Abstract
Plants are known to synthesize ethanolamine (Etn) moieties by decarboxylation of free serine (Ser), but there is also some evidence for phosphatidyl-Ser (Ptd-Ser) decarboxylation. Database searches identified diverse plant cDNAs and an Arabidopsis gene encoding 50-kD proteins homologous to yeast (Saccharomyces cerevisiae) and mammalian mitochondrial Ptd-Ser decarboxylases (PSDs). Like the latter, the plant proteins have putative mitochondrial targeting and inner membrane sorting sequences and contain near the C terminus a Glycine-Serine-Threonine motif corresponding to the site of proteolysis and catalytic pyruvoyl residue formation. A truncated tomato (Lycopersicon esculentum) cDNA lacking the targeting sequence and a chimeric construct in which the targeting and sorting sequences were replaced by those from yeast PSD1 both complemented the Etn requirement of a yeast psd1 psd2 mutant, and PSD activity was detected in the mitochondria of the complemented cells. Immunoblot analysis of potato (Solanum tuberosum) mitochondria demonstrated that PSD is located in mitochondrial membranes, and mRNA analysis in Arabidopsis showed that the mitochondrial PSD gene is expressed at low levels throughout the plant. An Arabidopsis knockup mutant grew normally but had 6- to 13-fold more mitochondrial PSD mRNA and 9-fold more mitochondrial PSD activity. Total membrane PSD activity was, however, unchanged in the mutant, showing mitochondrial activity to be a minor part of the total. These results establish that plants can synthesize Etn moieties via a phospholipid pathway and have both mitochondrial and extramitochondrial PSDs. They also indicate that mitochondrial PSD is an important housekeeping enzyme whose expression is strongly regulated at the transcriptional level.
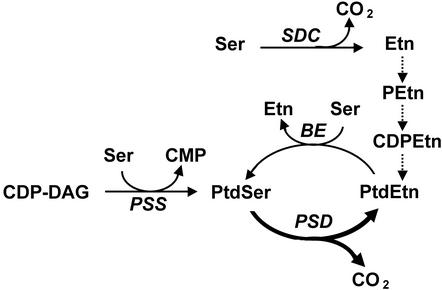
Ethanolamine (Etn) moieties can be synthesized by at least two routes in plants (Mudd and Datko, 1989). Currently, the most completely characterized route is decarboxylation of free Ser via Ser decarboxylase, a soluble enzyme that has been cloned from Arabidopsis and rapeseed (Brassica napus; Rontein et al., 2001; Fig. 1). A second possible route involves decarboxylation of phosphatidyl-Ser (Ptd-Ser) by a Ptd-Ser decarboxylase (PSD), yielding phosphatidylethanolamine (PtdEtn; Fig. 1). This latter pathway is well documented in yeast (Saccharomyces cerevisiae) and mammals, and the PSDs responsible have been shown to be membrane proteins (Voelker, 1997, 2000). In contrast, the presence of PSD in plants has not been clearly demonstrated. The only biochemical evidence comes from early reports of PSD activity in spinach (Spinacia oleracea) leaf extracts (Marshall and Kates, 1973, 1974). However, the activity was mainly in the soluble fraction rather than membranes, and the reaction product was not proven to be PtdEtn. These results could therefore be due to artifactual decarboxylation of the type that occurs in soluble plant extracts with Ser and other amino acids (Birecka et al., 1985; Rontein et al., 2001). The results of radiotracer kinetic studies with plant tissues fed [U-14C]Ser are consistent with the presence of PSD activity but do not unambiguously establish its existence (Marshall and Kates, 1974; Kinney and Moore, 1987a; Mudd and Datko, 1989).
Figure 1.
The origins of Etn moieties in plant phospholipids. Dotted arrows show the Kennedy pathway, which proceeds from Etn to PtdEtn via phospho and CDP derivatives of Etn. CDP-DAG, CDP diacylglycerol; CMP, cytidine 5′-phosphate; P, phospho; Ptd, phosphatidyl; SDC, Ser decarboxylase; PSD, Ptd-Ser decarboxylase; PSS, Ptd-Ser synthase; BE, base exchange. Note that when PSD and base exchange act together, they constitute a cycle.
Plant membranes are known to contain small amounts of Ptd-Ser (Mudd, 1980), and two different plant enzymes that synthesize Ptd-Ser have been identified. Ptd-Ser synthase has been cloned from wheat (Triticum aestivum; Delhaize et al., 1999) providing strong evidence that the yeast-type pathway (Carman and Zeimetz, 1996) for Ptd-Ser synthesis exists in plant tissues. Also, a Ser base-exchange reaction has been detected in castor bean (Ricinus communis) and leek (Allium porrum; Moore, 1975; Vincent et al., 1999), suggesting that the mammalian-type pathway (Kuge and Nishijima, 1997) occurs in plants. The base-exchange reaction requires Ser and PtdEtn as substrates. The PtdEtn could in principle be formed by PSD, but it could also come from Etn via phospho-Etn and CDP-Etn (the Kennedy pathway) because the enzyme activities required all occur in plants (Macher and Mudd, 1974, 1976; Tang and Moore, 1997; Fig. 1).
PSDs have been cloned from bacteria (Li and Dowhan, 1988; Matsumoto et al., 1998), yeast (Clancey et al., 1993; Trotter et al., 1993, 1995), and mammalian cells (Kuge et al., 1991). All of these PSDs belong to the group of pyruvoyl-dependent decarboxylase enzymes, which are autocatalytically cleaved at an internal Gly-Ser bond near the C terminus into α- and β-chains (Li and Dowhan, 1988). The Ser residue that gives rise to the pyruvoyl moiety is located in the conserved motif Glycine-Serine-Threonine (GST). Yeast PSD1 and Chinese hamster (Cricetulus griseus) PSD are mitochondrial proteins whose N-terminal regions have typical targeting peptides followed by an inner membrane sorting sequence. Yeast has a second PSD protein (PSD2) that is targeted to the Golgi/vacuole membrane (Trotter et al., 1995; Voelker, 1997). Double mutant psd1 psd2 yeast strains are Etn auxotrophs and have no detectable PSD activity (Trotter and Voelker, 1995). It has recently been shown that a critical minimum level of PtdEtn is required for yeast growth, although the biochemical basis for this requirement is not yet clear (Birner et al., 2001; Storey et al., 2001).
In view of the uncertain status of plant PSD, we first searched plant DNA databases for homologs of yeast and Chinese hamster PSDs. Having found a tomato (Lycopersicon esculentum) cDNA and an Arabidopsis gene encoding putative mitochondrial PSD proteins, we authenticated the former by complementation of a yeast psd1 psd2 mutant and by measuring PSD activity in complemented yeast cells. We further showed that the plant PSD protein is located in mitochondrial membranes and that its mRNA is expressed throughout the plant. Lastly, an Arabidopsis knockup mutant was used to show that mitochondrial PSD is active in planta and is subject to strong transcriptional control.
