Abstract
The Arabidopsis atmak3-1 mutant was identified on the basis of a decreased effective quantum yield of photosystem II. In atmak3-1, the synthesis of the plastome-encoded photosystem II core proteins D1 and CP47 is affected, resulting in a decrease in the abundance of thylakoid multiprotein complexes. DNA array–based mRNA analysis indicated that extraplastid functions also are altered. The mutation responsible was localized to AtMAK3, which encodes a homolog of the yeast protein Mak3p. In yeast, Mak3p, together with Mak10p and Mak31p, forms the N-terminal acetyltransferase complex C (NatC). The cytoplasmic AtMAK3 protein can functionally replace Mak3p, Mak10p, and Mak31p in acetylating N termini of endogenous proteins and the L-A virus Gag protein. This result, together with the finding that knockout of the Arabidopsis MAK10 homolog does not result in obvious physiological effects, indicates that AtMAK3 function does not require NatC complex formation, as it does in yeast. We suggest that N-acetylation of certain chloroplast precursor protein(s) is necessary for the efficient accumulation of the mature protein(s) in chloroplasts.
INTRODUCTION
Acetylation is the most common type of covalent modification found in proteins in eukaryotes, occurring either on the α-amino group (Nα) of N-terminal residues or on the ɛ-amino group of internal Lys residues (Polevoda and Sherman, 2002). Unlike the ɛ-Lys modification, N-terminal acetylation, catalyzed by N-terminal acetyltransferases (NATs), is irreversible and occurs cotranslationally on most eukaryotic proteins but rarely on prokaryotic or archaebacterial polypeptides (for review, see Polevoda and Sherman, 2000, 2003). The biological significance of cotranslational N-terminal acetylation varies from protein to protein: for tropomyosin (Urbancikova and Hitchcock-DeGregori, 1994), human fetal hemoglobin (Manning and Manning, 2001), yeast 20S proteasome subunits (Kimura et al., 2000), and NADP-specific glutamate dehydrogenase (Siddig et al., 1980), N-acetylation can modify protein function, protein–protein interaction, and/or thermal stability, whereas nonacetylated versions of the chaperonin Hsp10 (Ryan et al., 1995) and of alcohol dehydrogenase (Hoog et al., 1987) are fully functional.
Eukaryotic proteins subject to N-terminal acetylation have a variety of N-terminal sequences and no simple consensus motif. The complexity of acetylated termini is reflected in the existence of multiple NATs, each acting on different groups of N termini. In Saccharomyces cerevisiae, three NATs are known: NatA (Lee et al., 1989), NatB (Polevoda et al., 1999), and NatC (Tercero et al., 1992, 1993; Tercero and Wickner, 1992). NatA contains Nat1p and the catalytic subunit Ard1p (Mullen et al., 1989). Both nat1 and ard1 mutants grow slowly, do not enter the Go phase, and fail to acetylate in vivo the same subset of normally acetylated proteins (Mullen et al., 1989). Moreover, overexpression of both Ard1p and Nat1p is required for increased NAT activity in vivo (Mullen et al., 1989). In NatB, three proteins interact with the catalytic subunit Nat3p (Polevoda and Sherman, 2003).
Association of the catalytic Mak3p with Mak10p and Mak31p is a prerequisite for the formation of an active NatC complex (Rigaut et al., 1999; Uetz et al., 2000; Polevoda and Sherman, 2001). Loss of any of the three subunits results in identical abnormal phenotypes, including reduced growth on nonfermentable carbon sources and an inability to propagate the cytoplasmic L-A virus (Toh-e and Sahashi, 1985; Lee and Wickner, 1992; Tercero and Wickner, 1992; Tercero et al., 1993; Polevoda and Sherman, 2001). The latter effect is attributed to the lack of N-terminal acetylation of the viral Gag protein, whereas defective acetylation of one or more unidentified mitochondrial preproteins could be responsible for the growth phenotype (Tercero et al., 1993).
The characterization of mak3, nat3, and ard1 mutants has identified the substrate specificity of each of the NAT complexes (Tercero et al., 1993; Garrels et al., 1997; Arnold et al., 1999; Perrot et al., 1999; Polevoda et al., 1999; Kimura et al., 2000, 2003). Subclasses of proteins with N-terminal Ser, Ala, Thr, or Gly are not acetylated in ard1 cells; Met-Glu, Met-Asp, and some Met-Asn termini remain unacetylated in nat3; and proteins beginning with Met-Ile, Met-Leu, Met-Trp, or Met-Phe are not acetylated in mak3 mutants. Similar patterns of N-terminal acetylation also have been reported for mammalian proteins (Polevoda and Sherman, 2003). This finding, together with the presence of ARD1, NAT3, and MAK3 homologs in the genomes of human, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis, and other eukaryotes, indicates that similar N-acetylation mechanisms operate in all eukaryotes.
We have identified and characterized atmak3-1, an Arabidopsis mutant disrupted in a gene orthologous with MAK3 of S. cerevisiae. The absence of AtMAK3-mediated N-acetylation of cytoplasmic proteins is associated with defects in photosynthesis and growth, although the formation of a NatC complex like that in yeast is not required for AtMAK3 function.
RESULTS
Identification and Phenotype of the pam21 Mutant
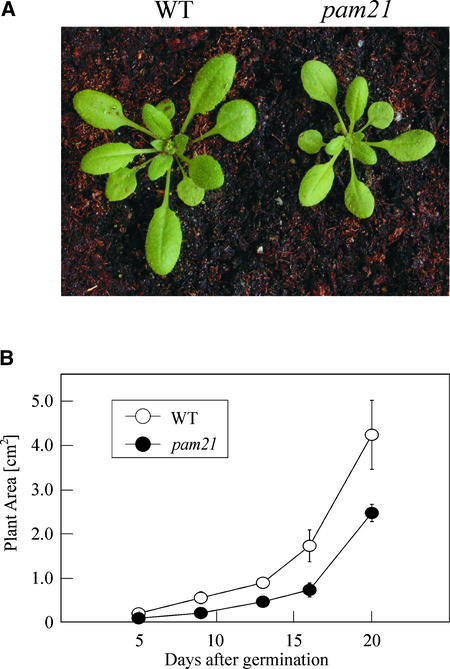
A screen for mutants with reduced effective quantum yields of photosystem II (ΦII) (Varotto et al., 2000a) led to the identification of photosynthesis altered mutant 21 (pam21). The mutant was identified among T-DNA–mutagenized Arabidopsis lines, and the photosynthetic lesion segregated as a monogenic, recessive trait. The mutation is associated with slightly paler leaves than the wild type and a reduction in plant size (Figure 1A). In pam21, growth rate, measured by noninvasive image analysis (Leister et al., 1999), was decreased by ∼40% relative to the wild type (Figure 1B). Additional phenotypic effects of the mutation were monitored by analyzing chlorophyll fluorescence. In addition to ΦII (pam21, 0.66 ± 0.04; wild type, 0.77 ± 0.01), FV/FM (the maximum quantum yield of photosystem II) was decreased (pam21, 0.73 ± 0.01; wild type, 0.83 ± 0.01), implying a defect in energy transfer within photosystem II (PSII).
Figure 1.
Phenotypes of pam21 and Wild-Type Plants.
(A) Four-week-old plants grown in the greenhouse. WT, wild type.
(B) Growth kinetics of pam21 (n = 100) compared with wild-type (n = 100) plants. Leaf area was measured in the period from 5 to 20 days after germination. Error bars indicate standard deviations.
HPLC analysis of pam21 leaf pigments revealed a decrease in chlorophyll concentration (chlorophyll a + b) (Table 1), as expected from the pale-green color of the leaves. Moreover, in mutant leaves, the reductions in chlorophyll a/b ratio and in β-carotene suggest a decrease in the concentration of the PSII core. In pam21, the disproportionate increase in the abundance of xanthophyll cycle pigments (violaxanthin + antheraxanthin + zeaxanthin) with respect to neoxanthin and lutein likely is the result of an increase in photooxidative stress. Interestingly, the abnormal pigment composition and chlorophyll fluorescence characteristics were observed in 3- to 4-week-old mutant leaves, whereas 5- to 6-week-old plants, although reduced in size, behaved like the wild type (data not shown).
Table 1.
Leaf Pigment and Thylakoid Protein Contents of 4-Week-Old pam21 and Wild-Type Plants
| Pigment/Protein | Wild Type | pam21 | Relative Level in pam21 (%) |
|---|---|---|---|
| Chlorophyll a/b | 3.78 ± 0.09 | 3.11 ± 0.01 | 82 |
| Chlorophyll a + b | 1050 ± 48 | 882 ± 35 | 84 |
| Neoxanthin | 42 ± 3 | 47 ± 4 | 112 |
| Lutein | 157 ± 13 | 149 ± 9 | 95 |
| β-Carotene | 66 ± 4 | 52 ± 3 | 79 |
| Violaxanthin + antheraxanthin + zeaxanthin |
52 ± 4 | 64 ± 4 | 123 |
| PSI core (PSI-A, PSI-B, PSI-C) | 1.36 ± 0.08 | 0.97 ± 0.09 | 71 |
| PSI-A/B | 7.90 ± 0.07 | 6.02 ± 0.08 | 76 |
| PSII core | 0.83 ± 0.06 | 0.52 ± 0.08 | 63 |
| Oxygen-evolving complex | 0.48 ± 0.06 | 0.39 ± 0.04 | 81 |
| PSII-D1 | 3.24 ± 0.10 | 1.99 ± 0.08 | 61 |
| ATPase (α + β) | 0.59 ± 0.05 | 0.34 ± 0.06 | 58 |
| LHCII | 2.27 ± 0.09 | 2.22 ± 0.10 | 98 |
| LHCII* | 3.7 ± 0.04 | 3.68 ± 0.05 | 99 |
The pigment content of pam21 (n = 3) and wild-type (n = 3) leaves determined by HPLC is expressed as nmol/g leaf fresh weight. Mean values (±sd) are shown. Values for proteins are average optical densities (±sd) measured from three independent two-dimensional protein gel analyses (PSII core, PSI core, ATPase [α + β], oxygen-evolving complex, and LHCII; see Figure 2A) or from three independent protein gel blot analyses (PSII-D1, PSI-A/B, and LHCII*; see Figure 2B). The relative values for the mutant genotype are percentages based on leaf fresh weight (g).
