Abstract
The NADPH/NADP-thioredoxin (Trx) reductase (NTR)/Trx system (NTS) is a redox system that plays a posttranslational regulatory role by reducing protein targets involved in crucial cellular processes in microorganisms and animals. In plants, the system includes several h type Trx isoforms and has been shown to intervene in reserve mobilization during early seedling growth of cereals. To determine whether NTS was operational during germination of legume seeds and which Trx h isoforms could be implicated, Trx h isoforms expression was monitored in germinating pea (Pisum sativum cv Baccara) seeds, together with the amount of NTR and NADPH. Two new isoforms were identified: Trx h3, similar to the two isoforms already described in pea but not expressed in seeds; and the more divergent isoform, Trx h4. Active recombinant proteins were produced in Escherichia coli and used to raise specific antibodies. The expression of new isoforms was analyzed at both mRNA and protein levels. The lack of correlation between mRNA and protein abundances suggests the occurrence of posttranscriptional regulation. Trx h3 protein amount remained constant in both axes and cotyledons of dry and imbibed seeds but then decreased 2 d after radicle protrusion. In contrast, Trx h4 was only expressed in axes of dry and imbibed seeds but not in germinated seeds or in seedlings, therefore appearing as closely linked to germination. The presence of NTR and NADPH in seeds suggests that NTS could be functional during germination. The possible role of Trx h3 and h4 in this context is discussed.
Thioredoxins (Trxs) constitute a family of small and ubiquitous proteins with two close and active Cys residues in a conserved motif: WCG/PPC. In their dithiol form, they are powerful disulfide reductases (Holmgren, 1985) that play a posttranslational regulatory role on protein targets involved in an ever-increasing number of cellular processes.
Plant cells have four different types of Trx isoforms, the well-known f and m types in the chloroplast (Buchanan, 1991), the recently described o type in the mitochondria (Laloi et al., 2001), and the cytosolic h type possibly associated with plasma membrane (Johnson et al., 1987; Florencio et al., 1988; Rivera-Madrid et al., 1995) and also found in the phloem sap (Ishiwatari et al., 1995; Schobert et al., 1998). The reduction of Trx h isoforms by NADPH is mediated by NADP-Trx reductase (NTR). These three components are referred to as the redox NTR/Trx system (NTS). Trx h is encoded by a multigenic family comprising eight members in the Arabidopsis genome (Rivera-Madrid et al., 1995; Sahrawy et al., 1996; Laloi et al., 2001) that differ by their level and cell-type expression, suggesting that they have specific functions (Reichheld et al., 2002). The best documented function of Trx h is its implication in reserve breakdown that sustains early seedling growth of germinated cereal seeds. The NTS was shown to promote the activation of α-amylase, pullulanase, and proteases and the reduction of storage proteins, resulting in the degradation of carbohydrate and protein reserves (Kobrehel et al., 1991, 1992; Jiao et al., 1992; Wong et al., 1995; Besse et al., 1996; Lozano et al., 1996; Cho et al., 1999; Marx et al., 2003). The recent finding that Trx h accumulates in aleurone and scutellum cells of germinated seeds of wheat (Triticum aestivum) is also consistent with a role in reserve mobilization (Serrato et al., 2001). However, considering the variety of functions reported for the counterpart system found in microorganisms and animal cells, including signal transduction, gene expression, and cell proliferation or apoptosis (for review, see Muller, 1995; Besse and Buchanan, 1997; Arrigo, 1999; Powis et al., 2000), it is likely that the multiple Trx h isoforms play diverse roles during germination. This process, which is a critical node in the life cycle of higher plants, incorporates events that begin with the uptake of water by the quiescent dry seed and ends with radicle protrusion (Bewley and Black, 1994). The profound changes in gene expression that are required to orientate metabolism toward a germination pattern are under the control of signal transduction pathways that might include redox regulation by NTS. Several potential targets of NTS, besides storage proteins, were identified in peanut (Arachis hypogaea) seeds and in germinating barley (Hordeum vulgare): peroxiredoxin, late-embryogenesis abundant or heat shock proteins, and dessication- and maturation-related proteins (Yano et al., 2001; Marx et al., 2003).
To find out whether the NTS was operational during germination of legume seeds, we characterized the expression of Trx h isoforms in germinating pea (Pisum sativum) seeds and brought to light two new isoforms. We also determined the relative amounts of NTR and the content in NADPH, the two components of the system that drive Trx h isoform reduction. Expression was investigated separately in cotyledons that mainly serve to reserve breakdown and in embryonic axes that prepare for cell elongation and cell division associated with radicle emergence. In this paper, we report for the first time, to our knowledge, about a NTS in germinating pea seeds that involves two new Trx h isoforms that are differentially expressed at protein levels.
RESULTS
Trx h Isoforms Expressed in Seeds
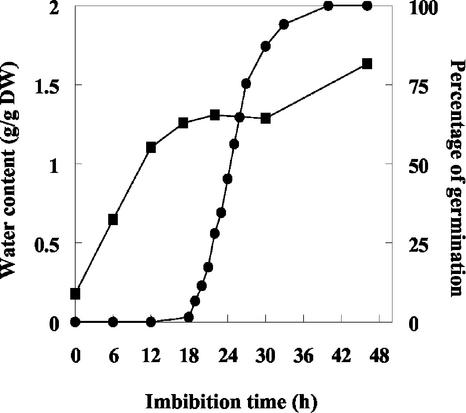
As a preliminary to the Trx h expression studies during germination, we established the germination rate and water uptake of pea seeds (Fig. 1). When pea seeds were imbibed at 20°C in the dark, 50% of the seeds had germinated after 25 h of imbibition. As expected, the water uptake curve shows the typical triphasic shape (Bewley, 1997; Obroucheva and Antipova, 1997). Initial imbibition (phase I) is characterized by a rapid water uptake from 0 to approximately 14 h, followed by a lag phase (phase II) between 14 and 30 h. A further increase in water uptake occurring after 30 h of imbibition (phase III) corresponds to post-germinative growth. Because the major metabolic events of germination take place during phase II to prepare for radicle emergence (Bewley and Black, 1994), seeds imbibed for 22 h were selected for initial investigations concerning the presence of Trx h isoforms related to germination.
Figure 1.
