Abstract
Because retrotransposons are the major component of plant genomes, analysis of the target site selection of retrotransposons is important for understanding the structure and evolution of plant genomes. Here, we examined the target site specificity of the rice retrotransposon Tos17, which can be activated by tissue culture. We have produced 47,196 Tos17-induced insertion mutants of rice. This mutant population carries ∼500,000 insertions. We analyzed >42,000 flanking sequences of newly transposed Tos17 copies from 4316 mutant lines. More than 20,000 unique loci were assigned on the rice genomic sequence. Analysis of these sequences showed that insertion events are three times more frequent in genic regions than in intergenic regions. Consistent with this result, Tos17 was shown to prefer gene-dense regions over centromeric heterochromatin regions. Analysis of insertion target sequences revealed a palindromic consensus sequence, ANGTT-TSD-AACNT, flanking the 5-bp target site duplication. Although insertion targets are distributed throughout the chromosomes, they tend to cluster, and 76% of the clusters are located in genic regions. The mechanisms of target site selection by Tos17, the utility of the mutant lines, and the knockout gene database are discussed.
INTRODUCTION
Transposable elements, especially retrotransposons, are one of the major components of the plant genome (Feschotte et al., 2002). For example, 80% of the maize genome and at least 40% of the fava bean genome are occupied by retrotransposons. At least 17% of the relatively small rice genome is estimated to consist of retrotransposons (McCarthy et al., 2002). During transposition, retrotransposons are reverse transcribed by a self-encoded reverse transcriptase, and the resulting cDNAs are integrated into the genome. This “copy-and-paste” mode of transposition contributes to the expansion of genome size. Recent study has shown that retrotransposon sequences are not eliminated readily from the genome and are maintained in an inactive form. For example, some retrotransposons in the maize genome have been maintained over a period of 6 million years (Walbot and Petrov, 2001). Currently, it is recognized that transposable elements influence genome evolution by increasing genome size and inducing various types of changes in higher eukaryotic genomes (Kidwell and Lisch, 1997; Bennetzen, 2000).
Genome-wide sequence analyses revealed that transposable elements contribute to changes in gene structure and expression. In the human and mouse genomes, many transposable elements are found in protein-coding genes. Two new genes in mouse were found to have been created by transposon insertion (Nekrutenko and Li, 2001). Almost 25% of the promoter regions of human genes contain transposable element–derived sequence, and some of them act as cis-regulatory elements (Jordan et al., 2003). In wheat, transcriptional activation of retrotransposons alters the expression of adjacent genes, and read-through transcription from retrotransposons is associated with the activation or silencing of flanking genes (Kashkush et al., 2003). Thus, transposable elements, including retrotransposons, have contributed more significantly to the evolution of genes and genomes than was thought previously.
An endogenous copia-like retrotransposon of rice, Tos17, is inactive under normal conditions. Tos17 is activated by tissue culture and inactivated again in regenerated plants (Hirochika et al., 1996). In contrast to other plant retrotransposons, the copy number of Tos17 is quite low: one to five copies depending on the rice cultivar. Nipponbare, which has been selected as a standard cultivar for the international rice sequencing project, carries only two copies. From 5 to 30 transposed copies are observed in plants regenerated from 3- to 16-month-old cultures. These features of Tos17 make it suitable not only for the analysis of interaction between retrotransposons and the host genome but also for the functional analysis of rice genes by gene disruption (Hirochika, 2001).
Here, we report the analysis of target site specificity of the retrotransposon Tos17 in the rice genome to understand the impact of Tos17 insertion on the genome and to evaluate the utility of Tos17 for the functional analysis of genes. We have produced 47,196 insertion mutant lines through tissue culture of cv Nipponbare. More than 42,000 Tos17-flanking sequences have been analyzed and mapped on rice genomic sequences. The results show that Tos17 prefers to integrate in genic regions and that hot spots for integration are distributed throughout the rice genome. There is a correlation between the preferred sites of Tos17 insertion and the locations of certain rapidly evolving gene classes. We discuss the interaction between the Tos17 retrotransposon and the rice genome and its utility for the functional analysis of rice genes.
RESULTS
Systematic Analysis of Tos17-Flanking Sequences and Detection of Disrupted Genes
To analyze the target site specificity and to develop a database of disrupted genes for the reverse-genetics analysis of gene function, we determined the 3′ flanking sequences of Tos17 insertion points. To amplify as many flanking sequences as possible, we adopted a thermal asymmetric interlaced PCR protocol and a suppression PCR protocol (Liu and Whittier, 1995; Siebert et al., 1995; Miyao et al., 1998). A total of 42,292 flanking sequences amplified by these methods from 4316 lines were determined. Because many of these single-run sequences were redundant, independent sequences were identified by comparing the sequences with each other. A total of 16,784 independent sequences were obtained. The efficiency of flanking sequence isolation was calculated by dividing the number of independent flanking sequences by the number of total insertions. To estimate the total number of insertions, we analyzed the number of insertions from 548 lines by DNA gel blot hybridization. Because each line was estimated to have, on average, 10 new insertions, the total number of insertions was 43,160. This simple calculation yields an estimated efficiency of flanking sequence isolation of 40%. However, this value might be an underestimate. Because the rice callus was cultured for 5 months, any Tos17 transposition events that occurred near the beginning of the culture period would have become widely distributed among the regenerated lines. To avoid the potential bias introduced by these events, we calculated the efficiency from each of 548 lines independently and obtained an average value. This efficiency, which represents the performance of the thermal asymmetric interlaced and suppression PCR methods, was estimated to be 75%. The difference between the two estimates of efficiency derives from the redundancy of Tos17 insertions in our mutant populations.
Analysis of the regions flanking Tos17 insertions in each line enables the identification and functional classification of the disrupted genes. Independent flanking sequences of sufficient length were subjected to a BLASTX (Basic Local Alignment Search Tool) search (Altschul et al., 1997) against the nonredundant data set from the National Center for Biotechnology Information. Because flanking sequences located at exon-intron junctions often show high E values even when there is significant identity, e-04 was chosen as the cutoff value for the BLASTX search. A total of 8495 of 16,784 independent sequences showed similarity to genes in the nonredundant data set. These were classified into functional categories based on similarity to known genes (Table 1). Of the 8495 candidates, 3536 sequences showed high similarity (E value < e-25) to known genes. Genes in the disease/defense and signal transduction categories, of which protein kinases were a major component, were disrupted by Tos17 insertion with high frequency.
