Abstract
Plant architecture results from the activity of the shoot apical meristem, which initiates leaves, internodes, and axillary meristems. KNOTTED1-like homeobox (KNOX) genes are expressed in specific patterns in the shoot apical meristem and play important roles in plant architecture. KNOX proteins interact with BEL1-like (BELL) homeodomain proteins and together bind a target sequence with high affinity. We have obtained a mutation in one of the Arabidopsis BELL genes, PENNYWISE (PNY), that appears phenotypically similar to the KNOX mutant brevipedicellus (bp). Both bp and pny have randomly shorter internodes and display a slight increase in the number of axillary branches. The double mutant shows a synergistic phenotype of extremely short internodes interspersed with long internodes and increased branching. PNY is expressed in inflorescence and floral meristems and overlaps with BP in a discrete domain of the inflorescence meristem where we propose the internode is patterned. The physical association of the PNY and BP proteins suggests that they participate in a complex that regulates early patterning events in the inflorescence meristem.
INTRODUCTION
The formation of the plant body is dependent on the activity of self-organizing groups of cells called meristems located at the shoot and root apices. The development of the shoot is controlled by the shoot apical meristem (SAM), which initiates leaf primordia repetitively from its flanks. Additional shoot meristems form in the axils of the leaf primordia. The axillary meristem may remain dormant, grow out with a similar body plan as the main stem, or develop into floral meristems, which are determinate meristems. The SAM also produces the internode, the stem portion between the leaves (Vaughn, 1955).
Arabidopsis has distinct vegetative and reproductive phases (Hempel and Feldman, 1994). The vegetative phase is characterized by a compact rosette with little or no internode elongation and the repression of axillary meristem development. Upon a signal to flower (Simpson and Dean, 2002), the SAM increases in size and axillary meristem growth is derepressed. Branches that mimic the growth pattern of the main shoot initiate in the axils of cauline leaves, whereas flowers initiate without visible subtending leaves. The branches (referred to as paraclades) initiate in a basipetal direction, toward the base, whereas the flowers initiate acropetally, toward the top (Hempel and Feldman, 1994).
The class-1 KNOTTED1-like homeobox (KNOX) genes are essential for proper meristem function (Reiser et al., 2000). Maize knotted1 (kn1) and its ortholog in Arabidopsis, SHOOT MERISTEMLESS (STM), are expressed in shoot meristems and stems but excluded from leaf primordia (Smith et al., 1992; Jackson et al., 1994; Long et al., 1996; Nishimura et al., 1999). Loss-of-function mutations in STM and kn1 produce a shootless phenotype in which the SAM is consumed during cotyledon development. These results indicate that kn1 and STM function in meristem maintenance (Endrizzi et al., 1996; Long et al., 1996; Kerstetter et al., 1997; Vollbrecht et al., 2000). Mutations in the Arabidopsis BREVIPEDICELLUS (BP) gene and its ortholog in rice, OSH15, have shortened internodes, suggesting that these genes regulate internode development (Sato et al., 1999; Douglas et al., 2002; Venglat et al., 2002). BP, OSH15, and the related maize gene rough sheath1 (rs1) are expressed in discrete domains of the SAM, possibly marking the position of an internode domain (Lincoln et al., 1994; Schneeberger et al., 1995; Sato et al., 1999).
KNOX proteins interact with members of the BEL1-like (BELL) family of homeodomain proteins in both monocots and dicots (Bellaoui et al., 2001; Muller et al., 2001; Smith et al., 2002). The cooperative interaction of KNOX/BELL complexes mediates high DNA binding affinity, indicating that this interaction is biochemically meaningful (Smith et al., 2002). Moreover, this interaction is selective, suggesting that KNOX proteins have a greater affinity for certain BELL proteins than for others (Smith et al., 2002). These findings indicate that specific KNOX/BELL heterodimers may function in distinct morphological events in the SAM. The combination of selective interactions between BELL and KNOX proteins and discrete expression patterns is likely to specify unique functions for the different KNOX/BELL heterodimers.
To gain more insight into KNOX function, we characterized a mutation in a BELL gene that we have named PENNYWISE (PNY). pny has similar defects in inflorescence architecture as bp. The double mutant of pny and bp shows a synergistic phenotype that severely affects internode patterning. The expression of PNY and BP overlaps in the meristem at the boundaries of developing floral primordia. In addition, PNY and BP interact physically, suggesting that they form a heterodimeric transcriptional unit required for inflorescence patterning.
RESULTS
Characterization of the pny Mutant Alleles
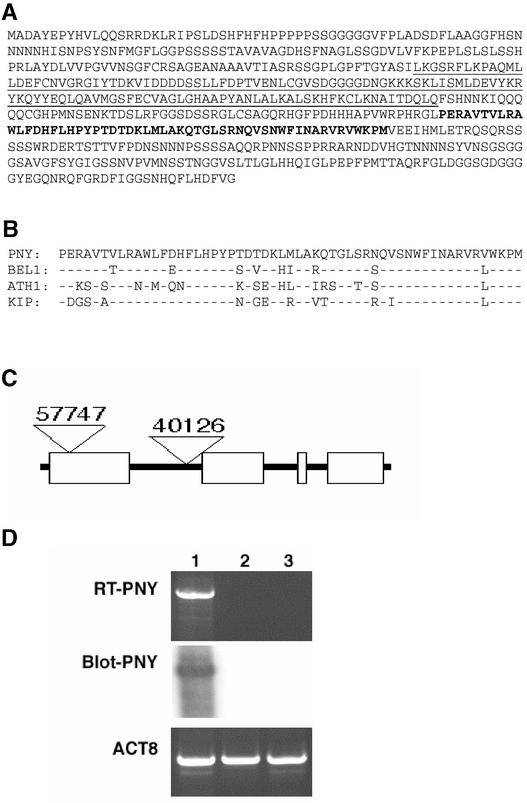
In Arabidopsis, the BELL gene family consists of 13 members, only one of which has been characterized by a mutant phenotype (Robinson-Beers et al., 1992; Modrusan et al., 1994; Reiser et al., 1995). We obtained two independent T-DNA insertion lines in the BELL gene At5g02030, which is located near the bottom of chromosome 5. This gene, renamed PENNYWISE (PNY), contains the conserved MEINOX-interacting domain near the N terminus and the three–amino acid loop extension homeodomain at the C terminus (Figure 1A) (Smith et al., 2002). The homeodomain of PNY is 66 to 84% identical to other BELL homeodomains (Figure 1B).
