Abstract
The function of the novel gene MSP1 (MULTIPLE SPOROCYTE), which controls early sporogenic development, was elucidated by characterizing a retrotransposon-tagged mutation of rice. The MSP1 gene encoded a Leu-rich repeat receptor–like protein kinase. The msp1 mutation gave rise to an excessive number of both male and female sporocytes. In addition, the formation of anther wall layers was disordered and the tapetum layer was lost completely. Although the mutation never affected homologous chromosome pairing and chiasma maintenance, the development of pollen mother cells was arrested at various stages of meiotic prophase I, which resulted in complete male sterility. Meanwhile, plural megaspore mother cells in a mutant ovule generated several megaspores, underwent gametogenesis, and produced germinable seeds when fertilized with wild-type pollen despite disorganized female gametophytes. In situ expression of MSP1 was detected in surrounding cells of male and female sporocytes and some flower tissues, but never in the sporocytes themselves. These results suggest that the MSP1 product plays crucial roles in restricting the number of cells entering into male and female sporogenesis and in initiating anther wall formation in rice.
INTRODUCTION
The life cycle of angiosperms alternates between diploid sporophyte and haploid gametophyte generation (Goldberg et al., 1993; Drews et al., 1998). In the majority of flowering plants, the dynamic phase transition of sporophyte to gametophyte occurs in the anther and ovule. Sporogenesis is characterized by the differentiation of hypodermal cells in anther and ovule primordia, termed primordial germ cells or archesporial cells, into microsporocytes and megasporocytes, respectively (Maheshwari, 1950). Sporocytes undergo meiosis to give rise to microspores and megaspores, in which a differential pattern of spore formation between male and female organs, called heterospory, usually is observed in flowering plants, including rice.
The plasticity of plant development and evidence from mosaic analysis indicate that cell fate generally is determined late in plant development as a result of positional information (Poethig, 1989; Dawe and Freeling, 1992; Huala and Sussex, 1993). That is, position-dependent communication processes probably occur between adjacent meristematic cells, and these signaling events influence floral organ cell specification pathways, including the establishment of germ cell fate. Indeed, cell signaling pathways at shoot and floral meristems have been studied by many researchers (reviewed by Hake and Char, 1997; van der Schoot and Rinne, 1999; Clark, 2001). However, it remains unclear when and how germ cells of angiosperms acquire their identity. Although several interesting mutants relating to the determination of germ cell fate have been isolated and characterized, such as spl/nzz (Schiefthaler et al., 1999; Yang et al., 1999), ems1 (Zhao et al., 2002), and exs (Canales et al., 2002; Sorensen et al., 2002) in Arabidopsis and mac1 in maize (Golubovskaya et al., 1992; Sheridan et al., 1996, 1999), this finding is not sufficient to determine the molecular basis regulating reproductive organ differentiation.
Tobacco stamen primordia and anther cell lineages have been inferred from histological analysis to be derived from three layers (Goldberg et al., 1993). The differentiation pattern of the stamen is mostly conserved in rice, in which the outermost layer gives rise to the epidermis, the second layer gives rise to archesporial cells, and the third and inner layers give rise to connective cells, vascular bundle cells, and a circular cell cluster adjacent to the stomium (Goldberg et al., 1993). The archesporial cells from the second layer undergo a series of mitotic periclinal divisions to differentiate the primary parietal cell (PPC) and the primary sporogenous cell (PSC). PSCs continue several mitotic divisions and differentiate into pollen mother cells (PMCs). Two subsequent periclinal divisions of the parietal layer give rise to the endothecium, the middle layer, and the tapetum layer in Gramineae species (Davis, 1966; Raghavan, 1988). Each of the four-walled layers expands with anticlinal divisions.
On the other hand, a female archesporial cell is differentiated from a single hypodermal cell at the top of the nucellar primordium in the majority of flowering plants. In Gramineae species, including rice, a female archesporial cell elongates longitudinally and directly differentiates the megasporocyte or megaspore mother cell (MMC) (Davis, 1966; Russell, 1979). The MMC then undergoes meiosis, resulting in the formation of four haploid spores. A chalazal spore becomes a functional megaspore, whereas three spores toward the micropyle undergo programmed cell death (Webb and Gunning, 1990).
Although early-developing stages of the anther and nucellus of rice were described previously by Kuwada (1910) and Raghavan (1988), few genetic studies have been performed to date. We attempted to elucidate the early steps of microsporogenesis and megasporogenesis in rice using the msp1 mutant tagged by retrotransposon Tos17. Here, we report that the MSP1 gene encodes a putative Leu-rich repeat (LRR) receptor–like protein kinase and plays important roles in restricting the number of cells entering into male and female sporogenesis and in initiating anther wall formation in rice flower development.
RESULTS
Screening of the msp1 Mutant
To study the processes of microsporogenesis and megasporogenesis in rice, we screened sterile mutant lines regenerated from the suspension culture of rice calli. It is known that the endogenous retrotransposon Tos17 is transposed in cultured calli and is expected to carry tagged mutations (Hirochika et al., 1996; Yamazaki et al., 2001). Among 600 primary selected partial and full sterile lines, we chose the mutant line ND0018, which segregated the plants with small collapsed anthers and full sterility. The final length of the mutant anthers was less than half the length of anthers in the heterozygous siblings (Figure 1A). Vegetative and reproductive organs of the mutants, except for the anthers, exhibited the same morphology as those of fertile siblings. The sterile phenotype was segregated as a single recessive mutation (fertile:sterile, 148:37, χ2 = 2.47 for 3:1). Heterozygous plants showed a completely normal phenotype. Observation of the mutant phenotype revealed that the number of PMCs and MMCs was multiplied abnormally in the anther and ovule, respectively. Therefore, we designated this mutation msp1 according to the phenotype of multiple sporocytes.
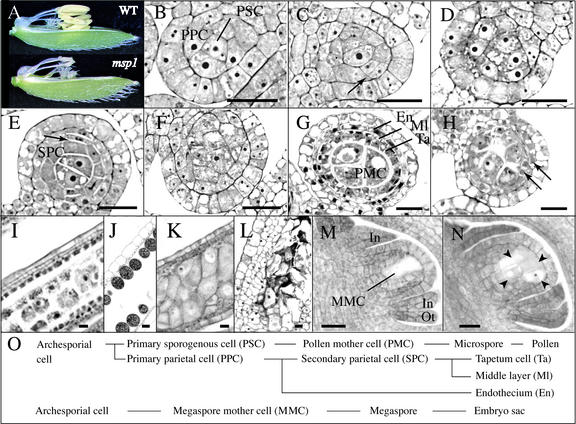
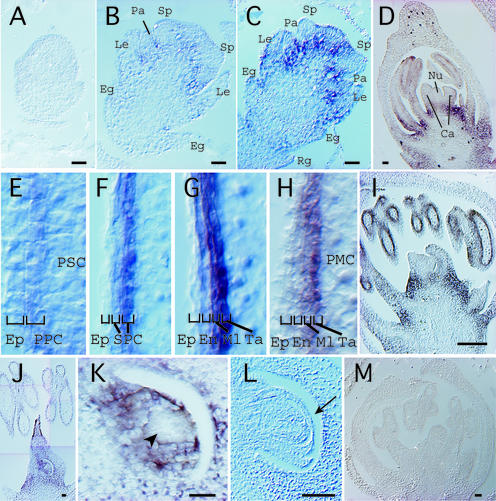
Figure 1.
Histological Features of Male and Female Reproductive Organ Development in the Wild Type and the msp1 Mutant.