Table 1.
Functional Classes of Disrupted Genes
| Category | Number (<e-4) |
Number (<e-25) |
Percent |
|---|---|---|---|
| Cell growth/division | 396 | 141 | 4.7 |
| Cell structure | 289 | 121 | 3.4 |
| Disease/defense | 1,175 | 661 | 13.8 |
| Energy | 36 | 12 | 0.4 |
| Intracellular trafficking | 32 | 9 | 0.4 |
| Metabolism | 598 | 231 | 7.0 |
| Protein synthesis | 69 | 35 | 0.8 |
| Protein targeting and storage | 138 | 36 | 1.6 |
| Secondary metabolism | 88 | 28 | 1.0 |
| Signal transduction | 768 | 357 | 9.0 |
| Transcription | 364 | 203 | 4.3 |
| Transporters | 364 | 180 | 4.3 |
| Transposons | 704 | 450 | 8.3 |
| Unclear classification | 3,474 | 1,072 | 40.9 |
| Total hits | 8,495 | 3,536 | |
| Total independent insertions | 16,784 |
A total of 42,292 sequences (from 4316 lines) were analyzed. The hit ratio in independent flanking sequences was 50.6%.
We evaluated the bias toward kinase/resistance genes for Tos17 insertion. The total length of annotated coding sequences (CDSs) was 54.5 Mbp in the whole rice genomic sequences used for this analysis. The total length of CDSs annotated with the term “kinase” or “disease resistance” was 2.8 Mbp. Thus, the proportion of the annotated CDSs that represent kinase and resistance genes was estimated to be 5%. Therefore, if Tos17 inserted into CDSs without bias, the number of insertions in kinase/resistance genes would be expected to be 5% of the total number of insertions in CDSs. Contrary to this expectation, 338 of 2900 insertions within the annotated CDSs, or 11.7%, were inserted in CDSs annotated with the term “kinase” or “disease resistance.” This result indicates that kinase and disease resistance genes are among the preferred targets for Tos17 insertion. Because resistance genes tend to be clustered and thus make gene-dense regions, this distribution of Tos17 may simply reflect its preference for inserting in gene-dense regions.
Some of the disrupted genes are listed in Table 2. Several lines showed the phenotypes expected for the genes disrupted. For example, mutants of Mg-chelatase and chlorophyll A oxygenase showed the albino and chlorina phenotypes, respectively. Some hot spots for Tos17 insertion also were detected. These include Tos17 itself (35 insertions) and genes for RNA polymerase largest subunit (104 insertions), sucrose synthase (23 insertions), Xa21 (18 insertions), and phytochrome A (9 insertions). Flanking sequence analysis also indicated that the termini of Tos17 are highly conserved, whereas those of integrated T-DNA often are rearranged, causing difficulty in flanking sequence analysis (Mayerhofer et al., 1991; Sessions et al., 2002).
Table 2.
Disrupted Genes with High Scores
| Accession Numbera | Entryb | Putative Identification | Organismc | Score | E Value |
|---|---|---|---|---|---|
| AG020730 | BAA24449.1 | 6-4 photolyase | Arabidopsis thaliana | 89.7 | 4e-31 |
| AG020828 | CAB41466.1 | Inositol 1,4,5-trisphosphate 5-phosphatase | Arabidopsis thaliana | 69.1 | 4e-11 |
| AG020899 | CAB88264.1 | Callose synthase catalytic subunit–like | Arabidopsis thaliana | 205 | 5e-53 |
| AG020942 | AAF74563.1 | Heat stress transcription factor A3 | Lycopersicon peruvianum | 119 | 3e-26 |
| AG020963 | AAF31730.1 | Phosphoribosylformylglycinamidine synthase–like | Arabidopsis thaliana | 159 | 1e-50 |
| AG020968 | BAA94795.1 | S-Adenosyl-l-Met:l-Met S-methyltransferase | Hordeum vulgare | 90.5 | 1e-27 |
| AG021106 | AAD37810.1 | NADP-specific isocitrate dehydrogenase | Oryza sativa | 77.6 | 5e-14 |
| AG021182 | BAB11335.1 | Eukaryotic release factor 1 | Arabidopsis thaliana | 181 | 2e-45 |
| AG021242 | JDMU1 | DNA-directed RNA polymerase II largest chain | Arabidopsis thaliana | 357 | 6e-98 |
| AG021377 | AAF23509.1 | Fructose-1,6-bisphosphatase | Porteresia coarctata | 101 | 2e-26 |
| AG021380 | BAA94509.1 | Protein kinase 1 | Populus nigra | 184 | 5e-46 |
| AG021400 | BAB08003.1 | Adenine phosphoribosyltransferase | Hordeum vulgare | 93.6 | 8e-19 |
| AG021445 | T03846 | Plasma membrane H+-ATPase | Oryza sativa | 102 | 3e-45 |
| AG021473 | T00020 | Bacterial blight resistance protein Xa1 | Oryza sativa | 289 | 1e-77 |
| AG021543 | T01661 | DNA (cytosine-5-)-methyltransferase | Zea mays | 90.1 | 2e-53 |
| AG021593 | CAA65053.1 | Pro transporter 2 | Arabidopsis thaliana | 116 | 2e-25 |
| AG021694 | BAA35120.1 | NADH-dependent glutamate synthase | Oryza sativa | 266 | 1e-70 |
| AG021741 | S64721 | Protoporphyrin IX magnesium chelatase precursor | Hordeum vulgare | 433 | e-121 |
| AG021763 | BAB10513.1 | NAM (no apical meristem)–like | Arabidopsis thaliana | 114 | 8e-25 |
| AG021795 | CAA94437.1 | PDR5-like ABC transporter | Spirodela polyrrhiza | 136 | 6e-52 |
| AG021958 | AAC39369.1 | Trehalose-6-phosphate phosphatase | Arabidopsis thaliana | 65.2 | 1e-24 |
| AG021996 | BAB03631.1 | Putative protein kinase Xa21 | Oryza sativa | 229 | 1e-59 |
| AG022284 | S47582 | High-affinity potassium uptake transporter | Triticum aestivum | 162 | 3e-39 |
| AG022440 | O23755 | Elongation factor 2 | Beta vulgaris | 161 | 2e-39 |
| AG022521 | P30298 | Sucrose synthase 1 | Oryza sativa | 85.8 | 6e-17 |
| AG022936 | P17801 | Putative receptor protein kinase zmpk1 precursor | Zea mays | 162 | 4e-61 |
| AG023034 | AAF26975.1 | Stelar K+ outward rectifying channel | Arabidopsis thaliana | 98.3 | 1e-25 |
| AG023207 | T09999 | Cytochrome P450 | Catharanthus roseus | 120 | 8e-40 |
| AG023219 | P10931 | Phytochrome A | Oryza sativa | 258 | 2e-68 |
| AG023602 | BAB08867.1 | DNA-3-methyladenine glycosylase | Arabidopsis thaliana | 65.2 | 6e-26 |
| AG023738 | AAD40979.1 | Peroxisomal copper-containing amine oxidase | Glycine max | 196 | 1e-49 |
| AG023869 | BAA34861.1 | Importin-β1 | Oryza sativa | 132 | 3e-30 |
| AG023905 | BAB10038.1 | Dihydropyrimidinase | Arabidopsis thaliana | 88.5 | 2e-25 |
| AG023996 | BAA90462.1 | Chlorophyll a oxygenase | Arabidopsis thaliana | 100 | 9e-31 |
| AG024163 | P52421 | Phosphoribosylamine-Gly ligase | Vigna unguiculata | 244 | 9e-64 |
| AG024164 | T04103 | Sucrose phosphate synthase | Oryza sativa | 311 | 4e-84 |
| AG024224 | AAC18440.1 | Argonaute protein | Arabidopsis thaliana | 68.7 | 1e-21 |
| AG024327 | AAC49302.1 | Erecta | Arabidopsis thaliana | 157 | 4e-38 |
| AG024330 | P52711 | Ser carboxypeptidase II-3 | Hordeum vulgare | 117 | 2e-25 |
Accession number of the flanking sequence.
