Abstract
Cellulose synthesis in the developing xylem vessels of Arabidopsis requires three members of the cellulose synthase (CesA) gene family. In young vessels, these three proteins localize within the cell, whereas in older vessels, all three CesA proteins colocalize with bands of cortical microtubules that mark the sites of secondary cell wall deposition. In the absence of one subunit, however, the remaining two subunits are retained in the cell, demonstrating that all three CesA proteins are required to assemble a functional complex. CesA proteins with altered catalytic activity localize normally, suggesting that cellulose synthase activity is not required for this localization. Cortical microtubule arrays are required continually to maintain normal CesA protein localization. By contrast, actin microfilaments do not colocalize with the CesA proteins and are unlikely to play a direct role in their localization. Green fluorescent protein–tagged CesA reveals a novel process in which the structure and/or local environment of the cellulose synthase complex is altered rapidly.
INTRODUCTION
Cellulose is composed of linear chains of β(1-4)–linked glucose. It is the major component of most plant cell walls and consequently the most abundant natural polymer. The orientation of cellulose microfibrils within the plant cell wall determines the direction of cell expansion; therefore, it is a major determinant of cell shape and plant morphology (reviewed by Emons et al., 1992; Baskin, 2001). In higher plants, cellulose is synthesized at the plasma membrane by a large membrane-bound complex (reviewed by Doblin et al., 2002). The complex has been visualized in freeze-fracture studies as a “rosette” structure composed of six distinct lobes (Herth, 1985; Haigler and Brown, 1986). The rosettes are thought to move through the plasma membrane synthesizing multiple individual cellulose chains that associate to form “crystallites,” which then may associate to form microfibrils (Delmer and Amor, 1995; Ha et al., 1998).
The only components of the cellulose synthase complex (CSC) that have been identified to date are the CesA (cellulose synthase) proteins, presumed to be the catalytic subunits (Arioli et al., 1998; Taylor et al., 1999, 2000; Fagard et al., 2000; Scheible et al., 2001). Based on measurements of elementary microfibril structure, the cellulose synthase rosette is likely to contain 18 to 36 CesA proteins (Delmer and Amor, 1995; Ha et al., 1998). At least three CesA genes are required for cellulose synthesis in the secondary cell walls of developing xylem vessels: IRX3 (AtCesA7) (Taylor et al., 1999), IRX1 (AtCesA8) (Taylor et al., 2000), and IRX5 (AtCesA4) (Taylor et al., 2003). These three proteins interact with each other, presumably forming part of the same complex (Taylor et al., 2000, 2003). A similar situation appears to occur in primary cell walls, where AtCesA1 (RSW1), AtCesA3 (IXR1), and AtCesA6 (PRC1/IXR2) all are required, supporting the idea that three CesA proteins are required in the primary wall and are a general requirement of cellulose synthesis in most, if not all, plant cell walls (Fagard et al., 2000; Scheible et al., 2001; Desprez et al., 2002; Ellis et al., 2002). To date, however, little information is available on the structure of the CesA proteins, how different CesA proteins are assembled within the complex, and how they interact with one another.
The orientation of cellulose microfibrils normally is parallel to the underlying cortical microtubules, and it has been hypothesized that these microtubules control microfibril orientation by constraining the movement of the CSC. Whether microtubules orientate microfibril deposition and how they might do so has been the source of much debate (reviewed by Baskin, 2001). A major obstacle to resolving these issues is that it has not been possible to visualize the CSC directly. Consequently, information about the role of microtubules in determining the movement of the CSC can be inferred only by examining the pattern of cellulose microfibrils produced rather than by visualizing the localization of the CSC directly.
The developing xylem has many advantages as a system for studying the assembly and targeting of the CSC. The secondary cell wall is deposited in a very characteristic pattern around the cell, and sites of secondary cell wall deposition are marked by dense bands of cortical microtubules (Hepler and Fosket, 1971; Brower and Hepler, 1976). Areas of secondary cell wall deposition are characterized by very high densities of the CSC, whereas adjacent areas of the same cell membrane not undergoing secondary cell wall thickening have very few if any CSCs (Herth, 1985; Haigler and Brown, 1986; Schneider and Herth, 1986).
In this study, cellulose synthase–specific antibodies and a green fluorescent protein (GFP) fusion were used to study the assembly and localization of the CSC in the developing xylem of Arabidopsis and to determine the role of the cytoskeleton in directing these processes. The results demonstrate that all three CesA proteins are required for localization to the plasma membrane, that microtubules assemble independently of the CSC, and that they are continuously required to maintain proper CSC localization.