Figure 1.
Characterization of the Loss-of-Function pny Alleles.
(A) The amino acid sequence of PNY showing the MEINOX-interacting domain (underlined) and the three–amino acid loop extension homeodomain (boldface).
(B) The homeodomain of PNY was aligned with three other BELL proteins, BEL1, ATH1, and KNOTTED INTERACTION PROTEIN (KIP).
(C) Scheme of the PNY gene. White boxes represent exons, and black lines represent introns. Triangles depict the approximate T-DNA insertion sites.
(D) RT-PCR analysis using PNY gene-specific primers. Amplification occurred only from wild-type tissue (lane 1) and not from the pny-40126 (lane 2) or pny-57747 (lane 3) tissue. Hybridization of a 32P-labeled PNY probe occurred in the wild-type sample only after PCR products were transferred to a nylon membrane (Blot-PNY). Primers to ACT8 show that approximately equal amounts were amplified.
The T-DNA insertion in pny-57747 is located in exon 1, whereas the T-DNA insertion in pny-40126 is located in the first intron (Figure 1C). Inflorescence mRNA was isolated from the wild type (Columbia), pny-40126, and pny-57747. Reverse transcription (RT) using PNY-specific primers showed that this gene was amplified in the wild type but not in either mutant. Signal was not detected in the mutant samples even when the RT-PCR products were transferred to nylon and probed with the radiolabeled PNY probe (Figure 1D). Primers to ACTIN8 (ACT8) were used as an internal control to demonstrate that relatively equal amounts of ACT8 could be amplified in the three mRNA samples (Figure 1D). These results suggest that both T-DNA insertions into PNY created mRNA null mutants.
Phenotypic Analysis of pny
pny-40126 and pny-57747 were backcrossed two times into Arabidopsis ecotype Columbia before the pny phenotype was characterized. Crosses between pny-40126 and pny-57747 produced F1 plants with the same phenotype, demonstrating that they were allelic (data not shown). Phenotypic analysis was performed with pny-40126. As shown in Figure 2A, the pny inflorescence was shorter in stature than the wild-type inflorescence. In addition, two to three more paraclades developed in this mutant than in wild-type plants (Figure 3B), indicating a partial loss of apical dominance. Internode patterning of the inflorescence also was affected in pny plants compared with wild-type plants (Figures 2B to 2E). The internode length was relatively constant in wild-type plants (Figure 2D), whereas close examination of the inflorescence in pny showed that internodes between the siliques often were absent or reduced severely (Figure 2E). This internode defect appears to be random, because normal-sized internodes were interspersed along the stem. Clusters of siliques often were visible on the inflorescences stems, indicating that the internode defect could span a large region of the shoot (Figure 2E).
Figure 2.
Characterization of the pny Phenotype.
(A) Wild-type (left) and pny (right) plants showing that pny plants were slightly shorter.
(B) Wild-type inflorescence with a regular pattern of internode development.
(C) pny inflorescence with short and long internodes.
(D) Close-up of a wild-type inflorescence.
(E) Close-up of a pny inflorescence showing a cluster of siliques.
(F) Paraclade in the axil of a cauline leaf on a wild-type plant.
(G) Paraclade in the axil of a cauline leaf on a pny plant. The paraclades produced cauline leaves with no internode elongation between them (arrowheads).
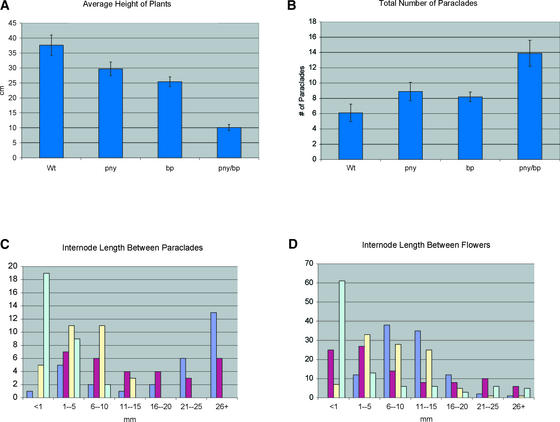
Figure 3.
Internode and Axillary Branch Defects.
Measurements were taken from 10 plants from each genotype of the F2 progeny that resulted from a cross between pny-40126 and bp-9.
(A) The average height of the wild type (Wt), pny, bp, and pny bp was determined, and the standard deviation was calculated.
(B) The average number of paraclades that grew >20 mm was measured, and the standard deviation was determined.
(C) Internode lengths between the first three paraclades (counting basipetally) were measured and grouped into 5-mm intervals. Wild type, dark blue; pny, red; bp, yellow; bp, pale blue.
(D) Ten internodes between the 1st and 11th siliques (counting acropetally) were measured and graphed. Color code same as in (C).
The paraclades had a highly penetrant defect that affected the first one or two internodes. These internodes were dramatically reduced in length, giving the appearance of leaves initiating in the axils of the cauline leaves (Figure 2G). The internodes on the main stem between the paraclades also often were reduced in size. In addition, the pedicels of flowers in pny were slightly shorter than those found in the wild type (Figures 2D and 2E). Finally, flowers occasionally were aberrant, producing small, curved, club-shaped siliques at a low penetrance (data not shown). Vegetative development appeared normal in pny plants (data not shown).