Entry name of the hit protein.
Source of the protein showing the highest similarity score.
Tos17 Insertions in Rice Genomic Sequences
To evaluate the distribution of Tos17 insertion sites in the rice genome, all flanking sequences were searched with the BLASTN program against the genomic sequences of rice (Table 3). A total of 20,458 loci were mapped onto 521 Mbp of genomic sequence, and the average insertion interval was estimated to be 22 kb. Because 16,784 flanking sequences were identified as independent by comparing the sequences with each other, 3674 insertions must be located on overlaps of P1 artificial chromosome (PAC)/BAC sequences or in duplicated genes. Insertions in duplicated genes were obtained from double-assigned flanking sequences on each PAC/BAC. Because there were 242 insertions in duplicated genes, the remaining 3432 insertions must be located on the overlaps. The best way to analyze insertion specificities on the rice genome would be to use 12 perfect sets of contigs without overlaps. The availability of data correlating the genetic map with the PAC/BAC clones of chromosome 1 (http://rgp.dna.affrc.go.jp) made it possible to construct high-quality contigs of chromosome 1 from which overlaps were eliminated. Although sequence data are available for the other chromosomes, high-quality contigs could not be constructed, because data correlating the genetic map with the PAC/BAC clones are limited and the quality of sequence data remains low. Thus, except for our in-depth analysis of chromosome 1 (see below), we performed most of our subsequent analyses using the data set of 20,450 sequences. Because there is no reason to assume that the overlaps of PAC/BAC clones are biased toward specific loci, the results are unlikely to be distorted.
Table 3.
Location of Tos17 Insertions in Published Rice Genomic Sequences
| Location | Number of Insertions |
Percent | Total Target Length (bp) |
Percent | Average Interval (bp) |
|---|---|---|---|---|---|
| Exon | 2,260 | 11.0 | 25,780,287 | 4.9 | 11,407 |
| Intron | 1,837 | 9.0 | 29,727,948 | 5.7 | 16,182 |
| Intergenic | 1,950 | 9.5 | 78,227,314 | 15.0 | 40,116 |
| Unknown | 14,412 | 70.4 | 387,279,814 | 74.3 | 26,872 |
| Total | 20,458 | 521,015,363 | 25,467 |
The average insertion interval in exon and intron DNA, shown in Table 3, is three times smaller than the interval in intergenic regions. In other words, Tos17 insertions were three times more likely to be found in genic regions containing exons and introns than in intergenic regions. Of 22,278 annotated genes in the rice genome, 794 genes were disrupted in exons and 1151 genes were disrupted in introns (638 were disrupted in both).
Target Site Specificity of Tos17
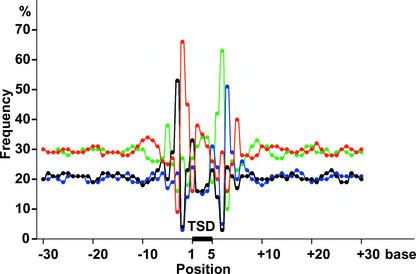
As described above, Tos17 insertions are not distributed randomly in the rice genome. To understand the molecular basis for target preference, the nucleotide composition of Tos17 insertion sites was analyzed. The nucleotide composition of the unique Tos17 insertion sites is shown in Figure 1. Relative to the 5-bp target site duplication sequence (TSD) (Hirochika et al., 1996), T (66%) at position −2 and A (63%) at position +2 are highly conserved. By contrast, adjacent T (9%) at position −3 and A (9%) at position +3 are avoided. G (53%) at position −3 and C (52%) at position +3 are preferred. A (38%) at position −5 and T (40%) at position +5 are somewhat preferred. The consensus sequence deduced from these conserved nucleotides is the palindrome ANGTT-TSD-AACNT. This consensus is relatively weak, like the target sequence requirements for T-DNA insertion (Brunaud et al., 2002), reflecting the ability of Tos17 to insert at highly divergent target sequences.
Figure 1.
Base Preferences of Tos17 Insertion Sites.
Average base preferences at each position were calculated based on flanking sequences at 20,458 loci. From position 1 to position 5 is the TSD sequence. Numbers with minus and plus signs are base numbers upstream and downstream, respectively, from the TSD. The percentages of A (green), C (blue), G (black), and T (red) at each position were plotted.
In the TSD, a relatively high number of Gs at position 1 and Cs at position 5 were observed. Positions 2, 3, and 4 show a slight bias against G and C. In the TSD, the base biases are too small to reveal any consensus, indicating that the sequence of the TSD is not a determinant of target preference. In the regions >10 bp away from the TSD (i.e., those from −30 to −10 and from +10 to +30), the frequency of A/T and G/C residues was 28 to 32% and 18 to 22%, respectively, at each base position. Because these values are equal to the average for all rice genomic sequences analyzed in this study, GC content within 25 bp of the target site consensus sequence is not a major factor determining target preference.