RESULTS
Identification of Plant PSD Homologs
Searches of the GenBank expressed sequence tag (EST) database identified a tomato cDNA (BE451597) encoding a protein homologous to mitochondrial PSDs from yeast and Chinese hamster. Complete sequencing of this cDNA demonstrated that the encoded 445-residue protein is approximately 37% identical to the yeast and hamster enzymes and has the characteristic GST motif (containing the Ser precursor of the active site pyruvoyl residue) close to the C terminus (Fig. 2). Like its yeast and mammalian counterparts, the N terminus of the deduced tomato protein has the features of a mitochondrial targeting peptide followed by an inner membrane sorting sequence (Fig. 2). The tomato protein was accordingly designated LePSD1. Searching GenBank with the LePSD1 sequence revealed homologous ESTs from nine other plants (including two monocots and a gymnosperm), and an Arabidopsis gene (At4g16700). The conceptual translation product of this gene (accession no. NP193403) lacks the C terminus due to a gene-prediction error but this region is present in the nucleotide sequence. The Arabidopsis cDNA encoding the NP193403 protein was cloned by reverse transcriptase (RT)-PCR, and sequenced. The deduced protein (AtPSD1) shows 63% identity with LePSD1 and diverges most from it at the N-terminal region, which is consistent with this being a mitochondrial targeting peptide (Fig. 2).
Figure 2.
Alignment of the tomato and Arabidopsis PSD homologs with yeast and Chinese hamster mitochondrial PSDs. Identical residues are shaded in black, similar residues are shaded in gray. Dashes are gaps introduced to maximize alignment. The arrowhead marks the conserved Ser residue implicated in autocatalytic cleavage into α- and β-subunits, and pyruvoyl prosthetic group formation. The bar indicates a hydrophobic inner membrane sorting sequence in the tomato and Arabidopsis proteins. LePSD1, tomato PSD1 (GenBank accession no. AY093689); AtPSD1, Arabidopsis PSD1 (AY189805); ScPSD1, yeast PSD1 (L20973); CgPSD, Chinese hamster mitochondrial PSD (M62722). The diamond shows where the LePSD1 sequence was truncated for expression in yeast, and the asterisk shows the junction between the parts of the chimeric yeast-plant enzyme.
Complementation of a Yeast psd1 psd2 Mutant
To test whether the LePSD1 protein is functional, full-length, truncated, and chimeric constructs were subcloned into the expression vector pVT103-U and introduced into yeast strain RYY51. This strain is a psd1 psd2 double disruptant that lacks PSD activity and is auxotrophic for Etn (Trotter and Voelker, 1995). The full-length LePSD1 was tested because some plant mitochondrial inner membrane proteins are correctly targeted and processed in yeast (Hamel et al., 1997). The truncated LePSD1 was constructed to delete residues 1 to 54, corresponding to the predicted mitochondrial targeting peptide (Fig. 2, diamond). In the chimeric construct, overlap extension PCR (Horton et al., 1993) was used to replace residues 1 to 117 of the plant protein with residues 1 to 139 of yeast PSD1 (Fig. 2, asterisk). This maneuver exchanged the putative mitochondrial targeting and inner membrane sorting sequences of LePSD1 for those of yeast.
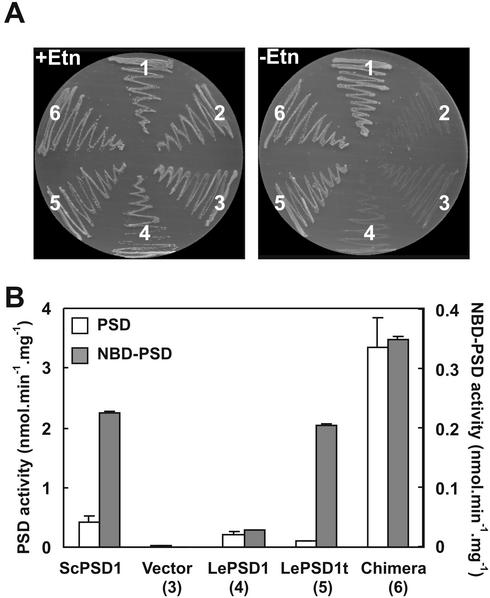
The chimeric and truncated constructs yielded Etn-independent transformants of the yeast mutant with high frequency, and their growth was similar to that of the wild-type strain (Fig. 3A). No complementation was observed with full-length LePSD1 or, as expected, with the vector alone (Fig. 3A). Retransformation of RYY51 with the chimeric and truncated constructs rescued from complemented cells restored Etn prototrophy, showing that complementation was due to the encoded plant protein.
Figure 3.
Complementation of a yeast psd1 psd2 mutant by LePSD1 constructs and PSD activities in complemented cells. A, Cells of wild-type yeast strain SEY6210 (segment 1), the psd1 psd2 mutant RYY51 (segment 2), RYY51 transformed with pVT103-U alone (segment 3) or containing full-length LePSD (segment 4), truncated LePSD (segment 5), and chimeric yeast PSD1/LePSD1 (segment 6) were plated on synthetic minimal medium plus or minus 5 mm Etn. B, PSD activities in homogenates of RYY51 cells harboring YCp50-PSD1 (ScPSD1), RYY51 cells complemented with pVT103-U alone (vector) or carrying full-length LePSD1, truncated LePSD1 (LePSD1t), and chimeric yeast PSD1/LePSD1 (chimera). Numbers in parentheses beneath the labels correspond to the segments in A. Cells were grown on minimal medium supplemented with Etn. White bars, PSD activity measured with Ptd[1′-14C]Ser; shaded bars, activity measured with NBD-Ptd-[1′-14C]Ser. Values are means of three replicates ± se.
Activity and Subcellular Localization of Recombinant LePSD1 in Yeast Cells
Total PSD activity in homogenates was assayed using as substrate either 1-acyl-2-[6-[(7-nitro-2–1, 3-benzoxadiazol-4-yl) amino]caproyl-Ptd-[1′-14C]Ser (NBD-Ptd-[1′-14C]Ser), which freely partitions into membranes in the absence of detergent (Trotter and Voelker, 1995), or Ptd[1′-14C]Ser dispersed in Triton X-100. Using either substrate, activity was detected in yeast cells expressing each of the LePSD constructs but not in cells harboring the vector alone (Fig. 3B). Activity was also found in RYY51 cells carrying a plasmid containing yeast PSD1, included as a positive control (Fig. 3B). With NBD-Ptd-[1′-14C]Ser as substrate, the activities in cells carrying the three LePSD1 constructs mirrored the Etn requirements for growth (Fig. 3A): The full-length construct that did not complement the RYY51 mutant exhibited much less enzyme activity than the other constructs. With Ptd[1′-14C]Ser as substrate, the PSD activity and Etn requirement data were mildly discrepant, inasmuch as cells expressing full-length LePSD1 had somewhat more activity than cells expressing the truncated version, although LePSD1 did not restore Etn prototrophy. This discrepancy may reflect differing detergent requirements for optimal enzymatic activities of the full-length and truncated proteins.