Synergistic Interactions between pny and bp
Given that bp plants also are shorter in stature with reduced internode length (Douglas et al., 2002; Venglat et al., 2002), we crossed pny-40126 to bp-9, both in the Columbia background, to determine the phenotype of the double mutant. Examination of the main inflorescences of the F2 progeny demonstrated that the short stature was enhanced in the double mutant (Figure 4A). Quantitative analysis showed that the average height of pny and bp plants was 7.9 and 12.2 cm shorter, respectively, than that of wild-type plants (Figure 3A). In pny bp double mutants, the average height was 27.5 cm shorter than that of the wild type. The loss of apical dominance also was enhanced in the double mutants. On average, pny and bp plants produced two or three more paraclades than wild-type plants, whereas the average number of paraclades that developed in pny bp more than doubled compared with that in the wild type (Figure 3B). The main axis terminated early in the double mutant, such that it quickly became difficult to distinguish the main axis from the paraclades.
Figure 4.
Phenotype of the pny bp Double Mutant.
(A) The inflorescence of the pny bp double mutant was very compact compared with those of the wild type (WT) and the bp and pny single mutants.
(B) Close-up of the inflorescence of pny bp.
(C) Paraclades from a pny bp plant were removed for photography. Clusters of siliques were visible on the main axis (m) and the paraclades (p).
(D) The short internodes between the paraclades and the siliques in pny bp produced an aerial rosette.
(E) Close-up view of a wild-type inflorescence.
(F) Close-up view of a pny inflorescence. Note the clusters of siliques.
(G) Close-up view of a bp inflorescence. Note the downward slant of the pedicels and the short internodes.
(H) Close-up view of a pny bp inflorescence. Note the exaggerated clustering of siliques and short pedicels.
A closer examination of the pny bp double mutant showed an enhanced clustering of siliques that was not found in either single mutant or the wild type (Figures 4E to 4H). This clustering occurred on the central axis as well as on the paraclades (Figure 4C). Often, a silique formed at the same position as a paraclade and more than one paraclade formed at the same node. This lack of internode elongation produced what appeared to be aerial rosettes on the main inflorescence (Figure 4D). The clusters of siliques in pny bp indicated that the internodes in these regions were absent or unelongated. In addition, internodes longer than those in the wild type were produced, demonstrating that internode patterning was not uniform (Figures 4E and 4H).
We measured internode lengths between the paraclades and between the first 11 siliques on the main axis. Internode lengths between the first three paraclades produced on the main axis tended to be very long in the wild type, >20 mm, whereas the same internodes in pny and bp were shorter, ranging from 1 to 25 mm and <1 to 10 mm, respectively (Figure 3C). In pny bp, more than half (63%) of the internodes between the first three paraclades were <1 mm (Figure 3C). The majority of internodes between siliques in the wild type ranged from 6 to 15 mm, whereas in bp and pny, the majority of internodes ranged in length from 1 to 15 mm and <1 to 10 mm, respectively (Figure 3D). In the double mutant, 61% of all internodes were <1 mm. Furthermore, only 12% of the internodes in pny bp were between 6 and 15 mm, the average internode length in the wild type (Figure 3D).
Cellular Organization of the Stem
To examine the histological organization of cells in the stem, we compared cross-sections through the internode located between the third and fourth siliques on the main axis. The internodes examined in this experiment were fully developed. As shown in Figures 5A and 5E, Columbia stems contained distinct cell types organized in a radial pattern, starting with the epidermis on the outside, followed by the cortex, and then the vascular bundles. These bundles were arranged an approximately equal distance apart separated by lignified interfascicular cells. The bundles contained phloem cells on the outside with xylem on the inside, separated by cambial cells. Large, vacuolated pith cells were found at the center of the stem. In pny and bp mutants, patterning defects in the organization of the bundles were apparent. Vascular bundles in bp varied in size and spacing but were somewhat regular in their organization (Figure 5B). Some bundles appeared to be undeveloped; they lacked lignin, and the xylem elements were small (Figure 5F, arrow). The pny mutants differed from the wild type and bp in having a thick continuous vascular ring with very little interfascicular space (Figures 5C and 5G). Instead of the 6 to 8 vascular bundles in a wild-type section, 8 to 10 bundles often were observed. There also was increased cambial activity between the phloem and the xylem cells (Figure 5G). In addition, cortex cells were larger and more densely packed in the single mutants than in the wild type (Figures 5E to 5G).
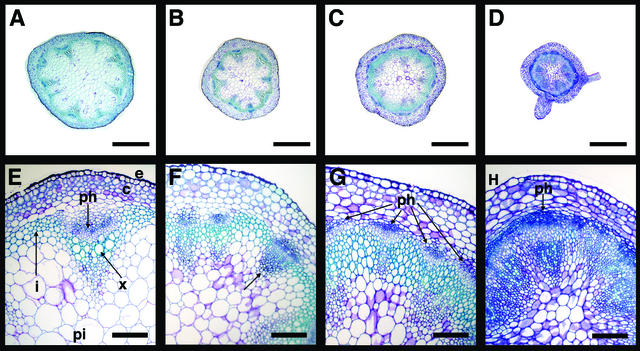
Figure 5.
Histological Analysis of the Stems.
(A) Cross-section through a wild-type stem.
(B) Cross-section through a bp stem.
(C) Cross section through a pny stem.
(D) Cross-section through a pny bp stem. A band of phloem (blue-stained cells) formed beneath the cortex layer.
(E) Close-up of a wild-type stem section. The vascular bundle, consisting of xylem and phloem, was detected inside of the cortex, protruding into the highly vacuolated pith cells. Cambial cells (opaque staining) were located between the xylem and phloem cells.