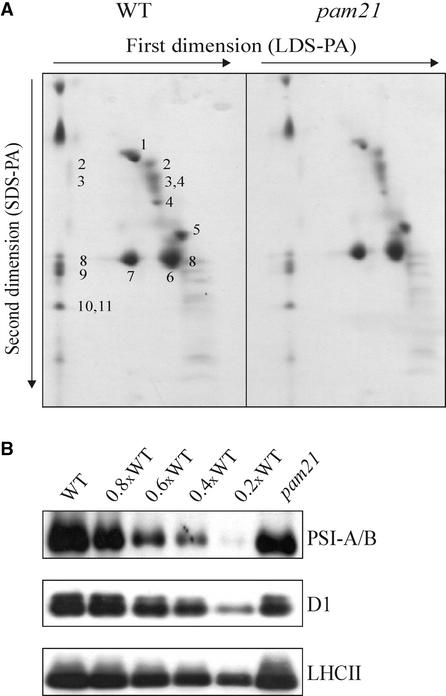
The Protein Composition of pam21 Thylakoids Is Altered
Thylakoid proteins were isolated from 4-week-old greenhouse-grown wild-type and mutant plants and subjected to two-dimensional gel electrophoresis (Figure 2A). Individual subunits of the photosynthetic apparatus were resolved and assigned to the core or to the oxygen-evolving complex of PSII, photosystem I (PSI), ATPase (α- and β-subunits), or the major light-harvesting complex of PSII (LHCII) as described by Pesaresi et al. (2001). Densitometric analyses of the two-dimensional protein gel after Coomassie blue staining revealed a reduction in the amounts of most of the proteins of the photosynthetic apparatus (Table 1). In particular, in mutant thylakoids, the abundance of proteins of the core of PSII and PSI and the α- and β-subunits of the ATPase was reduced by 30 to 40%. Levels of oxygen-evolving complex and LHCII were less affected (by 20% or less). Protein gel blot analysis of thylakoid proteins separated by denaturing one-dimensional PAGE with antibodies specific for PSI-A/B, PSII-D1, and LHCII confirmed the results of the two-dimensional gel analysis (Figure 2B, Table 1).
Figure 2.
Protein Composition of Thylakoid Membranes.
(A) Thylakoid proteins from identical amounts (fresh weight) of wild-type (WT) and pam21 leaves were fractionated first by electrophoresis on a nondenaturing lithium dodecyl sulfate polyacrylamide (LDS-PA) gel and then on a denaturing SDS-polyacrylamide (SDS-PA) gel. Positions of wild-type thylakoid proteins identified previously by protein gel blot analyses with appropriate antibodies are indicated: 1, α- and β-subunits of the ATPase complex; 2, D1-D2 dimer; 3, CP47; 4, CP43; 5, oxygen-evolving complex; 6, LHCII monomer; 7, LHCII trimer; 8, PSI-D; 9, PSI-F; 10, PSI-C; 11, PSI-H.
(B) Thylakoid proteins from identical amounts (fresh weight) of wild-type and pam21 leaves fractionated by one-dimensional PAGE. Decreasing levels of wild-type thylakoid proteins were loaded in the lanes marked 0.8×WT, 0.6×WT, 0.4×WT, and 0.2×WT. Three replicate filters were probed with antibodies raised against PSI-A/B, D1, and LHCII.
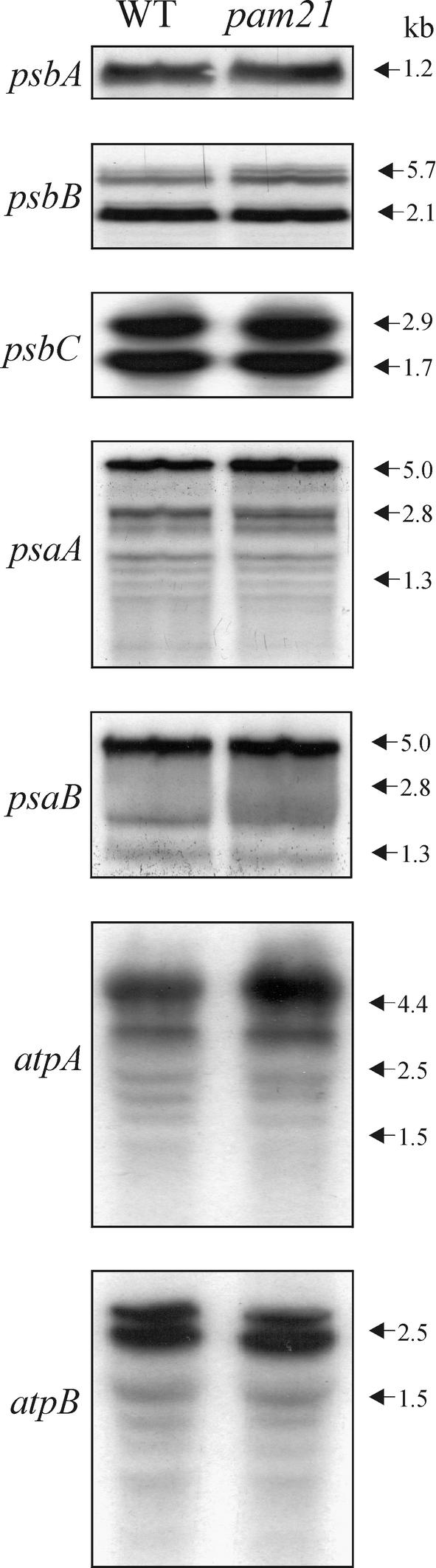
Translation of psbA Transcripts Is Affected in Mutant Chloroplasts
Transcription and translation patterns in mutant chloroplasts were characterized. RNA gel blot analysis indicated that identical amounts of psbA and psbC (encoding the D1 and CP43 subunits of PSII, respectively), psaA and psaB (encoding PSI-A and PSI-B, respectively), and atpB (encoding the β-subunit of ATPase) transcripts accumulated in pam21 and wild-type plants (Figure 3). The total amount of psbB transcript, encoding the CP47 protein, also was unchanged, although an accumulation of large transcripts was noted. The abundance of atpA mRNA, encoding the α-subunit of ATPase, actually was slightly higher in the mutant.
Figure 3.
mRNA Expression in Chloroplasts.
The numbers at right indicate RNA sizes as estimated by coelectrophoresis with denatured EcoRI-HindIII fragments of λDNA. Aliquots (30 μg) of total RNA from the wild type (WT) and pam21 were size-fractionated by agarose gel electrophoresis, transferred to a nitrocellulose filter, and probed with cDNA fragments specific for psbA, psbB, psbC, psaA, psaB, atpA, and atpB.
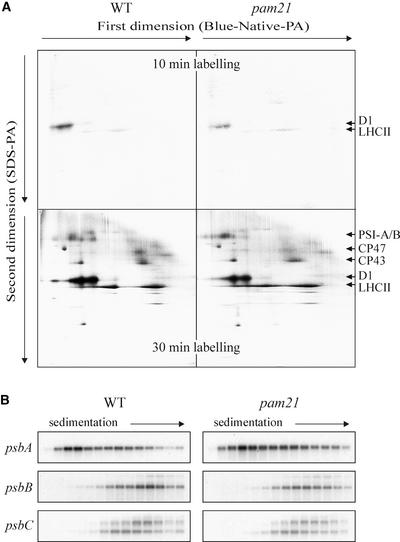
Because no marked decrease in the accumulation of plastome transcripts was observed in mutant plants, in vivo translation assays were performed to test for alterations in protein synthesis. Four-week-old leaves of wild-type and pam21 plants were incubated with 35S-Met and illuminated for 10 min. Subsequently, thylakoid membranes were isolated and polypeptides were fractionated by two-dimensional gel analysis using blue-native PAGE in the first dimension and denaturing SDS-PAGE in the second dimension. The accumulation of labeled D1 protein was reduced markedly in mutant leaves, whereas LHCII accumulated to a similar extent in wild-type and mutant plants (Figure 4A, top gel). To assess the rate of synthesis of other thylakoid proteins, labeling was performed for a longer period (30 min) (Figure 4A, bottom gel). Again, a marked decrease in the accumulation of labeled D1 and, to a lesser extent, of CP47 was observed, whereas all of the other proteins, including CP43, accumulated to wild-type levels.
Figure 4.
In Vivo Synthesis of Thylakoid Proteins and Polysome Accumulation.
(A) 35S-Met was applied for 10 min (top gels) or 30 min (bottom gels) to 4-week-old leaves under illumination. Thylakoid proteins from 100-mg portions of wild-type and pam21 leaves were separated by blue-native PAGE in the first dimension and by SDS-PAGE in the second dimension, electroblotted onto a nylon membrane, and analyzed by fluorography. Results from one of three independent experiments are shown, and the identities of the labeled bands are indicated at right according to Plucken et al. (2002).
(B) Total extracts from pam21 and wild-type leaves were fractionated on sucrose gradients. Fourteen fractions of equal volume were collected from the top to the bottom of the sucrose gradients. An equal proportion of the RNA purified from each fraction was analyzed by gel blot hybridization. Transcripts of psbA, psbB, and psbC were detected with gene-specific probes. Results from one of three independent experiments are shown.