RESULTS
Localization of the CesA Proteins IRX1, IRX3, and IRX5
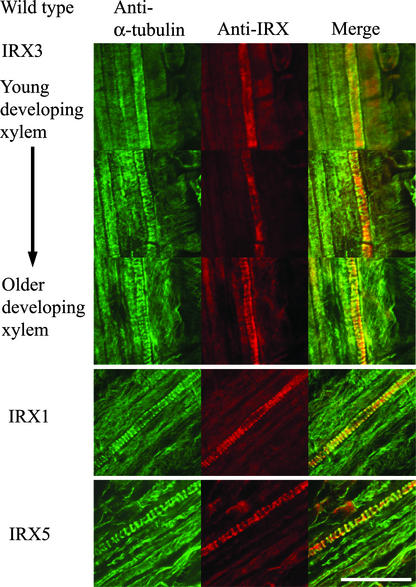
Early-forming protoxylem vessels in roots exhibit a characteristic annular or spiral pattern of cell wall thickening and develop in a highly predictable manner close to the apical meristem (Mahonen et al., 2000). In the youngest developing vessels, microtubules had not yet formed thick bands around the cell (Figure 1), and IRX3 staining was not localized to the plasma membrane (Figure 1). As the vessels matured, microtubules became localized in thick bands at the plasma membrane, and some IRX3 staining became associated with these microtubule bands and some remained within the cell (Figure 1). At late stages of vessel development, most of the IRX3 staining colocalized with the microtubule bands and little was present in the cell (Figure 1). IRX1 and IRX5 also exhibited a strong colocalization with microtubule bands in developing xylem vessels (Figure 1). Using the IRX antibodies, some faint fluorescence was visible in cells other than the xylem vessels. This staining represents some bleed-through from the different channels caused by overlap between the ranges of different filter sets used in the double labeling. In developing cotyledons, IRX proteins also were localized to developing xylem vessels, showing a banded pattern of distribution (data not shown).
Figure 1.
Localization of α-Tubulin and CesA Proteins in Developing Xylem Vessels.
Confocal images showing the localization of α-tubulin (left), CesA proteins (middle), and their overlap in distribution (right) in developing vessels from wild-type Arabidopsis roots. IRX3 images were taken from a single root and show early stages of xylem development (top) through the latter stages (bottom). IRX1 and IRX5 images were taken at the latter stages of vessel development. Bar = 25 μm.
Localization Requires All Three Proteins
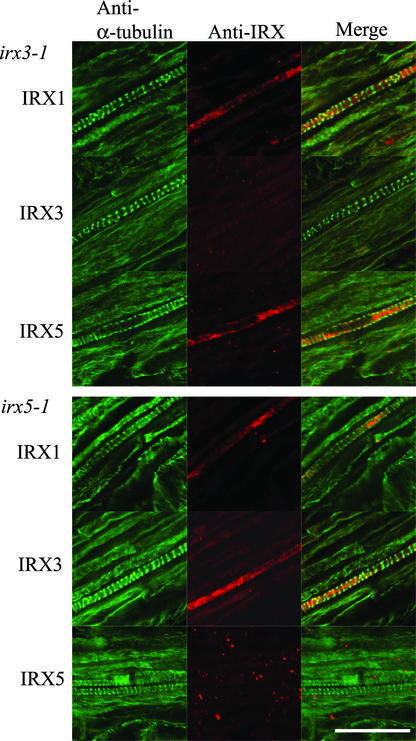
Plants homozygous for the irx3-1 mutation contain undetectable levels of the IRX3 protein (Taylor et al., 2000), and as expected, there was very little or no IRX3 labeling in developing xylem in these plants (Figure 2). The microtubules in these cells still exhibited the characteristic transverse banded pattern similar to that seen in wild-type roots (Figures 1 and 3). By contrast, IRX1 and IRX5 were not banded (Figure 2) and did not colocalize with microtubules (Figure 2), as seen in wild-type roots (Figure 1). The irx5-1 mutation is caused by a large T-DNA insertion in the gene, and no protein product was detectable by protein gel blot analysis (Taylor et al., 2003) or immunolabeling of whole roots (Figure 2). Although the cortical microtubules exhibited a banded distribution similar to that of the wild type (Figure 2), neither IRX1 nor IRX3 (Figure 2) colocalized with the cortical microtubules (Figure 2).
Figure 2.
Localization of α-Tubulin, IRX1, IRX3, and IRX5 in Developing Vessels of irx3-1 and irx5-1 Roots.
Confocal images showing the localization of α-tubulin (left), CesA proteins (middle), and their overlap in distribution (right) in developing vessels from irx3-1 (top) and irx5-1 (bottom) Arabidopsis roots. Bar = 25 μm.
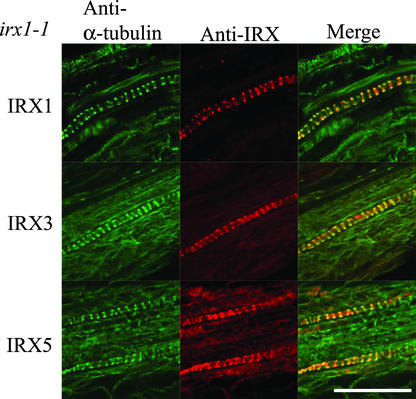
Figure 3.