(F) Close-up of a bp stem section. Bundles were closer together as a result of the overall reduction in size. The bundle indicated with an arrow contained small densely stained cells, suggesting that it was not fully differentiated.
(G) Close-up of a pny stem section. Note the increase in the number of vascular bundles, as depicted by the blue patches of phloem. In addition, there was an increase in lignification (green-stained cells).
(H) Close-up of a pny bp stem section. Note the thick continuous band of phloem cells. The cambium-like cells (opaque cells inside of the phloem) had random cell division patterns. Xylem elements were small and not lignified. Cortex and epidermal cells were larger, whereas pith cells were smaller.
c, cortex; e, epidermis; i, interfascicular cells; ph, phloem; pi, pith; x, xylem. Bars = 1 mm in (A) to (D) and 500 nm in (E) to (H).
The pny bp double mutants had extremely disrupted vascular patterning in the stem, displaying enhancement of characteristics observed in the single mutants (Figures 5D and 5H). The bundles in pny bp were similar to the undeveloped vascular bundles observed in bp. These bp-like bundles contained small, densely stained cells, and the xylem elements were not adjacent to each other, but scattered (Figures 5D and 5H). The pny characteristics also were enhanced in the double mutant. For example, the bundles were adjacent to each other, and a continuous band of phloem cells formed inside the large cortex cells. In addition, there also was an enhancement of disorganized cambial activity, with cell walls in random orientations (Figure 5H).
PNY Expression Overlaps with BP in the Inflorescence Meristem
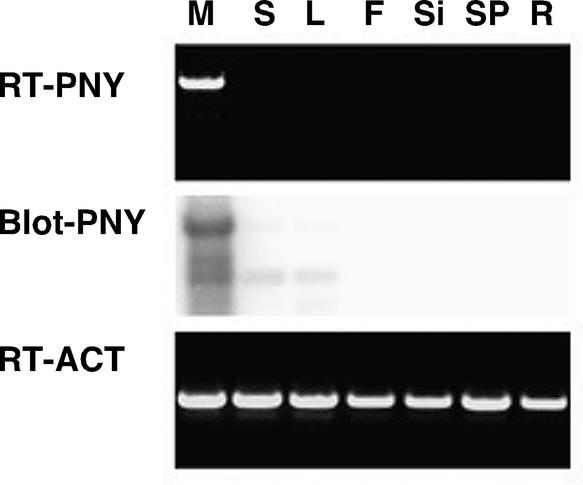
PNY transcripts were amplified using RT-PCR from wild-type inflorescences that contained meristems, developing flowers, internodes, and pedicles (Figure 1D). To determine the organs in which PNY was expressed, RT-PCR was performed on meristem, seedling, and root tissues as well as mature leaves, flowers, siliques, internodes, and pedicels. As shown in Figure 6, PNY could be amplified only from apetala1 cauliflower inflorescence meristem tissue, whereas little or no PNY transcript was amplified from other tissues (Figure 6). ACT8 was used as an internal control, and approximately equal amounts of this gene were amplified in all tissues examined.
Figure 6.
Expression Profile of PNY.
RT-PCR was performed on different plant tissues (RT-PNY), which then were blotted to nylon and probed with a 32P-labeled PNY probe (Blot-PNY). PNY was amplified from inflorescence meristem tissue (M) that was obtained from ap1 cal double mutants. Only minute amounts of PNY could be amplified from seedlings (S) and mature leaves (L). No PCR product was detected in mature flowers (F), siliques (Si), stems and pedicles (SP), or roots (R). Approximately equal amounts of ACT8 PCR product (RT-ACT) were amplified from the tissues examined.
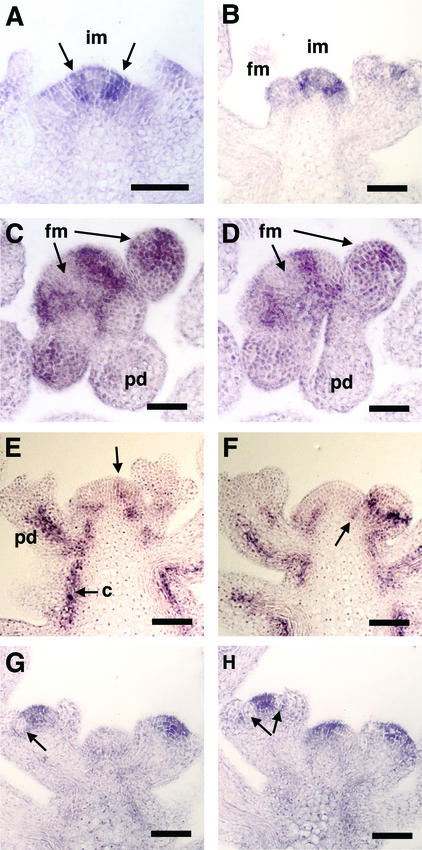
To localize PNY mRNA more precisely in the inflorescence, in situ hybridization was performed. PNY transcript localized to inflorescence and floral meristems. In the inflorescence meristem, PNY was observed in distinct zones at the apex. In a longitudinal section, the expression appeared as a stripe that extended from the epidermis to the fifth or sixth cell layer of the meristem (Figures 7A and 7B). Transverse consecutive sections confirmed that PNY was not expressed throughout the inflorescence meristem (Figures 7C and 7D). The expression pattern suggests that PNY may be excluded from the floral primordial cells of the inflorescence meristem, similar to the expression of STM (Long and Barton, 2000).
Figure 7.
In Situ Hybridization of PNY and BP in Wild-Type Inflorescence Tissues.
(A) and (B) Longitudinal cross-sections through the inflorescence meristem. PNY mRNA was localized in stripes of cells on the flanks of the meristem (arrows).
(C) and (D) Transverse sections through the inflorescence meristem. PNY localized to bands of cells that may encircle initiating floral primordia. Note that PNY was not expressed in early floral primordia but was expressed at a later stage of flower development.