The effect of the pam21 mutation on protein synthesis in the chloroplast was studied further by assessing polysome formation by sucrose density gradient fractionation (Figure 4B). Total leaf lysates were fractionated under conditions that maintain polysome integrity (Barkan, 1998), and RNA gel blots of RNA purified from gradient fractions were analyzed with probes specific for psbA, psbB, and psbC transcripts. As expected, an identical sedimentation profile between wild-type and pam21 plants was detected for psbC; however, no major differences were observed for the psbA and psbB genes as well.
Together, these results indicate that the photosynthetic lesion in pam21 is caused by the reduced synthesis of D1 and CP47, resulting in a substantial decrease in the abundance of PSII core proteins, and of other thylakoid multiprotein complexes. The concomitant decrease in the synthesis of D1 and CP47 is reminiscent of the “control by epistasy of synthesis” process described by Wollman and co-workers (Choquet and Wollman, 2002): in Chlamydomonas reinhardtii, a decrease in the synthesis of D1 downregulates the translation of CP47 by affecting the initiation step; similarly, the barley vir-115 mutant, which has a primary defect in D1 expression, fails to synthesize both CP47 and D1 (Gamble and Mullet, 1989; Kim et al., 1994). These findings suggest that in pam21 chloroplasts, the decrease in D1 expression could be the primary factor leading to the photosynthetic phenotype.
The PAM21 Gene Encodes a Homolog of NATs
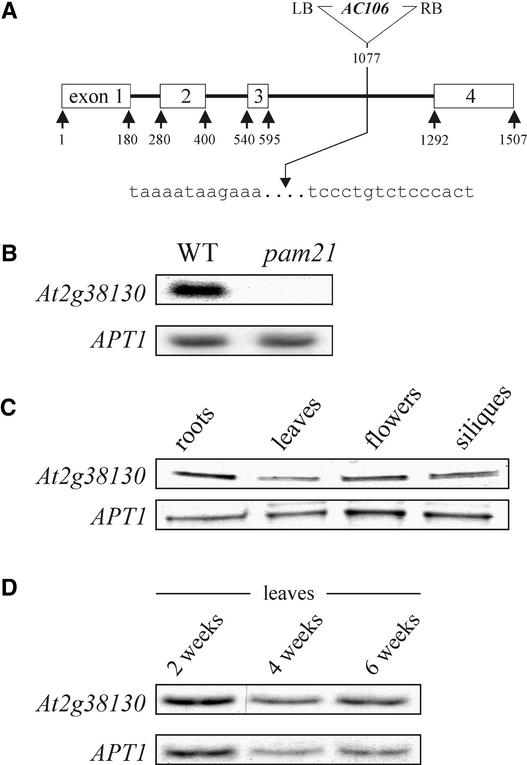
DNA gel blot analysis of a population of plants segregating for the pam21 allele, using the 5′ end of the AC106 T-DNA as a probe, revealed that only one T-DNA copy was present in the line, and this insertion cosegregated with the pam21 allele. Isolation of genomic sequences flanking the left border of the AC106 insertion was achieved by PCR after adapter ligation as described by Pesaresi et al. (2001). The AC106 T-DNA was found to be inserted in the third intron of At2g38130, at position +1077 relative to the start codon (Figure 5A). RNA gel blot analysis using an At2g38130 cDNA probe revealed that the T-DNA insertion drastically destabilized the At2g38130 transcript (Figure 5B). The wild-type phenotype could be restored fully by Agrobacterium tumefaciens–mediated transformation of homozygous mutant plants with At2g38130 genomic DNA, including 1 kb of its natural promoter. In all transformants, ΦII, pigmentation, and growth behavior were indistinguishable from those of wild-type plants (data not shown). Furthermore, in the progeny of all transformants, the wild-type phenotype cosegregated with the transgene.
Figure 5.
T-DNA Tagging and mRNA Expression of the At2g38130 Gene.
(A) At2g38130 was disrupted by an insertion of the 5.8-kb AC106 T-DNA in the third intron. The T-DNA insertion is not drawn to scale. Lowercase letters indicate plant DNA sequences flanking the T-DNA insertion. LB, left border; RB, right border.
(B) RNA gel blot analysis of the At2g38130 transcript in pam21 and wild-type (WT) leaves. Samples (30 μg) of total RNA were analyzed using At2g38130 full-length cDNA as a probe. To control for RNA loading, the blot was reprobed with a cDNA fragment derived from the APT1 gene, which is expressed at a low level in all tested tissues of Arabidopsis (Moffatt et al., 1994).
(C) Detection of At2g38130 transcripts in different plant organs by quantitative reverse transcriptase–mediated PCR. The analysis on 4.5% polyacrylamide gels was performed with 33P-labeled quantitative reverse transcriptase–mediated PCR products from roots, leaves, flowers, and siliques obtained after PCR for 20 cycles with At2g38130-specific primers (atmak3-137s and 33P-labeled primer atmak3-1494as) and control primers for the APT1 gene (Moffatt et al., 1994).
(D) The same analysis as in (C) was performed for leaves at different developmental stages.
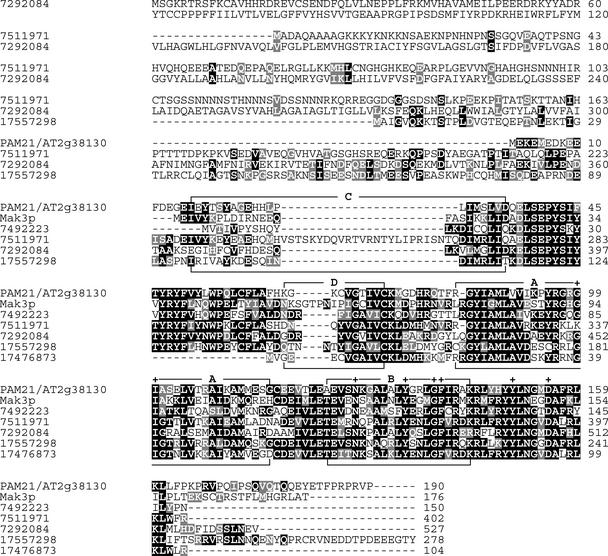
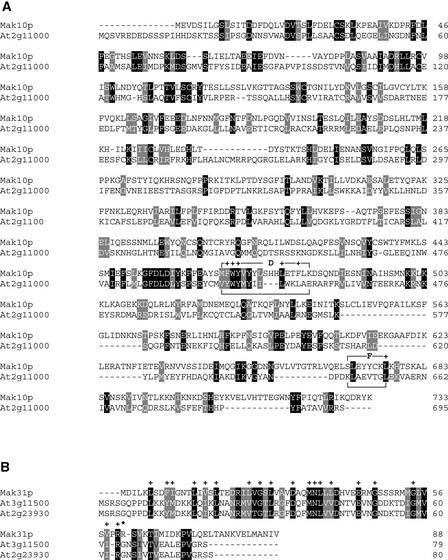
Database searches of the completely sequenced Arabidopsis genome revealed that At2g38130 is a single-copy gene. The predicted coding region is entirely covered by overlapping ESTs, and transcripts of the gene were detected in both green tissues and roots (Figure 5C) as well as during all leaf developmental stages tested (Figure 5D). The protein encoded by the At2g38130 gene contains 190 amino acid residues and shares significant sequence identity with two dozen proteins from diverse eukaryotes, including one from human (identity/similarity, 60/68%), two from Drosophila (60/66% [gene identifier: 7511971] and 55/62% [gene identifier: 7292084]), Mak3p of S. cerevisiae (52/62%), and proteins from Schizosaccharomyces pombe (50/63%) and C. elegans (48/59%). All of these proteins share four domains characteristic of the catalytic subunit of NATs, of which the A and B domains are predicted to be critical for acetyl CoA binding (Neuwald and Landsman, 1997) (Figure 6).
Figure 6.
Comparison of Mak3p-Like Sequences from Eukaryotes.
The amino acid sequence of the At2g38130 protein was compared with homologous sequences from S. cerevisiae (Mak3p), S. pombe (7492223), D. melanogaster (7511971 and 7292084), C. elegans (17557298), and human (17476873). Black boxes indicate strictly conserved amino acids, and gray boxes indicate closely related amino acids. Plus signs refer to positions of point mutations that abolish Mak3p function (Tercero et al., 1992). Conserved sequence stretches (motifs A to D) that are typical of NATs are indicated according to Neuwald and Landsman (1997).
The PAM21 Protein Is a Cytoplasmic NAT
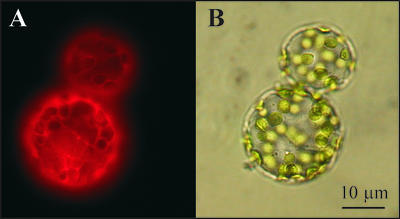
Analysis of the At2g38130 protein sequence with several predictors of subcellular targeting identified neither chloroplast- nor mitochondrion-targeting presequences, nor any other targeting signals, indicating that the protein resides in the cytoplasm. When the At2g38130 protein was fused to the red fluorescent protein dsRED (Jach et al., 2001) and transfected into tobacco protoplasts, cells harboring the At2g38130-dsRED fusion protein showed readily detectable red fluorescence in the cytoplasm (Figure 7).
Figure 7.
Intracellular Protein Localization.
(A) Fluorescence field of mesophyll tobacco protoplasts (SR1) transfected with the At2g38130-dsRED fusion protein.
(B) Bright-field view of the same section as in (A). Bar = 10 μm.