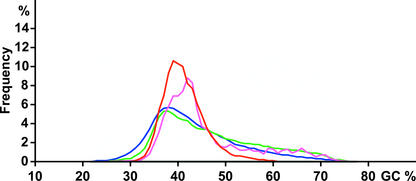
Tos17 Target Regions Have a Narrow GC Content Distribution
Figure 2 shows the frequency distribution of GC content in the regions within 500 bp of Tos17 insertion points (red line). For comparison, frequency distributions also were determined for total rice genomic sequence (blue line), CDS sequence (green line), and kinase and disease resistance genes (pink line). To determine the frequency distribution for all rice genomic sequences (521 Mbp), the sequences first were joined into one continuous sequence and then divided into 1-kb fragments. The GC content of these fragments then was calculated to generate a frequency distribution. Similarly, all rice CDS (55.5 Mbp) sequences and all rice sequences annotated with the terms “kinase” and/or “disease resistance” (2.8 Mbp) were joined and then analyzed in the same way. The frequency distributions of GC content were plotted using 1% intervals along the x axis.
Figure 2.
Frequency Distribution of the GC Content of Tos17-Inserted Regions and Published Rice Genomic Sequences.
The percentages of GC for 1-kb windows centered on 20,458 insertion points were calculated, and the frequency distribution was plotted in red. For continuous rice genomic sequences, derived from PAC or BAC clones with at least 70 kb of sequence, GC contents were calculated with a 1-kb sliding window, and the frequency distribution was plotted in blue. To determine the GC content distribution of CDS, all annotated CDS sequences were first joined into one large sequence. This sequence then was split into 1-kb pieces to determine the percentage of GC and the frequency distribution (green line). To generate a frequency distribution for protein kinase and defense-related genes (pink line), each 1-kb fragment of the rice genomic sequences was searched by BLASTX against an amino acid data set containing records of protein kinase and disease resistance genes. Fragments showing matches with E values of <e-10 were included in the frequency distribution. Each frequency distribution of GC content is plotted using 1% intervals.
The frequency distribution of GC content for all of the genomic sequences (blue line) is centered at 37% GC but is not symmetrical, showing a long tail toward high GC content. The frequency distribution for all CDS sequences (green line) also is centered at 37% GC and shows a similar pattern to that for all of the genomic sequences, although shifted toward higher GC content. This result agrees with the report that coding regions of the rice genome have high GC content (Sasaki et al., 2002).
The frequency distribution of GC content for Tos17 insertion points (red line) is centered at 39% GC and is narrower than that for the whole genome and CDS sequences. Surprisingly, the distribution plot for kinase/resistance genes is centered at 42% GC and overlaps the plot for Tos17 insertion points. By contrast, the plot for enzyme records annotated with “ase” but excluding “kinases” and “transcriptases” shows a pattern similar to the plot for all CDS sequences. This result strongly suggests that kinase/resistance genes reside in loci in the rice genome that have a specific GC content and that Tos17 prefers these loci.
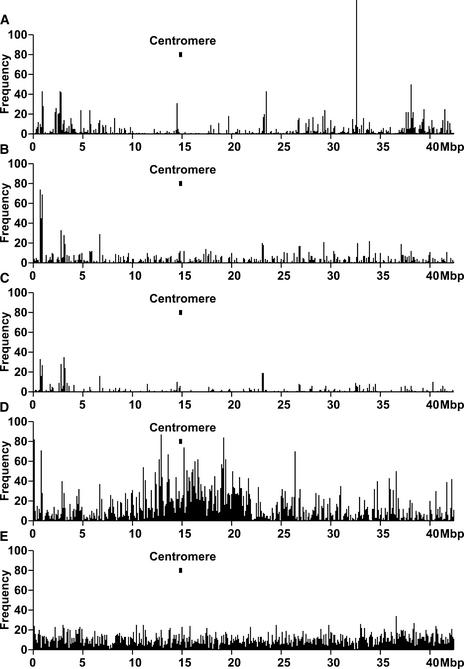
Tos17 Insertion Hot Spots in Rice Genomic Sequences
The number of Tos17 insertions was counted in each 100-kb interval of chromosome 1. Frequencies of protein kinases, resistance genes, retrotransposons, and ESTs in each 100 kb also were determined based on the following analysis. Genomic sequences of chromosome 1 were divided at 1-kb intervals, and then each 1-kb query was searched by BLASTX against data sets of protein kinase genes, resistance genes, and retrotransposons. Data annotated with “protein kinase,” excluding those labeled as “putative,” “similar,” “probable,” “homolog,” or “like,” were selected from the nonredundant data set to construct the protein kinase data set. The resistance gene and the retrotransposon data sets were built in the same way. These data sets were used to determine the distribution of these gene classes along chromosome 1.
Tos17 insertion sites are dispersed widely throughout the chromosome, but they are not distributed evenly. Insertions tend to be located in the distal regions of the chromosome. The most frequently disrupted region in chromosome 1 (the highest peak in Figure 3A) is a putative rust resistance gene. The density of retrotransposon-related sequences is high in the pericentromeric region (Figure 3D). However, the density of Tos17 insertions in the pericentromeric region is low. These results indicate that Tos17 avoids the retrotransposon-rich pericentromeric region.
Figure 3.
Tos17 Insertion Map on Contigs of Rice Chromosome 1.
Contigs were assembled through a combination of BLASTN searches and the EMBOSS megamerger program. Seven remaining gaps between the contigs were joined simply. The x axes represent the position on chromosome 1. The y axes show the frequency of Tos17 insertions, retrotransposons, and genes.
(A) Tos17 insertions.
(B) Protein kinase genes.
(C) Disease resistance–related genes.
(D) Retrotransposons.
(E) Rice ESTs.
Frequencies were calculated at 100-kb intervals along chromosome 1. The position of the centromere is shown with a box. For (B), (C), and (D), the joined sequence of chromosome 1 was split into 1-kb fragments. Each fragment was searched with BLASTX against an amino acid data set of protein kinase genes, disease resistance genes, and retrotransposons, respectively. Gene frequencies represent the number of 1-kb fragments that have matches with E values of <e-10 for BLASTX or <e-100 for BLASTN.