(E) and (F) BP was expressed in a band of cells on the flanks of the meristem (arrows). In addition, BP was expressed at the base of the floral primordia four to five cells from the epidermal layer. BP also localized to cortex cells adjacent to vascular tissue in elongating internodes and pedicels.
(G) and (H) PNY mRNA localized to cells in the floral meristem but not in developing floral organ primordia (arrows).
c, cortex; fm, floral meristem; im, inflorescence meristem; pd, pedicle. Bars = 100 nm.
In situ hybridization using the BP probe showed similar stripes of expression on the flanks of the inflorescence meristem (Figures 7E and 7F). BP expression also was detected at the bases of floral meristems and in the cortex cells. These results identify possible overlapping regions of PNY and BP expression. However, it should be noted that little or no PNY transcript was detected in the internodes adjacent to the vasculature, as seen with BP (cf. Figures 7E and 7G). We detected no difference in the expression of BP in the pny mutant compared with the wild type or in the expression of PNY in the bp mutant (data not shown), suggesting that BP and PNY do not regulate each other's transcription.
PNY also was expressed in a discrete domain in the first four to five cell layers of the floral meristem. PNY was not expressed in initiating sepals (Figures 7G and 7H). The zone of expression narrows in slightly older floral meristems, suggesting that it is downregulated as organ primordia initiate. No expression was seen in the pedicel, unlike the strong expression seen with BP (cf. Figures 7E and 7G). The antisense digoxigenin PNY mRNA probe did not hybridize to tissues in pny inflorescences (data not shown). Finally, no hybridization was detected with the sense digoxigenin PNY mRNA probe in wild-type tissue (data not shown).
PNY Interacts with BP and STM
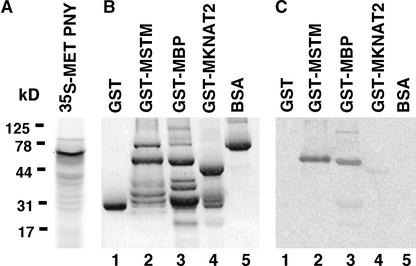
Ligand blot analysis in maize showed that KIP had an apparent higher affinity for the MEINOX domain of KN1 than RS1 and LIGULELESS3 (Smith et al., 2002). To determine the Arabidopsis class-1 KNOX binding partners for PNY, a ligand blot procedure was employed that used glutathione S-transferase (GST) fusion proteins containing the MEINOX domain of STM (GST-MSTM), BP (GST-MBP), and knotted1-like from Arabidopsis thaliana2 (KNAT2) (GST-MKNAT2). The GST-KNOX fusions, as well as GST and BSA, were purified, and approximately equal amounts of the full-length fusions were separated by SDS-PAGE (Figure 8B). After transfer to nitrocellulose, the membrane was hybridized with PNY protein (Figure 8C) that had been radiolabeled with 35S-Met in vitro (Figure 8A). PNY bound to GST-MSTM and GST-MBP, whereas little or no interaction was detected with GST-MKNAT2 (Figure 8C). In addition, PNY did not associate with the GST or BSA control.
Figure 8.
Ligand Blot Analysis of PNY.
(A) PNY was labeled in vitro with 35S-Met, separated by SDS-PAGE, and analyzed by autoradiography. The molecular mass of PNY is 67 kD (arrowhead).
(B) The MEINOX (M) domain of STM (lane 2), BP (lane 3), and KNAT2 (lane 4) was fused to GST, and ∼1 μg of full-length recombinant expressed and purified proteins was separated by SDS-PAGE. The molecular mass of the full-length GST fusions was 61 kD for GST-MSTM, 59 kD for GST-MBP, and 41 kD for GST-MKNAT2. Other protein products detected in each lane correspond to partially degraded fusion proteins or bacterial proteins that copurify with the GST fusion proteins. These protein products also serve as internal negative controls, because the interaction of KNOX and BELL proteins requires an intact MEINOX domain (our unpublished data). GST and BSA were used as negative controls.
(C) Ligand blots probed with radiolabeled PNY and analyzed by autoradiography demonstrated that PNY interacted with GST-MSTM and GST-MBP, whereas little or no binding was detected for GST-MKNAT2, GST, or BSA.
DISCUSSION
We identified a mutation in the Arabidopsis gene PNY with similar defects in internode patterning as BP. We propose that BP and PNY cooperatively regulate early events in internode patterning, based on the synergistic double mutant phenotype, the overlap in expression, and the interaction of the proteins in vitro.
Internode Patterning
Biochemical interactions of KNOX-BELL proteins have been described in many plant species (Bellaoui et al., 2001; Muller et al., 2001; Smith et al., 2002). In vitro binding and yeast-two hybrid studies show that the interactions are selective, suggesting that specific KNOX-BELL heterodimers regulate unique developmental pathways in the meristem. Moreover, cooperative interaction between KNOX-BELL heterodimers mediates high DNA binding affinity to the KN1 DNA motif, TGACAG(G/C)T (Smith et al., 2002). To address the biological significance of specific KNOX-BELL interactions, we initiated experiments to identify mutations in BELL genes.
Two KNOX orthologs, OSH15 (D6) and BP, are expressed in similar domains in the meristem and internode regions of rice and Arabidopsis, respectively (Sato et al., 1999; Douglas et al., 2002; Venglat et al., 2002). Plants that are mutant for BP and D6 are short in stature with reduced internodes. In d6, only the uppermost internodes of the inflorescence are affected, whereas in bp, short internodes appear randomly throughout the inflorescence. In addition, the cellular organization of the vasculature is affected in d6 (Sato et al., 1999).