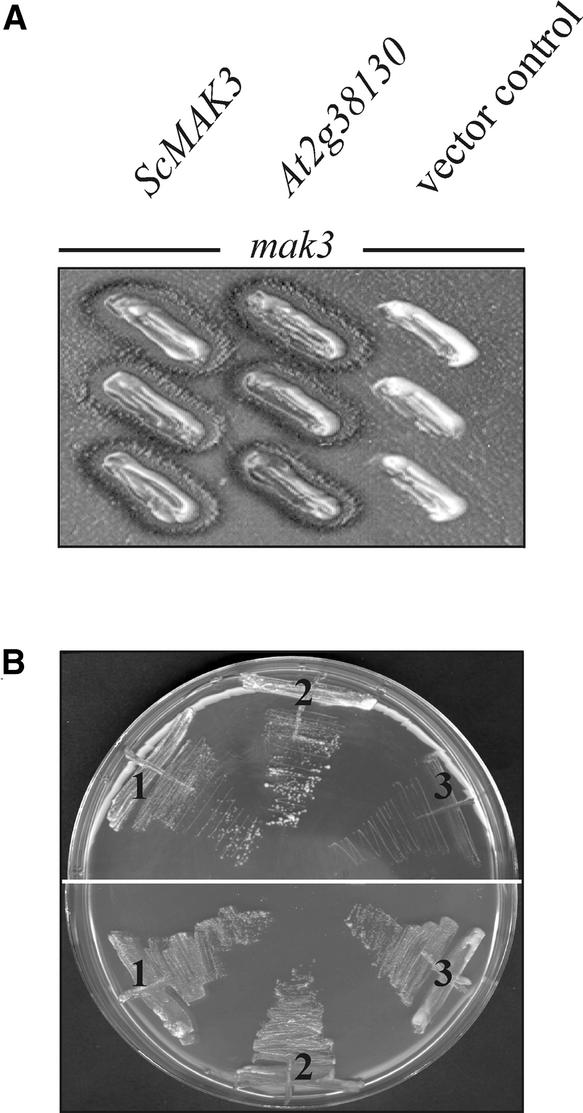
Moreover, transformation of a yeast mutant strain defective for Mak3p activity (mak3) (Sommer and Wickner, 1982) with the At2g38130 cDNA resulted in rescue of the two known phenotypes associated with the mak3 mutation: mak3 At2g38130 cells (1) support the propagation of the cytoplasmic double-stranded dsRNA virus L-A (Figure 8A) and (2) can grow on the nonfermentable YPG medium (1% yeast extract, 2% peptone, and 3% glycerol) (Figure 8B). This finding demonstrates that the At2g38130 protein can functionally replace the catalytic subunit of the yeast NatC. Because the PAM21/At2g38130 gene encodes a functional ortholog of Mak3p, the At2g38130 protein was designated AtMAK3 and the pam21 mutant was renamed atmak3-1.
Figure 8.
At2g38130 Complements the Yeast mak3 Mutation.
(A) The yeast mutant mak3 (Sommer and Wickner, 1982) was transformed with the Arabidopsis At2g38130 cDNA (At2g38130) and, as controls, with the S. cerevisiae MAK3 cDNA (ScMAK3) and with the empty vector alone (vector control). After cytoduction (Ridley et al., 1984), the three yeast strains were tested for their ability to propagate L-A-HNB M1 dsRNA virus particles. Both mak3 ScMAK3 and mak3 At2g38130 strains were able to propagate the virus, as detected by the presence of the killing zones surrounding the yeast colonies.
(B) The strains mak3 ScMAK3 (1), mak3 At2g38130 (2), and mak3 plus empty vector (3) were tested for their ability to grow on nonfermentable carbon sources (YPG) at 37°C (top half). As a control, strains were propagated on full medium (YPD; bottom half).
AtMAK3-Mediated N-Acetylation Does Not Require the Formation of a MAK3-MAK10-MAK31 Complex
To determine whether a NatC-like complex operates in Arabidopsis, the Arabidopsis genome sequence was searched for open reading frames that encode homologs of the other two known NatC subunits, Mak10p and Mak31p. The only MAK10 homolog in Arabidopsis corresponds to the At2g11000 gene, which encodes a protein with limited identity to yeast Mak10p (similarity/identity, 36/25%) (Figure 9A). Mak10p contains two sequence motifs that are common among a large number of T cell receptor α-subunits: region D ([LMF]FWY[LV].Y …L..[LF][LV]K) and the F region (S..YYC.[LV]) (Lee and Wickner, 1992). In the At2g11000 protein, motif D is partially conserved but no F domain is detectable.
Figure 9.
Comparison of Yeast Mak10p and Mak31p Sequences with Orthologous Sequences from Arabidopsis.
(A) Yeast Mak10p.
(B) Yeast Mak31p.
The D and F regions are motifs with similarity to domains of T cell receptor α-subunits (Lee and Wickner, 1992). Plus signs indicate amino acids conserved between Mak10p and T cell receptor α-subunits (A) and between Mak31p and Sm-like proteins (B). The asterisk in (B) indicates position 107 of the Sm-like protein consensus sequence according to Séraphin (1995). Black boxes indicate strictly conserved amino acids, and gray boxes indicate closely related amino acids.
Two Arabidopsis genes (At3g11500 and At2g23930) encode proteins related to Mak31p (At3g11500, 39/30% similarity/identity; At2g23930, 37/28% similarity/identity) (Figure 9B). Based on protein sequence similarity, Mak31p was predicted to be a Sm-like protein, although it lacks a Sm-typical Cys or Gly residue at the consensus position 107 defined by Séraphin (1995). Interestingly, both At3g11500 and At2g23930 contain a Gly residue at the corresponding position.
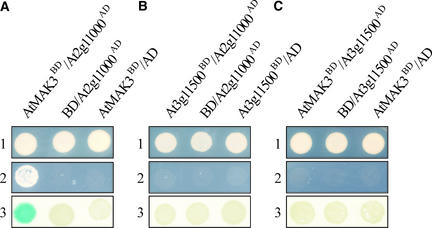
The copurification of yeast Mak3p, Mak10p, and Mak31p (Rigaut et al., 1999), in combination with protein–protein interactions detected between Mak3p and Mak10p and between Mak10p and Mak31p (Uetz et al., 2000), indicates that the three proteins form a ternary Mak3p-Mak10p-Mak31p complex. Therefore, we assayed for interactions among AtMAK3, At2g11000, At3g11500, and At2g23930 by yeast two-hybrid analysis. When AtMAK3 fused to the GAL4 DNA binding domain was coexpressed with a GAL4 activation domain fused to At2g11000 (the candidate Arabidopsis MAK10 protein), yeast cells grew robustly in medium lacking His and adenine and turned blue in the LacZ test, indicating that AtMAK3 and At2g11000 could interact (Figure 10A). No interaction was observed between At2g11000 and At3g11500 (the first Arabidopsis MAK31 candidate) (Figure 10B) or between At2g11000 and At2g23930 (the second Arabidopsis MAK31 candidate) (data not shown). Moreover, no interaction was detected between AtMAK3 and At3g11500 (Figure 10C) or between AtMAK3 and At2g23930 (data not shown).
Figure 10.
Yeast Two-Hybrid Analysis of Interactions between AtMAK3 and Candidate MAK10 and MAK31 Proteins from Arabidopsis.
(A) The AtMAK3 protein fused to the GAL4 binding domain (AtMAK3BD) was tested for interaction with At2g11000 fused to the GAL4 activation domain (At2g11000AD).
(B) The At3g11500 protein fused to the GAL4 binding domain (At3g11500BD) was tested for interaction with At2g11000 fused to the GAL4 activation domain (At2g11000AD).
(C) The AtMAK3 protein fused to the GAL4 binding domain (AtMAK3BD) was tested for interaction with At3g11500 fused to the GAL4 activation domain (At3g11500AD).
To detect interactions, transformed yeast strains were grown on either permissive (1) or selective (2) medium or subjected to the LacZ assay (3). AD, empty vector containing the GAL4 activation domain; BD, empty vector containing the GAL4 binding domain.
In yeast, functional Mak3p, Mak10p, and Mak31p are absolutely required for the acetylation of NatC-type substrates in vivo (Polevoda and Sherman, 2001). To determine whether At2g11000, the putative AtMAK10 protein in Arabidopsis, plays the same role as Mak10p in yeast, the insertion flanking database SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress) was searched for insertions in the gene. A line (Salk_015011) was identified that carries a copy of the 5.2-kb ROK2 T-DNA inserted in the eighth exon of At2g11000 at position +3670 relative to the start codon, leading to complete destabilization of At2g11000 transcripts (data not shown). However, in the mutant plants, photosynthesis was normal (FV/FM, 0.83 ± 0.01 [wild type], 0.83 ± 0.02 [at2g11000-1]; ΦII, 0.77 ± 0.01 [wild type], 0.78 ± 0.02 [at2g11000-1]), and growth rate and pigment composition were comparable to those of the wild type (data not shown), strongly suggesting that the At2g11000 protein is not required for the function of AtMAK3.