The subcellular location of the recombinant LePSD1 proteins was established by measuring PSD activity in mitochondrial, microsomal, and cytosolic fractions using NBD-Ptd-[1′-14C]Ser as substrate. The activity specified by all three constructs—including the truncated PSD lacking the putative mitochondrial targeting sequence—was present mainly in the mitochondria, which had the highest specific activity (Table I). There was some activity in the microsomes from the chimeric and truncated constructs, and there was also a trace in the cytosol from the truncated construct (Table I).
Table I.
Subcellular distribution of recombinant LePSD activity in yeast
PSD activity was measured using with NBD-Ptd-[1′-14C]Ser as substrate. Data are means of three replicates ± se.
| Construct
|
PSD Activity
|
||
|---|---|---|---|
| Mitochondria | Microsomes | Cytosol | |
| nmol min-1 mg-1 protein | |||
| Yeast PSD1 | 0.658 ± 0.063 | 0.02 ± 0.008 | <0.001 |
| Vector control | <0.001 | <0.001 | <0.001 |
| Full-length LePSD1 | 0.127 ± 0.048 | 0.003 ± 0.002 | <0.001 |
| Truncated LePSD1 | 0.550 ± 0.218 | 0.136 ± 0.033 | 0.004 ± 0.002 |
| Chimeric LePSD1 | 0.872 ± 0.222 | 0.150 ± 0.067 | 0.001 |
Localization of PSD1 in Plant Mitochondria
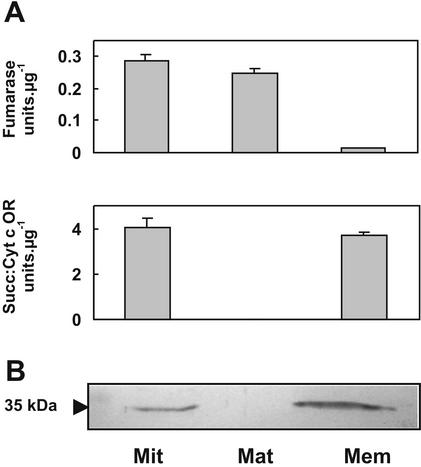
To confirm the location of PSD1 in planta, mitochondria were isolated from tubers of potato (Solanum tuberosum; a close relative of tomato) by a Percoll gradient procedure that yields highly pure mitochondria with about 95% integrity (De Leonardis et al., 1995). The mitochondria, and the membrane and matrix fractions derived from them, were then analyzed by SDS-PAGE and immunoblotting using antiserum raised against recombinant LePSD1. The distribution of enzyme markers for the matrix (fumarase) and inner mitochondrial membrane (succinate:cytochrome c oxidoreductase) validated the purity of these two submitochondrial fractions (Fig. 4A). A strong cross-reacting band was detected in entire mitochondria and their membranes but not in the matrix (Fig. 4B). This band had an apparent molecular mass of approximately 35 kD, which agrees with the value predicted for the entire LePSD1 protein (50 kD) from which the α-subunit (5.1 kD), the mitochondrial targeting peptide (approximately 6 kD), and the inner membrane sorting signal (approximately 4 kD) have been processed (Kuge et al., 1996; Voelker, 1997). The corresponding values for the potato protein are expected to be the same, because EST data indicate that it is approximately 97% identical to LePSD1.
Figure 4.
Evidence for the mitochondrial location of PSD1 in plants. Potato tuber mitochondria isolated by Percoll density gradient centrifugation were ruptured by freezing/thawing and separated into matrix and membrane fractions by ultracentrifugation. A, Specific activities (in micromoles per minute) of matrix (fumarase) and inner membrane (succinate:cytochrome c oxidoreductase) markers measured on mitochondria (Mit) and their matrix (Mat) and membrane (Mem) fractions. Data are means of three replicates ± se. B, The same fractions were analyzed by SDS-PAGE (5 μg protein lane–1) and immunoblotting with antibodies raised against recombinant LePSD1.
Expression of PSD1 mRNA in Arabidopsis Organs
The expression pattern of the AtPSD1 gene was analyzed by real-time quantitative RT-PCR rather than northern blotting because EST data indicated that this gene is not highly expressed, there being no cognate Arabidopsis sequences in the GenBank dbEST database and <30 sequences from plants as a whole (dbEST contains approximately 179,000 Arabidopsis ESTs and >2.5 × 106 plant ESTs). Because the predicted AtPSD1 gene in GenBank is incorrect (see above), introns and exons were identified by comparing the cDNA and genomic sequences, and the resulting map of the gene (Fig. 5A) was used to design a suitable amplicon (spanning two exons of the mature mRNA). The AtPSD1 mRNA levels measured in various organs of wild-type plants ranged from 94 to 228 fg 250 ng–1 total RNA (Table II). This corresponds to an mRNA frequency no higher than about 1 in 10,000, assuming mRNA makes up approximately 1% of total RNA. AtPSD1 is thus expressed at low levels throughout the plant.
Figure 5.
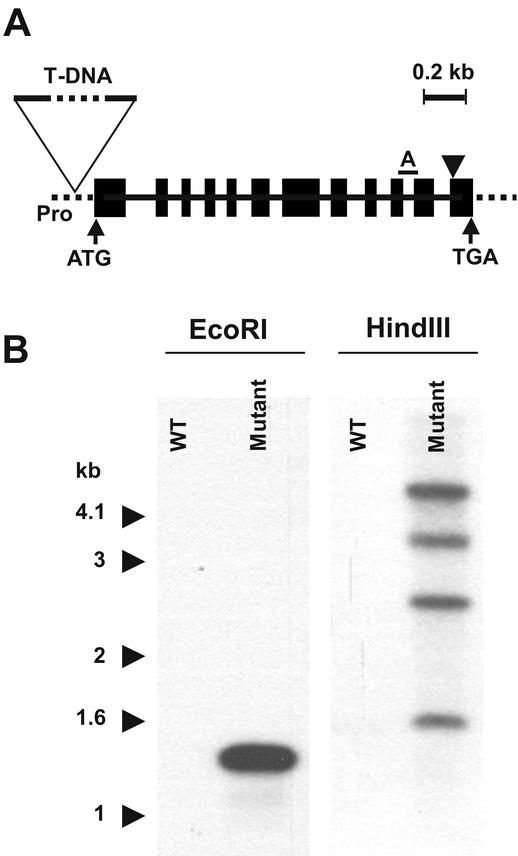
Analysis of the gene encoding AtPSD1 in wild-type and mutant Arabidopsis. A, Structure of the coding region of the gene. Black boxes represent exons, and solid lines represent introns. Dotted lines are outside the coding region; introns could not be identified here because the cDNA comprised the coding sequence only. The horizontal bar marked A is the position of the amplicon used in quantitative RT-PCR. The arrowhead shows the position of the codons for the GST consensus. The position of the T-DNA insertion in the probable promoter region is indicated. B, Southern-blot analysis of T-DNA mutants and their wild-type siblings. The 32P-labeled probe was specific for the BAR gene within the T-DNA. Genomic DNA was digested with EcoRI (which cuts each side of the probe) or HindIII (which cuts only at the 5′ end of the probe).