Water content and percentage of germination of pea seeds. Percentages of germination (•) and water content (▪) of seeds, allowed to germinate on wet filter paper at 20°C in the dark, are indicated for different times after the beginning of imbibition.
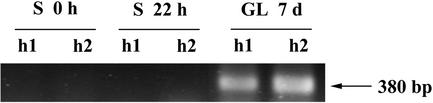
Two different isoforms of Trx h (GenBank accession nos. AJ319808 and AJ310990) have been identified previously in pea. Because both genes were given the same name, they will be referred to as Trx h1 and Trx h2. To investigate whether both genes were expressed in seeds, reverse transcription (RT)-PCR was performed using specific primers (Table I), using as templates cDNAs from dry or 22-h-imbibed seeds and cDNAs from 7-d-old green leaves. Figure 2 shows that Trx h1 and h2 were easily amplified in leaves but could not be detected in seeds even after 35 cycles of amplification. Cloning and sequencing of the PCR products amplified from leaf cDNAs confirmed that they actually encoded Trx h1 and h2, allowing the further use of these probes in northern experiments.
Table I.
Primers, annealing temperatures, and no. of cycles of PCR reactions
| Primers | Sequences | Annealing Temperatures × Cycle Nos. |
|---|---|---|
| h1-sens | acgcgtcgaca-ATGGCAGCAGAAAATGAG | 50°C × five cycles then 60°C × 30 cycles |
| h1-anti | acgcgtcgac-AGCAGTAGCAACAGTTGT | |
| h2-sens | acgcgtcgac-ATGGCAGGTTCATCAGAA | 50°C × five cycles then 60°C × 30 cycles |
| h2-anti | acgcgtcgac-AGCATTAGATGAAGCCACA | |
| Xs-1A | ggcggatccgccactgc-TGGTGYGGNCCNTGYM | 45°C × five cycles then 62°C × 30 cycles |
| or Xs-1B | ggcggatccgccactgc-TGGTGYCCNCCNTGYM | |
| Xa-anti2A | gcgactggatccgcgcg-RAANGTNGGCATNGCYTC | |
| or Xa-anti2B | gcgactggatccgcgcg-RAANGTNGGCATNGCYTG | |
| h3-5′1b | AGCTTTTTAGCAATCTCTGC | 65°C × five cycles then 56°C × 30 cycles |
| h3-5′2b | GTCAACCTTAAGGAAAGTG | 50°C × 30 cycles |
| h3-3′1b | TTTATTGCCCCAATTTTGGC | 65°C × five cycles then 50°C × 30 cycles |
| h3-3′2b | AAAAAGCTTACACATGTCAC | 50°C × 30 cycles |
| h4-5′1b | CGATCTTGATGAATTCAACG | 65°C × five cycles then 56°C × 30 cycles |
| h4-5′2b | CATAAACAATCGGTTCCATC | 50°C × 30 cycles |
| h4-3′1b | ATGGAACCGATTGTTTATGC | 65°C × five cycles then 50°C × 30 cycles |
| h4-3′2b | TTATGCCATGGCTAACGAAT | 50°C × 30 cycles |
| h3-sens | GAAGAAAGAGGAAAATG | 35°C × five cycles then 30°C × 30 cycles |
| h3-anti | CAAAGTTTTGTTCATGAC | |
| h4-sens | TATCAAGCAAGCAAGTCAACGAACC | 65°C × five cycles then 50°C × 30 cycles |
| h4-anti | CAAACAAATACATAAACATCATTCAACC | |
| h3pASK-sens | ATGGTAGGTCTCAAATGGCGGAAGAGGGACAAGTGATCG | 62°C × 20 cycles |
| h3pASK-anti | ATGGTAGGTCTCAGCGCTAGCTGCATGCTTGTCAATTTTCAAT | |
| h4pASK-sens | ATGGTAGGTCTCAAATGGGCTCAATTCTCTCTTCCCTCAT | 62°C × 20 cycles |
| h4pASK-anti | ATGGTAGGTCTCAGCGCTAGCTCTGTATTTTTTAATCTTGTTTTTG | |
| 18S rRNA-sens | CCAGGTCCAGACATAGTAAG | 55°C × 25 cycles |
| 18S rRNA-anti | GTACAAAGGGCAGGGACGTA | |
| h1-for | AGGCTATGCCAACCTTCTTGC | 60°C × 40 cycles |
| h1-rev | TGCATGCTTGGTTATTGCCA | |
| h2-for | GCGTTGATGCATGGAACGATAT | 60°C × 40 cycles |
| h2-rev | AGGAATGGTGCAATGAAACGG | |
| h3-for | GTGGATGCTTGGAAGGAACAGT | 60°C × 40 cycles |
| h3-rev | GGCAATAAAACGGCATGGAC | |
| h4-for | TTAGAATCCTCCGATCACGCC | 60°C × 40 cycles |
| h4-rev | TCAATCACAACGAGACGAGGC |
The part of the sequence that appears in lower case for several primers was arbitrarily defined to either insert a Sa/I restriction site (underlined) at both ends of the PCR product and then facilitate the subcloning of the PCR products or to increase the temperature in course of the PCR process according to Weighardt et al. (1993). b 5′1, 5′2, 3′1, or 3′2 primers were used for RACE reactions with the 5′, 5′-nested, 3′, or 3′-nested primers provided with the GeneRacer kit.
Figure 2.
Expression of Trx h1 and h2 in pea. Trx h1 and h2 mRNAs were amplified by RT-PCR using specific primers (h1-sens/h1-anti and h2-sens/h2-anti, respectively; see Table I) on different templates: cDNAs from either dry (S 0 h) or 22-h-imbibed seeds (S 22 h) and cDNAs from 7 d-old green leaves (GL 7 d). The PCR products were resolved in 1.5% (w/v) agarose gel and stained with etidium bromide.