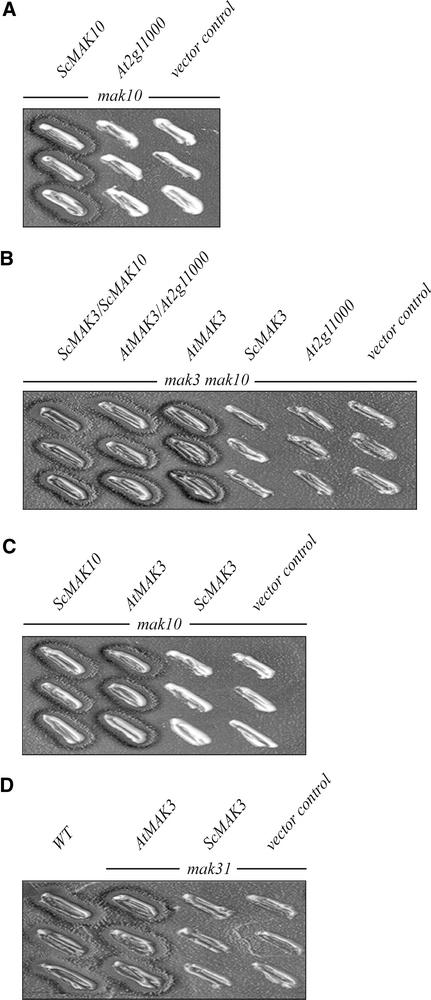
These results can be interpreted in two ways: either the Arabidopsis MAK3 protein does not require a functional MAK10 protein for its function, and/or the At2g11000 protein does not provide the MAK10 function. To determine whether At2g11000 can replace Mak10p in yeast, a mak10 strain (Sommer and Wickner, 1982) was transformed with the At2g11000 cDNA. The virus-killer assay showed that the At2g11000 protein was not able to replace Mak10p (Figure 11A), indicating that At2g11000 is not a functional Mak10p homolog.
Figure 11.
AtMAK3 Can Replace Mak3p, Mak10p, and Mak31p.
(A) The mutant yeast strain mak10 (Sommer and Wickner, 1982) was transformed with the S. cerevisiae MAK10 cDNA (ScMAK10), with the Arabidopsis At2g11000 cDNA (At2g11000), or with the empty vector (vector control). After cytoduction (Ridley et al., 1984), the three yeast strains were tested for the ability to propagate the L-A-HNB M1 dsRNA virus. Only ScMAK10 was able to propagate the virus, as detected by the presence of killing zones surrounding the yeast colonies.
(B) The double mutant yeast strain mak3 mak10 was transformed with both ScMAK3 and ScMAK10 cDNAs (ScMAK3 ScMAK10), with both AtMAK3 and At2g11000 cDNAs (AtMAK3 At2g11000), with the AtMAK3 cDNA (AtMAK3), with the ScMAK3 cDNA (ScMAK3), with the At2g11000 cDNA (At2g11000), or with the empty vector (vector control). Only ScMAK3 ScMAK10, AtMAK3 At2g11000, and AtMAK3 were able to complement the double mutation, enabling propagation of the L-A-HNB M1 dsRNA virus.
(C) The mutant yeast strain mak10 was transformed with the S. cerevisiae MAK10 cDNA (ScMAK10), with the Arabidopsis AtMAK3 cDNA (AtMAK3), with the S. cerevisiae MAK3 cDNA (ScMAK3), or with the empty vector (vector control). ScMAK10 and AtMAK3 were able to complement the mutation.
(D) The mutant yeast strain mak31 was transformed with the Arabidopsis AtMAK3 cDNA (AtMAK3), with the S. cerevisiae MAK3 cDNA (ScMAK3), or with the empty vector (vector control). As a control, an isogenic wild-type strain (WT) was used. Only AtMAK3 was able to complement the mutation.
The double mutant yeast strain mak3 mak10, which lacks both functional Mak3p and Mak10p, was transformed with the AtMAK3 and At2g11000 cDNAs together or with AtMAK3 cDNA alone. In both cases, the wild-type behavior in (1) propagation of the cytoplasmic dsRNA virus L-A (Figure 11B) and (2) growth on the nonfermentable YPG medium (data not shown) was restored. Additionally, AtMAK3 cDNA also complemented mak10 and mak31 single mutants (Figures 11C and 11D). The same mutant yeast strains were transformed with the ScMAK3 cDNA cloned into the same centromeric vector used for AtMAK3 cDNA under the control of the same ADH1 promoter (see Methods), but no wild-type phenotype could be obtained. Because both ScMAK3 and AtMAK3 cDNAs were able to express functional proteins, as shown by complementation of mak3, the data suggest that the AtMAK3 protein alone can replace the entire NatC complex. This particular functionality of the AtMAK3 protein is not reflected in the presence of evident additional domains related to Mak10p or Mak31p in its sequence. In fact, compared with yeast Mak3p, AtMAK3 contains at its N and C termini only 13 and 9 extra amino acids, respectively.
Effects of atmak3-1 on Mitochondrial Respiration and Nuclear Gene Expression
Yeast cells that lack NatC activity are not able to grow on the nonfermentable YPG medium, suggesting that their mitochondria are defective (Tercero et al., 1993). To clarify whether the atmak3-1 mutant has a similar defect, mitochondrial respiration was investigated in Arabidopsis by measuring oxygen uptake. Quantification of oxygen uptake in wild-type and mutant cells cultivated in the dark revealed no differences between the two genotypes (data not shown). However, because yeast mak3 mutants are affected only under specific growth conditions (37°C and nonfermentable carbon source), we cannot exclude the possibility that a mitochondrial defect exists in atmak3-1 that is not manifested under the specific conditions investigated here.
Previous studies have shown that the classification of Arabidopsis photosynthetic and other mutants based on physiological and biochemical data is reflected in characteristic patterns of gene expression (Wang et al., 2002; Richly et al., 2003). DNA arrays allow the quantification of gene expression on a large scale. Such an analysis was performed for the atmak3-1 mutant using a set of 3292 nuclear genes spotted on nylon membranes, 81% of which encode chloroplast proteins, and the mRNA expression pattern observed was compared with that of the wild type (Kurth et al., 2002; Richly et al., 2003). Differential gene expression values (atmak3-1 versus wild type) were compared with those obtained upon subjecting Arabidopsis plants to 34 different environmental and/or genetic conditions (Richly et al., 2003). In atmak3-1, 777 of the 3292 genes tested displayed significant differences in behavior relative to wild-type plants. In the mutant, moreover, the fraction of differentially regulated genes encoding nonchloroplast proteins was greater (200 different transcripts, or 26%) than was noted under any of the other 34 genetic or environmental conditions tested, suggesting that the function of AtMAK3 is not restricted to chloroplast processes.
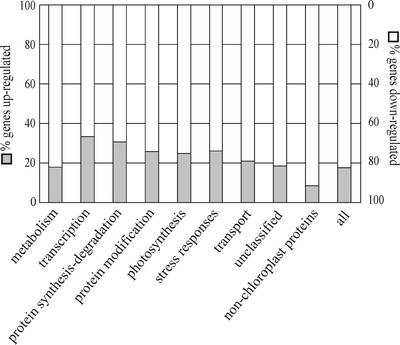
Among the 577 chloroplast protein–coding genes that were expressed differentially with respect to wild-type plants, 121 were upregulated and 456 were downregulated. The differentially expressed genes that encode chloroplast proteins could be grouped into seven major functional categories: metabolism, transcription, protein synthesis and degradation, protein modification, photosynthesis, stress response, and transport (Figure 12; see also supplemental data online). Less than others, genes for transcription and protein synthesis/degradation were downregulated, indicating that the plant may be able to monitor the change in plastid protein synthesis/accumulation caused by the atmak3-1 mutation and reacts—against the general trend in gene expression in this genotype—by upregulating appropriate nuclear genes. This finding supports the hypothesis that regulatory networks operate in plant cells that can sense the levels of key proteins, metabolites, and/or redox state in the chloroplast and activate gene expression in the nucleus (Surpin et al., 2002; Pfannschmidt, 2003).
Figure 12.
Effects of the atmak3-1 Mutation on the Accumulation of Nuclear Transcripts of Genes for Chloroplast Proteins.
Total RNA isolated from wild-type and atmak3-1 mutant plants was used to probe a nylon filter DNA array carrying 3292 gene sequence tags. In atmak3-1, 777 of the 3292 genes tested were significantly differentially expressed compared with wild-type plants. The 577 differentially expressed chloroplast protein–coding genes in atmak3-1 were grouped into seven major functional categories. A complete list is available in the supplemental data online.
The N-terminal protein sequences encoded by the 50 most upregulated or downregulated genes in atmak3-1 were compared with those of all proteins represented on the 3292–gene sequence tag array. No particular consensus motif at the protein level was identified for the most differentially expressed genes (data not shown). Moreover, the N-terminal sequences of proteins encoded by all of the 777 differentially regulated genes were compared with the sequences of proteins known to be acetylated by the NatC complex (Polevoda and Sherman, 2003). The At4g25250 protein, the transcript level of which was increased slightly in atmak3-1 (see supplemental data online), is annotated as a putative protein and possesses a N terminus (MLRFV) identical to that of the viral L-A Gag protein.
DISCUSSION
The mutant atmak3-1 is affected in the synthesis of the PSII core proteins D1 and CP47. This defect is associated with a decrease in the levels of most of the thylakoid multiprotein complexes; consequently, photosynthetic electron flow is disturbed. The reduction in both leaf pigmentation and growth rate can be related to the photosynthetic lesion. This phenotype predominates in young atmak3-1 leaves, indicating that AtMAK3 function is limiting in fast-growing tissues.
DNA array–based mRNA expression analysis indicates that putative extraplastid functions also are altered by the atmak3-1 mutation. Therefore, it was not surprising that the AtMAK3 gene encodes a nonchloroplast protein. AtMAK3 is located in the cytoplasm and exhibits high sequence identity with the catalytic subunit of eukaryotic NATs. Moreover, AtMAK3 can functionally replace the catalytic Mak3p subunit of the NatC complex in yeast, demonstrating that it is a genuine N-acetyltransferase. Because substrates of the yeast NatC complex are quite rare (Arnold et al., 1999; Polevoda et al., 1999), and because AtMAK3 is able to rescue the two known defects associated with the yeast mak3 mutation, propagation of killer virus particles and growth on nonfermentable carbon sources, it appears that the functional specificity of AtMAK3 is remarkably conserved. Nonetheless, significant differences between the yeast and Arabidopsis N-acetyltransferases are evident. In fact, AtMAK3 can replace ScMAK3, ScMAK10, and ScMAK31, and knockout of the putative plant AtMAK10 does not produce a phenotype. Thus, although two-hybrid analysis indicates that AtMAK3 and AtMAK10 can interact, the available results indicate that AtMAK3 does not function as an essential member of a multiprotein complex corresponding to the Mak3p-Mak10p-Mak31p complex of yeast. Whether AtMAK3 acts alone or as part of a different multiprotein complex remains to be determined. If an interaction between AtMAK3 and AtMAK10 occurs (as suggested by the two-hybrid data), this interaction is definitely not required for the effect of N-terminal acetylation on photosynthesis.