(A) Flower and anther morphology of the wild type (WT) and msp1 at the heading stage.
(B) to (H) Transverse sections of the anther.
(B) PSCs and PPCs were already differentiated in a wild-type anther at stage II.
(C) PPCs began to divide periclinally (arrow) to differentiate secondary parietal cells in a wild-type anther at stage IV.
(D) Cell mass was increased in the mutant microsporangia, but no layered structure was observed at stage IV.
(E) Secondary parietal cells (SPC) began to divide periclinally (arrow) in a wild-type anther at stage V.
(F) Cells of a similar size occupied the mutant microsporangia at stage V.
(G) PMCs, tapetum cells (Ta), the middle layer (Ml), and the endothecium (En) were established in a wild-type anther in meiotic prophase I.
(H) A layered structure, but with irregular proliferation, appeared in a mutant anther in meiotic prophase I (arrows).
(I) to (N) Longitudinal sections of the anther and the ovule.
(I) A densely stained tapetum layer located at the fourth inner layer was observed clearly in a longitudinal section of a wild-type anther in meiotic prophase I.
(J) Upon the completion of pollen grain maturation, inner wall layers were degraded in a wild-type anther.
(K) Tapetum cells were never observed in the longitudinal section of a mutant anther in meiotic prophase I.
(L) Cells of a similar size formed a layer-like structure. PMCs were degraded in the mutant at the stage corresponding to the pollen maturation in the wild type shown in (J).
(M) A single and elongated MMC was observed in a wild-type nucellus with developing inner integuments (In) and outer integuments (Ot).
(N) At least four MMCs (arrowheads) were observed in a mutant nucellus with normally developing inner and outer integuments.
(O) Wild-type cell lineage of male and female germ cells.
Bars = 20 μm.
Abnormal Anther and Ovule Development in the msp1 Mutant
Histological features were compared in the early developmental stages of anthers between wild-type cv Nipponbare and the msp1 homozygous mutant. In this study, we classified the normal development of young anthers into five stages. Stage I is the stage at which the anther primordium is ovoid in a transverse section and in which archesporial cells differentiate at four corners. In stage II, archesporial cells divide to form PSCs and PPCs (Figure 1B). Stage III is the stage at which PPCs continue to divide and enlarge. In stage IV, PPCs begin and continue to divide periclinally (Figure 1C). Stage V is the stage at which the formation of secondary parietal cell is completed in most anthers (Figure 1E).
In stages I and II, the composition of anther cells of the msp1 mutant was almost the same as that of the wild type (data not shown). A remarkable difference was first encountered in stage III. In the mutant anther, PSCs that contained larger nuclei and nucleoli than those of surrounding cells occupied microsporangia (Figure 1D) instead of the differentiated PPCs in the wild-type anther. Cells of a similar size occupied the mutant microsporangia at stage V (Figure 1F) instead of the differentiated secondary parietal cells in the wild-type anther. Even in the meiotic stages, callose was scarcely accumulated, and PMC-like larger cells of various shapes and sizes were packed tightly in the mutant microsporangia (Figures 1H and 1K). Most nuclei in these cells were enlarged and engaged in meiosis, indicating the PMC identity of the cells (data not shown). The number of PMCs per mutant microsporangium was increased abnormally (126 ± 21 in the wild type and 173 ± 14 in msp1). The anther wall of the mutant was composed of two or sometimes three cell layers in the meiotic stages, whereas four layers were observed clearly in the wild type (Figure 1I). The innermost tapetum cell layer was absent in the msp1 mutant (Figure 1K). In addition, none of the epidermal cells was elongated longitudinally (Figure 1K). In the pollen maturation stage, the inner wall layers of the wild-type anthers were degenerated (Figure 1J), whereas abnormal cell layers continued to proliferate and PMCs were collapsed in msp1 (Figure 1L). These observations indicated that the msp1 mutation affected the differentiation of PSCs and PPCs and induced an increased number of PMCs instead of the loss of inner wall cells in the anther.
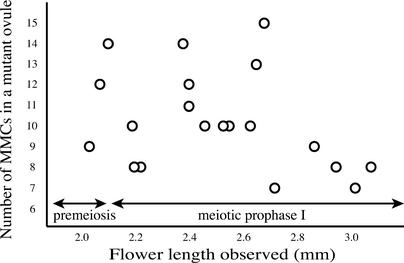
The effects of the msp1 mutation were found not only in male sporogenesis but also in female sporogenesis. In rice, a single MMC was formed just beneath the nucellar epidermis and was identified easily on the tissue section because of its larger size (Figure 1M). However, multiple MMC-like cells in meiotic prophase I were observed in the mutant nucellus (Figure 1N). Most of the cells entered into meiosis, suggesting the MMC identity of these cells (see Figure 3D). The influence of an excess number of MMCs seemed to increase nucellus width (Figure 1N). The mutant phenotype was observed only in the hypodermal nucellar cells and not in the epidermis. Various numbers of MMCs were counted in mutant flowers, with an average and a maximum of 9.4 and 15 MMCs per nucellus, respectively (Figure 2). If a single archesporial cell abnormally proliferates to produce supernumerary MMCs in the mutant, the number of mutant MMCs should increase according to their developmental progress. However, no correlation was detected between these parameters (Figure 2), suggesting that a discrete group of plural precursor cells was differentiated into archesporial cells in a mutant ovule and that each of them developed independently into MMCs. These results suggest that MSP1 function suppresses the surrounding cells converted into male and female sporocytes.
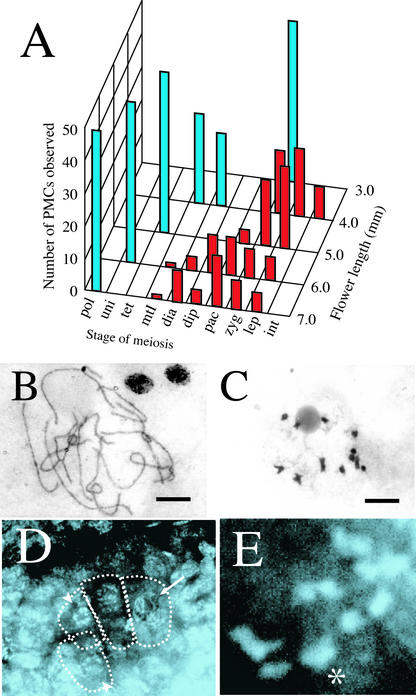
Figure 3.
msp1 PMCs and MMCs Underwent Normal Synapsis and Chiasma Maintenance in Meiosis.
(A) Histogram showing the number of PMCs in each meiotic stage investigated in flowers from 3.0 to 7.0 mm long. Blue and red bars indicate the number of PMCs in wild-type and mutant anthers, respectively. Stages of meiosis are from right to left: int, premeiotic interphase; lep, leptotene; zyg, zygotene; pac, pachytene; dip, diplotene; dia, diakinesis; mtI, metaphase I; uni, uninucleate; pol, matured pollen.
(B) A complete set of 12 homologous chromosome pairs observed in the mutant PMC in pachytene. Bar = 10 μm.
(C) Twelve bivalent chromosomes maintaining normal chiasmata in the mutant PMC in diakinesis. Bar = 10 μm.
(D) An optical and longitudinal section of the mutant ovule. Several meiotic nuclei in pachytene (arrow) and diakinesis (arrowhead) were observed in the nucellus.
(E) Magnified view of an MMC. A complete set of 12 homologous pairs in diakinesis was observed. The asterisk indicates the two overlapping pairs.