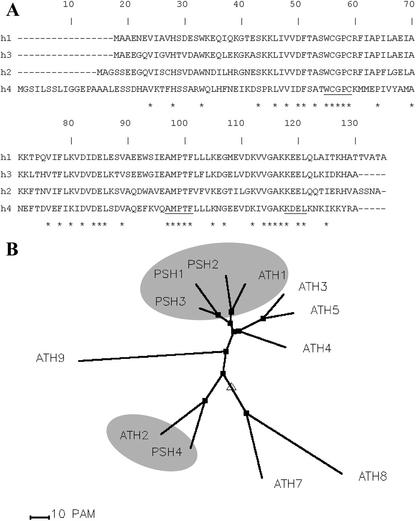
The lack of expression of these Trx h isoforms in pea seeds led us to investigate whether other unknown isoforms could be specifically expressed during germination. The presence of two conserved motives (WCG/PPC and Q/EAMPTF) in most plant Trx h allowed the design of degenerated oligonucleotide primers for RT-PCR experiments performed on cDNAs from 22-h-imbibed seeds (Table I). Two forward primers, Xs-1A and Xs-1B, were derived from the WCG/PPC sequence to take into account the presence of either a G (Xs-1A) or a P (Xs-1B) in the third position. Similarly, two reverse primers, Xa-anti2A and Xa-anti2B, were designed in the Q/EAMPTF consensus sequence, respectively. Of the four primer combinations, only those using forward primer Xs-1A and reverse primers Xa-anti2A or Xa-anti2B yielded products with an expected size of 170 bp (data not shown). Cloning and sequencing of the PCR products revealed two new Trx h cDNAs that will be referred to as Trx h3 and Trx h4. Among all the clones analyzed, Trx h3 and h4 were encountered several times, suggesting that they correspond to major transcripts of Trx h in germinating seeds. The full-length sequences of h3 and h4 cDNAs (647 and 533 bp) were obtained by RACE and found to encode proteins of 113 and 130 amino acids, respectively, with theoretical masses of 12,578 and 14,549 D (GenBank accession nos. AY170650 and AY170651). The protein sequences deduced from Trx h3 and h4 were aligned with those of Trx h1 and h2 (Fig. 3A). The sequence of Trx h3 appeared similar to those of h1 and h2 (72% and 64% identity, respectively), whereas that of Trx h4 was more divergent because it shared only 37% to 41% identity with the three other isoforms. In addition, Trx h4 exhibits an N-terminal 17-amino acid extension enriched in hydrophobic residues (Fig. 3A). A comparison of the four pea sequences with all the Trx h sequences in databases revealed that they were more similar to those of Arabidopsis than to any other species (data not shown). Thus, a phylogenetic tree was constructed with pea and Arabidopsis sequences (Fig. 3B). Pea Trx h1, h2, and h3 were found in the same subgroup as Arabidopsis Trx h1, whereas Trx h4 was clustered in another subgroup that included Trx h2 from Arabidopsis. This last cluster comprised so far only four isoforms: Trx h2 from Arabidopsis, Trx h1 and h2 from soybean (Glycine max; Shi and Bhattacharyya, 1996), and Trx h4 from pea.
Figure 3.
Primary sequences of Trx h1 to h4 from pea and comparison with those of Trx h isoforms from Arabidopsis. A, Pea Trx h protein sequences (GenBank accession nos.: h1, AJ319808; h2, AJ310990; h3, AY170650; and h4, AY170651) were aligned. Identical residues are indicated by an asterisk. Consensus sequences used to define degenerated primers and the possible endoplasmic reticulum retention motif KDEL present in h4 sequence are underlined. B, A phylogenetic tree was built using Trx h protein sequences from pea and Arabidopsis (h1, At3g51030; h2, At5g39950; h3, At5g42980; h4, At1g19730; h5, At1g45145; h7, At1g59730; h8, At1g69880; and h9, At3g08710 from The Institute for Genomic Research) according to Corpet (1988).
Overexpression in Escherichia coli of Trx h3 and Trx h4 and Production of Discriminating Antibodies
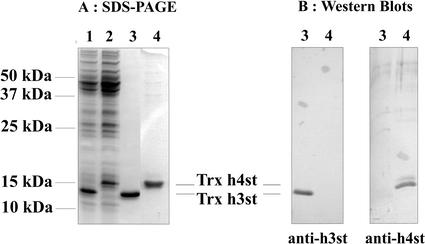
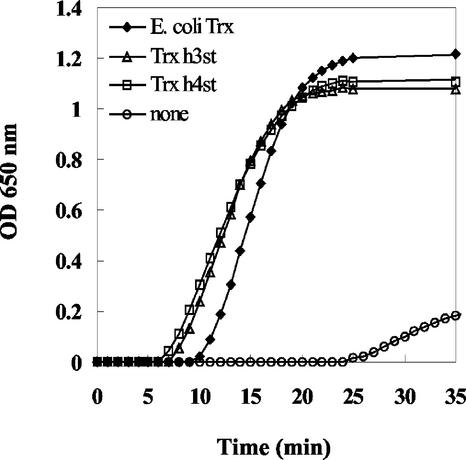
To investigate whether Trx h3 and Trx h4 encoded functional proteins, cDNAs were cloned into the expression vector pASK-IBA3 designed to produce recombinant proteins with a C-terminal Strep-tag. Recombinant Trx h3st and Trx h4st were found to be highly expressed in E. coli as soluble proteins and were purified using Strep-Tactin-Sepharose (Fig. 4A). The purified recombinant proteins were used to raise polyclonal antibodies in rabbits. The specificity of the two antisera (anti-Trx h3st and antiTrx h4st) was demonstrated by the absence of cross reactions using the recombinant proteins as antigens (Fig. 4B). Both purified Trx h3st and Trx h4st readily reduced insulin with similar kinetics. They appear to promote a faster insulin reduction than the E. coli Trx used as a control (Fig. 5).
Figure 4.
Purification and immunological characterization of Trx h3st and Trx h4st overexpressed in E. coli. A, Soluble extracts from E. coli transformed with pASK-IBA3-Trx h3st (lane 1) or pASK-IBA3-Trx h4st (lane 2) and 1 μg of respective proteins purified on StrepTactin (lanes 3 and 4) were resolved by 15% (w/v) SDS-PAGE, and then the gel was stained with Coomassie Blue. Molecular masses of standard proteins are indicated at the left of the figure. The respective apparent molecular masses determined for recombinant proteins Trx h3st or Trx h4st are 15 and 14 kD. B, Purified proteins (200 ng) resolved by SDS-PAGE were transferred onto nitrocellulose membranes that were probed with a 1:1,000 (v/v) dilution of antibodies raised against either Trx h3st or Trx h4st using the phosphatase alkaline assay.