As described above, Tos17 prefers protein kinase genes and disease resistance genes. Reflecting this preference, the locations of Tos17 hot spots (Figure 3A) correlate with those of clusters of protein kinase genes (Figure 3B) and disease resistance genes (Figure 3C).
Evaluation of the Aggregation of Tos17 Insertions in Rice Genomic Sequences
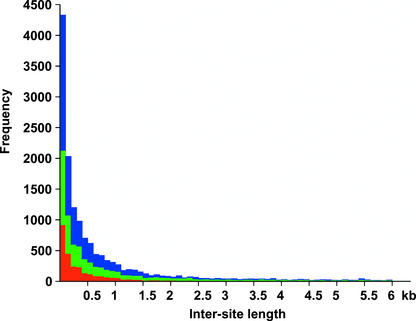
To further characterize Tos17 hot spots, the distance between neighboring Tos17 insertion points mapped on genomic sequences was determined. Figure 4 shows the distribution plot of distances between Tos17 insertion points in 100-bp increments. The intersite distances show an exponential distribution, which is a typical result for one-dimension, nearest-neighbor distances. However, the distribution is extremely biased toward short distances, indicating that there are hot spots for Tos17 insertion. We analyzed the distribution type using the Hopkins-Skellam index (Hopkins and Skellam, 1954). If Tos17 inserted at random, the mean value of the squares of the distances between adjacent Tos17 insertions (MI) would equal the mean value of the squares of the distances between Tos17 and randomly chosen points on the genome (MP). If Tos17 insertion points are aggregated, then MP would be greater than MI. The coefficient of aggregation is defined as MP divided by MI; thus, it will equal 1 for randomly distributed elements. From a total of 17,041 distances on PAC/BAC sequences longer than 70 kb, a coefficient of aggregation of 6.1 was obtained for Tos17 insertions, differing significantly from the value expected for random distribution (P < 0.001). This result shows that the Tos17 insertion points are highly aggregated.
Figure 4.
Estimation of the Window Sizes of Tos17 Hot Spots.
All insertion points were mapped onto public PAC and BAC sequences. The distances between adjacent Tos17 insertion points on the rice genomic sequences were obtained, and the frequency distribution of the distances was plotted in 100-bp increments. Each distance was calculated from continuous genomic sequences of at least 70 kb. Red, green, and blue bars indicate Tos17 insertions in coding, intergenic, and unknown regions, respectively.
The number of hot spots was examined by scanning with a 2.5-kb sliding window throughout rice genomic sequences. A hot spot was defined as a continuous group of insertion points within which each interinsertion distance was <2.5 kb. This window size was chosen based on the distribution plot of distances (Figure 4). From 20,458 insertions on the published genomic sequences, 1767 hot spots were detected, whereas the number of stand-alone insertions was 4490. This result indicates that 78% of the insertions are located in hot spots. The average insertion number per hot spot was 6.5, with a standard deviation of 12.
We also analyzed the locations of the hot spots on annotated PAC/BAC sequences (25% of published rice genomic sequences). In total, 519 hot spots were detected on annotated PAC/BAC sequences. Of these, 396 hot spots (76%) mapped to genic regions (i.e., CDS), whereas 123 (24%) were in intergenic regions. For comparison, 10% of the whole rice genome sequence is annotated as genic, whereas 15% is intergenic. Therefore, the hot spots of Tos17 insertion tended to be located in genic regions. This result indicates that insertion hot spots contribute to the preference of Tos17 for insertion into genic regions.
Insertion Hot Spots Are Not Determined By Nucleotide Sequence
As mentioned above, an RNA polymerase largest subunit gene was one of the hot spots of Tos17 insertion. In the published rice genomic sequences, RNA polymerase largest subunit genes are located on chromosomes 5 and 8. Identity between the exon regions of the two genes is 91%. Because the cutoff value for the identification of an independent Tos17 insertion point is 97%, we were able to compare the number of Tos17 insertions in the two copies of the RNA polymerase largest subunit gene. All 104 of the Tos17 insertions in the RNA polymerase largest subunit gene were mapped to chromosome 5. Considering the sequence identity between the two copies of the RNA polymerase largest subunit gene, this result clearly indicates that the determinant of Tos17 insertion hot spots is not the nucleotide sequence itself. Reverse transcriptase–mediated PCR analysis showed that only the gene copy on chromosome 8 is transcribed in callus, indicating that the transcriptional activity of the target regions also is not a determinant of hot spots.
DISCUSSION
The target site specificity of Tos17 was revealed clearly by large-scale analysis of target sequences. The insertion site consensus sequence is the palindrome ANGTT-TSD-AACNT. Interestingly, in vitro studies of retroviral integrase activity revealed that palindromic sequences that form cruciform structures are preferred targets for retroviral integrase (Katz et al., 1998). Indeed, for retroviral integrase, a cruciform structure is a more important determinant of target preference than the specific sequence of the site. The fact that the Tos17 target site consensus sequence is a palindrome suggests that the integrase encoded by Tos17 also may have a preference for cruciform structures. However, because the nucleotide sequence is well conserved at the target sites of Tos17, it is likely that the sequence itself also is a determinant of target specificity for this retrotransposon. The conserved sequence may act as a recognition sequence for Tos17 integrase. Interestingly, a small-scale analysis of target sites of the tobacco retrotransposon Tto1 in Arabidopsis revealed a preference for A or T at position −2 relative to the TSD, similar to the Tos17 consensus (Okamoto and Hirochika, 2000). This finding suggests that a common mechanism for target selection is conserved, at least among the copia-type retrotransposons of plants.
At the chromosome level, Tos17 insertions show an aggregated distribution pattern (Figure 4). Aggregation of retrotransposon insertions in host genomes is a commonly observed phenomenon. For example, the Saccharomyces cerevisiae retrotransposons Ty1 and Ty3 usually insert upstream of promoters recognized by RNA polymerase III (Chalker and Sandmeyer, 1992; Devine and Boeke, 1996), whereas Ty5 is targeted into silent chromatin regions. The fission yeast retrotransposon Tf1 usually inserts into regions between genes transcribed by RNA polymerase II (Behrens et al., 2000; Singleton and Levin, 2002). In plants, the miniature inverted-repeat transposable elements prefer to integrate into the 5′ regions of genes, including regulatory regions (Bennetzen, 2000). The S1 short interspersed nuclear element in the genus Brassica prefers matrix attachment regions (Tikhonov et al., 2001).
