic expression of VP1 confers ABA induction to a broad range of seed-specific genes in vegetative tissues. Moreover, altered ABA regulation of a large number of metabolism-related genes suggests a high potential for metabolic reprogramming. The analysis also reveals that VP1 and ABI3 share a potential to enhance ABA sensitivity in part through regulation of the ABI1/ABI2, appending another layer of feed-forward regulation of ABA signaling mediated by ABI3 (Soderman et al., 2000). ABI5-related bZIP proteins appear to specifically mediate the interaction of ABA signaling and VP1/ABI3 among all bZIPs. The analysis of promoters of genes in two major classes of the VP1/ABA-regulated genes identifies a subset of ABREs that are enriched elements in the 600-bp 5′ upstream region, further implicating ABI5-related bZIPs in their regulation. An asymmetric distribution of the Sph element in the two classes of coregulated genes indicates that B3 DNA-binding activity is an important determinant of VP1-mediated gene activation.
The microarray analysis revealed extensive VP1 regulation of known ABA-signaling components. VP1 activates ABI5 bZIP gene, a positive regulator of ABA signaling, in ABA-dependent manner. A similar response occurs in 35S::ABI3 transgenic Arabidopsis, leading to enhanced ABA sensitivity in vegetative tissues (Parcy et al., 1994; Finkelstein and Lynch, 2000). In this respect, VP1 and ABI3 function equivalently in a feed-forward pathway consistent with an enhancesome model (Soderman et al., 2000). In contrast to our results, other studies have reported that ABA alone can induce ABI5 in wild-type Arabidopsis (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2001; Kim et al., 2002). However, we note that the prior studies used either higher concentrations of ABA (50 or 100 μm versus 5 μm) or longer duration of ABA treatment with germinating seedlings (24 h with germinating seedlings versus 12 h with 2-week-old seedlings). Hence, the degree of VP1/ABI3 dependence of this response likely depends on ABA dosage and stage of plant growth. Despite the relatively low concentration of ABA in this study, strong activation by ABA treatment (as high as 800-fold) is detected in 35S::VP1 transgenic Arabidopsis (Supplemental Data 1). The dramatic effect is likely due to ABA hypersensitivity caused by overexpression of VP1 in vegetative tissues (Suzuki et al., 2001). The ABA hypersensitivity of ABI5 activation in 35S::VP1 might be a key mediator for the strong activation. It is noteworthy that overexpression of ABI5 enhances ABA sensitivity in germinating seeds and roots of seedlings (Lopez-Molina et al., 2001; Brocard et al., 2002).
Importantly, our results identify a second independent pathway of feed-forward regulation by VP1/ABI3 through repression of the two negative regulators of ABA signaling, ABI1 and ABI2. Both ABI1 and ABI2 are ABA inducible, consistent with the previous studies (Leung et al., 1997; Hoth et al., 2002). ABA induction of these genes is suppressed by VP1, suggesting that VP1 has a potential to suppress a negative feedback loop of ABA signaling. Consistent with that hypothesis, Gosti et al. (1999) and Merlot et al. (2001) have reported that loss-of-function or reduction-of-function mutations of ABI1 and ABI2, respectively, cause hypersensitivity to ABA at various developmental stages including roots. Overexpressed ABI3 and VP1 also cause hypersensitivity to ABA in Arabidopsis roots (Parcy and Giraudat, 1997; Suzuki et al., 2001) and that response is partly mediated by ABI1 (Parcy and Giraudat, 1997). These results suggest that feed-forward regulation mediated by VP1 through repression of ABI1 and ABI2 may be operating in roots. Our finding that transcript levels of ABI1 and ABI2 are elevated in abi3-6 seeds compared with Col wild type, suggests that this pathway is also active in normal seed development. Recently, Hoth et al. (2002) have reported genes possibly regulated by ABI1 in ABA signaling. These genes include AtHB-7 and AtHB-12, both of which belong to ABA-activated/VP1-suppressed class in our study. Interestingly, ABI1 and ABI2 are also members of ABA-activated/VP1-suppressed class, which makes up the second largest class of the VP1/ABA-regulated genes. VP1 functions as an activator or a repressor depending on the target gene (McCarty et al., 1991; Hattori et al., 1992; Hoecker et al., 1995; Nambara et al., 1995). An important although previously unrecognized role for the repressor function of VP1 may be to shut down part of ABA-induced expression. Both activator and repressor functions contribute to feed-forward regulation of ABA signaling through regulation of the ABI5 and ABI1/ABI2 gene families, respectively. Hence AtHB-7 and AtHB-12 may have a role in regulation of ABI1/ABI2 expression. Interaction of repression function of VP1 with the homeoprotein genes may affect their expression, altering ABA sensitivity including stomata response to ABA in 35S::ABI3 (Parcy and Giraudat, 1997).