Localization of α-Tubulin and IRX1, IRX3, and IRX5 in Developing Vessels of irx1-1 Roots.
Confocal images showing the localization of α-tubulin (left), CesA proteins (middle), and their overlap in distribution (right) in developing vessels from irx1-1 Arabidopsis roots. Bar = 25 μm.
The irx1-1 Arabidopsis mutant show reduced levels of cellulose as a result of a mutated Asp residue, which is thought to be crucial for the activity of cellulose synthase, but these plants contain approximately wild-type levels of the inactive protein (Taylor et al., 2000). In these plants, all three IRX proteins colocalized with the microtubules (Figure 3) in a manner identical to the wild type (Figure 1).
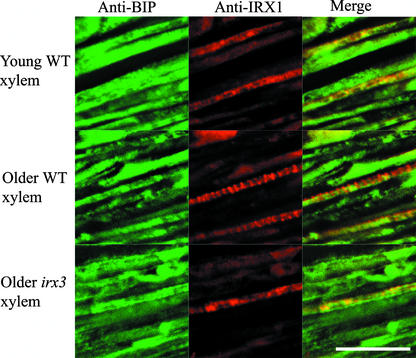
Anti–binding protein (BiP) staining in young developing xylem cells of wild-type Arabidopsis plants showed that it colocalizes with IRX1 protein (Figure 4). In older wild-type developing xylem, in which IRX proteins demonstrated a banded distribution, there was little or no colocalization between anti-BiP labeling and anti-IRX1 labeling (Figure 4). This finding demonstrates that there is no cross-reaction between the antibodies used for these double-labeling experiments. In older developing xylem in irx3-1 plants, there was strong colocalization between anti-BiP labeling and anti-IRX1 labeling (Figure 4).
Figure 4.
Localization of BiP and IRX1 in Developing Xylem Vessels.
Confocal images showing BiP (left), IRX1 (middle), and their degree of colocalization (right) in young (top) and older (middle and bottom) developing xylem vessels from wild-type (WT; top and middle) and irx3-1 (bottom) Arabidopsis roots. Bar = 25 μm.
CesA Protein Localization Requires Microtubules
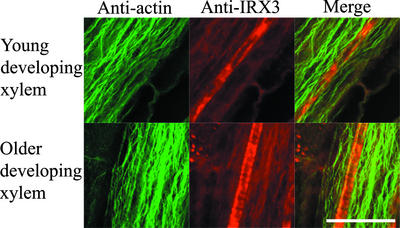
Actin cables in both younger and older developing xylem vessels run parallel to the long axis of cells throughout the stele (Figure 5). Consequently, actin did not colocalize with IRX3 in young developing xylem vessels or with banded IRX3 in older developing xylem vessels (Figure 5).
Figure 5.
Localization of Actin Microfilaments and IRX3 in Wild-Type Arabidopsis Roots.
Confocal images showing actin (left), IRX3 (middle), and their degree of colocalization (right) in developing xylem vessels from wild-type Arabidopsis roots. Bar = 25 μm.
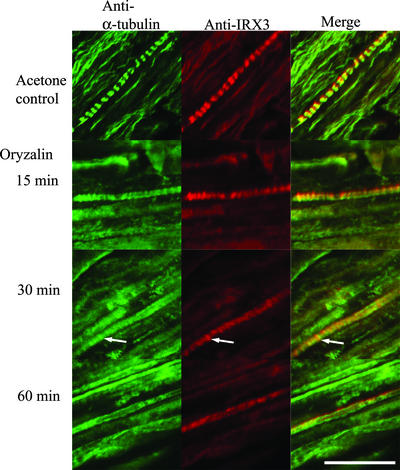
After 15 min, wild-type roots treated with 10 μM oryzalin showed reduced microtubule bands in developing xylem. After 30 min of oryzalin treatment, only vestigial microtubules remained in the developing xylem (Figure 6). During this time course, IRX3 distribution mirrored the pattern of microtubule organization. After 15 min, IRX3 started to lose its tightly banded distribution (Figure 6). By 30 min, only a few areas of IRX3 banding remained; these areas colocalized with the vestigial microtubules (Figure 6, arrows). After 60 min of oryzalin treatment, few microtubules were left and IRX3 staining was dispersed relatively evenly. Control roots treated with acetone for 1 h showed normal colocalization of both microtubules and IRX3 in developing xylem (Figure 6).
Figure 6.
Effect of Microtubule Depolymerization on IRX3 Localization.
Confocal images showing the localization of α-tubulin (left), IRX3 (middle), and their overlap in distribution (right) in developing vessels from wild-type Arabidopsis roots treated with 10 μM oryzalin for 15, 30, or 60 min or with 0.1% acetone for 60 min. Bar = 25 μm.