We determined the phenotype of pny, a BELL gene, through reverse genetics. Both pny and bp have increased numbers of paraclades and very short internodes mixed with long internodes. The phenotype of the double mutant is synergistic. Clusters of siliques are observed on the stems in pny bp with little or no internode elongation between the siliques. These clusters are bracketed by long internodes, occasionally longer than normal. The highly reduced internodes also are found between the paraclades and between cauline leaves on the paraclades. The resulting appearance of the double mutant is bushy.
Alterations to the vasculature also are enhanced in the double mutant. bp mutants have an occasional vascular bundle that lacks lignin and contains small xylem elements. pny mutants differ from the wild type in having fewer interfascicular cells and a concomitant increase in vascular bundles. The double mutant accentuates these individual defects. All of the bundles in the pny bp stems resemble the undeveloped bundles of bp, lacking lignin and containing small xylem elements. The vascular bundles form a continuous ring, leaving little or no space for interfascicular cells. The cellular disorganization occurs whether the internodes are very short or of normal length, suggesting that the defect in pny bp affects the entire stem.
Given the enhancement of the internode defect in the double mutant and the interaction of BP and PNY in vitro, we expect the two gene products to localize to the same tissues where they are required for proper internode development. PNY is expressed in inflorescence and floral meristems and is not detected in internodes using either in situ hybridization or RT-PCR. The mRNA expression patterns of BP and PNY overlap in a narrow region between the initiating floral primordia and the inflorescence meristem. We suggest that their coincident expression in the meristem establishes the proper patterning of the internode.
We propose that the PNY-BP protein complex may function in the inflorescence meristem to form a boundary between lateral organs and the inflorescence meristem. The PNY-BP boundary then would regulate patterning mechanisms to coordinate proper internode development. In the absence of just BP or PNY, the remaining protein interacts with another BELL or KNOX protein, respectively. These interactions partially compensate for the loss of BP or PNY; therefore, the single mutants are not as severe as the double mutant. For example, in a bp mutant, PNY may interact with STM, and in a pny mutant, BP may interact with another BELL protein. In the absence of both PNY and BP, the alternate partners do not interact; thus, the boundary is not specified properly. The result is mostly an absence of internode but the occasional long internode. The increase in vascular bundles also may result from the defect in patterning. Procambium is first detected below the initiating leaf primordium (Ye, 2002). If the patterning mechanisms are disrupted, it is possible that too many procambial initials form.
Histological analyses suggest that the internode of Arabidopsis arises from two zones of the meristem (Vaughn, 1955). The flank meristem zone (also called the peripheral or morphogenetic zone) produces lateral organs (leaves and flowers) and the cortex of the stem. The file meristem zone produces the pith of the stem. Both the file and flank meristem zones are produced from a central initiation zone (Vaughn, 1955). Clearly, initiation of the internode requires the coordination of file and flank meristem cells. Given that BP and PNY are expressed in the flank meristem zone and not the file meristem zone, they are likely to signal to the file meristem cells to coordinate internode patterning.
Studies in other systems strengthen the relationship of internode, leaf, and meristem. For example, clonal analyses in maize and sunflower suggest that the meristem uses the same population of cells to produce a leaf and its subtending internode (Johri and Coe, 1983; Poethig et al., 1986; McDaniel and Poethig, 1988; Jegla and Sussex, 1989). Mutations such as terminal ear, which increase leaf number, reduce internode length (Veit et al., 1998). Finally, comparative morphological studies also demonstrate the interrelationship of the leaf base and the subtending internodes (Kaplan, 2001).
Independent Functions of KNOX/BELL Heterodimers
Not all defects of bp are shared by pny, as seen in the pedicel orientation of bp (Douglas et al., 2002; Venglat et al., 2002). These phenotypic differences suggest that BP and PNY also have functions that are independent of each other. These independent functions likely result from expression patterns that do not overlap in specific tissues of the plant. For example, BP is expressed strongly in the pedicel and PNY is not. The expression of PNY in the floral meristem also differs from the expression of BP. In tissues in which there is no overlap, BP may interact with other BELL proteins that are expressed in the pedicle or internode, whereas PNY may interact with STM to control maintenance events in the floral meristem. Interestingly, in the inflorescence meristem, PNY expression overlaps with both BP and STM, and ligand blot analysis demonstrates that PNY can interact with these two KNOX proteins. Therefore, the independent functions of PNY may not depend only on its temporal and spatial expression patterns but also on whether it is bound to BP or STM. This observation suggests an important parameter for the function of all BELL and KNOX proteins.
Resemblance to Hormone Imbalance
Morphological changes that result from the interplay of KNOX gene expression and hormone homeostasis have been well documented. For example, ectopic KNOX gene expression can lead to an increase of cytokinin (Ori et al., 1999; Frugis et al., 2001) and the loss of apical dominance (Sinha et al., 1993). In addition, antagonistic interactions between gibberellin (GA) and KNOX genes are thought to contribute to the diversity in leaf morphology observed in higher plants (Tanaka-Ueguchi et al., 1998; Sakamoto et al., 2001; Hay et al., 2002) and may be required for meristem maintenance (Hay et al., 2002). Finally, both the semaphore and rough sheath2 mutants of maize misexpress KNOX genes and show altered auxin transport (Tsiantis et al., 1999; Scanlon et al., 2002).
Many of the phenotypic characteristics of pny bp are similar to defects in auxin and GA signaling. For example, the loss of apical dominance is typical of reduced auxin or increased cytokinin (Cline, 1994). The cellular defects in the stems of pny bp resemble the phenotype of increased GA and indoleacetic acid in cut stems (Digby and Wareing, 1966; Cutter, 1971). Finally, auxin and GA are thought to control internode elongation and development in rosette plants (Sachs, 1968). Further analysis will help determine exactly how PNY-BP integrates with hormones to establish proper patterning and cellular organization in the stem.