The feed-forward regulation by VP1 through ABI5 and ABI1/ABI2 is likely to be just a part of complex combinatorial regulation of ABA signaling (Soderman et al., 2000). EEL/AtDPBF4 is regulated by ABA and VP1 in a manner similar to ABI5. Bensmihen et al. (2002) have proposed that EEL inhibits full activation by ABI5 by competing for the same G-box element-binding site of AtEm1 gene. Hence, EEL/AtDPBF4 may be involved in VP1-mediated inhibition of ABA signaling by antagonistically interacting with ABI5. In another words, EEL/AtDPBF4 may even suppress the ABI5-mediated feed-forward regulation of ABA signaling. Although the eel mutant does not affect ABA sensitivity during seed germination, this may be due to functional redundancy of EEL/AtDPBF4 with other ABI5-related genes or/and little expression of the gene at late stage of seed development (Bensmihen et al., 2002). Interestingly, overexpression of ABI4 also enhances ABA induction of ABI5 and EEL/AtDPBF4 (Soderman et al., 2000; Brocard et al., 2002), suggesting that VP1/ABI3 and ABI4 may balance ABA sensitivity for induction of some genes such as AtEm1 through regulation of the ABI5-related bZIPs.
The capacity to interact with ABA-signaling components in Arabidopsis is evidently highly conserved in maize VP1. Like Arabidopsis ABI3, VP1 regulates other bZIPs related to ABI5. The Arabidopsis genome contains at least nine bZIPs closely related to ABI5 (Bensmihen et al., 2002). Four of these genes (ABF1, ABF2/AREB1, ABF3/DPBF5, and ABF4/AREB2) have been implicated in ABA signaling (Choi et al., 2000; Uno et al., 2000; Finkelstein et al., 2002; Kang et al., 2002). A rice homolog, TRAB1, physically interacts with OsVP1 and mediates ABA signaling. TRAB1 is more closely related to ABF/AREB subfamily than to ABI5 (Hobo et al., 1999b; Bensmihen et al., 2002), suggesting that VP1/ABI3 may physically interact with other ABI5-related proteins in Arabidopsis as well. An abi5 null mutant is milder in phenotype than null abi3 mutants (Finkelstein, 1994; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Finkelstein et al., 2002). These results strongly suggest that other ABI5-related bZIPs play major roles in interaction of ABI3/VP1 and ABA signaling (Finkelstein et al., 2002). The capacity of VP1 to regulate all of the ABI5-related genes in similar manner with ABI3 indicates strong functional conservation between VP1 and ABI3 at a level of transcriptional regulation of key ABA-signaling intermediates. Moreover, we find that the clade of ABI5-related genes is uniquely affected by VP1. The result reinforces the notion that ABI5-related bZIPs are key factors for the interaction of ABI3/VP1 and ABA signaling.
The regulation of the ABI5-clade bZIPs by VP1 may contribute to the complexity of responses that we observe at 12 h. G-box-related ABREs have been broadly implicated in ABA-regulated gene expression. Our analysis of the promoters of VP1/ABA-regulated genes clearly identified ABRE elements as highly enriched elements. Because the ABI5-related proteins are known to bind ABRE sequences (Choi et al., 2000; Uno et al., 2000; Bensmihen et al., 2002; Kim et al., 2002; Carles et al., 2002), many of the ABA response genes could also be affected by regulation of ABI5-related genes as a secondary cascade. Genes in the VP1 AND ABA-dependent class are strictly regulated by concerted action of VP1 and ABA. Two potential downstream factors, ABI5/DPBF1 and EEL/DPBF4, are themselves members of the VP1 AND ABA-dependent class. Hence, these genes may contribute to specification of this class. On the other hand, the ABF3/DPBF5 and ABF4/AREB2 bZIPs are members of the ABA-dependent class. Because both genes are ABA-inducible in the absence of VP1, their induction conceivably would condition a secondary response via protein-protein interaction with VP1. Conversely, bZIPs that are constitutively expressed, such as ABF1 and AREB3/DPBF3, encode candidate interacting proteins capable of mediating primary effects of VP1.