Figure 2.
Relationship between the Number of MMCs in a msp1 Ovule and the Progression of MMC Development.
The number of MMCs and their cell stages were observed using a series of longitudinal sections derived from a mutant ovule. From the very early stages of ovule development, the mutant nucellus contained multiple MMCs, suggesting that a discrete group of plural precursor cells was differentiated into archesporial cells in a mutant ovule, rather than a single archesporial cell proliferated abnormally to produce supernumerary MMCs. The correlation of meiosis progression to flower elongation was reported by Miyoshi et al. (2001).
PMCs, but not MMCs, Were Arrested during Meiosis in msp1
After the early development of sporogenous cells from stages I to V, the state of chromosome features in meiosis was observed in the PMCs and MMCs of the wild type and the msp1 mutant. All of the wild-type PMCs entered into diakinesis or metaphase I in the 4.0-mm flowers (Figure 3A). However, in flowers of the same length, the mutant PMCs continued through the leptotene or zygotene stages. In mutant flowers of 6.0 to 7.0 mm, the progression of meiosis was arrested at various stages from leptotene to diakinesis. In the PMCs of the msp1 mutant that reached pachytene or diakinesis, 12 pairs of homologous chromosomes were synapsed completely (Figure 3B) and chiasmata were maintained in all homologous pairs (Figure 3C). Normal synapsis and chiasma maintenance also were observed in mutant MMCs (Figures 3D and 3E). However, unlike PMCs, MMCs in the mutant completed meiosis and performed gametogenesis, as described below. These results indicate that this mutation does not directly affect meiotic events such as homologous chromosome pairing and chiasma formation but causes the arrest of male meiosis, resulting in complete male sterility.
Abnormal Megasporogenesis Frequently Yields Viable Gametophyte in msp1
To investigate whether a mutant embryo sac has the ability of fertilization, msp1 flowers were pollinated with wild-type pollen. Although all of the msp1 homozygous plants exhibited complete sterility in self-pollination, they exhibited ∼30% fertility in 206 flowers pollinated with wild-type pollen (Table 1). Interestingly, 7.7% of the hybrid seeds had twin embryos that germinated twin shoots and roots (Table 1, Figure 4B). These were not induced by apomictic development, because all of the hybrid plants, including twin plants, showed a heterozygous genotype on the MSP1 locus, a normal plant type, and normal seed fertility (data not shown). These results indicated that some of the multiple MMCs observed in the mutant ovule passed through meiosis, developed multiple sporocytes, underwent gametogenesis, and produced functional female gametophytes.
Table 1.
Pollination of Wild-Type Pollen to msp1 Flowers
| Cross Combination | No. of Flowers Pollinated | No. of Mature Seeds | No. Germinated (%) | No. of Twin Shoots (%) | |
|---|---|---|---|---|---|
| Female | Male | ||||
| msp1/msp1 | Nip | 206 | 62 | 35 (62.9) | 3 (7.7) |
| + / msp1 | Nip | 36 | 26 | 22 (84.6) | 0 (0.0) |
| msp1/msp1 | — | 129 | 0 | — | — |
Nip, Oryza sativa cv Nipponbare.
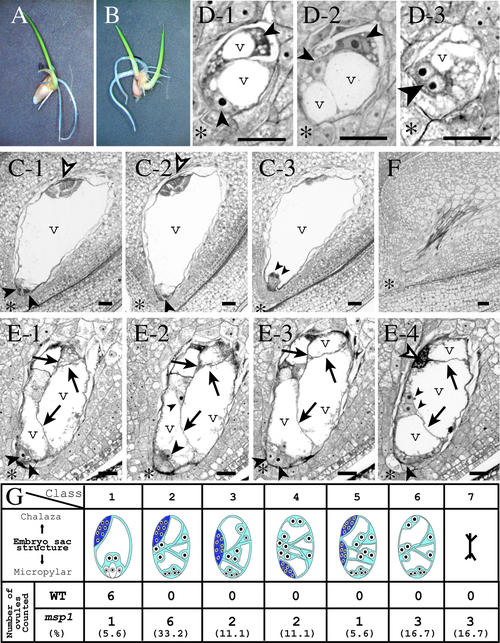
Figure 4.
Aberrant Megagametogenesis in the msp1 Ovule.
(A) A germinated hybrid seed with a single embryo derived from the msp1 mutant crossed with wild-type pollen.
(B) A germinated hybrid seed with twin embryos.
(C) to (E) Longitudinal sections of wild-type and mutant ovules.
(C) A series of three sections from a wild-type ovule. The egg apparatus cells (an egg cell and two synergids) appeared at the micropylar end (large arrowheads), polar nuclei were close to the egg apparatus (small arrowheads), and an antipodal tissue was at the chalazal end (open arrowheads). A central cell is mostly occupied by a vacuole (v). Asterisks indicate the micropylar end.
(D) A series of three sections from a single mutant ovule containing multiple megaspores. At least four megaspores were included in the ovule (small and large arrowheads). The binucleated spore (large arrowhead) suggested that the spore had just entered into the first mitotic division of megagametogenesis.
(E) A series of four sections from a mutant ovule. This ovule generated a single embryo sac but was compartmented by cell walls (arrows). Various numbers of cells were observed at the micropylar end (arrowheads). In this ovule, an antipodal tissue (open arrowheads) developed at a normal position.
(F) The mutant ovule, empty and collapsed.
(G) Embryo sac composition of the wild type (WT) and the mutant. The embryo sac morphology was classified into seven groups (from left to right): class 1 exhibited a normal configuration; classes 2 to 6 developed an embryo sac with aberrantly internal walls and various numbers of nuclei, but the position of the antipodal tissue developed differently as follows: class 2, at the normal position; class 3, at the middle portion; class 4, at the micropylar end; class 5, duplicated at the chalazal and middle portions; class 6, no antipodal tissue was developed. In class 7, the ovule was empty and collapsed. The frequency is presented at the bottom of each classification.
Bars = 20 μm.
In the wild-type ovules of many Gramineae species, a single MMC usually undergoes meiosis and produces four megaspores, among which only the chalazal-most megaspore survives while the other three degenerate (Huang and Sheridan, 1994). Consequently, the surviving megaspore enters into megagametogenesis, in which three subsequent nuclear divisions without cytokinesis yield an eight-nucleate embryo sac. This is followed by the simultaneous formation of cell wall compartments, two synergids, and an egg cell to occupy the micropylar pole and the central cell to occupy the center of the embryo sac (Figure 4C). Antipodal cells continue to divide and produce various numbers of cells at the chalazal pole (Figure 4C). In our case, in the msp1 mutant ovule, multiple megaspores survived and engaged in megagametogenesis (Figure 4D). In 14 of 18 msp1 ovules observed, the composition of the embryo sac frequently was disorganized, with internal cell walls positioned abnormally and various numbers of supernumerary nuclei (Figures 4E and 4G). In Figure 4E, at least six nuclei are observed at the mycropylar pole in the msp1 embryo sac. Despite the aberrant development of the msp1 embryo sac, 9 of 15 embryo sacs with internal walls formed antipodal tissue at almost normal positions (Figure 4G).