Figure 5.
Insulin reduction by Trx h3st and Trx h4st. The disulfide reductase activity of Trx h3st and Trx h4st (20 μg mL–1) was assayed by measuring the rate of insulin reduction, which was followed by turbidimetry at 650 nm. Control assays were performed in the absence of Trx (none) or in the presence of 20 μg mL–1 Trx from E. coli.
Differential Expression of Trx h3 and Trx h4 during Germination
The expression of the four Trx h isoforms during germination and early seedling growth of pea was first investigated by northern-blot analysis using DNA probes derived from the cDNA coding regions. Because cross-hybridization could occur between the different isoforms (Brugidou et al., 1993; Serrato et al., 2001), the specificity of the probes was tested with dot blots. No cross reaction was found between h2, h3, and h4, whereas h1 and h3 slightly hybridized with each other (results not shown). The cross reaction between h1 and h3 is most likely due to their high degree of identity at the nucleotide sequence level (72% in the coding regions).
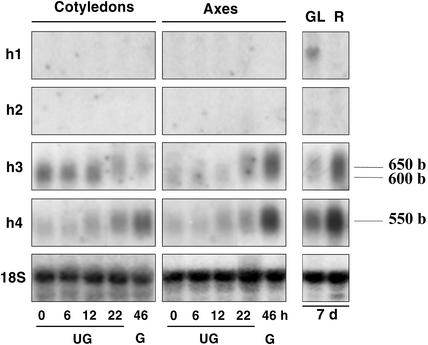
Transcripts of Trx h1 and h2 were not detected by northern blot in germinating seeds (Fig. 6), which confirmed the lack of detection by RT-PCR (Fig. 2). Thus, the signals obtained with the h3 probe in germinating seeds cannot be attributed to h1. Among the other organs tested during early seedling growth, h1 and h2 were only found in green leaves, albeit in a very low amount (Fig. 6). In contrast, Trx h3 and h4 were clearly detected in dry and imbibed seeds and in all other organs examined (Fig. 6). In dry seeds, Trx h3 and h4 transcripts were present both in embryo axes and cotyledons, transcript levels of h3 being higher than that of h4. Trx h3 and h4 were found to be differentially expressed during germination. In the course of imbibition, the content in h3 gradually decreased in cotyledons but increased in embryo axes before the onset of radicle protrusion (22 h) and afterward (Fig. 6). However, because a slight increase (600–650 b) in the size of detected band was systematically observed between early germination (0, 6, and 12 h) and latter stages (22 and 46 h), including 7-d-old seedlings, the presence of two very similar transcripts cannot be excluded. Trx h4 (550 b) amount started to increase during phase II of imbibition in both embryo axes and cotyledons (Fig. 6) and remained high during early seedling growth.
Figure 6.
Expression of Trx h isoforms in seeds and seedlings on northern blots. Total RNA from cotyledons and embryo axes from mature seeds imbibed for 0 to 46 h (UG, ungerminated seeds; G, germinated seeds) and from green leaves (GL) and roots (R) from 7-d-old seedlings were resolved per lane of 1.4% (w/v) agarose denaturing gels. They were transferred onto Hybond N membrane and probed with [32P]-labeled coding regions of Trx h1 to h4 cDNA or [32P]-labeled 18 S cDNA from Medicago truncatula. After hybridization and stringent washing (0.2× SSC and 0.02% [w/v] SDS at 65°C), membranes were autoradiographied for 3 d at –80°C.
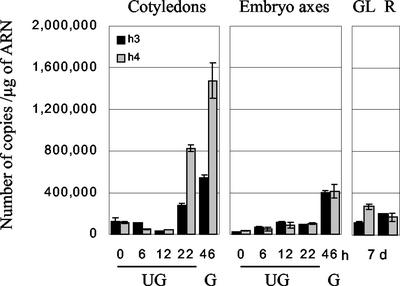
A further analysis of Trx h isoform expression was performed by quantitative RT-PCR. The lack of expression of Trx h1 and h2 during germination was further confirmed (data not shown). As for the northern blot, Trx h3 and h4 were found to be expressed in dry and imbibed seeds and in green leaves and roots (Fig. 7). In contrast, they showed similar expression profiles upon imbibition, with a strong induction of expression in axes after the radicle protrusion (46 h) and in cotyledons just before and after radicle protrusion (22 and 46 h). Concerning the isoform h4, this pattern is in accordance with that obtained in northern blot. Considering the expression of Trx h3 transcripts in cotyledons at early stages of germination, quantitative RT-PCR yielded different results because the amount of transcript was much lower than expected from the northern-blot signal (Figs. 6 and 7). Such a result might be explained by the presence of another cross-hybridizing isoform.
Figure 7.
Expression of Trx h3 and h4 isoforms in seeds and seedlings by quantitative PCR. cDNAs produced by RT of total RNAs from cotyledons and embryo axes from seeds imbibed for 0 to 46 h (UG, ungerminated seeds; G, germinated seeds) and from green leaves (GL) and roots (R) from 7-d-old seedlings were used as templates in quantitative PCR realized in the presence of Sybr Green. Copy number of each isoform was determined using a standard curve established with the recombinant plasmid containing the corresponding full-length sequence.
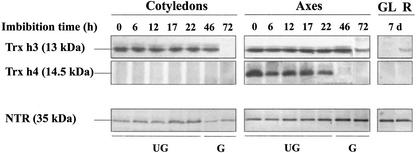
To investigate the expression of the protein isoforms Trx h3 and h4 in seeds during germination and early seedling growth, western blots were performed upon soluble proteins using the specific anti-Trx h3st and anti-Trx h4st antibodies (Fig. 8). Trx h4 exhibited an apparent higher molecular mass (14.5 kD) than Trx h3 (13 kD), which agrees well with the different theoretical molecular masses (14.549 and 12.578 kD, respectively) deduced from the cDNAs. Trx h3 was easily detected from 0 to 46 h of imbibition, both in cotyledons and embryo axes (Fig. 8). Although its level remained constant in both parts until 46 h, Trx h3 could not be detected anymore in cotyledons at 72 h, and its amount was markedly reduced in embryo axes at 72 h. It could be detected, albeit at low levels, in 7-d-old seedlings root tissues, but not in leaves (Fig. 8). The expression profile of Trx h4 was clearly different because the protein was only detected in embryo axes of dry or imbibed but ungerminated seeds (6–22 h), its amount decreasing slightly upon imbibition. The protein could not be detected in embryo axes and cotyledons of germinated seeds (46 and 72 h) or in the organs from 7-d-old seedlings examined. Thus, Trx h4 could be a seed-specific isoform.