Ectopic induction of seed-specific genes in vegetative tissues supports the idea that ABI3/VP1 is a key determinant of seed-specific gene expression (Parcy et al., 1994; Li et al., 1999; Suzuki et al., 2001). However, in this role, VP1/ABI3 activates genes in concert with interacting factors. Hence, the patterns of ectopic gene regulation may correlate with the presence of interacting factors. Bensmihen et al. (2002) report that ABI5 and the six related genes are expressed in developing siliques. Six of these genes including ABI5 show constitutive or inducible expression in vegetative tissues (Brocard et al., 2002; this study). This high degree of overlap may be one reason that ectopic activation of seed-specific genes shows a striking dependence on ABI3/VP1—for the most part the interacting factors are not limiting in vegetative cells. Conversely, expression of a seventh gene, AtbZIP67 (At3g44460), is not detected in vegetative tissues under any conditions (Brocard et al., 2002; this study). Thus, AtbZIP67 is a candidate-limiting determinant for expression of a handful of ABI3-regulated seed genes (e.g. M10 and PAP10) that are not ectopically activated by 35S::VP1. We cannot rule out the possibility that maize VP1 is incompletely interchangeable with Arabidopsis ABI3 (Suzuki et al., 2001), in this respect.
The present analysis also sheds light on VP1 and ABA functions in other developmental contexts. The anthocyanin 2 (AN2), dihydroflavonol 4-reductase (DFR), and leucoanthocyanidin dioxygenase (LDOX) genes, all of which are involved in flavonoid biosynthesis (Shirley et al., 1992; Huits et al., 1994), are responsive to VP1 and ABA (At1g56650, At5g42800, and At4g22880, respectively; Supplemental Data 1). The petunia AN2 gene has been shown to be a positive regulator for the DFR expression (Quattrocchio et al., 1999). Although direct binding of AN2 MYB proteins to the promoter of the DFR or LDOX has not been demonstrated, our data are consistent with the possibility that ABA induction of the AN2-like gene contributes to activation of the DFR and LDOX. In line with this view, overexpression of VP1 inhibits ABA induction of the AN2-like gene, coincident with reduced ABA activation of DFR and LDOX.
Although VP1/ABI3 has been thought to be a seed-specific factor, several studies reveal that ABI3-related factors have roles in vegetative tissue, especially in regulation of meristem activity. An ABI3 promoter-GUS fusion gene is expressed in meristems (Rohde et al., 1999), and in poplar (Populus spp.), an ABI3 homolog is implicated in arrest of bud development (Rohde et al., 2002). The abi3 mutant has an early flowering phenotype (Kurup et al., 2000). 35S::VP1 fully complements the early flowering phenotype in Arabidopsis, and overexpression delays time of flowering (Suzuki et al., 2001). Interestingly, in our chip experiment, VP1 and ABA together repress Twin Sister of FT, a positive regulator for flowering (Kardailsky et al., 1999; Kobayashi et al., 1999), indicating that VP1 and ABA are capable of regulating flowering timing through Twin Sister of FT.
Plant adaptation to cold is also partly mediated by ABA. Fowler and Thomashow (2002) have recently reported transcriptome analysis of cold acclimation in Arabidopsis. They identified five genes encoding structurally related AP2 domain proteins (RAP2) regulated by cold. Two of these genes, RAP2.1 and RAP2.6, are up-regulated by C-repeat/dehydration-responsive element-binding factor (CBF). They have proposed a subregulon of CBF-regulated genes mediated by RAP2.1 and RAP2.6. In our analysis, both genes are activated by ABA, suggesting that these proteins potentially integrate CBF and ABA signaling. In contrast to RAP2.1 and RAP2.6, the RAP2.9 gene is down-regulated by ABA, whereas it is activated by cold treatment. Interestingly, repression of RAP2.9 by ABA is completely abolished in 35S::VP1 plants, indicating that VP1 has a potential to alter the cold response as well as ABA signaling.