MSP1 Encodes a Putative LRR Receptor Ser/Thr Kinase
The mutant line ND0018 was derived from suspension culture of the calli of cv Nipponbare and was examined in the tagged Tos17 sequence linked to the MSP1 gene. In R2 and R3 progeny of the regenerated plant, a transposed Tos17 locus was confirmed to link completely with the msp1 phenotype by DNA gel blot analysis (Figure 5A). The flanking sequence to the Tos17 insertion was cloned and identified as the same sequence as that in the P1 artificial chromosome clone P0413C03, which was mapped at the 158.2-centimorgan position of chromosome 1L by the Rice Genome Research Program of Japan (http://rgp.dna.affrc.go.jp/). The flanking sequence was used as a probe to screen ∼70,000 cDNA clones from rice flowers, and 4 cDNA clones of an identical 4.8-kb sequence were isolated. The clones encoded a 1294–predicted amino acid peptide with a molecular mass of 141 kD (Figure 6A). Comparison of a genomic sequence with the cDNA sequence revealed that the coding region contained no intron, whereas the 5′ untranslated region contained two introns (Figure 5B). The coding sequence appeared as a single band in genomic DNA gel blot analysis (Figure 5C).
Figure 5.
Identification and Characterization of the MSP1 Gene.
(A) DNA gel blot analysis of some of 53 progeny of the msp1 heterozygotes. A Tos17 insertion induced a 4.0-kb polymorphic band, whereas the wild type showed a 4.3-kb band (lane P). The 4.0-kb bands were linked completely with the sterile phenotype (S). Homozygosity (W) or heterozygosity (H) of fertile plants was determined in the next generation.
(B) Genomic structure of the MSP1 gene. The sequences corresponding to the cDNA are shown as closed boxes. Vertical arrows indicate the Tos17 insert site found in three alleles. ATG and TAA indicate start and stop codons, respectively. A pair of horizontal arrows and three lines below the gene structure indicate the positions of primers for RT-PCR and probes for DNA gel blot analysis and the in situ hybridization experiment, respectively. Kp, KpnI; Xb, XbaI; Xh, XhoI.
(C) DNA gel blot analysis of cv Nipponbare genomic DNA using a probe shown in (B).
(D) Flowers of complemented msp1 with the 7.7-kb XbaI-BamHI fragment containing the entire MSP1 gene. The two flowers are from different transformations.
(E) An msp1 flower transformed with only a vector sequence having sterile anthers.
(F) MSP1 expression analyzed by RT-PCR with (+) and without (−) reverse transcriptase. Amplified fragments were provided for ethidium bromide staining (top gel) and for DNA gel blot analysis (middle gel). RT-PCR for a rice actin gene (RAc) is shown as a positive control (bottom gel). Lane C, a positive control using cDNA as a PCR template.
Figure 6.
The MSP1 Gene Encodes an LRR Receptor–Like Protein Kinase.
(A) The shaded bars (light and dark shading is used alternately) indicate 34 LRR units. The underlined sequences indicate (1) a predicted signal peptide, (2) a putative Leu zipper motif, (3) the islands of 21 and 13 amino acids separating LRRs, and (4) a putative transmembrane domain. The double underline indicates a putative kinase domain. Boldface letters indicate invariant amino acid residues conserved completely in all Ser/Thr kinases (Hanks and Quinn, 1991).
(B) Alignment of the kinase domains of OsMSP1, AtEMS1, and OsBRI1. Sequences were aligned with the CLUSTAL W program (http://www.ddbj.nig.ac.jp). Residues identical to those of OsMSP1 are highlighted in black.
A search for the coding sequence in the Tos17-flanking database (http://pc7080.abr.affrc.go.jp/miyao/pub/tos17/) revealed two allelic lines in addition to the ND0018 allele: one was found in ND0095, ND0116, and ND0122 (all possessed a completely identical insertion); another was found in NE0046. The allele identified first was designated msp1-1, and the additional two alleles were designated msp1-2 and msp1-3 (Figure 5B). All of the mutations were found to cause the phenotype of collapsed anthers and full sterility and to retain an in-frame stop codon that appeared in their inserted Tos17 sequence. A 5-bp target sequence was duplicated at both ends of all insertions, as described by Hirochika et al. (1996).
To confirm that the Tos17 insertion found in the MSP1 locus caused the mutant phenotype, we performed complementation analysis of the msp1 phenotype by introducing a 7.7-kb XbaI-BamHI fragment that contains the entire MSP1 coding region and a 2.3-kb upstream sequence (Figure 5B). The fragment proved to complement the mutant phenotype through the formation of normal anthers in seven of nine msp1-1 homozygous transformants (Figure 5D). The control vector had no effect on the phenotype (Figure 5E). Seed fertility also recovered, although it ranged from 60.9 to 2.2% among the complemented plants. This result confirmed that the msp1 mutation was caused by the insertion of the Tos17 sequence into the MSP1 coding region.
A database search revealed that the entire sequence of a putative MSP1 protein shows some similarity to several Ser/Thr kinases with an LRR domain as a receptor (Figure 6A). EMS1 and EXS of Arabidopsis, both of which are identified from allelic mutations in the same gene, show the highest identity, 63.8% (176 of 276), to MSP1 in the putative kinase domain (Figure 6B). This fact seems consistent with the phenotypic similarity in anther development observed in the Arabidopsis and rice mutants, with the excess number of microsporocytes and the abnormal formation of anther walls, including the lack of tapetal cells (Sorensen et al., 2002; Zhao et al., 2002). The MSP1 kinase domain also shows 54.6% identity (153 of 280) to OsBRI1 (Yamamuro et al., 2000) (Figure 6B) and 36.7% identity (80 of 218) to XA21 (Song et al., 1995) of rice and 54.6% identity (153 of 280) to BRI1 (Li and Chory, 1997), 41.8% identity (115 of 275) to ERECTA (Torii et al., 1996), and 41.3% identity (114 of 276) to CLV1 (Clark et al., 1997) of Arabidopsis.
Like other family members, the predicted MSP1 protein has several distinct domains: a signal peptide, a putative Leu zipper motif, an LRR domain, a transmembrane domain, and a cytoplasmic kinase domain (Figure 6A). The N-terminal 22 residues constitute a hydrophobic segment capable of acting as a signal peptide to transport the protein to the plasma membrane, as predicted by the computational program PSORT (Nakai and Kanehisa, 1992). A putative extracellular domain of 34 LRRs (Figure 6A) is separated by two small islands of 21 and 13 amino acids (Figure 6A), which probably are ligand binding domains, as expected in BRI1 (Li and Chory, 1997). The kinase domain of MSP1 completely conserves 15 invariant amino acid residues found in all Ser/Thr kinases (Hanks and Quinn, 1991) (Figure 6A). These results strongly suggest that MSP1 plays an important role in transferring an extracellular signal into the cytoplasm through the plasma membrane of reproductive organ cells.
MSP1 Is Expressed in the Early Development of Reproductive Organs
Total RNAs extracted from vegetative and reproductive organs were examined to detect MSP1 gene expression by reverse transcription–PCR (RT-PCR) with primers encompassing the splice sites in the 5′ untranslated region (Figure 5B). For the reproductive phase, total RNAs were extracted from whole panicles of five groups according to their length: panicles of <5 mm were classified as P1; those of 5 to 10 mm were classified as P2; those of 10 to 30 mm were classified as P3; those of 30 to 60 mm were classified as P4; and those of 60 to 100 mm were classified as P5. Although the stage progression was variable among plants or panicles, almost all panicles in the classified groups contained developing flowers at specific stages: P1 panicles developed a secondary branch or flower primordia; P2 panicles had flowers with anthers in stage I or II (Figure 1B); P3 panicles had flowers with anthers in stage III or IV (Figure 1C); P4 panicles had flowers with anthers in stage V (Figure 1E) or tapetum differentiation; and P5 panicles had anthers in meiosis (Figure 1G). Expression of the MSP1 mRNA was limited only in the reproductive phase of P1 to P4 panicles (Figure 5F), indicating that MSP1 specifically expresses itself from a very early stage of flower development to flowers with established tapeta.