Although, like other retrotransposons, Tos17 shows a tendency to aggregate, its distribution on chromosome 1 differs from that of other retrotransposon-related sequences (Figure 3). According to a BLASTX search throughout the chromosome, retrotransposon-related sequences are concentrated in the pericentromeric regions. This result is consistent with the finding that the intergene long terminal repeat retrotransposons, a class of high-copy-number retrotransposons located in intergenic regions (Bennetzen, 2000), might be concentrated in pericentromeric regions. Furthermore, many copies of Athila family retrotransposons are present in the heterochromatin knob on Arabidopsis chromosome 4 (Arabidopsis Genome Consortium, 2000), suggesting that Athila also may be targeted into silent heterochromatin regions. By contrast, Tos17, like T-DNA (Brunaud et al., 2002), seems to avoid the retrotransposon-rich pericentromeric region and to prefer genic regions. This target preference of Tos17 is consistent with the proposal that low-copy-number transposons prefer genic regions for insertion because they require more transcriptionally active loci to maintain their copy number than do the intergene long terminal repeat retrotransposons (Bennetzen, 2000).
The mammalian retrovirus HIV-1, which has integration machinery similar to that of the retrotransposons, also shows regional hot spots for integration (Schroder et al., 2002) and prefers genic regions that are transcriptionally active. Because Tos17 hot spots also tend to occur in genic regions, it seemed possible that transcriptional activity could be a determinant of target site preferences. However, of two nearly identical copies of the RNA polymerase largest subunit gene in the rice genome, only the untranscribed copy, on chromosome 5, is a hot spot for Tos17 insertion. This result suggests that transcriptional activity may not be a major determinant of Tos17 hot spots. Furthermore, in contrast to Ty1, Ty3, and Tf1, insertions in Tos17 hot spots are distributed evenly throughout the disrupted open reading frames and are not biased toward 5′ regulatory regions. This result suggests that Tos17 target selection is not mediated by interaction with transcription factors but may be encoded at the chromatin level. This mechanism of target selection is used by Ty5, whose integration was shown to be mediated by interactions between its integrase and the silent information regulator protein Sir4p (Xie et al., 2001).
Although expressed genes, detected by a BLAST search of ESTs, are distributed evenly throughout chromosome 1, defense-related and protein kinase genes tend to occur in clusters. The distribution of these clusters tends to correlate with hot spots for Tos17 insertion. Furthermore, BLASTX analysis shows that one-fourth of the Tos17 flanking sequences have similarities with protein kinase or disease/defense-related genes (Table 1). There are several possible explanations for this apparent concentration of Tos17 insertions in these two classes of genes. First, analysis of the frequency distribution of GC content (Figure 2) shows that Tos17 prefers target sites with a specific GC content. Because the narrow GC content distribution of Tos17 target sites overlaps that of the kinase and resistance genes, this may explain the preference of Tos17 for these genes. Second, the kinase and defense gene clusters are gene dense, and Tos17 prefers to insert in gene-dense regions of the genome.
It is tempting to speculate that the high number of Tos17 insertions in kinase and defense genes reflects the fundamental mechanisms of plant genome evolution. Comparative sequence analysis of a resistance gene complex in sorghum and maize shows that disease resistance gene clusters are unusually prone to frequent internal and adjacent chromosomal rearrangements (Ramakrishna et al., 2002). Such rearrangements may facilitate the rapid evolution of these genes and, in turn, help plants develop novel resistance mechanisms. Similarly, the rapid evolution of protein kinase genes might be needed to facilitate the adaptation of signal transduction pathways to environmental changes. Transposons are known to alter the genome through illegitimate recombination (Devos et al., 2002) and thus may contribute to the rapid evolution of resistance gene complexes. Because Tos17 has a significant preference for regions containing defense-related and protein kinase genes, this retrotransposon could be one agent driving the rapid evolution of these and other hot spots. The existence of different copy numbers of Tos17 among different japonica cultivars shows that Tos17 also was activated under natural conditions, although Tos17 is most active in tissue culture, and that Tos17 may have this potential. Protein kinase and disease/defense-related genes may reside in a surface stratum of the chromosome that is more accessible to modification and rapid evolution. Meanwhile, the redundancy within these classes of genes ensures that plants with Tos17-mediated gene disruptions will survive with high probability.
We have constructed 47,196 Tos17 insertion lines of rice, which will be useful for functional genomic studies of this important crop plant. Although T-DNA and many transposons have been used for gene tagging, there are several advantages of using Tos17: (1) Tos17 is an endogenous retrotransposon, so large-scale analysis of mutants can be performed in the field without the regulatory constraints applicable to recombinant plants; (2) the copy-and-paste mechanism of transposition used by retrotransposons means that Tos17 is not excised in regenerated rice and is inherited stably; (3) Tos17 has a low copy number, with only two copies in Nipponbare, making it suitable for flanking sequence and reverse-genetics analyses; and (4) the copy number depends on the duration of tissue culture, making it easy to control insertion number without the need for crossing, as in the Mutator and Ac/Ds systems. The disadvantages of Tos17 are (1) it is impossible to obtain revertants, as can be done with the Ac/Ds system; and (2) the insertion events are not random, and their target sites are selected.
Because of the multiple-copy nature of Tos17 and the background mutations that accompany prolonged tissue culture, it often is difficult to establish a correlation between disrupted genes and their corresponding mutant phenotypes in our mutant rice lines. Nevertheless, if two or more lines have independent insertions in the same gene and the same phenotype is observed in these lines, it is likely that the observed phenotype results from the disruption of that gene. For hot spots such as PHYA, these allelic mutants are obtained readily (Takano et al., 2001). Because 76% of the hot spots are located in CDS, this is a distinct advantage of using Tos17 for insertional mutagenesis.
T-DNA insertion lines are used mostly for flanking sequence analysis. However, it is reported that T-DNA often makes tandem or complex insertions (Mayerhofer et al., 1991; Brunaud et al., 2002; Sessions et al., 2002). The broken-end structure of T-DNA also makes analysis of flanking sequence difficult. By contrast, Tos17 inserts cleanly into target sites, with a 5-bp target site duplication. Because disruption of Tos17 end regions during insertion occurs with very low frequency, we were able to obtain >22,000 insertion points on the rice genome with a >97% base match.