The Arabidopsis atmak3-1 mutant was identified on the basis of its photosynthetic phenotype. This raises questions regarding the mechanism(s) by which the photosynthetic process is affected by a cytoplasmic N-acetyltransferase. It is noteworthy in this context that Mak3p, the orthologous yeast protein, also is required for function related to organelles. Thus, a simple explanation may involve the cotranslational modification of mitochondrial and/or chloroplast precursor proteins. Mitochondrial and chloroplast transit peptides are cleaved after import into the organelle. Thus, acetylation of protein N termini may be relevant for the stability and/or import competence of organellar precursor proteins and thus, indirectly, for organelle function. Two observations support this hypothesis. (1) Post-translational modification of preproteins can modulate their organelle import competence: phosphorylation of chloroplast transit peptides in the cytoplasm is necessary for the efficient import of precursor proteins into the chloroplast (Waegemann and Soll, 1996). (2) N-acetylation of the noncleavable mitochondrial targeting signal of rat chaperonin 10 increases N-terminal helix stability, thus facilitating mitochondrial import (Jarvis et al., 1995).
In S. cerevisiae, 10 years after the discovery of NATs, the substrates of the NatC complex that are instrumental in conferring the mutant growth phenotype on nonfermentable carbon sources still are unknown. Although N-terminal acetylation is a common protein modification, few substrates of NatC are known, and none was encountered among 55 abundant proteins (Polevoda et al., 1999) or among 68 ribosomal proteins (Arnold et al., 1999). The prediction of N-terminal acetylated substrates based on their amino acid sequences is difficult because no simple consensus sequence for N-acetylation has been defined (Polevoda and Sherman, 2000). Three mitochondrial proteins, however, have N-terminal sequences that are sufficient, in model experiments, to determine Mak3p-specific acetylation (Tercero et al., 1993). The situation is complicated because only a subset of acetylated proteins appears to require N-terminal acetylation for stability or enzymatic activity. Therefore, the identification of the proteins responsible for the photosynthetic phenotype in the Arabidopsis atmak3-1 mutant, or for the mitochondrial defect in the mak3, mak10, and mak31 mutants of yeast, will require extensive efforts. If the unknown substrates are in fact precursors of proteins targeted to mitochondria and chloroplasts, respectively, it might be difficult to identify them even using proteomics approaches, because the processed proteins would not have altered mobility on two-dimensional gels. On the other hand, the function of only a single protein, or very few proteins, requiring N-acetylation to be transported efficiently into the organelle may be sufficient to explain the phenotype(s) observed in mak3 mutants.
The viability of both the yeast mak3 and atmak3 mutants suggests a subtle role for protein acetylation, a modification that is not an absolute requirement for most proteins. The finding that Mak3p from the lower eukaryote yeast can be replaced by its higher plant homolog demonstrates that this type of cotranslational modification may be important, because it has been conserved during evolution. New substrates might have been acquired during the evolution of the plant lineage: AtMAK3, for example, probably acts on precursor protein(s) targeted to an organelle that does not exist in yeast. Because the functional specificity of the AtMAK3 proteins appears to be conserved (see above) and mitochondrial and chloroplast transit peptides show relatively wide variability in their primary sequence (Emanuelsson and von Heijne, 2001), it is plausible that evolutionary diversification of N-terminal protein sequences, rather than evolutionary diversification of the intrinsic specificity of N-acetyltransferases, is the driving force that alters the spectrum of N-acetylated proteins. In this context, to increase their stability and/or targeting competence, certain precursor proteins of mitochondrial or chloroplast organelles may have recruited N-terminal acetylation to extend the range of their molecular structures beyond the limits imposed by the size and amino acid composition of their transit peptides.
METHODS
Plant Lines, Propagation, and Growth Measurement
The atmak3-1 mutant was identified among a collection of AC106 T-DNA-mutagenized Arabidopsis thaliana (ecotype Columbia) lines on the basis of a decrease in the effective quantum yield of photosystem II (PSII). The insertion mutant for the At2g11000 gene was identified in the Salk collection (http://signal.salk.edu/), consisting of flank-tagged ROK2 T-DNA lines (ecotype Columbia), by searching the insertion flanking database SIGnAL (http://signal.salk.edu/cgi-bin/tdnaexpress).
Mutant and wild-type plants were grown on Minitray soil (Gebr. Patzer, Sinntal-Jossa, Germany) in an air-conditioned greenhouse (day period of 16 h at 20°C and PFD of 80 μmol·m−2·s−1; night period of 8 h at 15°C). Fertilization with Osmocote Plus (15% N, 11% P2O5, 13% K2O, and 2% MgO; Scotts Deutschland, Nordhorn, Germany) was performed according to the manufacturer's instructions. Plant growth was measured as described by Leister et al. (1999).
Chlorophyll Fluorescence and Pigment Analysis
Mutants that show alterations in ΦII, the effective quantum yield of PSII, were identified using an automatic pulse amplitude modulation fluorometer system as described by Varotto et al. (2000a). In vivo chlorophyll a fluorescence of single leaves was measured using the PAM 101/103 fluorometer (Walz, Effeltrich, Germany). Pulses (800 ms) of white light (6000 μmol·m−2·s−1) were used to determine the maximum fluorescence (FM) and the ratio (FM − F0)/FM = FV/FM. A 15-min illumination with actinic light (80 μmol·m−2·s−1) was supplied to drive electron transport between PSII and PSI before measuring ΦII. Pigment analysis was performed by reverse-phase HPLC as described by Färber et al. (1997).
Whole-Cell Mitochondrial Respiration Measurement
Arabidopsis cell lines from wild-type and mutant leaves generated according to Mathur and Koncz (1998) were cultivated in the dark at 26°C and 30% RH with gentle shaking (90 rpm). The culture medium was as described by Werhahn et al. (2001). Mitochondrial respiration was measured on intact cells in culture medium using a Clark-type oxygen electrode cuvette (Hansatech Oxygraph, Laborbedarf Helmut Sauer, Reutlingen, Germany) at 26°C. Cytochrome pathway capacity was defined as the portion of the oxygen consumption inhibited by 1 mM KCN, whereas alternative pathway capacity was the fraction of oxygen consumption inhibited by 2 mM salicylhydroxamic acid in the presence of 1 mM KCN (Maxwell et al., 2002). Residual respiration (in the presence of both salicylhydroxamic acid and KCN) was subtracted from all measurements.
Two-Dimensional PAGE and Protein Gel Blot Analyses
Leaves from 4-week-old plants were harvested in the middle of the light period, and thylakoids were prepared from mesophyll chloroplasts as described (Bassi et al., 1985). For two-dimensional gel analyses, thylakoid membrane samples equivalent to 100 mg of fresh leaf weight were first fractionated on nondenaturing lithium dodecyl sulfate polyacrylamide gradient gels (4 to 12% acrylamide) according to Peter and Thornber (1991) using lithium dodecyl sulfate instead of lauryl β-d-iminopropionidate in the upper electrophoresis buffer. Fractionation in the second dimension was performed on a denaturing SDS-PAGE gradient gel (10 to 16% acrylamide) as described by Schägger and von Jagow (1987). Densitometric analyses of protein gels after Coomassie Brilliant Blue R250 staining (Roth, Karlsruhe, Germany) were performed with Lumi Analyst 3.0 (Boehringer Mannheim/Roche, Basel, Switzerland). For immunoblot analyses, thylakoid proteins separated by one-dimensional SDS-PAGE were transferred to Immobilon-P membranes (Millipore, Eschborn, Germany) and incubated with antibodies specific for PSI-A/B, PSII-D1, and light-harvesting complex of PSII subunits. Signals were detected using the Enhanced Chemiluminescence Western Blotting Kit (Amersham Biosciences, Sunnyvale, CA).
In Vivo Translation Assay
Segments (100 mg) of 4-week-old wild-type and mutant leaves were harvested and vacuum-infiltrated (vacuum strength, 400 mm Hg) with 50 μL of reaction medium (1 mM KH2PO4, pH 6.3, 0.1% [w/v] Tween 20, 300 μCi of 35S-Met [specific activity > 1000 Ci/mmol; Amersham Buchler, Braunschweig, Germany]) for 5 s. After vacuum infiltration, leaves were illuminated (120 μmol·m−2·s−1) for different times (10 and 30 min) at 25°C, washed twice with 500 μL of wash buffer (20 mM Na2CO3 and 10 mM DTT), disrupted by grinding in the presence of 60 μL of 100 mM Na2CO3 buffer, and clarified by centrifugation (4°C for 15 min at 15,000 rpm; Sigma 2MK, Christ, Osterode, Germany). The green pellet was solubilized with 2% dodecyl β-d-maltoside, fractionated by blue-native PAGE (Schägger, 1994) in the first dimension and SDS-PAGE (Schägger and von Jagow, 1987) in the second dimension, and then transferred to Immobilon-P membranes (Millipore). Radioactive signals were detected using Fuji Bio imaging plates (Fuji Bio imaging analyzer, BAS 2000 software package, TINA software package version 2.08β; Fuji Photo Film, Tokyo, Japan).