In situ expression of the MSP1 gene was detected on a section of wild-type flowers. During flower development, an MSP1 signal was first visible at basal regions of developing stamen, carpel, palea, and lemma in flower primordia (Figures 7B to 7D) but not in younger primordia (Figure 7A) and in male and female archesporial cells (data not shown). Expression at the basal region of each organ continued until sporocytes entered into meiosis (Figure 7I), corresponding well with the RT-PCR results. However, no phenotypic change was observed in these regions of the msp1 mutant. In the anthers at stage II, in which the PSCs and PPCs were differentiated, the signal began to appear on parietal cells (Figures 7D and 7E). MSP1 was expressed in developing inner wall layers of wild-type anthers. The strongest signal was observed consistently on the innermost layer attached to the sporogenous cells or sporocytes (Figures 7E to 7I). On the other hand, the signal never was observed in female tissues while anthers expressed the MSP1 gene (Figures 7D and 7I). However, after PMCs entered into meiosis, an MSP1 probe stained a whole pistil, including an ovule (Figure 7J). Nucellar cells also were stained except for an MMC region (Figure 7K). These signals disappeared from the nucellus when the inner integuments wrapped the nucellus (Figure 7L). Through all stages of stamen and pistil development, the MSP1 gene was never expressed in male and female sporogenous cells or sporocytes; rather, it was expressed in their surrounding cells. The expression pattern of MSP1 together with the phenotype observed in the loss-of-function mutant indicated that the functional role of this gene in a signaling pathway is to suppress the surrounding cells entering into sporogenesis while simultaneously initiating anther wall formation in rice.
Figure 7.
In Situ Expression of the MSP1 Gene in Reproductive Organs of a Wild-Type Flower.
(A) A palea-primordium differentiation stage.
(B) An early differentiation stage of stamen and pistil primordia. The signal began to appear at the basal regions of each organ. Eg, empty glume; Le, lemma; Pa, palea; Sp, stamen primordium.
(C) A late differentiation stage of stamen and pistil primordia. Rg, rudimentary glume.
(D) A young flower at stage II. MSP1 signals began to appear in the inner wall layer of anthers, but no signal was observed in the female tissue. Ca, carpel; Nu, nucellus.
(E) to (H) Magnified views of the inner walls formed in developing anthers from earlier (E) to later (H) stages. The strongest MSP1 expression was detected in the innermost cell layer attached to the sporogenous cells or sporocytes. Ep, epidermis; En, endothecium; Ml, middle layer; SPC, secondary parietal cells; Ta, tapetum cells.
(I) A young flower at the stage of PMCs entering into meiotic prophase. No MSP1 signal was observed in the female tissue.
(J) MSP1 expression was observed in a whole pistil, whereas expression in the anther had already disappeared.
(K) A magnified view of the nucellus in (J). The nucellar cells were stained except for the MMC region indicated by the arrowhead.
(L) MSP1 expression disappeared in the nucellus wrapped by the inner integuments (arrow).
(M) Control experiment at the stage corresponding to (I) using a sense probe.
Bars = 20 μm.
DISCUSSION
The MSP1 Function in Sporogenesis
Gymnosperms, angiosperms, and some ferns are heterosporous plants. They develop two kinds of spore mother cells, PMCs and MMCs, which then grow into microspores and megaspores, respectively. It is generally accepted that heterospory evolved from the homospory found in lower plants, such as in most ferns. During this process, flowering plants acquire different developmental programs in sporogenesis and gametogenesis between the anther and the ovule. A number of male- or female-specific genes reported in many plant species might be acquired during and after heterospory establishment. Meanwhile, the gene or mutation that affects reproduction in both male and female organs might share a common role between homosporous and heterosporous plants that would be essential for germ cell initiation, differentiation, sporogenesis, and gametogenesis. The SPL/NZZ gene (Schiefthaler et al., 1999; Yang et al., 1999) of Arabidopsis, which encodes a nuclear protein that probably acts as a transcription factor, is the only gene isolated and with known function that affects both PMC and MMC development.
The spl/nzz mutation blocks the differentiation of primary sporogenous cells into sporocytes and the formation of the anther wall. SPL/NZZ expression is detected first in sporogenous cells and successively in sporocytes, but never in anther wall cells. Yang et al. (1999) proposed that SPL/NZZ functions in microsporocytes to regulate the expression of a subset of genes required for sporocyte formation and that microsporocytes regulate the differentiation and growth of parietal cell layers, and consequently anther wall development, through signal exchange or cell-to-cell interactions.
In the msp1 mutant, the supernumerary sporogenous cells were differentiated into sporocytes and engaged in meiosis, indicating that a major pathway of sporogenesis should be unaffected by the msp1 mutation. In the mutant anther, the number of hypodermal wall layers was decreased, whereas the number of PMCs was increased, suggesting that the cells originally programmed to be wall layers were converted to sporogenous cells. In addition, it was suggested that a discrete group of plural precursor cells developed into multiple MMCs in the msp1 mutant (Figure 2). These results indicate the existence of a genetic system common to both male and female sporogenic pathways to repress the surrounding cells of germ cells entering into ectopic sporogenesis. In situ expression observed in the basal region of developing flower organs and in the whole pistil (Figure 7) suggests a more global function of MSP1 for preventing cells of flower tissues from entering into ectopic sporogenesis. The reason that no phenotype is detected in those regions might involve the receptor kinase activity, which is dependent on a specific ligand distribution. The strongest signal of MSP1 expression was detected in the innermost layer touching the sporogenous cells (Figures 7E to 7H), suggesting that the ligand of MSP1 receptor kinase might have originated from sporogenous cells. The signal transferred by the MSP1 kinase also might be important to differentiate PPCs. However, even though parietal cells failed to differentiate in the msp1 mutant, an abnormal cell layer continued to grow in the mutant anther (Figure 1L), suggesting that many other genes function in the subsequent wall formation. Indeed, a number of genes and mutations related to anther wall formation have been reported in many higher plants (reviewed by Goldberg et al., 1993).
MSP1 expression was observed more significantly later in the wild-type pistil than in the anthers (Figure 7). This finding can be attributed to the difference in the initiation of organ development between the anther and the pistil. The rice OSH1 gene is a KNOTTED1-type homeobox gene and is expressed in the area of the floral shoot apical meristem (Sato et al., 1998; Sentoku et al., 1999), indicating that OSH1 expression should be a good molecular marker for identifying undifferentiated cells. During early flower development, OSH1 expression disappears in developing stamen primordia, whereas it remains in the area programmed for pistil formation and continues until the initiation of the carpel (H. Hirano, personal communication). These findings suggest that the pistil founder cells are maintained in an undifferentiated state even after the stamen is differentiated.
Why did MMCs pass through meiosis in spite of the fact that male meiosis was arrested in meiotic prophase I in the msp1 mutant? This difference could be caused by the difference in cell lineage between male and female nursery cells. In the wild-type anthers, a tapetum cell layer that plays an important role in meiosis progression (Chaubal et al., 2000) and pollen development (Chapman, 1987) is derived from archesporial cells, from which male sporocytes also originate (Maheshwari, 1950). On the other hand, in the ovule, it is known that a single archesporial cell is formed just beneath the nucellar epidermis and is differentiated directly into a megasporocyte (Davis, 1966; Russell, 1979) and that the majority of nucellar cells are consumed for embryo sac development (Bhojwani and Bhatnagar, 1992). In the msp1 mutant, the complete absence of the tapetum layer probably caused the arrest of male meiosis in prophase I. On the other hand, nucellar cells remained in the mutant ovule, even though some of them were converted to MMCs (Figure 1N), and they might be able to support female sporogenesis and gametogenesis.