Visualization of IRX3-Containing Organelles
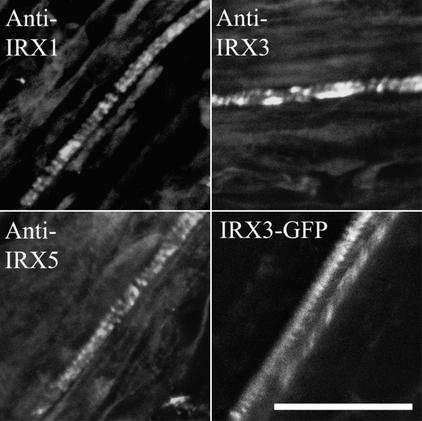
An IRX3-GFP fusion protein was constructed by inserting GFP close to the N terminus between amino acids 3 and 4. The irx3-1 line transformed with this construct exhibited a wild-type phenotype, indicating that the IRX3-GFP fusion protein was functional and able to complement the irx3-1 mutation (data not shown). Using whole roots of the IRX3-GFP plants, immunolabeling demonstrated that IRX1, IRX3-GFP, and IRX5 all showed a banded distribution characteristic of their distribution in the wild type (Figure 7). Direct imaging of IRX3-GFP by confocal microscopy also showed it was localized in a banded pattern within the developing xylem vessels (Figure 7).
Figure 7.
Localization of IRX3-GFP Fusion in Xylem Vessels.
Confocal images showing the distribution of IRX1, IRX5, and IRX3-GFP in roots of irx3-1 plants complemented with an IRX3-GFP construct. IRX1 and IRX5 were visualized using immunofluorescence, whereas IRX3 was visualized using immunofluorescence (IRX3) or directly using GFP fluorescence (IRX3-GFP). Bar = 25 μm.
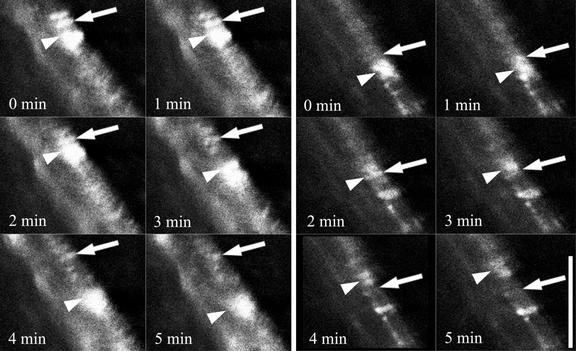
Time-course imaging of developing xylem revealed dynamic changes in GFP fluorescence. Frames taken at 1-min intervals revealed regions of much brighter GFP fluorescence that moved progressively along the cell (Figure 8, arrowheads). These brighter regions were in addition to the relatively stable banded pattern of IRX-GFP distribution that was visualized normally (Figure 7). The coincidence of these brighter regions with a stable band of fluorescence (Figure 8A, arrows) led to a transient increase in fluorescence from the stable band of IRX3-GFP. The signal from the stable band then diminished as the region of brightness moved along the cell (Figure 8A). This dynamic pattern of fluorescence was observed on several occasions and always appeared to cause a local increase in the intensity of the banded pattern of IRX3-GFP fluorescence (Figure 8B).
Figure 8.
Rapid Alteration in IRX3-GFP Fluorescence during Xylem Development.
Two series of successive confocal images taken at 1-min intervals showing IRX3-GFP fluorescence in a developing xylem vessel. Arrows indicate a banded area of IRX3-GFP fluorescence that transiently exhibits a dramatic increase in brightness. Arrowheads indicate a region of IRX3-GFP fluorescence moving along a cell that transiently associates with the stable band of IRX3-GFP fluorescence. Bar = 15 μm.
DISCUSSION
Localization in the Wild Type
Xylem development in the roots begins just behind the root apical meristem, where two characteristic xylem poles are formed. Early stages of protoxylem development can be identified by the expression of IRX proteins (Figure 1). In the very early stages of xylem formation, the IRX proteins exhibit a dispersed distribution throughout the cell (Figures 1 and 4). This distribution of the IRX proteins appears to overlap with that of the endoplasmic reticulum (ER) marker BiP (Figure 4). Given the very widespread staining with anti-BiP, it is possible that the IRX proteins also may localize to other regions of the cell. As these cells mature, the cortical microtubules form a banded pattern, and all three IRX proteins become progressively colocalized with these bands of cortical microtubules (Figure 1). In roots with more than two xylem elements, there appears to be greater density of IRX protein, which probably reflects the more complex pattern of secondary cell wall deposition in these cells (data not shown). These data are consistent with a number of studies that suggest that microtubule banding marks the site of, but precedes, secondary cell wall formation (Hepler and Fosket, 1971; Brower and Hepler, 1976; Haigler and Brown, 1986).