Nucleic Acid and Polysome Analyses
Arabidopsis DNA was isolated as described (Liu et al., 1995), and regions flanking the AC106 T-DNA insertion were identified according to Pesaresi et al. (2001).
Total RNA was extracted from fresh tissue using the RNeasy Plant System (Qiagen, Hilden, Germany), and RNA gel blot analyses were performed according to Sambrook et al. (1989). Probes complementary to the following genes from the chloroplast genome were used: psbA (positions 674 to 1129), psbB (72521 to 72880), psbC (33781 to 33800), psaA (41329 to 41782), psaB (39079 to 39509), atpA (10480 to 10951), and atpB (53287 to 53306). For the AtMAK3 gene, a full-length cDNA probe was amplified using the primers atmak3-22s (5′-CGGGATCTTGATCACTTCTTG-3′) and atmak3-1518as (5′-CGTTCATGGCCTTAAGGTAC-3′), whereas for the APT1 gene, RNA gel blot analysis was performed as described previously (Varotto et al., 2000b). For quantitative reverse transcriptase–mediated PCR of AtMAK3 expression, first-strand cDNA was synthesized using the SuperScript Preamplification System (Invitrogen, Karlsruhe, Germany). Primers specific for AtMAK3 (atmak3-137s, 5′-CTTACCGGTACTTCGTCTAC-3′; atmak3-1494as, 5′-GGCCTAGGAAAGGTCTCATA-3′) and APT1 (Moffatt et al., 1994) were used to amplify cDNA by PCR for 20 cycles, and the PCR products were separated on a 4.5% (w/v) polyacrylamide gel. Signals were quantified using a PhosphorImager (Storm 860; Molecular Dynamics, Sunnyvale, CA) and the program Image Quant for Macintosh (version 1.2; Molecular Dynamics).
Polysomes were isolated from leaf tissue of wild-type and mutant plants as described (Barkan, 1998). RNA was fractionated on 1.5% agarose-formaldehyde gels, blotted onto nylon membranes, and hybridized with 32P-labeled probes specific for psbA, psbB, and psbC.
Expression Profiling
The 3292–gene sequence tag nylon array, including 2661 nuclear chloroplast genes and 631 genes coding for nonchloroplast proteins, has been described (Richly et al., 2003). Total RNA was isolated from 5 g of leaf material obtained from plants at the eight-leaf rosette stage (∼4-week-old plants). Three independent experiments were performed with different filters and independent cDNA probes derived from plant material corresponding to pools of at least 50 individuals of wild-type or atmak3-1 plants, thus minimizing variation between individual plants, filters, or probes. cDNA probes were synthesized using as a primer a mixture of oligonucleotides matching the 3292 genes in the antisense orientation and hybridized to the gene sequence tag array as described (Kurth et al., 2002; Richly et al., 2003). Images were read using the Storm PhosphorImager (Molecular Dynamics). Hybridization images were imported into the ArrayVision program (version 6; Imaging Research, St. Catharines, Ontario, Canada), where artifacts were removed, background correction was performed, and resulting values were normalized with reference to the intensity of all spots on the array (Kurth et al., 2002). In the next step, those data were imported into the ArrayStat program (version 1.0, revision 2.0; Imaging Research) and a z test (nominal α set to 0.05) was performed to identify statistically significant differential expression values. Only differential expression values that fulfill the criteria of the z test are listed in the supplemental data online. Original data are available at http://www.mpiz-koeln.mpg.de/~leister/atmak3-1.html.
Intracellular Protein Localization
The red fluorescent protein from the reef coral Discosoma (dsRED) was used as a reporter to determine the intracellular localization of AtMAK3 (Jach et al., 2001). The AtMAK3 cDNA was fused 5′ to the dsRED gene, and this construct was used to transfect protoplasts. Mesophyll protoplasts were isolated from 6-week-old tobacco plants (Nicotiana tabacum cv Petit Havana SR1) (Maliga et al., 1975) according to the protocol of Negrutiu et al. (1987). For transient gene expression assays, 3.3 × 105 freshly isolated SR1 mesophyll protoplasts were transfected with 10 μg of plasmid DNA by polyethylene glycol–mediated DNA uptake (Walden et al., 1994). Protoplasts were cultured for 2 days at 26°C in the dark and in the presence of auxin (5 μM) and cytokinin (1 μM). The transfected protoplasts were analyzed using the Zeiss Axiophot fluorescence microscope (Jena, Germany) equipped with a filter set purchased from AF Analysentechnik (Tübingen, Germany). Photographs were taken using a video imaging system mounted on the microscope that consisted of a Hitachi charge-coupled device video camera (Tokyo, Japan) operated by the DISKUS software package (Technisches Büro Hilgers, Königswinter, Germany).
Complementation of the atmak3-1 Mutant
The AtMAK3 genomic DNA together with 1 kb of its natural promoter was ligated into the plant expression vector pP001-VS (constructed by Bernd Reiss, Max-Planck-Institute for Plant Breeding Research). Flowers of atmak3-1 mutant plants were transformed according to Clough and Bent (1998). Plants were transferred to the greenhouse, and seeds were collected after 3 weeks. Fifteen independent transgenic plants were selected on the basis of their resistance to the herbicide Basta. Successful complementation was confirmed by measurements of chlorophyll fluorescence, leaf pigmentation, and growth.
Media, Plasmids, and Yeast Strains
YPAD, YPG, SD (and appropriate dropout media), and 4.7MB media have been described previously (Ridley et al., 1984; Sherman, 1991). The yeast two-hybrid assay was performed using the yeast strain AH109 supplied by Clontech (Palo Alto, CA) (James et al., 1996). The pAS2-1 vector (Harper et al., 1993) carrying the GAL4 DNA binding domain was used to express the bait proteins, and the pGADT7 vector (Harper et al., 1993) carrying the GAL4 activation domain was used to express the prey proteins.
Saccharomyces cerevisiae strains 2967 (MATa leu2 ura3 mak10-1) (Lee and Wickner, 1992), ATCC#4005470 (MATa ura3 leu2 his3 met15 mak3::G418), ATCC#4003501 (MATa ura3 leu2 his3 met15 mak31::G418) (purchased from the American Type Culture Collection), and NY2 (MATa ura3 leu2 his3 met15 mak3::G418 mak10::HIS3) were used for AtMAK3 and At2g11000 complementation tests. Strains 3168 (MATα kar1 arg1 thr1 L-A-HNB M1) and 5X47 (MATa/MATα his1/+ trp1/+ ura3/+ [KIL-o]) (Ridley et al., 1984) were used for the cytoduction and killer test (see below). The wild-type strain used in the mak31 killer test was BY4741 (MATa ura3 leu2 his3 met15) from the American Type Culture Collection.
For the expression of AtMAK3 and ScMAK3 cDNAs, the vector pH124 was used. pH124 (kindly provided by H.K. Edskes, National Institutes of Health, Bethesda, MD) is a centromeric vector with the LEU2 marker gene and the ADH1 promoter upstream of a multiple cloning site (MCS). The vector was created by cloning the ADH1-MCS fragment into PvuII-digested pRS315 (Sikorski and Hieter, 1989) in the reverse orientation with respect to the LEU2 marker. For the expression of ScMAK10 and At2g11000 cDNAs, the vector pH270 (provided by H.K. Edskes) was used. pH270 is a centromeric plasmid with the URA3 marker gene and the ADH1 promoter upstream of a MCS. The vector was constructed by inserting the ADH1-MCS cassette between the two PvuII sites in pRS316 (Sikorski and Hieter, 1989) in the same orientation as the URA3 marker. Yeast transformation was performed using a lithium acetate procedure as described by Gietz et al. (1992). Cytoduction was performed as described by Ridley et al. (1984). Cytoductants were tested for the killer phenotype as reported by Ridley et al. (1984).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact D. Leister, leister@mpiz-koeln.mpg.de.
Accession Number
The GenBank accession number for the chloroplast genomic DNA of A. thaliana (ecotype Columbia) is AP000423.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutant. We are grateful to Tinka Eneva (Max-Planck-Institut für Züchtungsforschung) for in vitro cell culture, to Erik Richly (Max-Planck-Institut für Züchtungsforschung) for support with bioinformatics, and to Hans-Peter Braun (University of Hannover, Germany) for support with respiration measurements. Grateful acknowledgment is extended to Paul Hardy for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012377.
Footnotes
Online version contains Web-only data.