Comparison with Arabidopsis and Maize Mutants
The database search revealed that the MSP1 LRR receptor kinase of rice was homologous with the EMS1/EXS LRR receptor kinase of Arabidopsis, especially in the kinase domain (Figure 6B). Mutant phenotypes of both Arabidopsis and rice share some similarity in the disorganization of the anther, suggesting that the EMS1/EXS gene in Arabidopsis is an ortholog of the MSP1 gene in rice. Interestingly, however, there are some distinct differences. The mutation in the rice gene affects male and female plants, but that in Arabidopsis affects only male plants. In addition, MSP1 is expressed in the surrounding cells of sporogenous cells but not in sporogenous cells themselves. By contrast, EMS1/EXS is detected in both anther wall layers and sporogenous cells (Zhao et al., 2002). The exs mutation results in smaller embryonic cells, delayed embryo development, and smaller mature embryos (Canales et al., 2002), whereas no phenotype has been reported in female sporogenesis. On the contrary, the mac1 mutant of maize (Sheridan et al., 1996, 1999) exhibited an almost identical phenotype with the rice msp1 mutant in both male and female sporogenesis and gametogenesis. This finding suggests that the genetic regulation of macrosporogenesis differs between dicot and monocot plants. In the anther of the msp1 mutant, meiosis was arrested in prophase I, possibly because of the absence of the tapetum layer (Figure 1K), as shown in the maize mutants (Chaubal et al., 2000), in which the prospective tapetal layer divided into two layers and PMCs aborted after the onset of prophase I of meiosis. PMCs of the maize mac1 mutant also are arrested in prophase I (Sheridan et al., 1999), whereas those of the exs (Sorensen et al., 2002) and ems1 (Zhao et al., 2002) mutants of Arabidopsis are in the tetrad stage. Although the points of arrest were different among organisms, the arrest of meiosis in these mutants suggests a significant role of tapetum cells in all steps from meiosis to pollen maturation.
Aberrant Gametogenesis in the msp1 Ovule
In the msp1 mutant ovule, supernumerary MMCs underwent sporogenesis. Several megaspores were engaged in aberrant megagametogenesis, resulting in the development of an abnormal embryo sac structure with internal cell walls and various numbers of supernumerary nuclei (Figure 4). The female gametophyte of the indeterminate gametophyte (ig) mutant of maize is known to contain excessive numbers of cells and nuclei, resulting from asynchronous and extra cycles of nuclear divisions at the two poles (Kermicle, 1971; Lin, 1978; Huang and Sheridan, 1996). In addition, the 6% of the ig mutant embryo sacs govern sets of multiple embryos, suggesting that more than one cell functions as an egg cell (Kermicle, 1971). Phenotypic similarity in female gametophytes between the ig and msp1 mutants allows us to infer that the disorganization of the msp1 embryo sac is affected not by supernumerary megaspores but by the dysfunction of genes such as IG under the msp1 mutation. As another possibility, plural megaspores in an msp1 mutant ovule (Figure 4D) cooperate with each other to generate a single but chimeric embryo sac, as observed in the mac1 mutant of maize (Sheridan et al., 1996), or the multiplication of the megaspores may have an effect similar to that of the extra nuclear division of the ig mutant in female gametogenesis. However, it also was reported that two embryo sac units with almost normal cell composition occupied an ovule in mac1 (Sheridan et al., 1996), indicating that either process may be acceptable in aberrant megagametogenesis with multiple megaspores in Gramineae species.
Although egg apparatus or egg cells were difficult to discriminate from others in this study, at least one or two egg cells located at the micropylar pole were evident from the germination of the msp1 mutant seeds crossed with wild-type pollen (Table 1). This fact indicates that some of the disorganized embryo sacs in the msp1 mutant retain the fundamental and functional organization found in the wild-type embryo sac. More analyses of the msp1 phenotype combined with other gametogenesis mutations will be useful to elucidate the genetic processes and establish the megagametophyte function. However, the primary function of the MSP1 gene is thought to be limited to early sporogenesis, and the disorganized embryo sac would be a secondary effect, as suggested by the expression profile of the gene in early sporogenesis (Figures 5F and 7).
In conclusion, we have shown that the MSP1 gene participates in a signaling pathway to repress male and female nursery cells entering into sporogenesis and simultaneously to initiate anther wall formation. The in situ expression profile of MSP1 raises the possibility that Gramineae species have evolved a common system for both male and female organs to prevent ectopic sporogenesis during flower development. Bisexual effects of the msp1 mutant suggest that heterosporous higher plants might inherit this system from homosporous plants. Although the molecular mechanism at work in these reproductive events is scarcely known in the plant kingdom, analyses of mutants defective in each step of sporogenesis, like msp1, will contribute to our knowledge of the genetic network of the plant reproductive pathway.
METHODS
Plant Materials and Crossing Experiments
Calli induced from rice (Oryza sativa subsp japonica cv Nipponbare) were used for regeneration after 5 months of suspension culture as described by Hirochika et al. (1996). Of 27,998 regenerated plants, reduction of seed fertility was observed in 4032 lines in both primary regenerated plants (R1 generation) and their segregant progeny (R2 or R3 generation). From those sterile lines, 600 lines were chosen at random. Their young panicles were fixed with ethanol:acetic acid (3:1) and prepared for observation of the meiotic chromosome with a BX50 light microscope (Olympus, Tokyo, Japan). In the crossing experiments, the hot water emasculation method was used for all materials. Panicles were immersed in 42°C hot water for 7 min for emasculation, and the opened flowers were pollinated with foreign pollen. For the msp1 homozygous mutants, emasculation was effected by the mechanical exclusion of anthers using forceps because of the complete male sterility of the mutants. All materials were grown in a field in the city of Mishima, Shizuoka, Japan, or in a greenhouse at 30°C during the day and 24°C at night.
Isolation and Characterization of the MSP1 Gene
Genomic DNAs of 53 R3 plants segregating the msp1 phenotype were isolated according to Rogers and Bendich (1988), digested with XbaI at 37°C for 6 h, electrophoresed on a 0.8% agarose gel, blotted onto a nylon membrane (Biodyne B; Pall Corp., Port Washington, NY), and prepared for DNA gel blot hybridization with a 1.6-kb BamHI-XhoI fragment of Tos17 (Hirochika et al., 1996) as a probe. An ∼4.0-kb XbaI fragment tagged by Tos17, which was linked completely to the msp1 phenotype in DNA gel blot analysis, was sliced out from the gel, eluted using the GENECLEAN II kit (Qbiogene, Carlsbad, CA), and inserted into pBluescript SK− (Stratagene). Genomic sequences flanking Tos17 were amplified by PCR with the primers M13-20 (5′-GTAAAACGACGGCCAGT-3′) in the vector and T17LTR4MR (5′-CTGTATAGTTGGCCCATGTCCAG-3′) in the long terminal repeat of the Tos17 retrotransposon. The PCR product that was specific for the msp1 mutant was cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) and provided for sequencing and DNA gel blot analysis. A rice genomic sequence corresponding to the Tos17 flanking sequence was found using a Basic Local Alignment Search Tool (BLASTN) search against the DDBJ database. A cDNA library of rice flowers was constructed using the SuperScript Lambda system (Invitrogen) according to the manufacturer's instructions.