Tos17 insertions are distributed throughout the rice genome, and Tos17 prefers genic regions for integration. This characteristic is advantageous for the functional analysis of genes. However, there also are insertion hot spots for Tos17 in the rice genome, suggesting that chromatin structure is not homogeneous. Although >50% of the insertion mutants showed visible phenotypes, the contribution of the Tos17 insertions to these phenotypes was estimated to be <10%. This value indicates that many other genetic changes, such as insertions, deletions, and base exchanges, may be induced in cultured cells. Additional studies are required to understand the nature of the mutations induced in cultured cells and to fully use these mutations for the functional analysis of genes.
METHODS
Production of Tos17 Insertion Mutants
Conditions of callus induction, liquid culture, and regeneration were as described by Otsuki (1990), except that the concentration of casamino acids (acid-hydrolyzed casein) in the preregeneration medium was increased to 0.3%. The number of newly transposed Tos17 copies correlated approximately with the duration of tissue culture, suggesting that the number of primary sets for mutant screening could be reduced by prolonging the tissue culture period. However, prolonged culture leads to a reduction of the heterogeneity of mutations, possibly as a result of the selection of specific cell types that may have acquired a higher growth rate (Hirochika et al., 1996). As a compromise between these two factors, a 5-month culture period was adopted for the large-scale production of mutant lines. A total of 8885 NC (made in 1997), 9518 ND (made in 1998), 9593 NE (made in 1999), 9600 NF (made in 2000), and 9600 NG (made in 2001) mutant lines were obtained from rice (Oryza sativa cv Nipponbare) calli. Based on DNA gel blot analysis data, the average numbers of new insertions in the NC, ND, and NE lines were estimated to be 7.3, 10.7, and 12.2 per line, respectively. These estimates suggest that the 47,196 insertion lines produced carry ∼500,000 insertions.
Ten plants from the M2 generation of each insertion line were grown in the field to evaluate the frequency of visible mutations and the range of mutant phenotypes. From the analysis of 700 NE lines, 374 lines (53.4%) showed more than one visible phenotype. The most frequent phenotype was sterility (defined as <50% fertility), which occurred in 25.7% of all lines. The next most frequent phenotype was dwarf (14%). The frequencies of the albino, xantha, and chlorina phenotypes were 6.0, 0.6, and 7.1%, respectively. The frequent appearance of sterile and chlorotic phenotypes is consistent with a previous report of somaclonal mutations in plants derived from rice calli (Oono et al., 1984). The frequency of other minor phenotypes ranged from 0.1 to 3.9%. Similar mutation frequencies and ranges of mutant phenotypes were observed in mutant populations produced in different years. Because the mutant phenotypes and Tos17 insertions cosegregated in only 4 of 40 NE lines, the efficiency of tagging with Tos17 was estimated to be 10% or less. This value is consistent with the tagging efficiency estimated from large-scale screening of viviparous mutants (Agrawal et al., 2001). These results indicate that mutations other than Tos17 insertions were induced during tissue culture. Considering the relatively low efficiency of tagging with Tos17, a reverse-genetics approach is most suitable for functional analysis using Tos17 insertion mutants.
Isolation and Sequencing of Flanking Regions of Tos17 Insertions
Detailed methods for the isolation of Tos17 flanking regions and the direct sequencing of isolated fragments were described previously (Miyao et al., 1998). Both thermal asymmetric interlaced PCR (Liu and Whittier, 1995) and suppression PCR (Siebert et al., 1995) methods were used for the amplification of flanking regions. Additional adapter primers, 5′-GTNCGA(G/C)(A/T)CAN(A/T)AGC-3′, 5′-GTNCGA(G/C)(A/T)CNAA(A/T)GTT-3′, and 5′-CGTGNAG(A/T)ANCNAAG-3′, were used for thermal asymmetric interlaced PCR to increase efficiency. Isolated DNA fragments were used directly for BigDye terminator sequencing reactions. Single-run DNA sequences were obtained using ABI 377 and 3100 DNA sequencers (Applied Biosystems, Foster City, CA).
DNA Isolation and DNA Gel Blot Hybridization
Total DNA from each line was isolated using the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). DNA samples (500 ng) were digested completely with XbaI, which cuts Tos17 at one site. Digested samples were separated by electrophoresis on 0.8% agarose gels and then transferred onto positively charged nylon membranes. Tos17 XbaI-BamHI probe, which has a gag region, was used for hybridization at 65°C in 0.5 M phosphate buffer, pH 7.2, containing 1 mM Na2-EDTA, 7% SDS, and 200 μg/mL sonicated and autoclaved calf thymus DNA. After hybridization, the membrane was washed two times with 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) at 55°C.
Bioinformatic Analysis and Insertion Mutant Database
Genomic sequence data for rice were obtained from GenBank at the National Center for Biotechnology Information (NCBI) via a mirror site (ftp://bio-mirror.jp.apan.net/pub/biomirror/) and from the Rice Genome Research Program (http://rgp.dna.affrc.go.jp/). Personal computers running the FreeBSD operating system (http://www.freebsd.org/) were used for all data analysis and data storage. To identify independent flanking sequences, each sequence was compared using BLASTN (Basic Local Alignment Search Tool) with the other flanking sequences in the database. Flanking sequences were determined to be identical if they had >90% identity, expected values of <e-15, less than a 3-bp mismatch at their 5′ ends, and less than a 3-bp frameshift. For the BLASTX analysis, a nonredundant amino acid data set provided by NCBI was used. Disrupted gene products were categorized by manual inspection using the LIGAND database provided by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/). For the BLASTN analysis to determine insertion points in the genomic sequences, rice genomic sequences were obtained from GenBank Release 134 and daily updates. After the first screening by BLASTN, insertion points were reconfirmed with the EMBOSS (http://www.hgmp.mrc.ac.uk/Software/EMBOSS/) stretcher program. The criterion for identifying insertion points was >97% identity of the flanking sequence to the genomic sequence. EMBOSS megamerger was used to construct a local P1 artificial chromosome contig map. The pdflib_pl.pm module (http://www.pdflib.com/) was used to draw graphs. For primer construction, the Primer3 (Rozen and Skaletsky, 2000) program was incorporated into our common gateway interface (CGI) program.