METHODS
Plant Growth and Genetic Analysis
Arabidopsis thaliana (ecotype Columbia) plants were grown in a 16-h-light/8-h-dark cycle at 22°C. The two T-DNA insertion lines in PNY were obtained from the ABRC (Ohio State University, Columbus; stock numbers SALK_040126 and SALK_057747). The map positions of these T-DNA insertions were at 395,850 and 396,983 bp for SALK_057747 and SALK_040126, respectively. Both alleles were backcrossed two times into the Columbia ecotype. The SALK_040126 insertion line was used in our phenotypic and molecular analyses. Genotypes of pny-40126 and pny-57747 plants were determined using primers to the left border (Lba1) and a primer that was specific to PNY (Lba1, 5′-TGGTTCACGTAGTGGGCCATCG-3′; 40126-1, 5′-TGGAATTGGAGACAAAATGTGTTA-3′; 57747-1, 5′-GTTTCAAGAACCTTGATCC-3′). Only plants that were homozygous for the T-DNA displayed the pny phenotype (data not shown). bp-9 results from a dSpm insertion and was obtained by screening the SLAT lines for insertions (G. Mele, N. Ori, Y. Sato, and S. Hake, unpublished data). Quantitative analysis of the different genotypes—wild type, pny, bp, and pny bp—were performed on plants at equal developmental stages, 2 to 3 weeks after bolting.
Molecular in Situ and Biochemical Analyses
Reverse transcription PCR (RT-PCR) was performed as described (Hay et al., 2002) using the gene-specific primers AT5G02030-F and AT5G02030-R (see below). Transfer of PCR products to nylon membranes and hybridization with 32P-PNY probe was performed using standard procedures (Sambrook et al., 1989). A full-length PNY cDNA was obtained by RT-PCR using mRNA isolated from Columbia inflorescences with primers that were complementary to the 5′ and 3′ ends of the gene as follows: AT5G02030-F, 5′-ATCCGAATTCGAATGGCTGATGCATACGAGCCTTATC-3′; AT5G02030-R, 5′-TCCTGTCGACACCTACAAAATCATGTAGAAACTG-3′. The 5′ primer contained an EcoRI site and the 3′ primer contained a SalI site, which allowed for in-frame cloning with the T7 epitope tag in the pET-21d vector (Novagen, Madison, WI). The gene was sequenced to verify its integrity. As an internal control for RT-PCR, we used primers to the ACTIN8 gene: ACT8-F, 5′-ATGAAGATTAAGGTCGTGGCA-3′; ACT8-R, 5′-TCCGAGTTTGAAGAGGCTAC-3′.
To generate a probe for in situ hybridization, PNY was cleaved with XbaI (PNY has two internal XbaI restriction sites at 465 and 987 bp from the ATG start codon). This XbaI fragment was cloned into pBluescript SK− and KS+ plasmids (Stratagene, La Jolla, CA), and the T7 polymerase (Promega, Madison, WI) was used to synthesize sense and antisense UTP-digoxigenin–labeled RNA as described (Jackson, 1992). The BP probe was as described (Lincoln et al., 1994). Plant fixation and in situ hybridization were performed as described previously (Jackson, 1992).
To construct the GST-MEINOX fusion proteins, gene-specific primers were used to amplify the 5′ part of the genes that encoded the N-terminal region containing the MEINOX domain of STM, BP, and KNAT2. Pfu polymerase (Stratagene) was used to synthesize the gene fragments. Each PCR primer (forward and reverse) contained a specific restriction enzyme site that allowed the PCR products to be cloned in frame with GST. The 5′ fragments of STM1-703, BP1-747, and KNAT2162-310 were amplified using GSTMSTM-F (5′-CATTATGGATCCAGATGGAGAGTGGTTCCAACAGC-3′) and GSTMSTM-R (5′-GTATAGGATCCCTGCTGCTGTCTCTCCATAACCGGA-3′), GSTMBP-F (5′-CAGCTTAGGATCCAGATGGAAGAATACCAGCATGAC-3′) and GSTMBP-R (5′-TTATTGGATCCCATTGTCACTCTTCCCATCAGG-3′), and GSTMKNAT2-F (5′-GACAACGGATCCCGCGTATTCGAAAAGCTGAG-3′) and GSTMKNAT2-R (5′-GCAAGGTCGACCTAACCAGTGCACAAGTTCTGAAGCTG-3′). The restriction enzyme sites in the primers are underlined. STM1-703 and BP1-747 gene fragments then were cloned into pGEX-5X-2 (Pharmacia, Piscataway, NJ) using BamHI (Panvera/Takara, Madison, WI), whereas KNAT2162-310 was cloned into pGEX-5X-2 using BamHI and SalI (Panvera/Takara). All constructs were sequenced to verify the integrity of the clones. The ligand blot procedure was performed as described (Chen et al., 2000; Smith et al., 2002).
Histology
Tissues were fixed in 4% formaldehyde, 5% glacial acetic acid, 45% ethanol, and 1% Triton X-100 for 48 h and then dehydrated in an ethanol series. Dehydrated tissues were embedded in Technovit 7100 resin (Hereaus Kulzer, Wehrheim, Germany) according to the manufacturer's protocol. Sections were 8 μm thick and stained for 32 s in toluidine blue solution (0.1% toluidine blue and 0.1 M sodium phosphate buffer, pH 5.0). After destaining in running water, sections were dried and mounted.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact S. Hake, fax 510-559-5678, maizesh@nature.berkeley.edu.
Accession Numbers
The GenBank accession numbers for the sequences shown in Figure 1 are U39944 (BEL1), X80126 (ATH1), and AY082396 (KIP).
Acknowledgments
We thank the Salk Institute of Genomic Analysis Laboratory and the Salk/Stanford/PGEC Consortium for the T-DNA insertion lines SALK_040126 and SALK_057747. We thank George Chuck for technical advice on in situ hybridization experiments, George Chuck and Giovanni Mele for critical reading of the manuscript, Hans Holtan for graphics assistance, and David Hantz for greenhouse maintenance. H.M.S.S. was supported by National Institutes of Health postdoctoral fellowship GM20158-03. S.H. was supported by National Science Foundation Grant IBN-0131431 and the U.S. Department of Agriculture.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012856.