Table II.
Quantification of AtPSD1 mRNA in organs of wild-type Arabidopsis and a knockup mutant
Levels of mRNA were determined by real-time quantitative RT-PCR, using an amplicon spanning exons 10 and 11 of the AtPsd1 gene. Roots were either from hydroponically grown plants, or from in vitro cultures in Murashige and Skoog medium. Three independent RNA extracts were made of each organ, and triplicate mRNA determinations were made on each extract. Data are means of all nine determinations ± se. An internal AtPSD1 RNA standard (6.2 pg) was used to estimate recovery from RT-PCR of each sample; recovery values were 90% ± 4%. Data are corrected for recovery.
| Genotype | Organ | AtPSD1 mRNA |
|---|---|---|
| fg 250 ng-1 total RNA | ||
| Wild type | Young leaves | 121 ± 13 |
| Mature leaves | 94 ± 16 | |
| Stems + flowers | 106 ± 10 | |
| Roots (hydroponic) | 228 ± 12 | |
| Roots (Murashige and Skoog medium) | 143 ± 13 | |
| Homozygous mutant | Young leaves | 1605 ± 378 |
| Mature leaves | 740 ± 76 | |
| Roots (Murashige and Skoog medium) | 787 ± 29 |
Identification of an Arabidopsis T-DNA Knockup Mutant
A search of the Torrey Mesa database of sequences flanking T-DNA insertion sites (Sessions et al., 2002) using the AtPSD1 gene as query identified a line with an insertion positioned 66 bp before the start codon (Fig. 5A). After verifying this insertion site by sequencing genomic DNA, plants homozygous for the insert and their wild-type siblings were identified by PCR and were self-pollinated. Southern-blot analysis of the homozygous progeny demonstrated that only mutant plants possessed inserts (Fig. 5B), showing that the T-DNA is inserted only at the AtPSD1 locus. The multiple banding pattern in the HindIII-digested DNA (Fig. 5B) is indicative of concatenated insertion of several T-DNA copies at the same locus, which is not unusual (Waters and Guiney, 1993). To assess the impact of the T-DNA insertion on gene expression, real-time quantitative RT-PCR was performed on RNA from young leaves, mature leaves, and roots (Table II). The AtPSD1 mRNA level was elevated 6- to 13-fold in the plants homozygous for the T-DNA insert. Because the T-DNA insertional vector, pDAP101 (Sessions et al., 2002), contains no plant promoter besides that driving expression of the BAR marker gene, the knockup character of the mutation presumably reflects ablation of a negative regulatory element in the AtPSD1 gene or introduction of an adventitious promoter or enhancer element as a result of the insertion event.
Phenotype of the Knockup Mutant
There was no visual difference between homozygous mutant and wild-type plants under standard growth conditions, and their growth curves were indistinguishable (data not shown). However, mitochondrial PSD activity was 9-fold higher in the mutant when Ptd[1′-14C]Ser was used as substrate and was also much greater when assayed using NBD-Ptd-[1′-14C]Ser (Table III). This elevated activity was not accompanied by changes in mitochondrial PtdEtn content (Table IV) or whole-leaf PtdEtn content (Table V), indicating that the level of this lipid is tightly controlled. Despite the increase in mitochondrial PSD activity, there was no significant change in PSD activity measured in the total membrane fraction (Table III), which implies that Arabidopsis has extramitochondrial PSD activity and that this probably far exceeds that of AtPSD1. Although both of these inferences are based on comparing root mitochondria and leaf total lipids, the gene expression data of Table II show that AtPSD1 expression in roots and leaves is much the same.
Table III.
PSD activity in total membranes and purified mitochondria of wild-type Arabidopsis and a knockup mutant
PSD activity was measured in a total (80,000g) membrane pellet prepared from leaf extracts and in root mitochondria purified by Percoll density gradient centrifugation. The roots were cultured in Murashige and Skoog medium. Ptd[1-14C]Ser and NBD-Ptd-[1′-14C]Ser were used as substrates. Data are means of three replicates ± se.
| Substrate | Total Membranes
|
Purified Mitochondria
|
||
|---|---|---|---|---|
| Wild Type | Mutant | Wild Type | Mutant | |
| nmol min-1 mg-1 | ||||
| Ptd[1-14C]Ser | 0.412 ± 0.087 | 0.481 ± 0.072 | 0.034 ± 0.048 | 0.295 ± 0.021 |
| NBD-Ptd-[1′-14C]Ser | 0.112 ± 0.016 | 0.154 ± 0.028 | <0.015 | 0.134 ± 0.025 |
Table IV.
Phospholipid composition of mitochondria from wild-type Arabidopsis and a knockup mutant
Phospholipids were determined in root mitochondria purified by Percoll density gradient centrifugation. Data are average ± range for duplicate samples. Wild-type and mutant mitochondria had similar total phospholipid contents, as judged from their phospholipid phosphate/protein ratios.
| Phospholipid | Wild Type | Mutant |
|---|---|---|
| % | ||
| PtdEtn | 37.4 ± 0.3 | 39.4 ± 0.6 |
| Phosphatidylcholine | 38.2 ± 1.7 | 38.6 ± 0.2 |
| Phosphatidylinositol | 3.3 ± 0.2 | 2.5 ± 0.3 |
| Phosphatidylglycerol | 0.9 ± 0.5 | 0.3 ± 0.2 |
| Cardiolipin + Phosphatidate | 18.3 ± 0.5 | 19.3 ± 0 |
| Other | 2.0 ± 1.8 | 0.1 ± 0.1 |
Table V.
Phospholipid composition of leaves from wild-type Arabidopsis and a knockup mutant
Phospholipids were determined using total lipid extracts prepared from leaves. Data are the average ± range for three independent samples for wild type and two independent samples for mutant plants.
| Phospholipid | Wild Type | Mutant |
|---|---|---|
| % | ||
| PtdSer | 2.4 ± 1.2 | 2.8 ± 0.8 |
| PtdEtn | 25.5 ± 1.5 | 22.4 ± 1.2 |
| Phosphatidylcholine | 39.8 ± 2.3 | 37.6 ± 2.1 |
| Phosphatidylinositol | 3.0 ± 0.5 | 5.2 ± 0.6 |
| Phosphatidylglycerol | 22.3 ± 2.1 | 26.5 ± 0.5 |
| Other | 6.0 ± 2.9 | 4.8 ± 2.6 |
DISCUSSION
The data reported here show unambiguously that tomato and Arabidopsis have genes encoding PSDs, thereby substantiating earlier biochemical (Marshall and Kates, 1973, 1974) and in vivo radiotracer (Kinney and Moore, 1987b; Mudd and Datko, 1989) evidence for the presence of this enzyme in plants. The tomato and Arabidopsis proteins that we investigated— LePSD1 and AtPSD1—are homologous to mitochondrial PSDs from other eukaryotes and have characteristic mitochondrial targeting and inner membrane sorting sequences. Immunological evidence confirmed that the protein is located in plant mitochondrial membranes, and that the molecular mass of the mature plant protein is consistent with processing to remove the targeting and sorting sequences, as well as cleavage near the C terminus into α- and β-chains.