All Three CesAs Are Required for Proper Localization
In irx3-1 plants, it was possible to detect both IRX1 and IRX5 but not IRX3, consistent with IRX3 being absent from these plants (Figure 2). In contrast to the wild type, IRX1 and IRX5 were not localized to the plasma membrane but instead were retained within the ER (Figures 2 and 4). Likewise, in irx5-1 plants, both IRX1 and IRX3 were localized to the ER (Figures 2 and 4). The irx1-1 mutant is caused by the alteration of a highly conserved Asp residue that is assumed to result in the loss of catalytic activity, but the mutated protein is present at approximately wild-type levels (Taylor et al., 2000). Here, IRX1, IRX3, and IRX5 all colocalized with cortical microtubules as in the wild type (Figure 3). These results demonstrate that the presence of all three subunits is essential for the proper localization of these proteins to regions of the plasma membrane associated with cell wall thickening. The absence of catalytic activity, although clearly reducing cellulose deposition (Turner and Somerville, 1997; Taylor et al., 1999, 2000), has no effect on localization. These data support the ideas that all three IRX subunits are part of the CSC (Taylor et al., 2003) and that correct assembly of the CSC is required for its correct targeting to the plasma membrane. This study demonstrates that in the absence of one of these subunits, the two remaining subunits are retained in the ER.
Freeze-fracture studies have been used to visualize intact rosettes in Golgi vesicles (Haigler and Brown, 1986). The data presented here demonstrate that exit of the CSC from the ER, presumably to the Golgi secretory pathway, apparently occurs only when the entire complex is assembled. A similar situation has been seen for other oligomeric protein complexes destined for the plasma membrane. For example, the three subunits of epithelial sodium channels in Xenopus oocytes are retained in the ER until they are assembled into complexes (Valentijn et al., 1998), and a similar situation is observed with class-C L-type calcium channels, in which assembly of the two subunits is required to generate the signal for their correct targeting (Gao et al., 1999). What makes the situation with the IRX proteins unusual is the high level of identity between the three IRX subunits (Taylor et al., 2003). Although it remains possible that different CesA proteins may have different catalytic functions, these results support the idea that three CesAs are an essential structural requirement for the assembly of the complex. It is likely that different CesA subunits are required for the different intersubunit contacts that are necessary to form the elaborate rosette structure that constitutes the higher plant CSC.
The proteasomal degradation pathway is associated with the ER, and oligomeric membrane complexes are transported into the ER through the Sec61p translocation channel for degradation (Cresswell and Hughes, 1997). It is possible that cellulose synthase subunits retained in the ER are broken down subsequently by the proteasome.
Role of the Cytoskeleton in CesA Protein Organization
Several experiments have suggested that actin may be involved in secondary cell wall deposition. When secondary cell wall thickening first becomes apparent in differentiating Zinnia cells, the actin filaments change alignment parallel to the thickening (Kobayashi et al., 1987, 1988). It also has been reported that sucrose synthase, which may be involved in supplying UDP-glucose as a substrate to cellulose synthase (Amor et al., 1995) and localizes to regions of cell wall thickening in Zinnia culture cells (Salnikov et al., 2001), also binds actin (Winter et al., 1998). In both younger and older developing xylem vessels in Arabidopsis roots, large actin cables run longitudinally and do not colocalize with regions of cell wall thickening that are indicated by IRX3 localization (Figure 5). Some reports suggest that it can be technically difficult to entirely preserve plant actin. The experiments described here, however, were performed on whole mounts of Arabidopsis seedlings, which should minimize this problem. The data presented indicate that actin filaments of developing root xylem are unlikely to be involved in the localization of the CSC or other components to areas of cell wall thickening.
A body of literature suggests that microtubules are required for microfibril orientation (for a recent example, see Burk and Ye, 2002). However, work with cellulose synthase inhibitors has suggested that cellulose microfibrils might participate with microtubules in an information feedback loop and that cellulose synthase activity might be required for microtubule organization (Fisher and Cyr, 1998) or that CSC might stabilize microtubules. irx3-1 and irx5-1 plants represent an excellent opportunity to test this hypothesis, because none of the CesA subunits localize to the plasma membrane and there is little or no cellulose in the secondary cell wall. Microtubule organization in the developing xylem of these mutants was normal (Figure 2), demonstrating that here, cellulose synthesis, or the presence of the CSC, is not required for the correct localization of microtubules. This finding is consistent with the results seen in the AtCesA1 mutant rsw1 (Sugimoto et al., 2001) for the primary cell wall, in which a decrease in cellulose synthesis does not lead to a loss of microtubule organization.
