Abstract
We have developed a high-throughput T-DNA insertional mutagenesis program in tomato using activation tagging to identify genes that regulate metabolic pathways. One of the activation-tagged insertion lines (ant1) showed intense purple pigmentation from the very early stage of shoot formation in culture, reflecting activation of the biosynthetic pathway leading to anthocyanin accumulation. The purple coloration resulted from the overexpression of a gene that encodes a MYB transcription factor. Vegetative tissues of ant1 plants displayed intense purple color, and the fruit showed purple spotting on the epidermis and pericarp. The gene-to-trait relationship of ant1 was confirmed by the overexpression of ANT1 in transgenic tomato and in tobacco under the control of a constitutive promoter. Suppression subtractive hybridization and RNA hybridization analysis of the purple tomato plants indicated that the overexpression of ANT1 caused the upregulation of genes that encode proteins in both the early and later steps of anthocyanidin biosynthesis as well as genes involved in the glycosylation and transport of anthocyanins into the vacuole.
INTRODUCTION
The genomes of higher plants are composed of thousands of genes that control growth, physiology, reproduction, and pathogen interaction traits. Understanding the functions of these genes is a major challenge, and a variety of forward- and reverse-genetics methods are being applied to this challenge. A classic genetics approach to understanding the functions of uncharacterized genes is to identify loss-of-function mutations in these genes and to screen for associated phenotypes. The limitations of classic mutagenesis strategies include the difficulty of identifying genes that (1) act redundantly, (2) are required during multiple stages of the life cycle, (3) result in early embryonic or gametophytic lethality, or (4) occur in species not suited for classic mutagenesis approaches. We have used “activation tagging” in tomato to facilitate the rapid functional analysis of genes involved in metabolic pathways. Activation tagging produces dominant mutations by means of the overexpression of endogenous genes with transcriptional enhancers that cause the ectopic expression of genes in the vicinity of the T-DNA insertion site (Borevitz et al., 2000; Van der Fits and Memelink, 2000; Weigel et al., 2000; Huang et al., 2001; Zubuko et al., 2002).
Tomato is a well-established model organism for the study of many biological processes. Tomato offers several features that enable studies on the development and ripening of fleshy fruit and on many plant–pathogen interactions that affect economically important plants. Its moderately sized genome (950 Mb with hundreds of mapped traits and molecular markers), tolerance to inbreeding, amenability to genetic transformation, and diversity of secondary metabolism make tomato an excellent platform for genetic and molecular research (Tanksley, 1993). Compared with other commercial crop plants, a relatively large number of single-gene-determined traits have been described in tomato, with an estimate of 1200 available monogenic traits (Stevens and Rick, 1986; Hille et al., 1989). Map-based cloning recently resulted in the first cloned quantitative trait loci in tomato (Frary et al., 2000; Fridman et al., 2000). It is predicted that the tomato genome encodes ∼35,000 genes sequestered largely in the euchromatic regions, corresponding to less than one-quarter of the total DNA in the tomato nucleus (Van der Hoeven et al., 2002).
The tomato cultivar used in this study is Micro-Tom, a variety originally bred for home gardening purposes (Scott and Harbaugh, 1989). It is well suited for large-scale mutagenesis and thus functional genomics owing to its small size, rapid life cycle, and ease of transformation (Meissner et al., 1997, 2000). We have produced >10,000 independent activation-tagged transgenic lines in tomato and identified a large number of leaf and fruit color mutants. The biochemical and molecular characterization of one of these color mutants that displays an accumulation of anthocyanins is described here.
RESULTS
Generation of Activation-Tagged Lines of Micro-Tom Transgenic Lines
Several types of Micro-Tom tissues, including leaf, stem, hypocotyl, and shoot tip, were tested for their ability to be transformed by Agrobacterium tumefaciens strain GV3101 containing pAG3202, a binary vector with four copies of 35S enhancers and a selection cassette containing nptII. Although all of the tissues tested gave transformed plants, the hypocotyl segments were the explant of choice because of the ease of manipulation and the consistently high rate of transformation (40 to 60%).
Activation-tagged lines of Micro-Tom displayed variations from the wild-type untransformed plant in many phenotypic traits. From a population of 10,427 independent transgenic lines, 1338 transgenic lines (12.83%) exhibited variation from the wild type in one or more visually observable characteristics. The heritability of the observed variations was confirmed in a set of select mutants by evaluating T1 progeny (Table 1). The segregation ratio of the mutant phenotype in T1 indicated a dominant or semidominant trait. In addition to evaluating T1 progeny from plants observed to have a mutant phenotype in T0, we evaluated T1 progeny from 1014 randomly selected activation-tagged lines. Phenotypes not observed in the T0 generation were identified for 103 of these lines, indicating possible loss-of-function traits resulting from T-DNA integration. Ten percent of these lines produced T1 progeny with a visually observable variant phenotype (data not shown).
Table 1.
Confirmation of Mutant Phenotype in T1
| Mutant Phenotype in T0 | No. of T1 Plants Evaluated |
No. of Plants nptII Positive |
Percent of nptII Plants Displaying Mutant Phenotype |
|---|---|---|---|
| Anthocyanin color (ANT1) | 11 | 10 | 100 |
| “Budding” fruit | 7 | 6 | 83.33 |
| Chlorophyll/pale green leaves | 6 | 4 | 75 |
| Chlorophyll/pale green leaves | 17 | 14 | 57.14 |
| Dark green fruit | 18 | 10 | 100 |
| Dwarf stature | 7 | 5 | 80 |
| Dwarf, chlorophyll/pale green leaves | 14 | 10 | 100 |
| Increased numbers of flowers per inflorescence | 15 | 10 | 90 |
| Golden fruit | 13 | 10 | 90 |
| Golden fruit | 18 | 17 | 100 |
| Golden fruit | 11 | 10 | 100 |
| Golden fruit | 18 | 17 | 100 |
| Golden fruit | 16 | 16 | 62.5 |
| Golden fruit | 18 | 16 | 43.75 |
| Golden fruit | 14 | 10 | 100 |
| Golden fruit | 17 | 13 | 100 |
| Leaf margin/curly | 11 | 7 | 100 |
| Leaf margin/curly | 17 | 16 | 93.75 |
| Tall plant, approximately twice the normal height of Micro-Tom | 13 | 9 | 100 |
| Variegated (green and yellow striations on leaves) | 14 | 12 | 58.33 |
Some of the fruit mutants with varying color and shape are shown in Figure 1. The color phenotypes were of special interest in the present study as a means of finding genes involved in pigment biosynthesis. From among the various color mutants, an anthocyanin mutant (ant1) with prominent purple coloration was selected for complete characterization at the molecular, biochemical, and genetic levels. This anthocyanin mutant was identified at a very early stage of development in tissue culture as a purple-colored caulogenic callus of a hypocotyl explant. The T0 plant of the ant1 mutant had purplish leaves, stems and flowers with prominent purple striations, and fruit with purple cells on the epidermis, as shown in Figure 2.
Figure 1.
Fruit Color and Shape Mutants from Activation-Tagging Transgenic Lines.
(A) Wild-type fruit.
(B) Dark green fruit.
(C) Golden yellow fruit.
(D) Orange fruit.
(E) Salmon pink fruit.
(F) Cross-sections of wild-type and salmon pink fruit.
(G) Pear shape.
(H) Persimmon shape.
(I) Radish shape.
(J) Teardrop shape.
(K) Yeast-budding-type mutants.
All fruit variants were observed in the T0 generation except the salmon color mutant ([E] and [F]), which was observed in T1.
Figure 2.
ant1 Mutant.
(A) Wild-type and mutant plants of T1.
(B) Wild-type flower.
(C) Mutant flower of T0.
(D) Surface of a green fruit of the ant1 mutant from T0 at ×66 magnification.