Figure 8.
Expression of Trx h3, Trx h4, and NTR in seeds and seedlings. Soluble proteins from cotyledons and embryo axes of seeds imbibed for 0 to 72 h (UG, ungerminated seeds; G, germinated seeds) and those from green leaves (GL) and roots (R) from 7-d-old seedlings were resolved by SDS-PAGE. For respective expression analysis of Trx h3, Trx h4, and NTR, 15%, 15%, and 12% (w/v) acrylamide gels were used, and 25, 50, and 15 μg of proteins were loaded per lane. Proteins were then transferred onto nitrocellulose membranes that were probed with anti-Trx h3st, anti-Trx h4st, or anti-Arabidopsis NTR.
Expression of NTR during Imbibition and Content in NADPH
A functional NTS requires the presence of the two other components of the system, namely NTR and the co-enzyme NADPH. The amount in NTR in embryo axes and cotyledons was investigated by western blotting using antibodies raised against Arabidopsis NTR, which revealed a single polypeptide with the expected size of 35 kD. NTR, which was detected in both tissues, was more abundant in embryo axes than in cotyledons. Its content slightly increased in axes after radicle protrusion, whereas it remained almost constant in cotyledons (Fig. 8).
Because NADPH is an essential metabolite undoubtedly required for germination, we effectively measured NADPH amounts of 2 nmol g–1 dry weight in germinating seeds, which agrees well with other values reported in seeds and seedlings (Zhao et al., 1987).
DISCUSSION
Because the NTS is known to intervene in redox regulation of crucial molecular events in microorganisms and animal cells, similar functions in plant cells may be envisioned. The system has been extensively studied in germinated cereal seeds where its role in the mobilization of reserves was clearly established (for review, see Besse and Buchanan, 1997). We have addressed whether the NTS could be functional in legume seeds by monitoring the expression of Trx h isoforms and NTR during early germination and seedling growth of pea seeds.
All three components of the system, NADPH, NTR, and Trx h, were demonstrated to be present in dry pea seeds and in germinating seeds, indicating a role of the system during germination. Trx h being encoded by multiple genes in plants, we have dissected the system by analyzing the expression of Trx h isoforms in germinating seeds. Because the transcripts of already known h1 and h2 isoforms were not detected, we designed a strategy to clone other putative isoforms that could be expressed in seeds. This approach revealed that two new isoforms, Trx h3 and h4, were expressed both at mRNA and protein levels during germination.
The two new isoforms, h3 and h4, exhibit a primary structure highly similar with other Trx h and with the canonical disulfide active site (WCGPC). The analysis of the pea sequences indicated that Trx h1, h2, and h3, which were clustered together, differed more from Trx h4 than from Arabidopsis Trx h1. Such an observation is consistent with the hypothesis that the apparition of the numerous h isoforms had started before higher plant divergence (Sahrawy et al., 1996; Juttner et al., 2000; Laloi et al., 2001). Trx h1, h2, and h3 are also similar to the majority of h isoforms already isolated from different species and, likewise, they are also expected to be localized in the cytosol.
In contrast, Trx h4 belongs to the subgroup that includes Trx h2 from Arabidopsis and Trx h1 and h2 from soybean (Shi and Bhattacharyya, 1996). These isoforms exhibit a hydrophobic N-terminal extension of 15 or more amino acids, suggesting that they could be bound to membranes. They also contain a KDEL motif in the C-terminal region that was described to confer retention in the endoplasmic reticulum. It is noteworthy that because this motif is replaced by a similar one (KEEL) in the other pea isoforms expected to be cytosolic, it might not be actually related, therefore, to localization. In fact, divergent results were obtained concerning the localization of these isoforms in soybean (Shi and Bhattacharyya, 1996) and Arabidopsis (Rivera-Madrid et al., 1995). Soybean proteins were reported to be bound to the plasma membrane, whereas Arabidopsis h2 and pea Trx h4 were both extracted as soluble proteins. An important question is the subcellular localization of the numerous Trx h isoforms; we are currently investigating the expression of green fluorescent protein fusions transiently expressed in pea leaf protoplasts.
At the mRNA level, among the four known pea Trx h, only Trx h3 and h4 were found to be expressed in dry and germinating seeds, but also in seedlings. No transcript encoding Trx h1 and h2 could be detected, suggesting these isoforms do not intervene in germination. In addition, they were only slightly detected in green leaf tissue. The only clues regarding their function consist in their annotations in GenBank (accession nos. AJ319808 and AJ310990), with h2 being linked to oxidative stress and development. Because Trx h3 and h4 were the major Trx h in seeds, we produced recombinant proteins that proved to be active in terms of insulin reduction, thus confirming the Trx activity of the isoforms. We were then able to raise antibodies that were highly specific and, thus, allowed to discriminate Trx h3 and Trx h4 expression at the protein level. It is noteworthy that antibodies previously raised against Trx h generally cross-reacted with several isoforms (Lozano et al., 1996; Serrato et al., 2001; Marx et al., 2003).
Both Trx h3 and h4 were found in dry seeds either only in embryo axes (h4) or in both axes and cotyledons (h3), indicating that they were synthesized before germination. During germination, Trx h3 amount remained constant both in embryo axes and cotyledons, whereas h4 was found only in axes, its level decreasing slightly upon imbibition and the protein disappearing after radicle protrusion. After germination, Trx h3 amount decreased slowly but was still detected in 7-d-old seedlings. A decrease in the overall Trx h content was also reported in cereals during germination and early seedling growth (Lozano et al., 1996; Serrato et al., 2001; Marx et al., 2003). As a whole, our results point out the differential expression pattern between the two isoforms at protein level, Trx h4 being intimately linked to the germination process. However, no correlation could be found between mRNA amount, determined by quantitative PCR, and protein expression, with even an inverse relation in cotyledons where mRNAs for Trx h4 were abundant and the protein was not detected. Considering that quantitative PCR is a specific method for sequence detection, the only explanation is that in pea as in other higher organisms, posttranscriptional regulation intervenes in the control of Trx h genes, taking into account the low correlation coefficient between mRNA and protein abundances (Anderson and Seilhamer, 1997; Gygi et al., 1999).