Despite the similarity between plant and yeast PSDs, LePSD1 did not substitute for yeast PSD1 in complementation tests, although it did so when its targeting and sorting sequences were replaced by those of yeast PSD1. Together with the low PSD activity found in yeast cells expressing unmodified LePSD1, this result suggests that the targeting and sorting sequences of plant protein did not adequately convey it to the correct intramitochondrial site in yeast. Removing these sequences also resulted in complementation, but because the truncated enzyme was expressed at a high level and was the least specifically located, it may simply have overcome mistargeting by abundance.
Close homologs of LePSD1 and AtPSD1 were found among the ESTs from diverse dicots and monocots and a gymnosperm. Along with the substantial sequence conservation among the plant, fungal, and animal enzymes, this finding indicates that mitochondrial PSDs occur widely in living plants and have most probably been present in the plant lineage because plants, fungi, and animals diverged approximately 1.6 billion years ago (Wang et al., 1999). The evolutionary persistence of mitochondrial PSDs in plants argues that they retain an important function despite the presence of Ser decarboxylase, which provides an alternative way to generate Etn moieties and subsequently PtdEtn (Rontein et al., 2001). Another argument favoring an important function for the enzyme is the observation that the AtPSD1 mRNA was present in all organs tested.
In considering what this function might be, the situation in yeast is instructive. Yeast has two types of PSD—PSD1 in mitochondria and PSD2 in Golgi/vacuolar membranes (Voelker, 1997)—and can also produce PtdEtn via the Kennedy pathway using either exogenously supplied Etn or phospho-Etn derived from sphingolipid breakdown (Birner et al., 2001; Storey et al., 2001). But these three routes to PtdEtn in yeast are not redundant: Deletion of PSD1 results in PtdEtn-depleted mitochondria and in very poor growth on non-fermentable substrates (Birner et al., 2001; Storey et al., 2001). These findings indicate that the Kennedy pathway and extramitochondrial PSD2 are insufficient when mitochondrial function is required, and that transport of PtdEtn into mitochondria can become growth limiting. It may therefore be that plant mitochondria also have a requirement for PtdEtn that cannot be met by import of PtdEtn produced elsewhere in the cell.
The data on PSD activity in the total membrane fraction of wild-type and mutant plants (Table III) demonstrate that Arabidopsis, like yeast, has extramitochondrial PSD. Consistent with this result, searching the Arabidopsis genome revealed two genes (At4g25970 and At5g57190) that encode proteins with 40% identity to the α- and β-subunits of the yeast PSD2 enzyme. Supporting this genomic evidence, a cDNA corresponding to At4g25970 complemented a yeast psd1 psd2 mutant on Glc medium (D. Rontein and A.D. Hanson, unpublished data). Interestingly, both the Arabidopsis PSD2 homologs have potential chloroplastic targeting signals so that, although extramitochondrial, they may not share the Golgi/vacuolar location of their yeast counterpart.
The Arabidopsis T-DNA knockup mutant was informative in three ways, besides confirming that AtPSD1 encodes a mitochondrial PSD. First, the mutant indicated that mitochondrial PSD activity is far lower than extramitochondrial activity in plants, because the large increase in mitochondrial PSD activity in the mutant did not affect total membrane PSD activity. Second, the large and roughly equal increases in mRNA level and enzyme activity in the mutant point to strong transcriptional control over mitochondrial PSD protein level and suggest that the coresponse coefficient (the slope of a log-log plot of [protein] versus [mRNA]) is close to one, which is exceptionally high (Fell, 2001). Third, the increase in mitochondrial PSD enzyme level was not accompanied by a significant rise in mitochondrial PtdEtn content. This finding implies that the mitochondrial content of PtdEtn is tightly controlled, perhaps by regulating the import of Ptd-Ser into the mitochondria as well as the export of PtdEtn from the organelle. The findings may further indicate that PSD1 in plants is normally produced in excess of the amounts needed. No deleterious effects have been described for overexpression of either PSD1 or PSD2 in yeast (Trotter et al., 1993, 1995). Quantification of PtdEtn in yeast strains overexpressing PSD1 has not been reported, but strains overexpressing PSD2 10-fold do not have significantly altered cellular lipid content (Kitamura et al., 2002). This last finding contrasts with the results of overexpressing Ptd-Ser synthase (Fig. 1) in Arabidopsis, which led to overaccumulation of Ptd-Ser but not PtdEtn (Delhaize et al., 1999).
MATERIALS AND METHODS
Chemicals and Reagents
Ptd[1′-14C]Ser and NBD-Ptd-[1′-14C]Ser were synthesized as described (Trotter et al., 1995). [5,6-3H]UTP (46 Ci mmol–1) and [α-32P]UTP (800 Ci mmol–1) were from Amersham Biosciences (Uppsala). [α-32P]CTP (3,000 Ci mmol–1) was from PerkinElmer Life Sciences (Boston). Phospholipid standards for thin-layer chromatography were from Avanti Polar Lipids (Birmingham, AL) and Sigma-Aldrich (St. Louis). Thin layer Silica Gel H plates were from Analtech (Newark, DE). Protease inhibitor cocktail (I9599) and cytochrome c (C7752) were from Sigma-Aldrich. Bicinchoninic acid protein assay reagents were from Pierce (Rockford, IL).
Yeast Strains, Plasmids, and Growth Conditions
The Brewer's yeast (Saccharomyces cerevisiae) strains were RYY51 (ura3 his3 trp1 leu2 lys2 psd1-Δ1::TRP1 psd2-Δ1::HIS3) and the corresponding wild type, SEY6210 (ura3 his3 trp1 leu2 lys2; Trotter and Voelker, 1995). The vector for plant PSD constructs was pVT103-U, which contains the URA-3 gene for selection and the ADH1 promoter to drive gene expression (Vernet et al., 1987). Yeast PSD1 was expressed using plasmid YCp50-PSD1 (Trotter et al., 1995). Synthetic minimal media, supplements, and culture conditions were as described (Trotter and Voelker, 1995).