The ANT1 Gene Encodes a MYB-Factor Protein
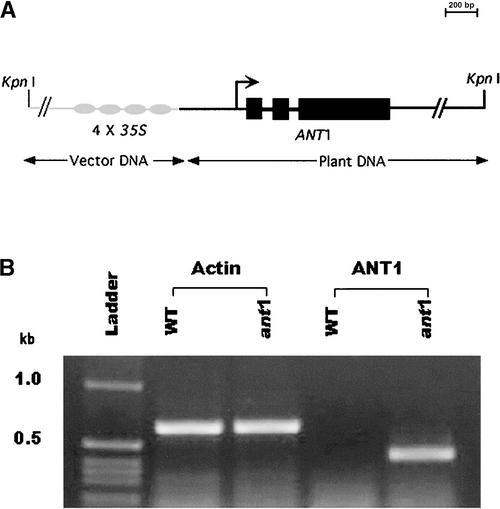
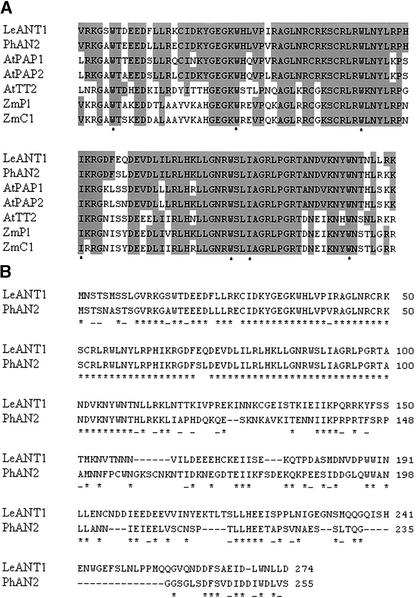
The inheritance of the ant1 mutant followed a typical Mendelian pattern for a dominant trait controlled by a single gene. Of 18 seeds that were sown, 11 germinated; 3 were sensitive to kanamycin (indicating the absence of the transgene), and the surviving 8 kanamycin-resistant plants displayed anthocyanin accumulation (Table 1). The presence of a single T-DNA insertion in the anthocyanin mutant was confirmed by T-DNA–specific PCR and DNA gel blot hybridization (data not shown). As shown in Figure 3A, a single predicted gene was identified in the 6.4-kb rescued genomic fragment adjacent to the 35S enhancers. The predicted gene has three exons that result in an 825-nucleotide coding region and a 274–amino acid protein. The translation start codon was 1810 bp away from the right T-DNA border. Reverse transcriptase (RT)–PCR confirmed that the transcriptional expression of the predicted ANT1 transcript is upregulated in the activation-tagged mutant relative to the wild-type background (Figure 3B), and sequencing of the product confirmed the predicted gene structure. There are no ESTs in the public databases that exactly match the predicted coding sequence of ANT1, which, in combination with the low abundance of this transcript in wild-type tomato, indicates that the expression of this gene normally is low or restricted to specific tissues and developmental stages. The ANT1 gene is predicted to encode a MYB-factor protein. An amino acid sequence alignment of the predicted ANT1 protein with other plant transcription factors known to function in the regulation of the anthocyanin biosynthetic pathway is shown in Figure 4.
Figure 3.
Structure and Expression of the Predicted Gene in the ant1 Mutant.
(A) Plasmid rescued from the anthocyanin mutant, and the predicted gene structure of ANT1 activated by 4× 35S enhancer.
(B) RT-PCR analysis of ANT1 expression. WT, wild type.
Figure 4.
Alignment of Predicted Protein Sequences of ANT1 and Related MYB Factors.
(A) Amino acid alignment of the R2R3 MYB domain of functionally characterized plant proteins involved in the regulation of flavonoid biosynthesis. The tomato MYB factor identified in this study (LeANT1) is compared with petunia AN2, Arabidopsis PAP1, Arabidopsis PAP2, Arabidopsis TT2, maize Pl, and maize C1. Residues that are identical to residues in the tomato ANT1 sequence are shaded. The positions of the conserved W/I residues implicated in DNA binding are indicated below the alignment with asterisks.
(B) Amino acid sequence alignment of the full-length tomato ANT1 and petunia AN2 proteins showing identical (asterisks) and similar (dashes) residues (CLUSTAL W, version 1.4: open gap penalty of 10.0; extended gap penalty of 0.1; similarity matrix of blosum62).
Recapitulation of the ant1 Mutant Phenotype
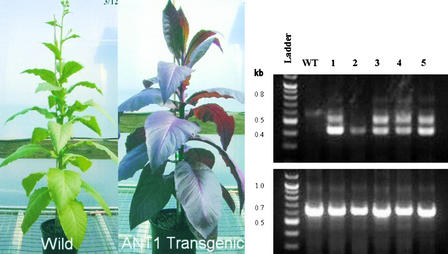
To demonstrate a direct relationship between ANT1 overexpression and the pigmented phenotype, we transformed tomato cv Micro-Tom with the ANT1 genomic DNA expressed under a strong constitutive promoter, cassava vein mosaic promoter (Verdaguer et al., 1996). As shown in Figure 5, the transgenic plants of tomato demonstrated a range of purple phenotypes—partial, weak, or strong accumulation of anthocyanin—confirming that overexpression of the gene ANT1 is responsible for the accumulation of anthocyanin. From 64 independent ANT1 transgenic events confirmed by genomic PCR, 54 displayed visibly evident purple coloration and 10 plants were green with no observable difference in color from the wild type. RT-PCR was performed to confirm the expression of the ANT1 gene in six of these transgenic plants, five that displayed purple coloration and one that appeared to exhibit a wild-type–like phenotype (Figure 6). RT-PCR confirmed the expression of transcript in the lines displaying purple coloration, whereas the green plant showed no detectable ANT1 gene expression (Figure 6, lane 3).
Figure 5.
ANT1 Recapitulated Tomato Plants AX, BK, and AW in the Greenhouse.
The degree of purpleness in flowers and fruit was correlated directly to the increased levels of transcript and anthocyanin pigment, respectively, in the seedling analyses of the various events.
Figure 6.
RT-PCR of ANT1 Transgenic Plants of Micro-Tom.
Plants 1 to 6 were transgenic plants. The Actin gene was used as a positive control. ANT1 was overexpressed in all plants that were purple in appearance (lanes 1, 2, 4, 5, and 6). Lane 3 contained RNA from a transgenic plant that was positive for ANT1 DNA (data not shown) but showed no purple color. WT, wild type.
To determine whether the ANT1 gene product could function in other solanaceous plants, tobacco cv Wisconsin was transformed with the same ANT1 construct described above. Transgenic plants of tobacco expressing the ANT1 gene also gave a range of phenotypes, from green to slightly or intensely purple plants. Figure 7 shows an intensely purple tobacco plant expressing the tomato ANT1 gene. Of 89 independent transgenic events confirmed by genomic PCR, 56 had purple coloration in tobacco. RT-PCR analysis showed the expression of ANT1 transcript in all of the samples with purple color (Figure 7).
Figure 7.
Recapitulation of Tobacco with ANT1 Gene.
At left, wild-type and ANT1 transgenic plants of tobacco cv Wisconsin. At right, RT-PCR data. All of the plants of ANT1 transgenic lines of tobacco were positive for the transcript (lanes 1 to 5). WT, wild type.