Plastic and Paraffin Sections of the Tissue
Observation of flower development was performed on standard paraffin and plastic sections as described by Hong et al. (1995). Young panicles or flowers were fixed with 3% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 N sodium phosphate buffer, pH 7.5, for 20 h at 4°C, rinsed with 0.1 M phosphate buffer, pH 7.0, and dehydrated in an ethanol series. For the plastic sections, the samples were embedded in Technovit 7100 resin (Hereaus Kulzer, Wehrheim, Germany), polymerized at 45°C, and divided into 2-μm sections using a rotary microtome. For the paraffin sections, the samples were substituted with xylene, embedded in Paraplast Plus (Oxford Labware, St. Louis, MO), and divided into 7-μm sections. All sections were stained with toluidine blue (Chroma Gesellshaft Shaud, Münster, Germany) and photographed using a BX50 light microscope and a charge-coupled device camera system (DP50; Olympus). For counting the number of sporocytes in anthers and ovules, 7-μm sections embedded in paraffin or 4-μm plastic sections were used. We paid careful attention to cell shape to prevent counting the same cells twice, and in some cases, we counted the nuclei or nucleoli that were larger in the sporocytes than in the other cells (Raghavan, 1988).
In Situ Hybridization
Three DNA fragments were amplified by PCR with MSP1 cDNA as a template (Figure 5B), cloned into a pCRII-TOPO dual promoter system (Invitrogen) and transcribed in vitro from the T7 or SP6 promoter with RNA polymerases using the DIG RNA labeling kit (Roche, Indianapolis, IN), and this mixture was prepared for hybridization. Young panicles of cv Nipponbare showing lengths of 5 to 20 mm were fixed and embedded in Paraplast Plus as described above. Microtome sections, 7 μm thick, were applied to glass slides coated with 3-amino-propyltriethoxy-silane (Shinetsu Chemical, Tokyo, Japan). In situ hybridization was performed according to the methods of Kouchi and Hata (1993). Tissue sections were deparaffinized in xylene, dehydrated through a graded ethanol series, and air-dried. The sections were incubated with 5 μg/mL proteinase K, 100 mM Tris-HCl, pH 7.5, and 50 mM EDTA at 37°C for 10 min, with 4% paraformaldehyde at room temperature for 10 min, and with 0.1 M triethanolamine and 0.5% acetic anhydride at room temperature for 10 min. Then, they were rinsed twice with double-distilled water for 5 min after each treatment. The sections were washed twice in 2× SSPE (1× SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4) for 5 min, dehydrated through a graded ethanol series, and hybridized with 1 μg/mL RNA probes in 50% deionized formamide, 0.3 M NaCl, 10% dextran sulfate, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 100 mM DTT, and 500 ng/μL poly(A)+ RNA (Wako Pure Chemical, Osaka, Japan) at 50°C for 16 h.
The sections were washed three times with 4× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 50°C for 5 min each and treated with 50 μg/mL RNase A at 37°C for 30 min. They were then rinsed twice with 0.5× SSC at 50°C for 20 min, rinsed twice with buffer 1 (150 mM NaCl and 100 mM Tris-HCl, pH 7.5) at room temperature for 5 min each, and incubated in blocking reagent of 20% normal rabbit serum (Wako Pure Chemical). Immunological detection of the hybridized probes was performed with anti-digoxigenin alkaline phosphatase (Roche) according to the manufacturer's recommendations. The sections were washed three times with buffer 1 for 10 min each, washed with buffer 3 (100 mM NaCl, 50 mM MgCl2, and 100 mM Tris-HCl, pH 9.5) at room temperature for 5 min, and incubated with 0.34 mg/mL nitroblue tetrazolium salt and 0.175 mg/mL 5-bromo-4-chloro-3-indolyl phosphate in buffer 3 for 6 h. They were rinsed with double-distilled water, dehydrated through a graded ethanol series, mounted in Eukitt (O. Kindler, Freiburg, Germany), and photographed using a differential interference contrast microscope and a DP50 system (Olympus). Images were enhanced with Photoshop 7.0 (Adobe, Mountain View, CA).
Chromosome Observation in Meiosis
Chromosome behavior in meiosis was observed in two ways. To observe the organization of sporangia and chromosome pairing simultaneously, whole-mounted stamens and pistils were prepared for the observation using confocal laser-scanning microscopy according to Clark et al. (1993) and Motamayor et al. (2000) with slight modifications. The flowers in meiosis were fixed in ethanol:acetic acid (3:1), rehydrated through an ethanol series, incubated in 100 mg/mL RNase A at 37°C for 30 min to 1 h, stained overnight in 1.0 mg/mL propidium iodide and 0.1 M l-Arg, pH 12.4, and destained two times for 1 h each in 0.1 M l-Arg, pH 8.0. The stamens and pistils were dissected gently by needle, mounted on a glass slide with a drop of Vectorshield (Vector Laboratories, Burlingame, CA), and viewed with a helium-neon laser (543-nm excitation wavelength) using the Olympus FLUOVIEW confocal laser-scanning microscopy system. Images were enhanced and pseudo-colored using Photoshop 7.0 (Adobe).
To observe the chromosome spreads of the pollen mother cells, a flame-drying method described by Kurata et al. (1981) was applied. Young flowers or anthers in meiotic stages were fixed, incubated in 2% cellulase Onozuka RS (Yakult Honsha, Tokyo, Japan), 0.3% pectolyase Y-23 (Kikkoman, Chiba, Japan), 1.5% macerozyme R200 (Yakult Honsha), 1 mM EDTA, and 75 mM KCl, pH 4.2, at 37°C for 40 min, rinsed with distilled water, squashed in several drops of fixative, flame-dried on a slide glass, stained with Giemsa solution (Sigma) diluted 30-fold with 33 mM phosphate buffer, pH 6.8, for 30 min, and photographed using a BX50 light microscope with a DP50 system (Olympus).
Reverse Transcription–PCR
Total RNA was isolated from root, shoot, adult leaf, and young panicle according to Sambrook et al. (1989) with slight modifications. One microgram of total RNA was reverse-transcribed using Superscript II RNase H− reverse transcriptase (Invitrogen). For the reverse transcription reaction, the MSP1-specific primer 5′-AGCACCATTTCCTTCAGCATCTTC-3′ was designed downstream of the 5′ untranslated region. The two specific primers for the MSP1 gene, encompassing the first and second spliced sites, were used for PCR: 5′-ATCTCCAGGTTTTTAGGCTTTACG-3′ and 5′-CTAGCAGGATGAAAAGCCAGAAAC-3′ (Figure 5B). The PCR products were loaded on an agarose gel, and DNA gel blot analysis was performed using a nested PCR probe to confirm precise amplification from the MSP1 mRNA. As a control, total RNA was reverse-transcribed with an oligo(dT)20 primer and provided for PCR using primers in the rice actin (RAc): 5′-AACTGGGATGATATGGAGAA-3′ and 5′-CCTCCAATCCAGACACTGTA-3′. The PCR conditions involved a 94°C incubation for 2 min followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min.