Plant Materials
Arabidopsis (ecotype Columbia) plants were grown in Super Fine Germination Mix (Fafard, Agawam, MA) at 22°C in 12-h d (80–150 μE m–2 s–1) and irrigated with water. Young leaves were harvested at d 14, mature leaves at d 21, and flowering stems at d 25; roots were from plants cultured hydroponically as described (Gibeaut et al., 1997). Roots were also grown from sterilized seeds in darkness at 22°C to 24°C in 1-L flasks containing 300 mL of liquid Murashige and Skoog medium (Murashige and Skoog, 1962) plus 30 g L–1 Suc and 1 μm indolebutyric acid, with rotary shaking (100 rpm). At 3 weeks, the medium was renewed and roots were harvested 1 week later. Potato (Solanum tuberosum) tubers were purchased locally.
cDNA Generation, Sequencing, and Sequence Analysis
A tomato (Lycopersicon esculentum Mill.) EST (GenBank accession no. BE451597) encoding LePSD1 was supplied by Clemson University Genomics Institute (Clemson, SC). An Arabidopsis cDNA encoding AtPSD1 was cloned by RT-PCR using RNA extracted from leaves with an RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA), the SuperScript preamplification system (Invitrogen, Carlsbad, CA), and Taq DNA polymerase (Invitrogen). The forward primer was 5′-ATGAAACCTCGTTTTCCTCAAAATG-3′, and the reverse primer was 5′-TCATTCCTCTTTCCATCTTCCCAA-3′. The PCR product was cloned into pGEM-T Easy (Promega, Madison, WI). DNA sequencing and sequence analysis were as described (Bourgis et al., 1999). Subcell location and transmembrane helices were predicted with TargetP (Emanuelsson et al., 2000) and TMHMM (Krogh et al., 2001) programs, respectively.
cDNA Expression in Yeast
DNA sequences were amplified using Pfu DNA polymerase (Stratagene, La Jolla, CA). The reverse primer for all LePSD1 sequences was 5′-PCTAGGAATCATGCCACCT-3′. Forward primers were: for full-length LePSD1, 5′-ATATGAGCTCATGAAATTTAGGGCTTCT-3′; for truncated LePSD1, 5′-GAGCTCAACAACATGTCTCAGGGTAACACTCTTTTG-3′, which includes an AACAAC sequence to enhance translation initiation (Miyasaka, 1999) and a start codon preceding codons 55 to 61 of LePSD1. The chimeric yeast PSD1/LePSD1 was constructed using gene splicing by overlap extension (Horton et al., 1993). Yeast PSD1 codons 1 to 139 were amplified using the primers 5′-TGACGAGCTCAACAACATGTCAATTATGCCAGTTA-3′ (forward) and 5′-TACCCCAGAATCGAGACATCGCATTCAGCGG-3′ (reverse). LePSD1 codons 118 to 445 were amplified using the forward primer 5′-ATGTCTCGATTCTGGGGTACCTTGACCAAT-3′ and the reverse primer above. The two amplified fragments were mixed 1:1 and were joined by PCR using the yeast PSD1 forward primer and the LePSD1 reverse primer. Constructs were ligated between the SstI and PvuII sites of pVT103-U and electroporated into Escherichia coli strain DH10B. The sequence-verified constructs were used to transform strain RYY51 as described (Trotter and Voelker, 1995).
cDNA Expression in E. coli
The LePSD1 coding sequence was PCR-amplified using the primers 5′-GGAATTCCATATGAAATTTAGGGCTTCTCAGAGA-3′ (forward) and 5′-TGACGAGCTCCTAGGAATCATGCCACCT-3′ (reverse) and ligated between the NdeI and SacI sites of pET28b (Novagen, Madison, WI). This procedure added an N-terminal hexa-His tag. The construct was electroporated into E. coli strain DH10B and then, after sequence verification, into E. coli strain BL21 CodonPlus (DE3). For protein production, 100-mL cultures were grown at 37°C to an A600 of 0.6 in Luria-Bertani medium containing 100 μg mL–1 kanamycin; isopropyl β-d-1-thiogalactopyranoside was then added (1 mm final concentration), and incubation was continued at 30°C for 4 h.
Isolation of His-Tagged LePSD1 and Antibody Production
Operations were at 4°C. Cells were harvested by centrifugation, washed in buffer A (0.1 m Tris-HCl, pH 7.2, 1 mm K2EDTA, and 10% [v/v] glycerol), resuspended in 1.5 mL of buffer B (buffer A plus 0.2% [w/v] Nonidet P-40), lysed with a Mini-BeadBeater (Biospec Products, Bartlesville, OK) using 0.1-mm zirconia/silica beads at maximum speed for 3 × 30 s, and centrifuged at 16,000g for 5 min. The beads and cell residues were washed with 6 × 1 mL of buffer B and then (at room temperature) 10 × 1 mL of buffer C (8 m urea, 0.1 m sodium phosphate, and 0.01 m Tris-HCl, pH 8). Each wash was analyzed by SDS-PAGE. LePSD was recovered almost pure from the buffer C washes; the band was excised from the gel and was used to raise antibodies in rabbits (Cocalico Biologicals, Reamstown, PA).
Subcellular Fractionation and Ptd-Ser Decarboxylase Assays in Yeast
Cells were grown to an A600 value of 1.5 to 2.0 in synthetic complete medium containing 0.05% (w/v) Glc and 4% (w/v) lactate as carbon source and 2 mm Etn. Mitochondrial, microsomal, and soluble supernatant fractions were prepared by differential centrifugation as described (Glick and Pon, 1995; Trotter and Voelker, 1995). PSD activity was measured by a 14CO2 trapping method using either Ptd[1′-14C]Ser or NBD-Ptd-[1′-14C]Ser as described (Trotter et al., 1995).
Isolation, Fractionation, and Immunoblot Analysis of Potato Mitochondria
Mitochondria were isolated and then purified on a Percoll density gradient as described (De Leonardis et al., 1995) except that 1-kg batches of tubers (cut in 1-cm cubes) were homogenized with a Waring blender for 30 s at maximum speed. The purified mitochondria were suspended in 10 mm potassium phosphate buffer, pH 7.2, disrupted by six freeze/thaw cycles, and centrifuged at 80,000g for 90 min to obtain matrix and membrane fractions. Proteins were separated by SDS-PAGE and transferred to nitrocellulose (Protran BA 85, Schleicher & Schuell, Dassel, Germany) for immunoblot analysis as described (Nuccio and Thomas, 1999) using antiserum to recombinant LePSD1.
Isolation of Arabidopsis Root Mitochondria
Mitochondria were prepared using a Percoll/Suc step gradient essentially as described (Klein et al., 1998). Roots were disrupted first in a Waring blender 5 s at maximum speed) and then in a Polytron (two 5-s bursts, setting 5). The 16,000g (crude mitochondria) pellet was thoroughly resuspended and deposited on the three-layer (18%/23%/40% [w/v]) Percoll gradient. Centrifugation was at 4°C for 45 min at 12,000g in an SW41Ti rotor (Beckman Coulter, Fullerton, CA). Mitochondria were collected from the 23%/40% (w/v) Percoll interface, washed, frozen in liquid N2, and kept at –80°C until analysis. Markers for the chloroplast stroma (NADP-linked glyceraldehyde 3-phosphate dehydrogenase), mitochondrial matrix (fumarase), and mitochondrial inner membrane (succinate:cytochrome c oxidoreductase) were assayed as described (Douce et al., 1973; Trossat et al., 1996).