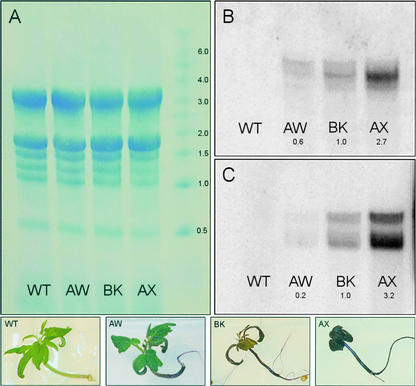
Quantitation of Anthocyanin Content and ANT1 Expression Levels in ANT1 Transgenic Tomato
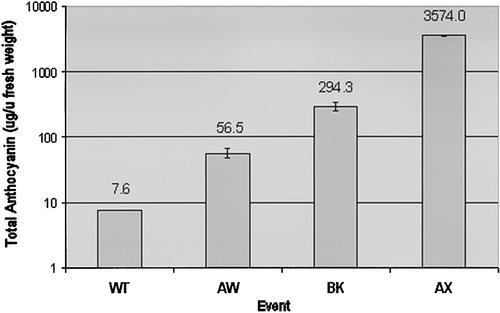
HPLC analysis with photodiode array detection was used to quantitate the anthocyanin content in three recapitulated ANT1 Micro-Tom lines designated BK, AW, and AX as well as in wild-type plants. All lines were grown in vitro and sampled in triplicate 3 weeks after seeds were sown. As shown in Figure 8, among the recapitulated lines, AX was by far the highest pigment accumulator, averaging 3574 μg of anthocyanin per gram fresh weight, an almost 500-fold increase over wild-type levels. BK had 294 μg/g fresh weight, and AW had 57 μg/g fresh weight. The amounts of anthocyanins detected in these lines correlated well with the levels of transcript expressed by these lines, as measured by RNA gel blot analysis (Figure 9). RNA gel blot analysis also showed the upregulation of a downstream gene that encodes glutathione S-transferase (GST) along with the increased expression of ANT1 (Figure 9C).
Figure 8.
Average Anthocyanin Content of Transgenic and Wild-Type Micro-Tom Plants (3-Week-Old Whole Seedlings).
Error bars indicate standard errors for n = 4. F.Wt, fresh weight.
Figure 9.
ANT1 Expression Levels in Transgenic Tomato Lines AW, BK, and AX.
(A) Equal RNA loading.
(B) and (C) Blots probed with radiolabeled DNA fragments corresponding to ANT1 or MTP4 encoding GST.
WT, wild type.
Further analysis of ant1 Micro-Tom leaf extracts by liquid chromatography/photodiode array tandem mass spectrometry indicated the presence of nine discrete anthocyanins. All had UV absorbance maxima near 520 nm, and their molecular weights in order of retention time were 773, 787, 801, 935, 949, 963, 919, 933, and 947. Fragmentation of the nine molecular ions yielded one of three daughter ions of molecular weight 303, 317, or 331, indicating the presence of delphinidin-, petunidin-, or malvidin-type anthocyanidins (aglycones), respectively. Comparison of the molecular weights and mass spectrometry fragmentation patterns of the tomato anthocyanins with common anthocyanin glycosylation and acylation motifs led us to conclude that ant1 Micro-Tom produces the 3-rutinoside-5-glucoside of delphinidin, petunidin, and malvidin. Further acylation of these pigments with p-coumaric acid or caffeic acid led to the production of six additional red to violet pigments (Table 2). All are known anthocyanins, and three have been reported previously in light-stressed tomatoes (Bovy et al., 2002).
Table 2.
Anthocyanin Molecular Ions Detected by Liquid Chromatography/Tandem Mass Spectrometry in ANT1 Micro-Tom
| Anthocyanin | Molecular Ion (M+) |
|---|---|
| Delphinidin 3-rutinoside-5-glucoside | 773.4 |
| Delphinidin 3-(coumaroyl)rutinoside-5-glucoside | 919.6 |
| Delphinidin 3-(caffeoyl)rutinoside-5-glucoside | 935.6 |
| Petunidin 3-rutinoside-5-glucoside | 787.4 |
| Petunidin 3-(coumaroyl)rutinoside-5-glucoside | 933.6 |
| Petunidin 3-(caffeoyl)rutinoside-5-glucoside | 949.6 |
| Malvidin 3-rutinoside-5-glucoside | 801.4 |
| Malvidin 3-(coumaroyl)rutinoside-5-glucoside | 947.6 |
| Malvidin 3-(caffeoyl)rutinoside-5-glucoside | 963.6 |
ANT1 Regulates a Variety of Genes Involved in Anthocyanin Accumulation
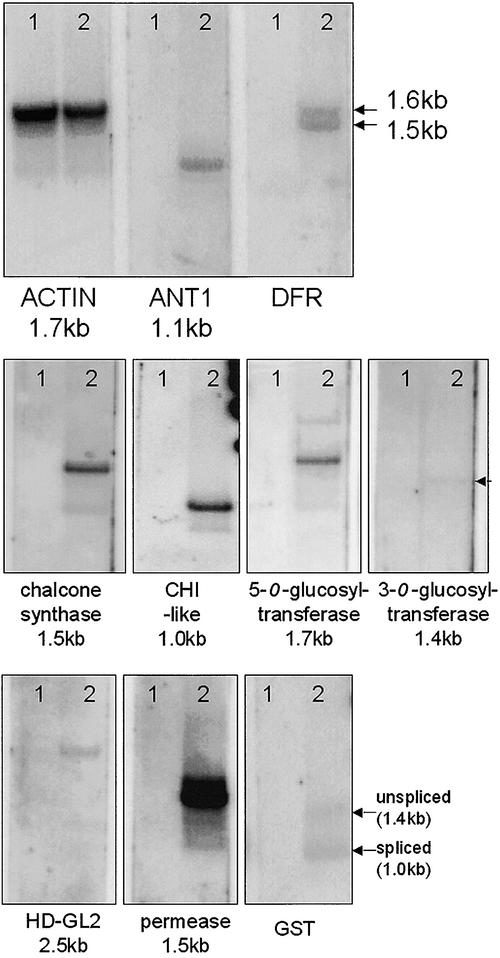
Validation of gene expression via SMART cDNA gel blot hybridization is similar to RNA gel blot hybridization and can resolve the presence of different splice forms of differentially expressed transcripts. As shown in Figure 10, the overexpression of ANT1 resulted in the upregulation of genes that encode proteins in the early (chalcone synthase [CHS]) and late (dihydroflavonol reductase [DFR]) steps of anthocyanin biosynthesis. In addition, several other genes involved in anthocyanin molecule decoration and transportation were identified by suppression subtractive hybridization (SSH). A summary of the validated differentially expressed transcripts in the ANT1 transgenic tomato is presented in Table 3. The EST contig with the highest match to the SSH fragment was assigned a putative identity based on a BLASTX (Basic Local Alignment Search Tool) search against the nonredundant protein database in GenBank (Altschul et al., 1997). The genes likely to encode the “decorating” enzymes 3-O-glucosyltransferase and 5-O-glucosyltransferase as well as a type-I GST, a flavonoid binding protein required for vacuolar transport (similar to petunia AN9), were upregulated by ANT1. The methodology also revealed substantial upregulation of three novel genes in the ANT1 transgenic line: a gene similar to chalcone isomerase (CHI-like), a homeodomain-GLABRA2 (HD-GL2) gene similar to Arabidopsis HD-GL2 protein, and a putative permease similar to proteins with ∼10 transmembrane helices required for the vacuolar transport of proanthocyanidins such as the Arabidopsis TT12 gene. The amino acid sequence alignment performed using the default parameters of CLUSTAL W showed the relative similarity of putative CHI and anthocyanin permease from tomato and other genera (Figures 11 and 12).
Figure 10.
SMART cDNA Gel Blot Hybridization.
SMART cDNA from 3-week-old seedlings of wild-type Micro-Tom (lanes 1) or ANT1 transgenic Micro-Tom, event BK (lanes 2), was separated, transferred, and probed with a radiolabeled DNA fragment corresponding to actin and each labeled gene. In all cases, the transcripts that were upregulated in the ANT1 transgenic tomato were undetectable in the untransformed control. Note the putative alternate polyadenylated forms of tomato DFR transcript. The numbers (in kb) indicate the estimated molecular masses of the hybridizing bands.