The main result inferred from this work is the expression in seeds of two new isoforms Trx h3 and h4, which raises the question of their functions. The growing body of evidence that Trx h are involved in reserve mobilization in cereals (Besse and Buchanan, 1997; Wong et al., 2002) speaks in favor of a similar role in legume seeds that harbor starch and protein reserves. We have identified in the soluble proteins of germinating pea seeds (22 h of imbibition) several proteolytic peptides derived from storage proteins (data not shown), which confirms that solubilization and proteolysis were already engaged. To demonstrate the potential role of Trx h3 or h4 in reserve mobilization, the characterization of their redox status together with the proteolysis and redox status of storage proteins during germination will be required, using for instance the monobromobimane approach (Lozano et al., 1996). Another general role that could be envisioned is the protection against reactive oxygen species that are highly produced with the resumption of metabolism (Leprince et al., 1990; Puntarulo et al., 1991; Cakmak et al., 1993; Aalen, 1999). Trx h has been suggested to act as hydrogen donor to 1Cys-peroxiredoxin, which protects embryo macromolecules from oxidation during early imbibition (Aalen, 1999). However, such a role for Trx h3 and h4 is questionable because their Arabidopsis orthologs could not confer hydrogen peroxide tolerance by complementation of a yeast (Saccharomyces cerevisiae) mutant deficient in Trx and that exhibited hydrogen peroxide hypersensitivity, Met auxotrophy, inability to reduce Met sulfoxide, and a perturbed cell cycle (Muller, 1995; Mouaheb et al., 1998). Interestingly, although some redundancy (complementation of cell cycle defect) was observed in the complementation experiments performed with Trx h1 to h5 from Arabidopsis, only the ortholog of pea Trx h4 was able to both restore Met prototrophy and allow growth on Met sulfoxide (Mouaheb et al., 1998). In connection with these observations, pea Trx h4 could play a role in sulfate assimilation and in the repair of protein damaged by oxidative stress. Trx h4 expression is restricted to embryonic axes, where the assimilation of sulfate and the reduction of Met sulfoxide into Met could be crucial processes to provide sulfur-containing amino acids that are necessary in embryo axes to sustain early protein synthesis.
The results presented here provide the first characterization, to our knowledge, of the NTS in legume seeds. The system, which appeared operational, involves new Trx h isoforms differentially expressed in seeds and, thus, expected to carry different roles during germination. Further understanding of the function of the system with regard to germination of legume seeds will be addressed through the identification of Trx h isoforms targets using mutated proteins as ligands in affinity chromatography or as baits in yeast double-hybrid screening (Verdoucq et al., 1999).
MATERIALS AND METHODS
Materials
Pea (Pisum sativum L. cv Baccara) seeds harvested in 2000 were obtained locally from an agronomical research institute (Fédération Nationale des Agriculteurs Multiplicateurs de Semences, Brain-sur-l'Authion, France) and stored in sealed plastic bags at 5°C (70% relative humidity).
Mature seeds were allowed to germinate on folded filter paper moistened with water (0.18 mL cm2) at 20°C in the dark or in the light (16 h of light/8 h of dark). Water content was determined by measuring the fresh and dry weight of the seeds (the latter was determined after heating at 100°C for 3 d).
Total RNA Extraction, RACE, and RT-PCR
Total RNAs were extracted according to Verwoerd et al. (1989). Full-length 5′- and 3′-adapted ends cDNAs from dry or 22-h-imbibed pea seeds, obtained as already reported (Duval et al., 2002) using the GeneRacer kit from Invitrogen (Groningen, The Netherlands), or 7-d-old green leaves cDNAs, obtained by classical RT (Huang et al., 1996), were used as templates in RACE and nonquantitative RT-PCR experiments. PCR reactions were performed using oligonucleotides and conditions (annealing temperature and number of cycles) described in Table I, with a preliminary denaturation step of 4 min at 94°C, followed by one or two sets of cycles (94°C × 1 min, annealing temperature × 1 min, and 72°C × 1 min) and then a final elongation step of 15 min at 72°C.
The two already known sequences of pea Trx h cDNAs (GenBank accession nos. AJ319808 and AJ310990, respectively, numbered h1 and h2 for convenience in this paper) were amplified by RT-PCR using specific primers designed at the 5′ and 3′ ends of their coding sequence (Table I). To amplify cDNAs from Trx h that could be expressed during seed germination, four degenerated primers (Xs-1A, Xs-1B, Xa-anti2A, and Xa-anti2B) were derived from two motifs conserved in protein sequences of Trx h from different sources. PCR products were then cloned into the PCR4-TOPO vector of the TOPO TA Cloning Kit for Sequencing (Invitrogen) according to the manufacturer's instructions. To obtain the corresponding full-length cDNAs, 3′1, 3′2, 5′1, and 5′2 primers (Table I) were designed to perform RACE according to the GeneRacer kit's instructions. DNA sequencing was performed by MWG-Biotech AG (Ebersberg, Germany).
For quantitative RT-PCR experiments, cDNAs—obtained from 2 μg of total RNAs by classical RT (Huang et al., 1996) in a total volume of 50 μL—were used as templates using forward and reverse primers (-for and -rev, Table I) designed with the Primer Express software (PE-Applied Biosystems, Foster City, CA). Quantitative PCR was performed in a total volume of 25 μL containing 2 μL of cDNAs, 0.3 μm each primer, and 12.5 μL of 2× Sybr Green master mix (Applera, Courtaboeuf, France), with a preliminary step of 5 min at 94°C, followed by 40 cycles of 94°C × 15 s and 60°C × 1 min (ABI Prism 7000 SDS, PE-Applied Biosystems). For quantification, standard curves were realized with 5,000 to 100,000 copies of recombinant plasmids containing full-length sequences of Trx h1 to h4.