Since microtubules were first discovered in plants, it has been suggested that they control the orientation of cellulose microfibril deposition (Ledbetter and Porter, 1963; reviewed by Gunning and Hardham, 1982; Giddings and Staehelin, 1991; Baskin, 2001), although the direct link between the cytoskeleton and control of the direction of cellulose synthesis has not been established. The dinitroaniline herbicide oryzalin specifically binds plant tubulin, causing microtubules to depolymerize (Vaughn, 1986; Anthony et al., 1998). When microtubules are removed from developing xylem vessels using colchicine, subsequent vessel development does not exhibit the characteristic localized pattern of secondary cell wall deposition but is deposited over the entire cell (Hepler and Fosket, 1971; Brower and Hepler, 1976; Taylor et al., 1992). In plants treated with oryzalin, IRX3 localization was altered rapidly coincident with the loss of microtubule organization. Changes in the pattern of IRX3 and microtubule localization were visible within 15 min, and by 60 min, IRX3 appeared to be distributed evenly around the developing vessel, demonstrating that microtubules are required continuously to maintain the localization of CSCs (Figure 6). The rapid manner in which the IRX3 distribution was altered suggests that this altered localization of IRX3 occurs, at least partially, by the movement of existing rosettes. Consequently, the data support a model in which the CSC is attached, directly or indirectly, to the microtubules (Hepler and Fosket, 1971; Heath, 1974; Seagull and Heath, 1980), but it is unlikely that a cell wall component is involved in the localization of the cellulose synthase complexes, as has been suggested (Baskin, 2001).
Organelles Containing IRX3
The irx3-1 mutant was complemented using an IRX3-GFP construct. This finding demonstrates that the IRX3-GFP fusion protein is functional as a component of the CSC. In addition, in IRX3-GFP plants, both IRX1 and IRX5 proteins exhibited a banded distribution characteristic of a wild-type localization, confirming that the CSC was assembled correctly (Figure 7). Using confocal microscopy, it is possible to observe the IRX3-GFP protein directly in living cells. Although some IRX3-GFP protein was localized within the cell, possibly to the ER, IRX3-GFP also exhibited the characteristic banded pattern found in mature cells undergoing secondary cell wall formation. The majority of GFP fluorescence located in the bands generally was very weak. Typically, the laser was used at high power (30%) to view the fluorescence. It was possible, however, to view bright regions of fluorescence that moved rapidly along the cell. These areas of bright fluorescence appeared to correspond to a transient increase in the banded IRX3-GFP fluorescence (Figure 8). This region presumably reflects an alteration in the structure and/or the environment of the IRX3-GFP. It is unclear whether the increase in fluorescence reflects a cellular process peculiar to the cellulose synthesis complex. Although the bright fluorescence may result from the movement of some organelle within the cell, the fact that it appears to give an increase in the banded pattern of fluorescence suggests that it in some way associates with regions of cell wall thickening. It is possible that these regions of bright fluorescence represent specific organelles, or possibly specific regions of the ER, dedicated to supplying CSCs to, or recycling them from, regions of secondary cell wall deposition.
Conclusions
The work described here presents an analysis of the localization and targeting of components of the CSC. It demonstrates that all three subunits of the CSC of developing xylem vessels in Arabidopsis are required for correct localization and that the lack of one subunit results in the other two subunits being retained within the cell, possibly within the ER. The incorporation of IRX3-GFP into functional CSCs reveals a dynamic process in which the GFP fluorescence is altered dramatically but transiently, probably as a result of alteration in the structure and/or the local environment of the CSC. One explanation is that a specific organelle, possibly a region of the ER, may be responsible specifically for supplying CSC components to, or recycling them from, regions of cell wall thickening. Microtubules are necessary to correctly localize IRX proteins to regions of cell wall thickening, but actin filaments appear unlikely to play a major role in IRX protein localization.
Defining the processes involved in the assembly and targeting of the CSC, in addition to the identification of other components of this complex, will allow us to further unravel the details of cellulose synthesis. The discoveries that all three cellulose synthase catalytic subunits are required to be present and of the relationship with microtubule organization represent a major step toward these goals.
METHODS
Immunofluorescence of Arabidopsis Roots for Microtubules and ER
All chemicals used were from Sigma (Poole, UK) unless stated otherwise. Immunofluorescent staining of Arabidopsis thaliana roots was performed as described by Harper et al. (1996) except for the digestion step, in which we used 0.5% (w/v) pectolyase and 0.5% (w/v) cellulase for 25 min.
Prepared seedlings on slides were incubated with a 1:2000 dilution of monoclonal antibody B512 against α-tubulin in antibody dilution buffer, or rabbit anti-BiP at a dilution of 1:600 (a generous gift from R. Boston, North Carolina State University, Raleigh), and anti-IRX1 (1:100) (Taylor et al., 2000), anti-IRX3 (1:200) (Taylor et al., 2000), or anti-IRX5 (1:200) (Taylor et al., 2003). Secondary antibodies used were donkey anti-sheep Alexa Fluor 568 (1:200; Molecular Probes, Eugene, OR), sheep anti-mouse fluorescein isothiocyanate (FITC) diluted 1:125, and goat anti-rabbit FITC diluted 1:80. Where more than one secondary antibody was required, they were added sequentially to avoid cross-reaction. Samples were viewed using a Nikon Eclipse TE300 fluorescence microscope equipped with a ×40 Nikon Plan Fluor oil-immersion lens (Tokyo, Japan) and a Bio-Rad MRC1024 MP confocal scanning head with a krypton/argon laser. Excitation lines of 488 and 568 nm were used to view FITC and Alexa Fluor 568, respectively.