Table 3.
Genes That Are Upregulated in ANT1 Transgenic Tomato
| SSH Clone Name |
SSH Insert Size (bp) |
EST Contig Size (bp) |
ESTs in the Contig (GenBank Accession Number) |
BLASTN Score (P Value) |
Putative Identity |
|---|---|---|---|---|---|
| MT13 | 1004 | 498 | BE462282, BE462229, AW626121 | 1232 (4.9e-51) | MYB factor, similar to petunia AN2 (=ANT1) |
| MT16 | 480 | 1410 | BM411645 | 2382 (2.0e-103) | CHS |
| MT2 | 546 | 1667 |
BE461567, BE460511, BE459234, BE436963, BE436886, BE436794, BE436417, BE436385, BE436300, BE436296, BE436116, BE435816, BE435361, BE435208, BE435015, BE434398, BE434141, BE434084, BE433520, BE433099, BE432679, BE432054, BE431801, AW933199, AW932099, AW931633, AW931609, AW930557, AW651280, AW651250, AW650795, AW648641, AW442216, AW442098, AW216889, AW220654, AW220655, AW220656, AW220874, AW221860, AW222352 |
941 (2.0e-38) | 5-O-Glucosyltransferase |
| MT11 | 281 | 673 | BI209061 AI775693 BG628982 BG631865 BG630259 | 1400 (9.3e-59) | 3-O-Glucosyltransferase |
| MTP4 | 400 | 521 | AI778224 | 730 (2.2e-28) | Type-I GST, similar to petunia AN9 |
| MTP96 | 970 | 831 | BQ505699 | 1753 (1.1e-72) | Similar to CHI |
| MTP2 | 301 | 362 | AI896332 | 1463 (2.5e-61) | HD-GL2, similar to ANL2 |
| MTP77 | 390 | 620 | BE354224 | 816 (2.4e-32) | Permease, similar to TT12 (and family) |
SSH fragments were cloned and sequenced. The SSH fragment sequence was compared with a database containing tomato ESTs assembled into the least number of contigs (BLASTN). The EST contig with the greatest match to the SSH fragment was assigned a putative identity based on a BLASTX search against the nonredundant protein database in GenBank (Altschul et al., 1997).
Figure 11.
Amino Acid Sequence Alignments of CHI from Various Species.
Sequences are from tomato MTP96 (this study), Arabidopsis At3g6317, citrus CHI, rice CHI, alfalfa CHI A and CHI B, and petunia CHI1 and CHI2. Identical amino acid residues conserved across species are indicated below the alignment with asterisks, and similar amino acids are indicated with dashes.
Figure 12.
Amino Acid Sequence Alignments of the Putative Anthocyanin Permease from Various Species.
Sequences are from tomato MTP77 (this study), Arabidopsis TT12, the At4g00350 gene product, and the At4g25640 gene product. Identical amino acid residues conserved across species are indicated below the alignment with asterisks, and similar amino acids are indicated with dashes.
DISCUSSION
We have demonstrated the utility of an activation-tagging approach to induce genetic mutants in tomato using a high-throughput tissue culture transformation system. From a population of 10,427 T0 transgenic plants, 1338 showed visible phenotypic variations, and a subset of these tested in T1 confirmed the heritability of the observed traits (Table 1). In addition, analysis of the T1 plants from randomly selected activation-tagged lines gave novel variations in 103 of the 1014 lines analyzed in T1. These observed variation rates indicate the potential of activation-tagging technology as an effective mutagen in tomato. Based on recent estimates of the number, organization, and evolution of genes in the tomato genome (Van der Hoeven et al., 2002), the present collection of >10,000 activation-tagged lines may represent more than one-quarter of the “activation” gene space of the tomato genome.
Of the many observed pigmentation mutants, we have characterized one, ant1, in which the purple phenotype resulted from the overexpression of a predicted gene that encodes a MYB transcription factor. The gene was cloned and overexpressed in tomato and tobacco, demonstrating that the upregulation of ANT1 MYB factor was responsible for the observed color phenotype. The overexpression of this single transcription factor caused the heavy accumulation of purple color, whereas the simultaneous expression of two heterologous genes (transcription factors LC and C1 of maize) were necessary to produce high-flavonol tomatoes (Bovy et al., 2002).
Characterization of anthocyanin mutants in a variety of plant species has led to the identification of genes that encode not only the enzymes of the anthocyanin biosynthetic pathways but also the regulatory elements that confer tissue-specific accumulation of anthocyanin (Dooner et al., 1991; Holton and Cornish, 1995; Mol et al., 1998; Sakamoto et al., 2001; Winkel-Shirley, 2001). Although many steps in anthocyanin biosynthesis are shared among plant species, the regulatory elements that underlie the expression levels are diverse. The ANT1 predicted protein shows the strongest identity with the AN2 gene product from Petunia hybrida and with a number of other plant transcription factors that are known to function in the regulation of the anthocyanin biosynthetic pathway, such as PAP1, PAP2, and TT2 from Arabidopsis and the PL and C1 proteins from maize (Quattrocchio et al., 1998, 1999; Borevitz et al., 2000). These proteins all contain a conserved N-terminal DNA binding motif called a MYB domain after its similarity to the proto-oncogene c-MYB (Martin and Paz-Ares, 1997). MYB-related proteins from plants generally contain two related helix-loop-helix motifs, the R2 and R3 repeats (Kranz et al., 1998; Nesi et al., 2001). These proteins are known to control a wide range of processes, including morphology, development, environmental responses, and secondary metabolism.
ANT1 has a highly conserved R2R3 MYB domain from residues 11 to 116. When this region is aligned with the same region from other functionally characterized plant proteins involved in flavonoid accumulation, a high degree of conservation is evident (Figure 4A). The C termini among these proteins generally have little identity. Outside of the R2R3 domain, ANT1 displays amino acid sequence similarity only to AN2 (Figure 4B). The two sequences show 53% identity and 62% similarity over the length of the protein. Functional domains in these regulatory proteins have been identified. For example, the conserved Trp (or Ile) residues in the MYB repeats have been demonstrated to be involved in DNA binding (Li and Parish, 1995). Transcriptional activation requires the presence of the C-terminal domain (Quattrocchio et al., 1999), but DNA and cofactor binding specificity is determined by residues in the MYB repeats (Quattrocchio et al., 1999; Grotewold et al., 2000). Some of the MYB domain proteins (C1 and AN2) are known to interact physically with the basic/helix-loop-helix domain regulatory proteins (R and JAF13) to control anthocyanin production in a combinatorial interaction (Quattrocchio et al., 1998, 1999). Amino acid residues that define the specificity of this interaction have been identified in the R3 domain (Grotewold et al., 2000). Based on its sequence similarity to the petunia AN2 protein, it is predicted that ANT1 may have similar functions and interactions.
SSH and SMART cDNA hybridization have revealed a suite of genes upregulated in plants overexpressing the ANT1 transcript. The ANT1 transgene itself was isolated by SSH as clone MT13, validating the experimental approach. The genes that encode proteins in the early and late biosynthetic pathways of anthocyanin synthesis, CHS and DFR, were upregulated in ANT1-overexpressing plants. DFR, a single-copy gene in tomato, is known to be represented by two transcript sizes resulting from alternating polyadenylation signals (Bongue-Bartelsman et al., 1994). The SMART cDNA gel blot resolved the two cDNA sizes, which differed by ∼100 bp (Figure 10). DFR is one of the structural enzymes in the anthocyanin pathway, and its transcription is activated by AN1, which in turn is regulated by AN2 in petunia (Spelt et al., 2002).