Several of the affected metabolic pathways potentially alter hormone and sugar signaling in 35S::VP1 plants. Notably, expression of GA4 (At1g15550), which encodes a 3β-hydroxylase for gibberellin biosynthesis (Yamaguchi et al., 1998), was activated 8-fold by VP1 and ABA. Because GA is well known as an antagonist of ABA signaling in several contexts (for review, see Bethke et al., 1997; White et al., 2000), activation of GA4 and GA synthesis is a potential feedback mechanism maintaining a balance of hormonal responses between ABA and GA. If so, it is intriguing that this response is dependent on VP1. Whereas feedback regulation of GA4 by GA is known, ABA has not been implicated in this response; nor is it known whether ABI3 has a similar potential for regulation of GA synthesis in the seed. We have previously shown that ABA and VP1 have a potential to interact with auxin-regulated gene expression and development (Suzuki et al., 2001). In our chip experiment, VP1 and ABA affected expression of several auxin-regulated genes (e.g. GH3-like gene; At4g27260), possibly altering development. Another intriguing example is the activation of trehalose-6-phosphatase (At4g12430) by VP1 and ABA. Trehalose-6-phosphatase is in the trehalose biosynthesis pathway. Although plants accumulate minute amounts of trehalose, recent studies have revealed an essential role of trehalose synthesis in embryo development in Arabidopsis (Eastmond et al., 2002). VP1-dependent activation of this gene in vegetative cells suggests that ABI3 may affect regulation of trehalose synthesis in seeds. We cannot rule out the possibility that overexpression of VP1 in vegetative tissues causes ectopic interactions leading to changes in expression of genes that are not regulated by VP1/ABI3 in seeds. Therefore, these intriguing possibilities require further confirmation and functional tests.
The picture that emerges from these data is that the complex hierarchy of VP1 and ABA response classes shown in Figure 1 arises from a combination of (a) primary interactions between VP1- and ABA-regulated transcription factors and (b) secondary interactions induced by feed-forward regulation of ABA-signaling components. Because VP1 and ABI3 directly regulate their interaction partners, there is not necessarily a clear boundary between primary and secondary gene regulation. However, we predict that ultimately the specificity of these interactions must be resolved by cis-acting regulatory sequences of affected genes. Our quantitative analysis of enriched sequence motifs that distinguish two classes of VP1-regulated promoters reveals several patterns as potential determinants of VP1 action. The pronounced enrichment of Sph elements in promoters of VP1 AND ABA-activated class relative to the ABA-dependent-activated class suggests that DNA contacts of the B3 domain are an important determinant of the former class. Moreover, within the VP1 AND ABA-dependent class we observe a consistent pairing of Sph elements with ABRE motifs in the highly activated genes. However, presence of strong consensus Sph elements is not a universal requirement in this class especially in the less strongly activated genes within that class.
Three ABRE sequences (ACGTGTC, ACGTGGC, and ACGTGTT) are highly enriched in both the VP1 AND ABA-dependent and ABA-dependent classes. Two of these elements, ACGTGTC and ACGTGGC, correspond to sequences that have the highest activity of mediating ABA signaling in a functional analysis of the Osem ABRE (Hattori et al., 2002). Interestingly, one of the ABRE motifs (ACGTGTC) shows significant enrichment in the VP1 AND ABA class relative to the ABA-dependent class. Hence, variants of the core ABRE motif may discriminate binding of ABA-signaling components that are capable of mediating positive interactions with VP1.
Although our analysis identified candidates for determinants of VP1 activation, we find less evidence for specific determinants of VP1-mediated repression (Fig. 5; Supplemental Data 2). We find substantially less evidence of enriched motifs shared within groups of negatively regulated promoters (data not shown). For instance, we did not detect any significant enriched element in ABI1 promoter (At4g26080). There are at least three possibilities: (a) Repression is the default condition mediated by physical interactions that have very broad specificity (e.g. chromatinbased repression), whereas activation requires specific determinants. (b) Repression is mediated by specific interactions that occur off of the DNA. (c) Specific determinants of repression are localized elsewhere in the gene, i.e. in introns or 3′ to the coding sequence. In addition to known VP1 and ABRE response elements, our analysis reveals several other enriched motifs that may contribute to specification of this class (data not shown; see Supplemental Data 2). Confirmation of the biological relevance of these candidate sequences will require functional testing.
MATERIALS AND METHODS
Plant Growth
Seeds of Col, abi3-6, and 35S::VP1 (G4; Suzuki et al., 2001) were sterilized and sown on plates containing germination media (Huang and Ma, 1992). Seedlings were grown for 12 d at 22°C under continuous light. The seedlings were transferred to plates containing media with or without 5 μm ABA (catalog no. A-1049, Sigma-Aldrich, St. Louis). The treated seedlings were grown for 12 h under continuous light and then harvested for the total RNA isolation.