Northern-Blot Analysis
Total RNA aliquots (7.5 μg) were run on 1.4% (w/v) agarose denaturing gel as described by Lehrach et al. (1977), transferred onto Hybond N membrane (Amersham Biosciences, Piscataway, NJ), and UV cross linked. Prehybridization and hybridization of the membranes were then performed according to the manufacturer's instructions. Membranes were probed with [32P]-labeled coding regions of Trx h1 to h4 cDNA (using [32P]dCTP and the Rediprime II kit, Amersham Biosciences). Then, they were washed (0.2× SSC and 0.02% [w/v] SDS for 1 h at 65°C) and subsequently exposed to film (Hyperfilm MP, Amersham Bioscience) for 2 to 3 d at –80°C. Equal loading of RNA per lane was checked by using Medicago truncatula 18 S cDNA (amplified with 18 S primers, Table I) as a control probe.
Specificity of the Trx h probes was ascertained by a dot-blot analysis. One nanogram of each plasmid containing a pea Trx h sequence was denaturated by 0.4 m NaOH, dotted in the edge of each blot, and UV cross linked.
Overexpression of Pea Trx h3 and h4 in Escherichia coli
The coding sequences of pea Trx h3 and h4 were subcloned in the BsaI site of the pASK-IBA3 plasmid (IBA, Gottingen, Germany) using the primers h3pASK-sens/h3pASK-anti and h4pASK-sens/h4pASK-anti, respectively (Table I), which were designed with the Primer D'Signer software (IBA, Gottingen, Germany). The pASK-IBA3 plasmid was designed to express target proteins fused to a C-terminal tag, allowing the purification of recombinant on Strep-Tactin resin (IBA).
The recombinant plasmids were introduced into competent E. coli XL2 blue (Hanahan, 1983). Overexpression and purification of the recombinant protein were carried out according to the manufacturer's instructions. The activities of recombinant Trx h (Trx h3st or Trx h4st) were assayed by following, at optical density 650 nm, the reduction of insulin according to Holmgren (1979) in the reaction mixture: 100 mm potassium phosphate buffer (pH 7), 2 mm EDTA, 0.8 mg mL–1 insulin, 1 mm dithiothreitol, and 20 μg mL–1 Trx h3st or Trx h4st. Control assays were performed in the absence of Trx (none) or in the presence of 20 μg mL–1 Trx from E. coli.
Western-Blot Analysis
For soluble protein extraction, embryo axes and cotyledons from pea seeds imbibed for 0 to 46 h were ground in liquid nitrogen and 5 mL g fresh weight–1 (or 10 mL for tissues from dry seeds) of 50 mm Tris-HCl (pH 7.8), 5 mm EDTA, and 5 mm 2-mercaptoethanol was added. The resulting homogenates were centrifuged (50,000g for 20 min at 4°C), and the soluble proteins were stored at –20°C. Protein contents were determined by using bovine serum albumin as a standard (Bradford, 1976).
For western-blot analysis, 15 to 50 μg of proteins was resolved per lane on 12% or 15% (w/v) acrylamide gels by SDS-PAGE (Laemmli, 1970) and transferred onto nitrocellulose membranes (Schleicher & Schull, Dassel, Germany) as previously described (Duval et al., 2002). Membranes were probed with a 1:1,000 (v/v) dilution of antibodies raised against pea Trx h3st, Trx h4st, or Arabidopsis NTR, and immunodetection was performed using the phosphatase alkaline assay in the presence of 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Rabbit antibodies raised against pea Trx h3 and h4 were produced by Davids Biotechnologie (Regensburg, Germany). Rabbit antibodies raised against Arabidopsis NTR were a gift of Y. Meyer (Unité Mixte de Recherche 5096, Centre National de la Recherche Scientifique, University of Perpignan, France).
Measurement of NADPH Amounts
Freshly separated embryo axes and cotyledons from pea seeds imbibed for 0 to 46 h were ground at room temperature in 0.1 m NaOH for the extraction of NADPH (16 mL g–1 dry seeds or 12 mL g–1 imbibed seeds) according to Zhao et al. (1987). Measurements of NADPH amounts were performed using Glc-6-P dehydrogenase of baker's yeast (Saccharomyces cerevisiae) in the presence of phenazine ethosulfate and methylthiazoletetrazolium (Delumeau et al., 2000).
Acknowledgments
We thank Dr Julia Buitink for stimulating discussions and suggestions with the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.019562.
This work was supported by the Conseil Régional Pays de Ra Loire, France (CER Semences 2000–2003).