The diverse group of transcriptional regulators controls not only anthocyanin biosynthesis but also their modification and transport into the vacuole. Overexpression of ANT1 upregulated genes likely encodes the decorating enzymes 3-O-glucosyltransferase and 5-O-glucosyltransferase as well as a type-I GST, a flavonoid binding protein required for vacuolar transport (similar to petunia AN9). Anthocyanins frequently are glycosylated, and glycosyltransferases are upregulated coordinately with other anthocyanin biosynthetic enzymes (Bovy et al., 2002; Yamazaki et al., 2002). In maize and tomato, glucosyltransferases are controlled by the MYB class of transcription factors (Dooner et al., 1991; Bovy et al., 2002). Finally, vacuolar pH, a determinant of pigment color and stability, also is controlled by a coordinated effort of the transcription factors in petunia mentioned above (Spelt et al., 2002).
A gene with strong similarity to the petunia AN9 gene that encodes GST, which is required for efficient vacuolar sequestration (Mueller et al., 2000), was upregulated in the ANT1 transgenic line. Anthocyanins are cytotoxic and unstable in the neutral pH of the cytoplasm. Therefore, sequestration of anthocyanins into the acidic vacuole is an important component of the pathway leading to anthocyanin accumulation. The transport of anthocyanins into the vacuole was long believed to involve the transport of an anthocyanin-glutathione conjugate by a GS-X pump (Marrs et al., 1995). However, more recent studies dispute the formation of an anthocyanin-glutathione conjugate (Mueller et al., 2000) and suggest, instead, that GST acts as an anthocyanin binding protein that may serve as a chaperone. SMART cDNA gel blot analysis corroborates the RNA gel blot analysis of the GST transcript, which was shown to be present in two forms, corresponding to the spliced and unspliced forms. Approximately 40% of the total transcript is unspliced in the ANT1 transgenic line, independent of the GST expression level. GST is represented as both spliced and unspliced forms, a possible point of regulation in the pathway (Figure 10). Splicing regulation has not been reported previously for this pathway, and it may result from the high level of expression of the transgene that controls the phenotype in the transgenic tomato. Unspliced GST transcript was not detected in the pap1-D Arabidopsis mutant, for example (Borevitz et al., 2000).
Three new genes in tomato anthocyanin synthesis and accumulation were identified in ANT1 transgenic tomato by SSH: a gene similar to CHI (CHI-like), a HD-GL2 gene similar to Arabidopsis HD-GL2, and a putative permease similar to proteins with ∼10 transmembrane helices required for the vacuolar transport of proanthocyanidins (Arabidopsis TT12 and family). The SSH clone MTP2 may encode a protein with similarity to the HD-GL2 class of transcription factors. The MTP2 fragment and the corresponding EST contig represent only a partial coding region. A similar gene product from Arabidopsis, ANTHOCYANINLESS2, was shown to be required for the accumulation of anthocyanins in subepidermal cells of vegetative tissues but had no effect on proanthocyanidin accumulation in the seed coat (Kubo et al., 1999). In the ANT1 transgenic tomato leaves, anthocyanins accumulated primarily in the epidermal cells (data not shown); therefore, although the tomato MTP2 cDNA and Arabidopsis ANL2 gene products both might function in regulating the tissue-specific accumulation of anthocyanins, their roles may be confined to different cell types.
The MTP96 transcript, which is upregulated in the ANT1 transgenic line, encodes a protein with similarity to CHI. The CHI-like gene product encoded by MTP96 is most similar to the Arabidopsis At3g63170 gene product and is only 17% identical (32% similar) to the petunia CHI-A gene product (Figure 12). However, the MTP96 product lacks the conserved residues reported to be involved in (2S)-naringenin binding and substrate preference determination (Jez et al., 2000), suggesting that the substrate for this enzyme may be modified. There is no report of a CHI gene from tomato in the public databases, although a CHI cDNA clone was reported to have been isolated recently from tomato (Bovy et al., 2002). However, the reported tomato CHI transcript is not regulated by the heterologous expression of maize transcription factors that regulate other enzymatic steps in the flavonoid pathway (Bovy et al., 2002). It is tempting to speculate that the recently reported CHI transcript and the MTP96-encoded CHI-like protein isolated in this study function in separate tissues or even separate steps of flavonoid biosynthesis, with the CHI-like transcript involved directly in anthocyanin biosynthesis and accumulation in the leaves of tomato.
Finally, the MTP77 clone that encodes a putative anthocyanin permease was characterized. The complete MTP77 cDNA sequence was assembled, translated, and compared with the Arabidopsis TT12 gene product and two other related Arabidopsis gene products (Figure 12). The MTP77 cDNA encodes a protein that is 36% identical (56% similar) to TT12 but is even more like At4g00350 (53% identical and 68% similar) and At4g25640 (61% identical and 71% similar). TT12 resembles multidrug secondary transporters in the MATE family and is likely to mediate the vacuolar sequestration of proanthocyanidins in the seed coat of Arabidopsis (Debeaujon et al., 2001). The similarity of the MTP77 permease to TT12 and its coregulation with the ANT1 transcription factor suggest that the gene product functions as an anthocyanin vacuolar transporter in tomato leaves. The high degree of amino acid sequence similarity between the tomato MTP77 permease and the Arabidopsis At4g00350 and At4g25640 gene products further suggests that these Arabidopsis genes may play a role in anthocyanin sequestration in vegetative tissues.
Mutant analysis in Arabidopsis also has led to the identification of genes involved in the cell type–specific accumulation of anthocyanins (ANL2; Kubo et al., 1999) and the vacuolar accumulation of proanthocyanidins (TT12; Debeaujon et al., 2001). Although the roles of genes that encode HD-GL2 and TT12-like transporter proteins in flavonoid accumulation have been shown in Arabidopsis, the evidence has been restricted to a role in the seed coat, and their regulation by a MYB factor (ANT1) has not been reported previously. It has been shown that anthocyanin accumulation is controlled differently in the seed coat and in vegetative tissues (Kubo et al., 1999; Borevitz et al., 2000). TT12 is unlikely to play a role in the accumulation of anthocyanins in the leaves, but the other closely related Arabidopsis genes may. Likewise, we predict that the permease encoded by MTP77 functions as a major vacuolar transporter of anthocyanins in the leaves of tomato and that a similar gene product is likely to be found in petunia and maize.
Genetic mutants have always played a central role as tools for research into the developmental processes of plants. A number of genes have been isolated in tomato by insertional mutagenesis using targeted tagging of Ac/Ds elements (Jones et al., 1994; Bishop et al., 1996; Keddie et al., 1996; Van der Biezen et al., 1996; Takken et al., 1998). Meissner's group (Meissner et al., 2000) reported the production of mutants in Micro-Tom with stabilized Ds element insertions and gene trapping with β-glucuronidase and luciferase genes. Micro-Tom mutants with altered pigments and modified leaves and fruits were recovered by screening 9000 M1 and 20,000 M2 plants of the ethyl methanesulfonate–mutagenized population (Meissner et al., 1997). The identification of ANT1 demonstrates the utility of activation-tagging in micro-tomato. Its regulatory role in the flavonoid pathway indicates the possibility of the metabolic engineering of tomato for nutritionally important compounds.
METHODS
Activation-Tagging Construct
The activation-tagging construct pSKI015 (Weigel et al., 2000) was modified to contain a kanamycin resistance selection cassette. The selection cassette in pAG3202 uses the RE4 promoter (U.S. patent 6,054,635, issued April 25, 2000; inventors: R.K. Bestwick, J.A. Kellogg, and H.V. Mathews; assignee: Agritope, Inc., Portland, OR), the nptII gene conferring kanamycin resistance, and the Agrobacterium tumefaciens gene 7 termination sequence adjacent to the T-DNA right border (Beck et al., 1982; Velten and Schell, 1985) in place of the bialaphos resistance selection cassette.