Preparation of a Total Membrane Fraction from Arabidopsis Leaves
The procedure was based on that of Macher and Mudd (1974). Leaf samples (1 g) were disrupted using a Polytron (three 10-s bursts at setting 7) in 6 mL of 100 mm Tris-HCl, pH 7.5, containing 0.25 m Suc, 1 mm EDTA, 2 mm dithiothreitol, 15 mm sodium ascorbate, 1.5% (w/v) polyvinylpolypyrrolidone, and 50 μL of protease inhibitor cocktail. The brei was centrifuged at 5,000g for 15 min, the pellet was discarded, and the supernatant was centrifuged at 80,000g for 90 min in a Beckman Coulter 75-Ti rotor. The pellet was frozen in liquid N2 and was kept at –80°C.
Phospholipid Analysis
Frozen leaves (0.6 g) were transferred to a 50-mL polypropylene tube containing liquid N2 and 6 g of 4-mm glass beads and were vortexed repeatedly in 10-s bursts to produce a fine powder. The tubes were cooled in liquid N2 after each burst of vortexing. Lipids were extracted by the addition of 6 mL of chloroform:methanol:formic acid (1:1:0.1, v/v) and continued vortex mixing and cooling. The solution was transferred to a 20-mL glass tube, and the beads were washed once with an additional 2 mL of the same solvent mixture. The resultant 8-mL extract was partitioned into two phases with the addition of 4 mL of 1 m KCl. The lower organic phase was separated by centrifugation and washed with upper phase one additional time. Organic solvents were removed from the extracts, which were subsequently resuspended in 9:1 (v/v) chloroform:methanol and spotted on Silica gel 60 plates. The lipids were separated by two-dimensional thin-layer chromatography—first, 65:25:10 (v/v) chloroform:methanol:acetic acid and then 65:25:10 (v/v) chloroform:methanol:formic acid—and visualized by iodine staining. Lipid phosphorus was determined by the method of Rouser et al. (1966). Mitochondrial lipids were extracted using the procedure of Bligh and Dyer (1959); the phospholipids were separated and quantified as described for leaves.
Real-Time Quantitative RT-PCR
Total RNA was extracted from three samples of each tissue using a Qiagen RNeasy Plant Mini Kit. Real-time quantitative RT-PCR was performed on 250 ng of RNA in 25-μL reactions using Taq-Man One-Step RT-PCR Master Mix Reagents (Applied Biosystems, Foster City, CA) and an Applied Biosystems GeneAmp 5700 sequence-detection system. The primers and Taq-Man probe (designed with Applied Biosystems Primer Express software) were as follows: forward primer, 5′-TGGAAAGAAGGTTTTATGGCACTT-3′; reverse primer, 5′-GGTTCAATGAAAAGCTCAATGGA-3′; and probe, 5′-CTGCTGTAGGCGCGACCAACATTG-3′ with the fluorescent reporter dye 6-carboxyfluorescein and the quencher dye 6-carboxytetramethylrhodamine bonded to the 5′ and 3′ end, respectively. The reverse primer spans exons 10 and 11 of the AtPSD1 gene, to avoid amplifying contaminating genomic DNA. The amplicon length was 74 bp. RT-PCR conditions were as follows: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard RNA was synthesized from AtPSD1 cDNA (see below); the standard curve was linear from 5 fg to 16 pg. As internal standard, 6.2 pg of AtPSD1 RNA was mixed with each sample before the RT-PCR reaction. A Ct threshold value was determined from amplification curves by selecting an optimal ΔRn (emission of the reporter dye over starting background fluorescence) in the exponential part of the plots.
AtPSD1 RNA Synthesis and Quantification
The in vitro transcription (MAXIscript, Ambion, Austin, TX) used 5.4 μm [5,6-3H]UTP or 3.1 μm [α-32P]UTP as limiting nucleotide. The efficiency of incorporation of [5,6-3H]UTP into RNA was 60% as evaluated by TCA precipitation of an aliquot of the in vitro transcription product. The 3H- and 32P-labeled RNAs were run on a 5% (w/v) polyacrylamide gel (16 × 20 cm) containing 8 m urea. The full-length [32P]RNA was localized by autoradiography, thus indicating the position of corresponding 3H-RNA. The [3H]RNA band was excised, eluted overnight at 37°C with DEPC-treated water, and quantified by scintillation counting. The overall [3H]RNA yield was 15%.
Insertional Arabidopsis Mutant
Searching the Torrey Mesa Research Institute Arabidopsis (ecotype Columbia) T-DNA insertion database (Sessions et al., 2002) for insertions in the AtPSD1 gene region identified line SAIL_508_C12, mutated with the pDAP101 vector. The location of the insert was verified by sequencing the PCR product obtained using the forward gene-specific primer 5′-TGAAGAGCTCCAAGCATAAGG-3′ and the reverse T-DNA left border primer 5′-GCATCTGAATTTCATAACCAATC-3′. Plants homozygous for the T-DNA insert and their wild-type siblings were selfed; the progeny were PCR-screened to check the genotype and used for analyses.
Southern-Blot Analysis
Genomic DNA was isolated from 2-g batches of leaves pooled from 25 Arabidopsis plants 21 d of age. Isolated DNA (5 μg) was digested, separated by 0.8% (w/v) agarose gel, and transferred to nitrocellulose (Protran BA 85, Schleicher & Schuell). Hybridization was at 65°C in 6 × SSC, 5 × Denhardt's solution, 1% (w/v) SDS, and 1 mm EDTA. The probe was the PCR-amplified BAR gene from the T-DNA insert, labeled with [α-32P]CTP by random priming. The final wash was at 65°C in 0.1 × SSC containing 0.1% (w/v) SDS.
Acknowledgments
We thank the Torrey Mesa Research Institute for the Arabidopsis insertional mutant and Michael J. Ziemak for help with cloning and yeast transformation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023242.
This work was supported in part by the Florida Agricultural Experiment Station, by an endowment from the C.V. Griffin, Sr. Foundation, by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 2001–35100–10620 to A.D.H.), and by the National Institutes of Health (grant no. GM 32453 to D.R.V.), and has been approved for publication as journal series no. R–09358.