Information about seed stocks, the phenotype of each mutant line, the nucleotide sequence of the flanking region of each Tos17 insertion, and similarity search data are stored in the relational database management system PostgreSQL (http://www.postgresql.org/). Data are accessed via the World Wide Web server program Apache (http://www.apache.org/) and a CGI of perl script with the CGI::SpeedyCGI module (http://daemoninc.com/speedycgi/).
Our database stores not only data about disruption loci but also information regarding the phenotypes of insertion lines, along with >40,000 photographic images. Users can search for insertion lines for genes of interest by BLAST search on our World Wide Web site (http://tos.nias.affrc.go.jp/). Phenotype data with a photographic image of each insertion line also are available via links from the BLAST search result. These phenotype data provide an efficient means of predicting the function of disrupted genes. Seeds of mutant lines are freely available to the scientific community. A PDF file of request forms with barcode tags can be accessed by following the link from the phenotype table. Once request forms with the user's signature are received, 20 grains in each line will be distributed. The primer design page for the Tos17 3′ end primer and target gene primer also can be accessed via links from the phenotype table. Mutant lines also are searchable via the BLAST search service at NCBI using the Genome Survey Sequence data set.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact A. Miyao, miyao@nias.affrc.go.jp.
Accession Numbers
The nucleotide sequence data, together with each insertion's position on the rice genomic sequence, the BLASTX results, and the original line name, were uploaded to the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession numbers AG020727 to AG025611 and AG205093 to AG215049.
Acknowledgments
We thank Yumiko Yamashita, Yuki Machida, and Ai Miyazaki for technical assistance. This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan, the Enhancement of Center-of-Excellence, Special Coordination Funds for Promoting Science and Technology in Japan, and the Program for the Promotion of Basic Research Activities for Innovative Biosciences.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012559.
References
- Agrawal, G.K., Yamazaki, M., Kobayashi, M., Hirochika, R., Miyao, A., and Hirochika, H. (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion: Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 125, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Consortium (2000). The complete sequence of heterochromatic island from higher eukaryote. Cell 100, 377–386. [PubMed] [Google Scholar]
- Behrens, R., Hayles, J., and Nurse, P. (2000). Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 28, 4709–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42, 251–269. [PubMed] [Google Scholar]
- Brunaud, V., et al. (2002). T-DNA integration into the Arabidopsis genome depends on sequences of pre-insertion sites. EMBO Rep. 3, 1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker, D.L., and Sandmeyer, S.B. (1992). Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 6, 117–128. [DOI] [PubMed] [Google Scholar]
- Devine, S.E., and Boeke, J.D. (1996). Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10, 620–633. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., Brown, J.K.M., and Bennetzen, J.L. (2002). Genome size reduction through illegitimate recombination counteracts ge-nome expansion in Arabidopsis. Genome Res. 12, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., Jiang, N., and Wessler, S.R. (2002). Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (2001). Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 4, 118–122. [DOI] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, B., and Skellam, J.G. (1954). A new method for determining the type of distribution of plant individuals. Ann. Bot. N.S. 18, 213–227. [Google Scholar]
- Jordan, I.K., Rogozin, I.B., Glazko, G.V., and Koonin, E.V. (2003). Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72. [DOI] [PubMed] [Google Scholar]
- Kashkush, K., Feldman, M., and Levy, A.A. (2003). Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33, 102–106. [DOI] [PubMed] [Google Scholar]
- Katz, R.A., Gravuer, K., and Skalka, A.M. (1998). A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem. 273, 24190–24195. [DOI] [PubMed] [Google Scholar]
- Kidwell, M.G., and Lisch, D. (1997). Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Bakkeren, G., Crameri, A., Angelis, K., Redei, G.P., Schell, J., Hohn, B., and Koncz, C. (1991). T-DNA integration: A mode of illegitimate recombination in plants. EMBO J. 10, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, E.M., Liu, J., Lizhi, G., and McDonald, J.F. (2002). Long terminal repeat retrotransposons of Oryza sativa. Genome Biol. 3, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao, A., Yamazaki, M., and Hirochika, H. (1998). Systematic screening of mutants of rice by sequencing retrotransposon-insertion sites. Plant Biotech. 15, 253–256. [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko, A., and Li, W. (2001). Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 17, 619–621. [DOI] [PubMed] [Google Scholar]
- Okamoto, H., and Hirochika, H. (2000). Efficient insertion mutagenesis of Arabidopsis by tissue culture-induced activation of the tobacco retrotransposon Tto1. Plant J. 23, 291–304. [DOI] [PubMed] [Google Scholar]
- Oono, K., Okuno, K., and Kawai, T. (1984). High frequency of somaclonal mutations in callus culture of rice. In Gamma Field Symposia, Vol. 23. (Ohmiya, Ibaraki, Japan: Institute of Radiation Breeding), pp. 71–94.
- Otsuki, Y. (1990). A Visual Manual for the Protoplast Culture System of Rice. (Tokyo: Food and Agriculture Research Development Association).
- Ramakrishna, W., Emberton, J., SanMiguel, P., Ogden, M., Llaca, V., Messing, J., and Bennetzen, J.L. (2002). Comparative sequence analysis of the sorghum Rph region and the maize Rp1 resistance gene complex. Plant Physiol. 130, 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420, 312–316. [DOI] [PubMed] [Google Scholar]
- Schroder, A.R.W., Shinn, P., Chen, H., Berry, C., Ecker, J.R., and Bushman, F. (2002). HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellogg, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23, 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, T.L., and Levin, H.L. (2002). A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of insertion. Eucaryot. Cell 1, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, M., Kanegae, H., Shinomura, T., Miyao, A., Hirochika, H., and Furuya, M. (2001). Isolation and characterization of rice phytochrome A mutants. Plant Cell 13, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonov, A.P., Lavie, L., Tatout, C., Bennetzen, J.L., Avramova, Z., and Deragon, J.M. (2001). Target sites for SINE integration in Brassica genomes display nuclear matrix binding activity. Chromosome Res. 9, 325–337. [DOI] [PubMed] [Google Scholar]
- Walbot, V., and Petrov, D.A. (2001). Gene galaxies in the maize genome. Proc. Natl. Acad. Sci. USA 98, 8163–8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W., Gai, X., Zhu, Y., Zappulla, D.C., Sternglanz, R., and Voytas, D.F. (2001). Targeting of the yeast Ty5 retrotransposon to silent chromatin is mediated by interactions between integrase and Sir4p. Mol. Cell. Biol. 21, 6606–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]