Plant Transformation
Tomato seeds (Lycopersicon esculentum cv Micro-Tom; Tomato Growers Supply Company, Fort Meyers, FL) were surface-sterilized in 25% bleach with Tween 20 for 10 min and rinsed with sterile water before plating on seed germination medium (MS salts [Murashige and Skoog, 1962], Nitsch vitamins [Nitsch and Nitsch, 1969], 3% sucrose, 0 to 0.5 mg/L benzyladenine, and 0.7% agar, pH 5.8). Hypocotyls and shoot tip explants of 7- to 8-day-old seedlings were used for transformation experiments. In addition, leaf segments of sterile plants 30 to 40 days old in culture also were used as explants.
Agrobacterium strain GV3101 containing the binary vector plasmid mentioned above was streaked on MGL medium (5 mg/L tryptone, 2.5 mg/L yeast extract, 5 mg/L NaCl, 5 mg/L mannitol, 1.17 mg/L sodium glutamate, 0.33 mg/L K2HPO4·3H2O, 0.1 mg/L MgSO4, and 0.02 mg/L biotin at pH 5.4) with 100 mg/L carbenicillin and grown at 28°C for 2 to 3 days. A single colony was picked from the plate and grown in 30 to 40 mL of MGL medium with 50 μM acetosyringone and 50 mg/L carbenicillin overnight. The bacteria were centrifuged at 4500 relative centrifugal force for 15 min to remove antibiotics and then suspended in MGL or liquid plant cocultivation medium (MS salts, Linsmaier and Skoog [1965] vitamins, 3% sucrose, 200 mg/L KH2PO4, 0.2 mg/L 2,4-D, 0.1 mg/L kinetin, and 0 to 100 mg/L acetosyringone, pH 5.4) to a working concentration of 5 × 108 cells/mL (OD600 = 0.200).
Hypocotyls and stems were segmented into 3- to 5-mm sections. Leaves were cut into 4-mm cross-sections exclusive of the shoot tip and petiole. Shoot tips were excised from the seedlings, leaving 2 to 3 mm of hypocotyl for ease of handling. Cotyledons were removed and the shoot tips were cut in half longitudinally, being careful to bisect the meristem. The explants were immersed in Agrobacterium suspension for 15 min, blotted onto sterile filter paper, and plated on cocultivation medium (MS salts, Linsmaier and Skoog [1965] vitamins, 3% sucrose, 200 mg/L KH2PO4, 0.2 mg/L 2,4-D, 0.1 mg/L kinetin, 0 to 100 μM acetosyringone, and 0.7% agar, pH 5.4). After a 2-day cocultivation period, the explants were transferred to regeneration medium (MS salts, Nitsch vitamins, 3% sucrose, 0.5 to 2 mg/L zeatin, 0 to 1 mg/L indoleacetic acid, 0 to 750 mg/L carbenicillin, 0 to 300 mg/L timentin, and 0.7% agar, pH 5.8) with 75 mg/L kanamycin for selection. Explants were transferred to fresh medium every 2 weeks. The selection was increased gradually over an 8-week period to 90 mg/L kanamycin. Shoots developing from the explants were excised and transferred to rooting medium (MS salts, Nitsch vitamins, 3% sucrose, 0.5 to 1 mg/L indolebutyric acid, 100 mg/L carbenicillin/timentin, 50 mg/L kanamycin, and 0.7% agar, pH 5.8) when they were ∼1 cm tall. Rooted shoots were outplanted to soil in the greenhouse for further evaluation.
Greenhouse Evaluation of Activation-Tagged Micro-Tom Transgenic Plants
The transgenic plants were maintained in a cool, shady environment for 2 days at 20°C for hardening before transfer to a greenhouse for further growth and observation. The greenhouse maintained a daytime temperature of 25 to 28°C and a night temperature of 20 to 22°C. Supplemental lighting was provided by 400-W high-pressure sodium lights to extend the daylength to 18 h.
Morphological observations were made throughout the development of the T0 plants for phenotypes such as plant stature; growth habit; leaf size, shape, texture, and color; flower size, shape, and color; fruit size, shape, and color; and seed set. Digital images were captured for plants with interesting morphological phenotypic modifications. Fruit was collected at maturity and allowed to ferment in a plastic bag before seed collection. The collected seeds were dried and cataloged in a temperature- and humidity-controlled chamber for future use. Leaf and fruit tissues were frozen in liquid nitrogen and stored at −80°C for biochemical and molecular analyses.
T1-generation plants were grown to evaluate genetic segregation, confirm the mutant phenotypes observed in the T0 generation, and screen for possible loss-of-function mutations. The emerging seedlings were sprayed with kanamycin at 1.0 mg/L three times at 2-day intervals to identify plants that were nulls without the transgene. Approximately 1 week after the last spraying, the lines were scored for germination rate as well as seedling resistance to kanamycin. The kanamycin-resistant seedlings were given individual identifier codes and transplanted to 4-inch pots for further observations like those used for T0 transplants.
Plasmid Rescue of DNA Flanking the T-DNA in the ant1 Mutant
Plasmid rescue was performed to recover genomic DNA flanking the T-DNA insertion (Weigel et al., 2000). A 6.4-kb genomic fragment was rescued after KpnI digestion of DNA from the ANT1 line. A primer-walking strategy was used to obtain the complete sequence of the rescued fragment. Sequencing was performed using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems) and the ABI Prism 310 Genetic Analyzer (Perkin-Elmer).
Reverse Transcriptase–PCR
Total RNA was extracted from leaves of mutant and wild-type tomato using the SV Total RNA Isolation System (Promega, Madison, WI). Reverse transcriptase–PCR then was performed using the Access RT-PCR system (Promega) according to the protocol supplied by the manufacturer. One microgram of RNA was used as a template in each reaction with gene-specific primers (ANT1 F2, 5′-TTCATTGGGAGTGAGAAAAGGTTC-3′; ANT1 R2, 5′-CATTGTGCTTGAGAAATACTTGCG-3′). Primers specific for actin were used in control reactions (Actin 5F, 5′-ATGACTCAAATCATGTTTGAGACCTTC-3′; Actin 3R, 5′-ACCTTAATCTTCATGCTGCTTGGAGC-3′).
Recapitulation of the ant1 Phenotype in Tomato and Tobacco
A tomato genomic fragment containing only the predicted coding region of ANT1 was amplified from the rescued plasmid using primers 5′-TCCCCCGGGATGAACAGTACATCTATGTCTTCAT-3′ and 5′-GGACTAGTTTAATCAAGTAGATTCCATAAGTCA-3′ and subcloned into a plant expression construct. The construct uses a pBIN19 vector, and expression of the ANT1 coding region is controlled by the cassava vein mosaic promoter (Verdaguer et al., 1996) and the nopaline synthase terminator (Depicker et al., 1982). The construct also contains the kanamycin-resistant selection marker as described for the activation-tagging construct. This ANT1 expression construct was introduced into Agrobacterium strains GV3101 and EHA105 for the transformation of tomato and tobacco. The transformation protocol in tomato followed the same procedure described above for the generation of ACTTAG lines; for tobacco, we followed our earlier published protocol (Viegas et al., 1987).
RNA Gel Blot Hybridization
Untransformed Micro-Tom (wild type) was compared with transgenic Micro-Tom overexpressing the ANT1 gene via a strong constitutive promoter (event BK). T1 seedlings were raised in vitro. Only transgenic plants showing the pigmented phenotype were analyzed. At least six plants per sample were pooled before RNA extraction. Total RNA was extracted from 3-week-old seedlings using TriReagent according to the supplied protocol (Sigma). For each sample, 20 μg of total RNA was separated on a 1.2% agarose formaldehyde gel and transferred to a Nytran Plus membrane (Schleicher & Schuell, Keene, NH) as described by Sambrook et al. (1989). Equal RNA loading was confirmed by methylene blue staining of the RNA on the membrane.