Immunofluorescence of Arabidopsis Roots for Actin
The procedure for immunolabeling actin in Arabidopsis roots was modified from D. Collings and G.O. Wasteneys (unpublished data). Five-day-old seedlings were incubated in 50 mM Pipes, 5 mM MgSO4, and 5 mM EGTA, pH 7.0 (PME), containing 400 μM maleimidobenzoyl-N-hydroxy- succinimide ester for 10 min. Seedlings then were fixed for 40 min in PME buffer containing 3.7% formaldehyde, 1% glutaraldehyde, 2 mM phenylmethylsulfonyl fluoride, 400 μM maleimidobenzoyl-N-hydroxy- succinimide ester, and 0.1% Triton X-100 before seedlings were washed twice in PME containing 0.1% Triton X-100 (PMET) for 5 min. Seedlings then were extracted for 10 min in PME containing 1% Triton X-100 before being washed twice for 5 min in PMET. This step was followed by digestion for 25 min in PMET, 1% BSA, 0.5% cellulase, 0.5% pectolyase, 0.4 M mannitol, and 2 mM phenylmethylsulfonyl fluoride. Seedlings then were washed three times for 5 min in PME buffer before they were permeabilized for 10 min in methanol at −20°C. Then, they were rehydrated in PBS for 10 min, transferred to 1 mg/mL NaBH4 for 20 min, and washed in PBS three times for 10 min. Seedlings were next blocked for 30 min in incubation buffer (PBS containing 1% BSA and 50 mM Gly). Antibodies were applied as described by Harper et al. (1996), except for the incubation buffer, which contained 50 mM Gly. Primary antibodies used were sheep anti-IRX1 antibody diluted 1:100 and mouse anti-chicken actin C4 antibody (ICN Biomedicals, Basingstoke, UK) diluted 1:200. Secondary antibodies used and microscopy were as described above.
GFP Construct Assembly
Soluble modified GFP (Davis and Vierstra, 1998) from plasmid pCD3-326 was digested with BamHI and EcoRI and cloned into similarly cut pBluescript KS+ (Stratagene, Amsterdam, The Netherlands) to give a clone carrying the GFP coding sequence and the nopaline synthase terminator. This clone (pKS-326) was used to amplify the coding region of GFP, appending an NheI site at either terminus using Pfu polymerase (Promega, Madison, WI) with primers N-termGFP-F (5′-ACTACTGCAGCTAGCATGAGTAAAGGAGAAGAACTTTT-3′) and N-termGFP-R (5′-GCCTCGAGCTAGCTTTGTATAGTTCATCCATGCC-3′). The resulting 750-bp product was cloned into pBluescript KS cut with EcoRV. After digestion with NheI, the GFP fragment was cloned into NheI-cut pCS12 (pCB2300 carrying an 8.3-kb XhoI-MunI genomic DNA fragment containing the entire IRX3 coding region and 1.7 kb of promoter sequence). This construct was transformed into irx3 plants by vacuum infiltration (Clough and Bent, 1998). GFP plants were observed using a ×40 Nikon C Apo water-immersion lens on the confocal microscope described above.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact S.R. Turner, simon.turner@man.ac.uk.
Acknowledgments
We thank David Collings (Australian National University, Canberra) for providing the actin immunolabeling protocol and for helpful discussions, Rebecca Boston (North Carolina State University, Raleigh) for her generous gift of the polyclonal anti-BiP antibody, and Tony Wade and Sarah Kirk (University of Manchester) for their assistance with confocal imaging. J.C.G. and N.G.T. were supported by Biotechnology and Biological Science Research Council Grants P15838 and P13259, respectively.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012815.