Total RNA Preparation
The total RNA isolation was prepared independently from two replicate experiments using the RNeasy Plant mini kit (Qiagen USA, Valencia, CA). The sampled tissues included all aerial vegetative parts. The total RNA from the siliques was isolated as described (Chang et al., 1993).
DNA Microarray Expression Analysis
A sample containing 8 μg of the total RNA was used for the cDNA synthesis. The cDNA synthesis and the in vitro transcription reactions were performed according to manufacturer's instructions (Affymetrix; Enzo Biochem, New York). The hybridization to the Arabidopsis GeneChip (Affymetrix) was performed at the University of Florida Interdisciplinary Center for Biotechnology Research Core facility. Elements with an absolute difference above 1,000 in at least one treatment were chosen for the further analysis. The complete raw dataset is available for download by email request to drm@ufl.edu in the form of a MySQL database.
Northern-Blot Analysis
The total RNA was prepared from wild-type (Col), abi3, and 35S::VP1 seedlings for expression analysis for ABI1, ABI2, and ABI5-related genes. The 12.5 μg of the total RNA was resolved in a 1.2% (w/v) agarose gel and was transferred onto a nylon membrane. Hybridization was performed as previously described (Suzuki et al., 2001). The probes for the northern analysis were prepared by PCR. To obtain gene-specific probes, divergent regions of the genes were chosen for the PCR amplification. The primer sequences for each specific probe are listed in Supplemental Data 3. The amplified DNA fragments were cloned into pCR4-TOPO (Invitrogen, Carlsbad, CA) and sequenced for confirmation before probe preparations.
Statistical Analysis of the Promoters of the VP1/ABA-Regulated Genes
We extracted 5′ upstream sequences (600 nucleotide upstream of the annotated coding sequence) of almost all of the 7,402 nuclear genes on the Affymetrix GeneChip by automated parsing of the XML format chromosome assemblies (http://www.tigr.org). We then constructed a dictionary containing a complete, nonredundant set of 8-mer sequences containing two degenerate bases. The frequency of each 8-mer was compared between a control set of 1,000 randomly extracted promoters and a set of coregulated promoters (ABA-dependent-activated class and VP1 AND ABA-dependent-activated class) using a simple chi-square test. Eight-mer motifs with P < 0.0001 were chosen as enriched motifs in each coregulated class. To compare the enriched motifs between the two classes, each motif was mapped on the promoters of the genes from both classes. Motifs from ABA-dependent class were labeled in blue, and those from VP1 AND ABA-dependent class were labeled in orange (Supplemental Data 2). Motifs that were identified from both classes were consequently labeled in purple. The Sph and ABREs were searched and counted if they were identified in a stretch of at least eight contiguous significant bases.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Dr. Michael Popp and Joint Shands Cancer Center-Interdisciplinary Center for Biotechnology Research at University of Florida for assistance with performing the microarray experiment. We also thank Dr. Eiji Nambara (Riken Institute, Yokohama) for abi3-6 mutant seeds.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022475.
This work was supported by the National Science Foundation (grant no. 0080175 to D.R.M.) and by the Florida Agricultural Experiment Station (journal series R-09523).
The online version of this article contains Web-only data. The supplemental material is available at http://www.plantphysiol.org.