DNA fragments used as probes were amplified by PCR from tomato SMART cDNA using oligonucleotide primers designed for DFR (DFR-F, 5′-GCTTGTCATGAGACTCCTTGAACG-3′; DFR-R, 5′-CGCACCATCTTAGCCACATCG-3′) and for ANT1 (ANT1-F, 5′-GGATCCTATATTATCAAATTATTATGAACA-3′; ANT1-R, 5′-ACTAGTACATATCTTTTAATCAAGTAGATT-3′). The probes used to validate the differentially ex-pressed transcript were amplified by PCR from the pCR2.1 vector with oligonucleotide primers complementary to the vector flanking the TA cloning site (M13F, 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′; M13R, 5′-AGCGGATAACAATTTCACACAGGA-3′). Amplification of all probe fragments was performed for 30 cycles in a Perkin-Elmer 480 thermal cycler using a 60°C annealing temperature and a 1-min extension.
The amplified probe fragments (∼50 ng) were labeled with 32P-dCTP (DuPont–New England Nuclear, Boston, MA) using the Ready-To-Go DNA labeling kit (Amersham Pharmacia). Hybridization conditions were as described by Church and Gilbert (1984). High-stringency washes were performed under the following conditions: 65°C in 1% SDS, 40 mM sodium phosphate buffer, pH 7, and 1 mM EDTA. RNA gel blot hybridization signal was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Suppression Subtractive Hybridization
Two micrograms of total RNA from 3-week-old seedlings of transgenic line BK were used to synthesize SMART cDNA according to the protocol supplied with Clontech's Super SMART PCR cDNA Synthesis Kit (K1054-1). Triplicate 100-μL amplification reactions were set up for each of the cDNAs; 17 cycles were determined to be optimal for the amplification of the double-stranded cDNA. After amplification, the triplicate reactions were pooled and phenol chloroform was extracted and ethanol precipitated. The precipitated double-stranded SMART cDNA was resuspended in TE buffer (10 mM Tris, 1 mM EDTA at pH 7.4).
SMART cDNA was digested with RsaI, and adaptors were ligated according to the protocol supplied with the Clontech PCR-Select cDNA Subtraction Kit (K1804). Two rounds of subtractive hybridization were performed with the wild-type and ANT1 transgenic SMART cDNA samples, including both forward (MTP) and reverse (MTC) subtractions. Primary and secondary PCR amplifications were performed using 27 and 12 cycles, respectively. The resulting pools of differentially expressed fragments were cloned into a TA cloning kit (pCR2.1; Invitrogen, Carlsbad, CA). The ligation reactions were purified over a G-50 column before transformation into INVαF' competent cells (Invitrogen, Carlsbad, CA). Each transformation (MTC and MTP) was plated on selective medium (ampicillin).
To estimate the relative transcript abundance in each pool of cloned fragments, 48 colonies per transformation were chosen for PCR colony screening with primers flanking the TA cloning site (M13F and M13R as described above). The 96 reaction products (amplified insert) were separated on duplicate agarose gels, transferred to nylon membranes with 0.4 M NaOH, and probed separately with SMART cDNA from either wild-type or ANT1 transgenic Micro-Tom. Probe labeling, hybridization, and signal detection were performed as described previously. The average signal intensity of the 48 cloned fragments from the forward subtraction (MTP) was twofold greater than that from the reverse subtraction (MTC), indicating that the MTP pool was enriched in upregulated transcript fragments (data not shown).
The clones showing the highest fold change in expression between the wild-type and transgenic samples were selected for validation by DNA gel blot hybridization and for DNA sequencing. Plasmid DNA templates were sequenced using the M13F and M13R primers on an ABI3100 DNA sequencer. Vector, primer, and poly(A) sequences were removed from the output before Basic Local Alignment Search Tool (BLASTN) analysis against the tomato EST collection in GenBank, assembled into the least number of contigs.
For DNA gel blot hybridization, SMART cDNA (3 μg/lane) was separated, transferred to nylon membranes (0.4 M NaOH), and hybridized with labeled PCR fragments corresponding to candidate regulated transcript fragments. Hybridization with probes to ANT1, DFR, and GST verified that these genes were upregulated in the ANT1 transgenic plants. The results also confirmed that the SMART cDNA gel blot results were similar to results from RNA gel blot hybridization, even including the ability to resolve different splice and polyadenylated forms of the GST and DFR transcripts.
The 5′ and 3′ ends of the MTP77 cDNA were amplified from SMART cDNA using nested sequence-specific primers and primers complementary to the adaptors on the ends of the SMART cDNA fragments. Reactions were set up according to the protocol supplied with the Universal GenomeWalker Kit (Clontech, K1807-1) and the Advantage Genomic Polymerase Mix (Clontech) using a Perkin-Elmer 480 thermal cycler. Primary amplifications consisted of 7 cycles (94°C for 25 s and 70°C for 3 min), 32 cycles (94°C for 25 s and 65°C for 3 min), and 1 cycle (67°C for 7 min). Secondary amplifications consisted of 5 cycles (94°C for 25 s and 68°C for 3 min), 20 cycles (94°C for 25 s and 63°C for 3 min), and 1 cycle (67°C for 7 min). The 5′ end of the MTP77 cDNA was amplified in the first round with SMART PCRIIA (5′-AAGCAGTGGTATCAACGCAGAGT-3′) and MTP77_F1 (5′-GAAAGACACAAGTAGCTAGCAGGATTA-3′) and in the second round with PCRIIA-UP (5′-GTGGTATCAACGCAGAGTACGCGG-3′) and MTP77_F2 (5′-CCATATGCCTGGCCACATAGTGTCTC-3′). The 3′ end of MTP77 cDNA was amplified in the first round with SMART PCRIIA and MTP77_R1 (5′-CTTCAAGCTCAGAGTAAAGTGGATGT-3′) and in the second round with PCRIIA-DOWN (5′-GTGGTATCAACGCAGAGTACTTTTT-3′) and MTP77_R2 (5′-GGCTCTTTATATATACACTTGAATGGGG-3′). A full-length MTP77 cDNA clone then was amplified from SMART cDNA using sequence-specific primers based on the sequences of the 3′ and 5′ ends. The 1.7-kb fragment was cloned and sequenced.
Biochemical Analysis
Extraction
In vitro–grown, 3-week-old wild-type and transgenic Micro-Tom plants were harvested, weighed, and extracted exhaustively with 1-mL aliquots of 5% HOAc. Each extract was derived from three plants, and three separate extracts were made for each line. The soluble portion of each extract was transferred to a centrifuge tube and spun at 3000 rpm under vacuum until dry. The solid pellet was dissolved in 1 mL of 5% HOAc, passed through a 0.45-μm acid-compatible filter, and transferred to a 12- × 32-mm vial with a conical insert for HPLC analysis.
HPLC Conditions
Five microliters of anthocyanin extract was injected onto a Waters 2795 HPLC system (Milford, MA) equipped with a Waters RP-C18 Symmetry-Shield column (3.5 μm, 4.6 × 150 mm) and a guard column (3.9 × 20 mm). Extracts were eluted with a 30-min mobile phase gradient of 5 to 35% CH3CN in 1.5% aqueous H3PO4 at a flow rate of 1 mL/min. Compounds were detected using a Waters 996 photodiode array detector. Samples were injected in triplicate, and their results were averaged.
Quantitation
Total anthocyanin content was determined by photodiode array detector response at 520 nm based on the external calibration standard cyanin chloride (Indofine, Hillsborough, NJ). A calibration curve was constructed by triplicate injection of a series of the standards (15.6, 31.3, 62.5, 125, 250, and 500 μg/μL), and the calibration curve had a correlation coefficient of 0.987. Total anthocyanin concentration of the unknown extract was expressed as cyanin chloride equivalents by summing the peak areas at 520 nm and comparing them with the standard curve.