References
- Bellaoui, M., Pidkowich, M.S., Samach, A., Kushalappa, K., Kohalmi, S.E., Modrusan, Z., Crosby, W.L., and Haughn, G.W. (2001). The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 11, 2455–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.H., Sheng, J., Hind, G., Handa, A.K., and Citovsky, V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, M.G. (1994). The role of hormones in apical dominance: New approaches to an old problem in plant development. Physiol. Plant. 90, 230–237. [Google Scholar]
- Cutter, E.G. (1971). Plant Anatomy: Experiment and Interpretation. (London: Edward Arnold).
- Digby, J., and Wareing, P.F. (1966). The effect of applied growth hormones on cambial activity and the differentiation of the cambial derivatives. Ann. Bot. 30, 539–548. [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10, 101–113. [DOI] [PubMed] [Google Scholar]
- Frugis, G., Giannino, D., Mele, G., Nicolodi, C., Chiappetta, A., Bitonti, M.B., Innocenti, A.M., Dewitte, W., Van Onckelen, H., and Mariotti, D. (2001). Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126, 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type ho-meobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hempel, F.D., and Feldman, L.J. (1994). Bi-directional inflorescence development in Arabidopsis thaliana: Acropetal initiation of flowers and basipetal initiation of paraclades. Planta 192, 276–286. [Google Scholar]
- Jackson, D. (1992). In situ hybridization in plants. In Plant Molecular Pathology: A Practical Approach, D.J. Bowles, ed (Oxford, UK: Oxford University Press), pp. 163–174.
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jegla, D.E., and Sussex, I.M. (1989). Cell lineage patterns in the shoot meristem of the sunflower embryo in the dry seed. Dev. Biol. 131, 215–225. [DOI] [PubMed] [Google Scholar]
- Johri, M.M., and Coe, E.H. (1983). Clonal analysis of corn plant development. I. The development of the tassel and the ear shoot. Dev. Biol. 97, 154–172. [DOI] [PubMed] [Google Scholar]
- Kaplan, D.R. (2001). The science of plant morphology: Definition, history, and role in modern biology. Am. J. Bot. 88, 1711–1741. [PubMed] [Google Scholar]
- Kerstetter, R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. (1997). Loss of function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124, 3045–3054. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A Knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6, 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218, 341–353. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- McDaniel, C.N., and Poethig, R.S. (1988). Cell-lineage patterns in the shoot apical meristem of the germinating maize embryo. Planta 175, 13–22. [DOI] [PubMed] [Google Scholar]
- Modrusan, Z., Reiser, L., Feldmann, K.A., Fischer, R.L., and Haughn, G.W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27, 13–23. [DOI] [PubMed] [Google Scholar]
- Nishimura, A., Tamaoki, M., Sato, Y., and Matsuoka, M. (1999). The expression of tobacco knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J. 18, 337–347. [DOI] [PubMed] [Google Scholar]
- Ori, N., Juarez, M.T., Jackson, D., Yamaguchi, J., Banowetz, G.M., and Hake, S. (1999). Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 11, 1073–1080. [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S., Coe, E.H., Jr., and Johri, M.M. (1986). Cell lineage patterns in maize embryogenesis: A clonal analysis. Dev. Biol. 117, 392–404. [Google Scholar]
- Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G.W., and Fischer, R.L. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83, 735–742. [DOI] [PubMed] [Google Scholar]
- Reiser, L., Sanchez-Baracaldo, P., and Hake, S. (2000). Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Biol. 42, 151–166. [PubMed] [Google Scholar]
- Robinson-Beers, K., Pruitt, R.E., and Gasser, C.S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, R.M. (1968). Control of intercalary growth in the scape of Gerbera by auxin and gibberellic acid. Am. J. Bot. 55, 62–68. [Google Scholar]
- Sakamoto, T., Kamiya, N., Ueguehi-Tanaka, M., Iwahori, S., and Matsuoka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sato, Y., Sentoku, N., Miura, Y., Hirochika, H., Kitano, H., and Matsuoka, M. (1999). Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18, 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon, M.J., Henderson, D.C., and Bernstein, B. (2002). SEMAPHORE1 functions during the regulation of ancestrally duplicated knox genes and polar auxin transport in maize. Development 129, 2663–2673. [DOI] [PubMed] [Google Scholar]
- Schneeberger, R.G., Becraft, P.W., Hake, S., and Freeling, M. (1995). Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 9, 2292–2304. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7, 787–795. [DOI] [PubMed] [Google Scholar]
- Smith, H.M., Boschke, I., and Hake, S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. USA 99, 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.G., Greene, B., Veit, B., and Hake, S. (1992). A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30. [DOI] [PubMed] [Google Scholar]
- Tanaka-Ueguchi, M., Itoh, H., Oyama, N., Koshioka, M., and Matsuoka, M. (1998). Over-expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. Plant J. 15, 391–400. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Brown, M.I., Skibinski, G., and Langdale, J.A. (1999). Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol. 121, 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn, J.G. (1955). The morphology and growth of the vegetative and reproductive apices of Arabidopsis thaliana (L.) Heynh, Capsella bursa-pastoris (L.) Medic. and Anagallis arvensis. J. Linn. Soc. Lond. LV, 279–301.
- Veit, B., Briggs, S., Schmidt, R.J., Yanofsky, M.F., and Hake, S. (1998). Regulation of leaf initiation by the terminal ear1 gene of maize. Nature 393, 166–168. [DOI] [PubMed] [Google Scholar]
- Venglat, S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53, 183–202. [DOI] [PubMed] [Google Scholar]