References
- Aalen RB (1999) Peroxiredoxin antioxidants in seed physiology. Seed Sci Res 9: 285–295 [Google Scholar]
- Anderson L, Seilhamer J (1997) A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18: 533–537 [DOI] [PubMed] [Google Scholar]
- Arrigo AP (1999) Gene expression and the thiol redox state. Free Radic Biol Med 27: 936–944 [DOI] [PubMed] [Google Scholar]
- Besse I, Buchanan BB (1997) Thioredoxin-linked plant and animal processes: the new generation. Bot Bull Acad Sin 38: 1–11 [Google Scholar]
- Besse I, Wong JH, Kobrehel K, Buchanan BB (1996) Thiocalsin: a thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Nat Acad Sci USA 93: 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD, Black M (1994) Seeds Physiology of development and germination. Ed 2. Plenum Press, New York
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantity of protein utilizing the principle of protein dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brugidou C, Marty I, Chartier Y, Meyer Y (1993) The Nicotiana tabacum genome encodes two cytoplasmic thioredoxin genes which are differently expressed. Mol Gen Genet 238: 285–293 [DOI] [PubMed] [Google Scholar]
- Buchanan BB (1991) Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys 288: 1–9 [DOI] [PubMed] [Google Scholar]
- Cakmak I, Dragana S, Marschner H (1993) Activities of hydrogen peroxide-scavenging enzymes in germinating wheat seeds. J Exp Bot 44: 127–132 [Google Scholar]
- Cho MJ, Wong JH, Marx C, Jiang W, Lemaux PG, Buchanan BB (1999) Overexpression of thioredoxin h leads to enhanced activity of starch debranching enzyme (pullulanase) in barley grain. Proc Nat Acad Sci USA 96: 14641–14646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delumeau O, Morère-Le Paven M-C, Montrichard F, Laval-Martin DL (2000) Effects of short-term NaCl stress on calmodulin transcript levels and calmodulin-dependent NAD kinase in two species of tomato. Plant Cell Environ 23: 329–336 [Google Scholar]
- Duval FD, Renard M, Jaquinod M, Biou V, Montrichard F, Macherel D (2002) Differential expression and functional analysis of three calmodulin isoforms in germinating pea (Pisum sativum L.) seeds. Plant J 32: 481–493 [DOI] [PubMed] [Google Scholar]
- Florencio FJ, Yee BC, Johnson TC, Buchanan BB (1988) An NADP/thioredoxin system in leaves: purification and characterization of NADP-thioredoxin reductase and thioredoxin h from spinach. Arch Biochem Biophys 266: 496–507 [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R (1999) Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254: 9627–9632 [PubMed] [Google Scholar]
- Holmgren A (1985) Thioredoxin. Annu Rev Biochem 54: 237–271 [DOI] [PubMed] [Google Scholar]
- Huang Z, Fasco MJ, Kaminsky LS (1996) Optimization of Dnase I removal of contaminating DNA from RNA for use in quantitative RNA-PCR. BioTechniques 20: 1012–1020 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195: 456–463 [DOI] [PubMed] [Google Scholar]
- Jiao J-A, Yee BC, Kobrehel K, Buchanan BB (1992) Effect of thioredoxin-linked reduction on the activity and stability of the Kunitz and Bowman-Birk soybean inhibitor proteins. J Agric Food Chem 40: 2333–2336 [Google Scholar]
- Johnson TC, Cao RQ, Kung JE, Buchanan BB (1987) Thioredoxin and NADP-thioredoxin reductase from cultured carrot cells. Planta 171: 321–331 [PubMed] [Google Scholar]
- Juttner J, Olde D, Langridge P, Baumann U (2000) Cloning and expression of a distinct subclass of plant thioredoxins. Eur J Biochem 267: 7109–7117 [DOI] [PubMed] [Google Scholar]
- Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB (1992) Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol 99: 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrehel K, Yee BC, Buchanan BB (1991) Role of the NADP/thioredoxin system in the reduction of alpha-amylase and trypsin inhibitor proteins. J Biol Chem 266: 16135–16140 [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Laloi C, Rayapuram N, Chartier Y, Grienenberger JM, Bonnard G, Meyer Y (2001) Identification and characterization of a mitochondrial thioredoxin system in plants. Proc Nat Acad Sci USA 98: 14144–14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H, Diamond D, Wozney JM, Boedtker H (1977) RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry 16: 4743–4751 [DOI] [PubMed] [Google Scholar]
- Leprince O, Deltour R, Thorpe P, Atherton N, Hendry G (1990) The role of free radicals and radical processing systems in loss of dessication tolerance in germinating maize (Zea mais L.). New Phytol 116: 573–580 [Google Scholar]
- Lozano RM, Wong JH, Yee BC, Peters A, Kobrehel K, Buchanan BB (1996) New evidence for a role for thioredoxin h in germination and seedling development. Planta 200: 100–106 [Google Scholar]
- Marx C, Wong J-H, Buchanan BB (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216: 454–460 [DOI] [PubMed] [Google Scholar]
- Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y (1998) In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast (Saccharomyces cerevisiae). Proc Nat Acad Sci USA 95: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E (1995) A redox-dependent function of thioredoxin is necessary to sustain a rapid rate of DNA synthesis in yeast. Arch Biochem Biophys 318: 356–361 [DOI] [PubMed] [Google Scholar]
- Obroucheva NV, Antipova OV (1997) Physiology of the initiation of seed germination. Russ J Plant Physiol 44: 250–264 [Google Scholar]
- Powis G, Mustacich D, Coon A (2000) The role of the redox protein thioredoxin in cell growth and cancer. Free Rad Biol Med 29: 312–322 [DOI] [PubMed] [Google Scholar]
- Puntarulo S, Galleano M, Sanchez R, Boveris A (1991) Superoxide anion and hydrogen peroxide metabolism in soybean embryonic axes during germination. Biochim Biophys Acta 1074: 277–283 [DOI] [PubMed] [Google Scholar]
- Reichheld J-P, Mestres-Ortega D, Laloi C, Meyer Y (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol Biochem 40: 685–690 [Google Scholar]
- Rivera-Madrid R, Mestres D, Marinho P, Jacquot JP, Decottignies P, Miginiac-Maslow M, Meyer Y (1995) Evidence for five divergent thioredoxin h sequences in Arabidopsis thaliana. Proc Nat Acad Sci USA 92: 5620–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y (1996) Intron position as an evolutionary marker of thioredoxins and thioredoxin domains. J Mol Evol 42: 422–431 [DOI] [PubMed] [Google Scholar]
- Schobert C, Baker L, Szederkenyi J, Grossmann P, Komor E, Hayashi H, Chino M, Lucas WJ (1998) Identification of immunologically related proteins in sieve-tube exudate collected from monocotyledonous and dicotyledonous plants. Planta 206: 245–252 [Google Scholar]
- Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ (2001) Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol 46: 361–371 [DOI] [PubMed] [Google Scholar]
- Shi J, Bhattacharyya MK (1996) A novel plasma membrane-bound thioredoxin from soybean. Plant Mol Biol 32: 653–662 [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Vignols F, Jacquot JP, Chartier Y, Meyer Y (1999) In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem 274: 19714–19722 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S (1993) A simple procedure for enhancing PCR specifivity. PCR Methods Applic 3: 77–80 [DOI] [PubMed] [Google Scholar]
- Wong JH, Kim Y-B, Ren P-H, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Nat Acad Sci USA 99: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Kobrehel K, Buchanan BB (1995) Thioredoxin and seed proteins. Methods Enzymol 252: 228–240 [DOI] [PubMed] [Google Scholar]
- Yano H, Wong JH, Lee YM, Cho MJ, Buchanan BB (2001) A strategy for the identification of proteins targeted by thioredoxin. Proc Nat Acad Sci USA 98: 4794–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Hu X, Ross C (1987) Comparison of tissue preparation methods for assay of nicotinamide coenzymes. Plant Physiol 84: 987–988 [DOI] [PMC free article] [PubMed] [Google Scholar]