Liquid Chromatography/Tandem Mass Spectrometry
Anthocyanin structures were elucidated by liquid chromatography/tandem mass spectrometry. Extracts (10 μL) were resolved on a C18 Symmetry column (Waters) with a mobile phase gradient of 10 to 50% CH3CN in water with 0.1% aqueous formic acid running at 0.3 mL/min and detected with a Micromass Quattro Micro triple quadrupole mass spectrometer with an ES+ source (a positive electro-spray source; Milford, MA).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact D. Ry Wagner, rwagner@exelixis.com.
Accession Numbers
The accession numbers for the sequences shown in the figures are petunia AN2 (PhAN2; gi|7673084), Arabidopsis PAP1 (AtPAP1; gi|11935171), Arabidopsis PAP2 (AtPAP2; gi|11935173), Arabidopsis TT2 (AtTT2; gi|14272363), Zea mays Pl (ZmPl; gi|7438350), Z. mays C1 (ZmC1; gi|127585), Arabidopsis At3g63170 (gi|18412649), Citrus sinensis CHI (gi|4126399), Oryza sativa CHI (gi|20152984), Medicago sativa CHI1 (gi|116134), M. sativa CHI2 (gi|116135), petunia CHI A (gi|7331150), petunia CHI B (gi|68483), Arabidopsis TT12 (gi|27151710), the At4g00350 gene product (gi|18411304), and the At4g25640 gene product (gi|15235172).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012963.
References
- Altschul, S.F., Madden, T.L., Schäffe, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, E., Ludwig, G., Auerswald, E., Reiss, B., and Schaller, H. (1982). Nucleotide sequence and exact location of the neomycin phosphotransferase gene from transposon Tn5. Gene 19, 327–336. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., Harrison, K., and Jones, J.D.G. (1996). The tomato Dwarf gene isolated by heterologous transposon tagging encodes the first member of a new cytochrome P450 family. Plant Cell 8, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongue-Bartelsman, M., O'Neill, S.D., Tong, Y., and Yoder, J.I. (1994). Characterization of the gene encoding dihydroflavonol 4-reductase in tomato. Gene 138, 153–157. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovy, A., de Vos, R., Kemper, M., Schijlen, E., Pertejo, M.A., Muir, S., Collins, G., Robinson, S., Verhoeyen, M., Hughes, S., Santos-Buelga, C., and van Tunen, A. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14, 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Leon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depicker, A., Stachel, S., Dhaese, P., Zambryski, P., and Goodman, H.M. (1982). Nopaline synthase: Transcript mapping and DNA sequence. J. Mol. Appl. Genet. 1, 561–573. [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25, 173–199. [DOI] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Grandillo, S., Knapp, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. [DOI] [PubMed] [Google Scholar]
- Fridman, E., Pleban, T., and Zamir, D. (2000). A recombinant hotspot delimits a wild QTL for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 97, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., Sainz, M.B., Tagliani, L., Hernandez, J.M., Bowen, B., and Chandler, V.L. (2000). Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 97, 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, J., Koornneef, M., Ramanna, M.S., and Zabel, P. (1989). Tomato: A crop species amenable to improvement by cellular and molecular methods. Euphytica 42, 1–23. [Google Scholar]
- Holton, T.A., and Cornish, E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Cerny, R.E., Bhat, D.S., and Brown, S.M. (2001). Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol. 125, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez, J.M., Bowman, M.E., Dixon, R.A., and Noel, J.P. (2000). Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 7, 786–791. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–792. [DOI] [PubMed] [Google Scholar]
- Keddie, J.S., Carroll, B., Jones, J.D.G., and Gruissem, W. (1996). The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 15, 4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. [DOI] [PubMed] [Google Scholar]
- Kubo, H., Peeters, A.J., Aarts, M.G., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.F., and Parish, R.W. (1995). Isolation of two novel myb-like genes from Arabidopsis and studies on the DNA-binding properties of their products. Plant J. 8, 963–972. [DOI] [PubMed] [Google Scholar]
- Linsmaier, E.M., and Skoog, F. (1965). Organic growth factor requirements of tobacco tissue cultures. Physiol. Plant. 18, 100–127. [Google Scholar]
- Marrs, K.A., Alfenito, M.R., Lloyd, A.M., and Walbot, V. (1995). A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature 375, 397–400. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Chague, V., Zhu, Q., Emmanuel, E., Elkind, V., and Levy, A.A. (2000). A high throughput system for transposon tagging and promoter trapping in tomato. Plant J. 22, 265–274. [DOI] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A. (1997). A new model system for tomato genetics. Plant J. 12, 1465–1472. [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Mueller, L.A., Goodman, C.D., Silady, R.A., and Walbot, V. (2000). AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid binding protein. Plant Physiol. 123, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch, J.P., and Nitsch, C. (1969). Haploid plants from pollen grains. Science 163, 85–87. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., and Koes, R. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., van der Woude, K., Mol, J.N., and Koes, R. (1998). Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Ohmori, T., Kageyama, K., Miyazaki, C., Saito, A., Murata, M., Noda, K., and Maekawa, M. (2001). The Purple leaf (Pl) locus of rice: The Plw allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42, 982–991. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scott, J.W., and Harbaugh, B.K. (1989). Micro-Tom: A miniature dwarf tomato. Fla. Agric. Exp. Stn. Circ. 370, 1–6. [Google Scholar]
- Spelt, C., Quattrocchio, F., Mo, J., and Koes, R. (2002). ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 14, 2121–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, M.A., and Rick, C.M. (1986). Genetics and breeding. In The Tomato Crop, J.G. Atherton and J. Rudich, eds (London: Chapman and Hall), pp. 35–109.
- Takken, F., Schipper, D., Nijkamp, H., and Hille, J. (1998). Identification and Ds-tagged isolation of a new gene at the Cf-4 locus of tomato involved in disease resistance to Cladosporium fulvum race 5. Plant J. 14, 401–411. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D. (1993). Linkage map of tomato (Lycopersicon esculentum) (2N=24). In Genetic Maps: Locus Maps of Complex Genomes, J. O'Brien, ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 6.39–6.60.
- Van der Biezen, E., Brandwagt, B., Leeuwen, V.W., Nijkamp, H., and Hilles, J. (1996). Identification and isolation of the FEEBLY gene from tomato by transposon tagging. Mol. Gen. Genet. 12, 267–280. [DOI] [PubMed] [Google Scholar]
- Van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten, J., and Schell, J. (1985). Selection-expression plasmid vectors for use in genetic transformation of higher plants. Nucleic Acids Res. 13, 6981–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer, B., de Kochko, A., Beachy, R.N., and Fauquet, C. (1996). Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol. Biol. 6, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Viegas, P., Mathews, H., Bhatia, C.R., and Notani, N.K. (1987). Monohybrid and dihybrid segregations in the progenies of tobacco transformed for kanamycin resistance with Ti-vector system. J. Genet. 66, 25–31. [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2001). Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M., Yamagishi, E., Gong, Z., Fukuchi-Mizutani, M., Fukui, Y., Tanaka, Y., Kusumi, T., Yamaguchi, M., and Saito, K. (2002). Two flavonoid glucosyltransferases from Petunia hybrida: Molecular cloning, biochemical properties and developmentally regulated ex-pression. Plant Mol. Biol. 48, 401–411. [DOI] [PubMed] [Google Scholar]
- Zubuko, E., Adams, C.J., Machaekova, I., Malbeck, J., Scollan, C., and Meyer, P. (2002). Activation tagging identifies a gene from Petunia hybrida responsible for the production of active cytokinins in plants. Plant J. 29, 797–808. [DOI] [PubMed] [Google Scholar]