Complementation of the msp1 Mutant
A chimeric clone of the genomic and cDNA fragments was constructed for the complementation test of the msp1 mutant. A genomic XbaI-KpnI fragment including 5′ upstream of MSP1 (Figure 5B) was inserted into the binary vector pPZP2H-lac (Fuse et al., 2001), and subsequently, a KpnI-BamHI fragment including 3′ downstream of the MSP1 cDNA was inserted into the KpnI-BamHI site. This MSP1 gene construct or the control pPZP2H-lac vector was introduced into the msp1 homozygous calli by Agrobacterium tumefaciens–mediated transformation and regenerated as described by Hiei et al. (1994).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact K. Nonomura, knonomur@lab.nig.ac.jp. The mutant line described in this article will be made available at the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan (http://www.nias.affrc.go.jp/).
Accession Numbers
The MSP1 cDNA sequence described in this article has been deposited in DDBJ under accession number AB103395. Accession numbers for the other sequences mentioned are DDBJ AP003451 (P1 artificial chromosome clone P0413C03), GenBank AJ488154 (EMS1), and GenBank AJ496433 (EXS).
Acknowledgments
We thank H. Mochizuki and M. Nakano for technical assistance. We also thank M. Yano (National Institute of Agrobiological Sciences) for providing vectors for rice transformation and H. Hirano (University of Tokyo) for useful comments on flower development. This research was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences, Japan, and in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Rice Genome Project MP-2112).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012401.
References
- Bhojwani, S.S., and Bhatnagar, S.P. (1992). The Embryology of Angiosperms. (New Delhi: Vikas Publishing House).
- Canales, C., Bhatt, A.M., Scott, R., and Dickinson, H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12, 1718–1727. [DOI] [PubMed] [Google Scholar]
- Chapman, G.P. (1987). The tapetum. Int. Rev. Cytol. 107, 111–125. [Google Scholar]
- Chaubal, R., Zanella, C., Trimnell, M.R., Fox, T.W., Albertsen, M.C., and Bedinger, P. (2000). Two male-sterile mutants of Zea mays (Poaceae) with an extra cell division in the anther wall. Am. J. Bot. 87, 1193–1201. [PubMed] [Google Scholar]
- Clark, S.E. (2001). Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2, 276–284. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Davis, G.L. (1966). Systematic Embryology of the Angiosperms. (New York: John Wiley & Sons).
- Dawe, R.K., and Freeling, M. (1992). The role of initial cells in maize anther morphogenesis. Development 116, 1077–1085. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse, T., Sasaki, T., and Yano, M. (2001). Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol. 18, 219–222. [Google Scholar]
- Goldberg, R.B., Beals, T.P., and Sanders, P.M. (1993). Anther development: Basic principles and practical applications. Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya, I., Avalkina, N.A., and Sheridan, W.F. (1992). Effects of several meiotic mutations on female meiosis in maize. Dev. Genet. 13, 411–424. [Google Scholar]
- Hake, S., and Char, B.R. (1997). Cell-cell interactions during plant development. Genes Dev. 11, 1087–1097. [DOI] [PubMed] [Google Scholar]
- Hanks, S.K., and Quinn, A.M. (1991). Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 200, 38–62. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.K., Aoki, T., Kitano, H., Satoh, H., and Nagato, Y. (1995). Phenotypic diversity of 188 rice embryo mutants. Dev. Genet. 16, 298–310. [Google Scholar]
- Huala, E., and Sussex, I.M. (1993). Determination and cell interactions in reproductive meristems. Plant Cell 5, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.Q., and Sheridan, W.F. (1994). Female gametophyte development in maize: Microtubular organization and embryo sac polarity. Plant Cell 6, 845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.Q., and Sheridan, W.F. (1996). Embryo sac development in the maize indeterminate gametophyte1 mutant: Abnormal nuclear behavior and defective microtubule organization. Plant Cell 8, 1391–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J.L. (1971). Pleiotropic effects on seed development of the indeterminate gametophyte gene in maize. Am. J. Bot. 58, 1–7. [Google Scholar]
- Kouchi, H., and Hata, S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238, 106–119. [DOI] [PubMed] [Google Scholar]
- Kurata, N., Omura, T., and Iwata, N. (1981). Studies on centromere, chromomere and nucleolus in pachytene nuclei of rice, Oryza sativa, microsporocytes. Cytologia 46, 791–800. [Google Scholar]
- Kuwada, Y. (1910). A cytological study of Oryza sativa L. Bot. Mag. 24, 267–281. [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Lin, B.Y. (1978). Structural modifications of the female gametophyte associated with the indeterminate gametophyte (ig) mutant in maize. Can. J. Genet. Cytol. 20, 249–257. [Google Scholar]
- Maheshwari, P. (1950). An Introduction to the Embryology of An-giosperms. (New York: McGraw-Hill).
- Miyoshi, K., Kurata, N., and Nonomura, K.I. (2001). Detection of pre-meiotic DNA synthesis in pollen mother cells by immunofluorescence technique in rice. Rice Genet. Newsl. 18, 82–84. [Google Scholar]
- Motamayor, J.C., Vezon, D., Bajon, C., Sauvanet, A., Grandjean, O., Marchand, M., Bechtold, N., Pelletier, G., and Horlow, C. (2000). switch (swi1), an Arabidopsis thaliana mutant affected in the female meiotic switch. Sex. Plant Reprod. 12, 209–218. [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, S. (1989). Genetic mosaics and cell lineage analysis in plants. Trends Genet. 5, 273–277. [DOI] [PubMed] [Google Scholar]
- Raghavan, V. (1988). Anther and pollen development in rice (Oryza sativa). Am. J. Bot. 75, 183–196. [Google Scholar]
- Rogers, S.O., and Bendich, A.J. (1988). Extraction of DNA from plant tissue. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–10.
- Russell, S.D. (1979). Fine structure of megagametophyte development in Zea mays. Can. J. Bot. 57, 1093–1110. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sato, Y., Sentoku, N., Nagato, Y., and Matsuoka, M. (1998). Isolation and characterization of a rice homeobox gene, OSH15. Plant Mol. Biol. 38, 983–998. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H., and Matsuoka, M. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell 11, 1651–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan, W.F., Avalkina, N.A., Shamrov, I.I., Batygina, T.B., and Golubovskaya, I.N. (1996). The mac1 gene: Controlling the commitment to the meiotic pathway in maize. Genetics 142, 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan, W.F., Golubeva, E.A., Abrhamova, L.I., and Golubovskaya, I.N. (1999). The mac1 mutation alters the developmental fate of the hypodermal cells and their cellular progeny in the maize anther. Genetics 153, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Wang, G.L., Chen, L.L., Kim, H.S., Pi, L.Y., Holsten, T., Gardner, J., Wang, B., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sorensen, A., Guerineau, F., Canales-Holzeis, C., Dickinson, H.G., and Scott, R.J. (2002). A novel extinction screen in Arabidopsis thaliana identifies mutant plants defective in early microsporangial development. Plant J. 29, 581–594. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Schoot, C., and Rinne, P. (1999). Networks for shoot design. Trends Plant Sci. 4, 31–37. [DOI] [PubMed] [Google Scholar]
- Webb, M.C., and Gunning, E.S. (1990). Embryo sac development in Arabidopsis thaliana. I. Megasporogenesis, including the microtubular cytoskeleton. Sex. Plant Reprod. 3, 244–256. [Google Scholar]
- Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., Ashikari, M., Kitano, H., and Matsuoka, M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M., Tsugawa, H., Miyao, A., Yano, M., Wu, J., Yamamoto, S., Matsumoto, T., Sasaki, T., and Hirochika, H. (2001). The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol. Genet. Genomics 265, 336–344. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D.Z., Wang, G.F., Speal, B., and Ma, H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]