References
- Birecka H, Bitonti AJ, McCann PP (1985) Assaying ornithine and arginine decarboxylases in some plant-species. Plant Physiol 79: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R, Burgermeister M, Schneiter R, Daum G (2001) Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol Biol Cell 12: 997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, Fisher DB, Tarczynski MC, Li C, Herschbach C, Rennenberg H, Pimenta MJ, Shen TL et al. (1999) S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 11: 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Zeimetz GM (1996) Regulation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. J Biol Chem 271: 13293–13296 [DOI] [PubMed] [Google Scholar]
- Clancey CJ, Chang SC, Dowhan W (1993) Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant. J Biol Chem 268: 24580–24590 [PubMed] [Google Scholar]
- De Leonardis S, De Lorenzo G, Borraccino G, Dipierro S (1995) A specific ascorbate free radical reductase isozyme participates in the regeneration of ascorbate for scavenging toxic oxygen species in potato tuber mitochondria. Plant Physiol 109: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Hebb DM, Richards KD, Lin JM, Ryan PR, Gardner RC (1999) Cloning and expression of a wheat (Triticum aestivum L.) phosphatidylserine synthase cDNA: overexpression in plants alters the composition of phospholipids. J Biol Chem 274: 7082–7088 [DOI] [PubMed] [Google Scholar]
- Douce R, Mannella CA, Bonner WD (1973) The external NADH dehydrogenases of intact plant mitochondria. Biochim Biophys Acta 292: 105–116 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Fell DA (2001) Beyond genomics. Trends Genet 17: 680–682 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Pon LA (1995) Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol 260: 213–223 [DOI] [PubMed] [Google Scholar]
- Hamel P, Sakamoto W, Wintz H, Dujardin G (1997) Functional complementation of an oxa1-yeast mutation identifies an Arabidopsis thaliana cDNA involved in the assembly of respiratory complexes. Plant J 12: 1319–1327 [DOI] [PubMed] [Google Scholar]
- Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR (1993) Gene splicing by overlap extension. Methods Enzymol 217: 270–279 [DOI] [PubMed] [Google Scholar]
- Kinney AJ, Moore TS (1987a) Phosphatidylcholine synthesis in castor bean endosperm: 1. Metabolism of l-serine. Plant Physiol 84: 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ, Moore TS (1987b) Phosphatidylcholine synthesis in castor bean endosperm: the localization and control of CTP:choline-phosphate cytidylyltransferase activity. Arch Biochem Biophys 259: 15–21 [DOI] [PubMed] [Google Scholar]
- Kitamura H, Wu W, Voelker DR (2002) The C2 domain of phosphatidylserine decarboxylase 2 is not required for catalysis but is essential for in vivo function. J Biol Chem 277: 33720–33726 [DOI] [PubMed] [Google Scholar]
- Klein M, Binder S, Brennicke A (1998) Purification of mitochondria from Arabidopsis. Methods Mol Biol 82: 49–53 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kuge O, Nishijima M (1997) Phosphatidylserine synthase I and II of mammalian cells. Biochim Biophys Acta 1348: 151–156 [DOI] [PubMed] [Google Scholar]
- Kuge O, Nishijima M, Akamatsu Y (1991) A cloned gene encoding phosphatidylserine decarboxylase complements the phosphatidylserine biosynthetic defect of a Chinese hamster ovary cell mutant. J Biol Chem 266: 6370–6376 [PubMed] [Google Scholar]
- Kuge O, Saito K, Kojima M, Akamatsu Y, Nishijima M (1996) Posttranslational processing of the phosphatidylserine decarboxylase gene product in Chinese hamster ovary cells. Biochem J 319: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QX, Dowhan W (1988) Structural characterization of Escherichia coli phosphatidylserine decarboxylase. J Biol Chem 263: 11516–11522 [PubMed] [Google Scholar]
- Macher BA, Mudd JB (1974) Biosynthesis of phosphatidylethanolamine by enzyme preparations from plant tissues. Plant Physiol 53: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macher BA, Mudd JB (1976) Partial purification and properties of ethanolamine kinase from spinach leaf. Arch Biochem Biophys 177: 24–30 [DOI] [PubMed] [Google Scholar]
- Marshall MO, Kates M (1973) Biosynthesis of phosphatidyl ethanolamine and phosphatidyl choline in spinach leaves. FEBS Lett 31: 199–202 [DOI] [PubMed] [Google Scholar]
- Marshall MO, Kates M (1974) Biosynthesis of nitrogenous phospholipids in spinach leaves. Can J Biochem Cell B 52: 469–482 [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Okada M, Horikoshi Y, Matsuzaki H, Kishi T, Itaya M, Shibuya I (1998) Cloning, sequencing, and disruption of the Bacillus subtilis psd gene coding for phosphatidylserine decarboxylase. J Bacteriol 180: 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka H (1999) The positive relationship between codon usage bias and translation initiation AUG context in Saccharomyces cerevisiae. Yeast 15: 633–637 [DOI] [PubMed] [Google Scholar]
- Moore TS (1975) Phosphatidylserine synthesis in castor bean endosperm. Plant Physiol 56: 177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd JB (1980) Phospholipid biosynthesis. In PK Stumpf, EE Conn, eds, The Biochemistry of Plants, Vol 4. Academic Press, New York, pp 249–282 [Google Scholar]
- Mudd SH, Datko AH (1989) Synthesis of ethanolamine and its regulation in Lemna paucicostata. Plant Physiol 91: 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nuccio ML, Thomas TL (1999) ATS1 and ATS3: two novel embryo-specific genes in Arabidopsis thaliana. Plant Mol Biol 39: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Rontein D, Nishida, Tashiro G, Yoshioka K, Wu W-I, Voelker DR, Basset G, Hanson AD (2001) Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J Biol Chem 276: 35523–35529 [DOI] [PubMed] [Google Scholar]
- Rouser G, Siakotos AN, Fleische S (1966) Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids 1: 85–86 [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey MK, Clay KL, Kutateladze T, Murphy RC, Overduin M, Voelker DR (2001) Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J Biol Chem 276: 48539–48548 [DOI] [PubMed] [Google Scholar]
- Tang F, Moore TS (1997) Enzymes of the primary phosphatidylethanolamine biosynthetic pathway in postgermination castor bean endosperm. Plant Physiol 115: 1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trossat C, Nolte KD, Hanson AD (1996) Evidence that the pathway of dimethylsulfoniopropionate biosynthesis begins in the cytosol and ends in the chloroplast. Plant Physiol 111: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Voelker DR (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae: isolation of mutants, cloning of the gene, and creation of a null allele. J Biol Chem 268: 21416–21424 [PubMed] [Google Scholar]
- Trotter PJ, Pedretti J, Yates R, Voelker DR (1995) Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae: cloning and mapping of the gene, heterologous expression, and creation of the null allele. J Biol Chem 270: 6071–6080 [DOI] [PubMed] [Google Scholar]
- Trotter PJ, Voelker DR (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J Biol Chem 270: 6062–6070 [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52: 225–233 [DOI] [PubMed] [Google Scholar]
- Vincent P, Maneta-Peyret L, Sturbois-Balcerzak B, Duvert M, Cassagne C, Moreau P (1999) One of the origins of plasma membrane phosphatidylserine in plant cells is a local synthesis by a serine exchange activity. FEBS Lett 464: 80–84 [DOI] [PubMed] [Google Scholar]
- Voelker DR (1997) Phosphatidylserine decarboxylase. Biochim Biophys Acta 1348: 236–244 [DOI] [PubMed] [Google Scholar]
- Voelker DR (2000) Interorganelle transport of aminoglycerophospholipids. Biochim Biophys Acta 1486: 97–107 [DOI] [PubMed] [Google Scholar]
- Wang DY, Kumar S, Hedges SB (1999) Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc R Soc Lond B Biol Sci 266: 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters VL, Guiney DG (1993) Processes at the nick region link conjugation, T-DNA transfer and rolling circle replication. Mol Microbiol 9: 1123–1130 [DOI] [PubMed] [Google Scholar]