References
- Amor, Y., Haigler, C.H., Johnson, S., Wainscott, M., and Delmer, D.P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92, 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, R.G., Waldin, T.R., Ray, J.A., Bright, S.W.J., and Hussey, P.J. (1998). Herbicide resistance caused by spontaneous mutation of the cytoskeletal protein tubulin. Nature 393, 260–263. [DOI] [PubMed] [Google Scholar]
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Brower, D.L., and Hepler, P.K. (1976). Microtubules and secondary wall deposition in xylem: The effects of isopropyl N-phenylcarbamate. Protoplasma 87, 91–111. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., and Ye, Z.H. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell 14, 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cresswell, P., and Hughes, E.A. (1997). Protein degradation: The ins and outs of the matter. Curr. Biol. 7, R552–R555. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Delmer, D.P., and Amor, Y. (1995). Cellulose biosynthesis. Plant Cell 7, 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refregier, G., Desnos, T., Aletti, E., Py, N., Pelletier, S., and Hofte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin, M.S., Kurek, I., Jacob-Wilk, D., and Delmer, D.P. (2002). Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 43, 1407–1420. [DOI] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons, A.M.C., Derksen, J., and Sassen, M.M.A. (1992). Do microtubules orient plant-cell wall microfibrils? Physiol. Plant. 84, 486–493. [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refregier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Hofte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.D., and Cyr, R.J. (1998). Extending the microtubule/microfibril paradigm: Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 116, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T.Y., Chien, A.J., and Hosey, M.M. (1999). Complexes of the alpha(1C) and beta subunits generate the necessary signal for membrane targeting of class C L-type calcium channels. J. Biol. Chem. 274, 2137–2144. [DOI] [PubMed] [Google Scholar]
- Giddings, T.H.J., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (London: Academic Press), pp. 85–99.
- Gunning, B.E.S., and Hardham, A.R. (1982). Microtubules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 33, 651–698. [Google Scholar]
- Ha, M.A., Apperley, D.C., Evans, B.W., Huxham, M., Jardine, W.G., Vietor, R.J., Reis, D., Vian, B., and Jarvis, M.C. (1998). Fine structure in cellulose microfibrils: NMR evidence from onion and quince. Plant J. 16, 183–190. [DOI] [PubMed] [Google Scholar]
- Haigler, C.H., and Brown, R.M. (1986). Transport of rosettes from the Golgi apparatus to the plasma membrane in isolated mesophyll cells of Zinnia elegans during differentiation to tracheary elements in suspension culture. Protoplasma 134, 111–120. [Google Scholar]
- Harper, J.D.I., Holdaway, N.J., Brecknock, S.L., Busby, C.H., and Overall, R.L. (1996). A simple and rapid technique for the immunofluorescence confocal microscopy of intact Arabidopsis root tips. Cytobios 87, 71–78. [PubMed] [Google Scholar]
- Heath, I.B. (1974). A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J. Theor. Biol. 48, 445–449. [DOI] [PubMed] [Google Scholar]
- Hepler, P.K., and Fosket, D.E. (1971). The role of microtubules in vessel member differentiation in Coleus. Protoplasma 72, 213–236. [Google Scholar]
- Herth, W. (1985). Plasma membrane rosettes involved in localized wall thickening during xylem vessel formation of Lepidium sativum L. Planta 164, 12–21. [DOI] [PubMed] [Google Scholar]
- Kobayashi, H., Fukuda, H., and Shibaoka, H. (1987). Reorganization of actin filaments associated with the differentiation of tracheary elements in Zinnia mesophyll cells. Protoplasma 138, 69–71. [Google Scholar]
- Kobayashi, H., Fukuda, H., and Shibaoka, H. (1988). Interrelation between the spatial disposition of actin filaments and microtubules during the differentiation of tracheary elements in cultured Zinnia cells. Protoplasma 143, 29–37. [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahonen, A.P., Bonke, M., Kauppinen, L., Riikonen, M., Benfey, P.N., and Helariutta, Y. (2000). A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salnikov, V.V., Grimson, M.J., Delmar, D.P., and Haigler, C.H. (2001). Sucrose synthase localizes to cellulose synthesis sites in tracheary elements. Phytochemistry 57, 823–833. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R., Eshed, R., Richmond, T., Delmer, D., and Somerville, C. (2001). Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. USA 98, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, B., and Herth, W. (1986). Distribution of plasma membrane rosettes and kinetics of cellulose formation in xylem development of higher plants. Protoplasma 131, 142–152. [Google Scholar]
- Seagull, R.W., and Heath, I.B. (1980). The organisation of cortical microtubule arrays in the radish root hair. Protoplasma 103, 205–229. [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2001). Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215, 172–183. [DOI] [PubMed] [Google Scholar]
- Taylor, J.G., Owen, T.P., Koonce, L.T., and Haigler, C.H. (1992). Dispersed lignin in tracheary elements treated with cellulose synthesis inhibitors provides evidence that molecules of the secondary cell wall mediate wall patterning. Plant J. 2, 959–970. [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100, 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn, J.A., Fyfe, G.K., and Canessa, C.M. (1998). Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J. Biol. Chem. 273, 30344–30351. [DOI] [PubMed] [Google Scholar]
- Vaughn, K.C. (1986). Cytological studies of dinitroaniline-resistant eleusine. Pestic. Biochem. Physiol. 26, 66–74. [Google Scholar]
- Winter, H., Huber, J.L., and Huber, S.C. (1998). Identification of sucrose synthase as an actin-binding protein. FEBS Lett. 430, 205–208. [DOI] [PubMed] [Google Scholar]