References
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14: 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Schuurink R, Jones RL (1997) Hormonal signalling in cereal aleurone. J Exp Bot 48: 1337–1356 [Google Scholar]
- Bobb AJ, Eiben HG, Bustos MM (1995) PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. Plant J 8: 331–343 [DOI] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol 131: 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Leon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Carson CB, Hattori T, Rosenkrans L, Vasil V, Vasil IK, Peterson PA, McCarty DR (1997) The quiescent/colorless alleles of viviparous1 show that the conserved B3 domain of VP1 is not essential for ABA-regulated gene expression in the seed. Plant J 12: 1231–1240 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Bartels D (1997) Structure and function of the vp1 gene homologue from the resurrection plant Craterostigma plantagineum Hochst. Mol Gen Genet 256: 539–546 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Crowe AJ, Abenes M, Plant A, Moloney MM (2000) The seed-specific transactivator, ABI3, induces oleosin gene expression. Plant Sci 151: 171–181 [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJ, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JD, Smeekens SC, Graham IA (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L (2000) Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J 24: 57–66 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765–771 [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville C (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94: 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Zhang C, Chen X, Gong Z, Zhu J-K (2002) Constitutive activation and transgenic evaluation of the function of an Arabidopsis PKS protein kinase. J Biol Chem 277: 42088–42096 [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu J-K (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna ST (1994) Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol Biol 24: 805–810 [DOI] [PubMed] [Google Scholar]
- Hattori T, Terada T, Hamasuna S (1995) Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J 7: 913–925 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK (1992) The Viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Hobo T, Asada M, Kowyama Y, Hattori T (1999a) ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J 19: 679–689 [DOI] [PubMed] [Google Scholar]
- Hobo T, Kowyama Y, Hattori T (1999b) A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc Natl Acad Sci USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR (1995) Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes Dev 9: 2459–2469 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1–1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Huang H, Ma H (1992) An improved procedure for transforming Arabidopsis thaliana (Landsberg erecta). Plant Mol Biol Rep 10: 372–383 [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Huits HS, Gerats AG, Kreike MM, Mol JN, Koes RE (1994) Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J 6: 295–310 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK (2002) The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130: 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hobo T, Murata M, Ban A, Hattori T (2002) Abscisic acidinduced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 14: 3177–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ma J, Perret P, Li Z, Thomas TL (2002) Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol 130: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis. Physiol Plant 6: 377–383 [Google Scholar]
- Kurkela S, Franck M (1990) Cloning and characterization of a cold- and ABA-inducible Arabidopsis gene. Plant Mol Biol 15: 137–144 [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones H, Holdsworth M (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Bishop KJ, Chandrasekharan MB, Hall TC (1999) β-Phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc Natl Acad Sci USA 96: 7104–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chandrasekharan MB, Wolffe AP, Hall TC (2001) Chromatin structure and phaseolin gene regulation. Plant Mol Biol 46: 121–129 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G, Paul AL, McCarty DR, Ferl RJ (1996) Transcription factor veracity: Is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Marcotte WR Jr, Russell SH, Quatrano RS (1989) Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell 1: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte WR Jr, Quatrano RS (1993) ABA-regulated gene expression: cisacting sequences. In TJ Close, EA Bray, eds, Plant Response to Cellular Dehydration during Environmental Stresses. American Society of Plant Physiologists, Rockville, MD, pp 185–192
- McCarty DR, Carson CB, Stinard PS, Robertson DS (1989) Molecular analysis of viviparous-1: an abscisic acid-insensitive mutant of maize. Plant Cell 1: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258: 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87: 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S (2000) The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220: 412–423 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121: 629–636 [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1994) Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol 35: 509–513 [PubMed] [Google Scholar]
- Nambara E, Naito S, McCourt P (1992) A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J 2: 435–441 [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P (2002) A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Giraudat J (1997) Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J 11: 693–702 [DOI] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R (1999) Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E (1998a) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Leube MP, Grill E (1998b) Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol Biol 38: 879–883 [DOI] [PubMed] [Google Scholar]
- Rohde A, Ardiles-Diaz W, Van Montagu M, Boerjan W (1998) Isolation and expression analysis of an ABSCISIC ACID-INSENSITIVE 3 (AB13) homologue from Populus trichocarpa. J Exp Bot 49: 1059–1060 [Google Scholar]
- Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W (2002) PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14: 1885–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Van Montagu M, Boerjan W (1999) The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ 22: 261–270 [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 301. [DOI] [PubMed] [Google Scholar]
- Shen Q, Zhang P, Ho TH (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8: 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota H, Satoh R, Watabe K, Harada H, Kamada H (1998) C-ABI3, the carrot homologue of the Arabidopsis ABI3, is expressed during both zygotic and somatic embryogenesis and functions in the regulation of embryo-specific ABA-inducible genes. Plant Cell Physiol 39: 1184–1193 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Hanley S, Goodman HM (1992) Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 4: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, Cocciolone S, McCarty DR (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28: 409–418 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil V, Marcotte WR Jr, Rosenkrans L, Cocciolone SM, Vasil IK, Quatrano RS, McCarty DR (1995) Overlap of Viviparous1 (VP1) and abscisic acid response elements in the Em promoter: G-box elements are sufficient but not necessary for VP1 transactivation. Plant Cell 7: 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Bies-Etheve N, Delseny M (2000) Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J Exp Bot 51: 995–1003 [DOI] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CN, Proebsting WM, Hedden P, Rivin CJ (2000) Gibberellins and seed development in maize: I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol 122: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001a) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1: 771–781 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee BH, Ishitani M, Lee H, Zhang C, Zhu JK (2001b) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (1998) Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993a) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236: 331–340 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1993b) The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet 238: 17–25 [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.