References
- Arnold, R.J., Polevoda, B., Reilly, J.P., and Sherman, F. (1999). The action of N-terminal acetyltransferases on yeast ribosomal proteins. J. Biol. Chem. 274, 37035–37040. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297, 38–57. [Google Scholar]
- Bassi, R., dal Belin Peruffo, A., Barbato, R., and Ghisi, R. (1985). Differences in chlorophyll-protein complexes and composition of polypeptides between thylakoids from bundle sheaths and mesophyll cells in maize. Eur. J. Biochem. 146, 589–595. [DOI] [PubMed] [Google Scholar]
- Choquet, Y., and Wollman, F.A. (2002). Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 529, 39–42. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., and von Heijne, G. (2001). Prediction of organellar targeting signals. Biochim. Biophys. Acta 1541, 114–119. [DOI] [PubMed] [Google Scholar]
- Färber, A., Young, A.J., Ruban, A.V., Horton, P., and Jahns, P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants: The relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching. Plant Physiol. 115, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, P.E., and Mullet, J.E. (1989). Translation and stability of proteins encoded by the plastid psbA and psbB genes are regulated by a nuclear gene during light-induced chloroplast development in barley. J. Biol. Chem. 264, 7236–7243. [PubMed] [Google Scholar]
- Garrels, J.I., McLaughlin, C.S., Warner, J.R., Futcher, B., Latter, G.I., Kobayashi, R., Schwender, B., Volpe, T., Anderson, D.S., Mesquita-Fuentes, R., and Payne, W.E. (1997). Proteome studies of Saccharomyces cerevisiae: Identification and characterization of abundant proteins. Electrophoresis 18, 1347–1360. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Sciestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wie, N., Keyomarsi, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hoog, J.O., Weis, M., Zeppezaue, M., Jornvall, H., and von Bahr-Lindstrom, H. (1987). Expression in Escherichia coli of active human alcohol dehydrogenase lacking N-terminal acetylation. Biosci. Rep. 7, 969–974. [DOI] [PubMed] [Google Scholar]
- Jach, G., Binot, E., Frings, S., Luxa, K., and Schell, J. (2001). Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J. 28, 483–491. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, J.A., Ryan, M.T., Hoogenraad, N.J., Craik, D.J., and Hoj, P.B. (1995). Solution structure of the acetylated and noncleavable mitochondrial targeting signal of rat chaperonin 10. J. Biol. Chem. 270, 1323–1331. [DOI] [PubMed] [Google Scholar]
- Kim, J., Klein, P.G., and Mullet, J.E. (1994). Vir-115 gene product is required to stabilize D1 translation intermediates in chloroplasts. Plant Mol. Biol. 25, 459–467. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., Saeki, Y., Yokosawa, H., Polevoda, B., Sherman, F., and Hirano, H. (2003). N-terminal modifications of the 19S regulatory particle subunits of the yeast proteasome. Arch. Biochem. Biophys. 409, 341–348. [DOI] [PubMed] [Google Scholar]
- Kimura, Y., Takaoka, M., Tanaka, S., Sassa, H., Tanaka, K., Polevoda, B., Sherman, F., and Hirano, H. (2000). Nα-acetylation and proteolytic activity of the yeast 20S proteasome. J. Biol. Chem. 275, 4635–4639. [DOI] [PubMed] [Google Scholar]
- Kurth, J., Varotto, C., Pesaresi, P., Biehl, A., Richly, E., Salamini, F., and Leister, D. (2002). Gene-sequence-tag expression analyses of 1,800 genes related to chloroplast functions. Planta 215, 101–109. [DOI] [PubMed] [Google Scholar]
- Lee, F.J., Lin, L.W., and Smith, J.A. (1989). Nα-acetyltransferase deficiency alters protein synthesis in Saccharomyces cerevisiae. FEBS Lett. 256, 139–142. [DOI] [PubMed] [Google Scholar]
- Lee, Y.J., and Wickner, R.B. (1992). MAK10, a glucose-repressible gene necessary for replication of a dsRNA virus of Saccharomyces cerevisiae, has T-cell receptor alpha-subunit motifs. Genetics 132, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister, D., Varotto, C., Pesaresi, P., Niwergall, A., and Salamini, F. (1999). Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol. Biochem. 37, 671–678. [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Maliga, P., Sz-Bresnovits, A., Marton, L., and Joo, F. (1975). Non-mendelian streptomycin-resistant tobacco mutant with altered chloroplasts and mitochondria. Nature 255, 401–402. [DOI] [PubMed] [Google Scholar]
- Manning, L.R., and Manning, J.M. (2001). The acetylation state of human fetal hemoglobin modulates the strength of its subunit interactions: Long-range effects and implications for histone interactions in the nucleosome. Biochemistry 40, 1635–1639. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Koncz, C. (1998). Establishment and maintenance of cell suspension cultures. In Methods in Molecular Biology, Vol. 82: Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 31–34. [DOI] [PubMed]
- Maxwell, D.P., Nickels, R., and McIntosh, L. (2002). Evidence of mitochondrial involvement in the transduction of signals required for the induction of genes associated with pathogen attack and senescence. Plant J. 29, 269–279. [DOI] [PubMed] [Google Scholar]
- Moffatt, B.A., McWhinnie, E.A., Agarwal, S.K., and Schaff, D.A. (1994). The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143, 211–216. [DOI] [PubMed] [Google Scholar]
- Mullen, J.R., Kayne, P.S., Moerschell, R.P., Tsunasawa, S., Gribskov, M., Colavito-Shepanski, M., Grunstein, M., Sherman, F., and Sternglanz, R. (1989). Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu, I., Shillito, R., Potrykus, I., Biasini, G., and Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. Plant Mol. Biol. 8, 363–373. [DOI] [PubMed] [Google Scholar]
- Neuwald, A.F., and Landsman, D. (1997). GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155. [DOI] [PubMed] [Google Scholar]
- Perrot, M., Sagliocco, F., Mini, T., Monribot, C., Schneider, U., Shevchenko, A., Mann, M., Jeno, P., and Boucherie, H. (1999). Two-dimensional gel protein database of Saccharomyces cerevisiae (update 1999). Electrophoresis 20, 2280–2298. [DOI] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., and Thornber, J.P. (1991). Biochemical composition and organisation of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266, 16745–16754. [PubMed] [Google Scholar]
- Pfannschmidt, T. (2003). Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 8, 33–41. [DOI] [PubMed] [Google Scholar]
- Plucken, H., Müller, B., Grohmann, D., Westhoff, P., and Eichacker, L.A. (2002). The HCF136 protein is essential for assembly of the photosystem II reaction center in Arabidopsis thaliana. FEBS Lett. 532, 85–90. [DOI] [PubMed] [Google Scholar]
- Polevoda, B., Norbeck, J., Takakura, H., Blomberg, A., and Sherman, F. (1999). Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 18, 6155–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda, B., and Sherman, F. (2000). Nα-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 275, 36479–36482. [DOI] [PubMed] [Google Scholar]
- Polevoda, B., and Sherman, F. (2001). NatC Nα-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J. Biol. Chem. 276, 20154–20159. [DOI] [PubMed] [Google Scholar]
- Polevoda, B., and Sherman, F. (2002). The diversity of acetylated proteins. Genome Biol. 3, REVIEWS0006. [DOI] [PMC free article] [PubMed]
- Polevoda, B., and Sherman, F. (2003). N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J. Mol. Biol. 325, 595–622. [DOI] [PubMed] [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Co-variation in the nuclear chloroplast transcriptome reveals a regulatory master switch. EMBO Rep. 4, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley, S.P., Sommer, S.S., and Wickner, R.B. (1984). Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Ryan, M.T., Naylor, D.J., Hoogenraad, N.J., and Hoj, P.B. (1995). Affinity purification, overexpression, and characterization of chaperonin 10 homologues synthesized with and without N-terminal acetylation. J. Biol. Chem. 270, 22037–22043. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schägger, H. (1994). Electrophoretic isolation of membrane proteins from acrylamide gels. Appl. Biochem. Biotechnol. 48, 185–203. [Google Scholar]
- Schägger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Séraphin, B. (1995). Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Siddig, M.A., Kinsey, J.A., Fincham, J.R., and Keighren, M. (1980). Frameshift mutations affecting the N-terminal sequence of Neurospora NADP-specific glutamate dehydrogenase. J. Mol. Biol. 137, 125–135. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, S.S., and Wickner, R.B. (1982). Yeast L dsRNA consists of at least three distinct RNAs: Evidence that the non-Mendelian genes [HOK], [NEX] and [EXL] are on one of these dsRNAs. Cell 31, 429–441. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Larkin, R.M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14 (suppl.), S327.–S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero, J.C., Dinman, J.D., and Wickner, R.B. (1993). Yeast MAK3 N-acetyltransferase recognizes the N-terminal four amino acids of the major coat protein (gag) of the L-A double-stranded RNA virus. J. Bacteriol. 175, 3192–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero, J.C., Riles, L.E., and Wickner, R.B. (1992). Localized mutagenesis and evidence for post-transcriptional regulation of MAK3: A putative N-acetyltransferase required for double-stranded RNA virus propagation in Saccharomyces cerevisiae. J. Biol. Chem. 267, 20270–20276. [PubMed] [Google Scholar]
- Tercero, J.C., and Wickner, R.B. (1992). MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267, 20277–20281. [PubMed] [Google Scholar]
- Toh-e, A., and Sahashi, Y. (1985). The PET18 locus of Saccharomyces cerevisiae: A complex locus containing multiple genes. Yeast 1, 159–172. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Urbancikova, M., and Hitchcock-DeGregori, S.E. (1994). Requirement of amino-terminal modification for striated muscle α-tropomyosin function. J. Biol. Chem. 269, 24310–24315. [PubMed] [Google Scholar]
- Varotto, C., Pesaresi, P., Maiwald, D., Kurth, J., Salamini, F., and Leister, D. (2000. a). Identification of photosynthetic mutants of Arabidopsis by automatic screening for altered effective quantum yield of photosystem 2. Photosynthetica 38, 497–504. [Google Scholar]
- Varotto, C., Pesaresi, P., Meurer, J., Oelmüller, R., Steiner-Lange, S., Salamini, F., and Leister, D. (2000. b). Disruption of the Arabidopsis photosystem I gene psae1 affects photosynthesis and impairs growth. Plant J. 22, 115–124. [DOI] [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1996). Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem. 271, 6545–6554. [DOI] [PubMed] [Google Scholar]
- Walden, R., Fritze, K., Hayashi, H., Miklashevichs, E., Harling, H., and Schell, J. (1994). Activation tagging: A means of isolating genes implicated as playing a role in plant growth and development. Plant Mol. Biol. 26, 1521–1528. [DOI] [PubMed] [Google Scholar]
- Wang, H., Ma, L., Habashi, J., Li, J., Zhao, H., and Deng, X.W. (2002). Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 32, 723–733. [DOI] [PubMed] [Google Scholar]
- Werhahn, W., Niemeyer, A., Jansch, L., Kruft, V.V., Schmitz, U.K., and Braun, H.P. (2001). Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis: Identification of multiple forms of TOM20. Plant Physiol. 125, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.