Abstract
Although calcium is a critical component in the signal transduction pathways that lead to stress gene expression in higher plants, little is known about the molecular mechanism underlying calcium function. It is believed that cellular calcium changes are perceived by sensor molecules, including calcium binding proteins. The calcineurin B–like (CBL) protein family represents a unique group of calcium sensors in plants. A member of the family, CBL1, is highly inducible by multiple stress signals, implicating CBL1 in stress response pathways. When the CBL1 protein level was increased in transgenic Arabidopsis plants, it altered the stress response pathways in these plants. Although drought-induced gene expression was enhanced, gene induction by cold was inhibited. In addition, CBL1-overexpressing plants showed enhanced tolerance to salt and drought but reduced tolerance to freezing. By contrast, cbl1 null mutant plants showed enhanced cold induction and reduced drought induction of stress genes. The mutant plants displayed less tolerance to salt and drought but enhanced tolerance to freezing. These studies suggest that CBL1 functions as a positive regulator of salt and drought responses and a negative regulator of cold response in plants.
INTRODUCTION
Plants are constantly bombarded with environmental signals, some of which cause stress and limit plant growth and development. In response to the stress signals, plants activate a number of defense responses that increase tolerance to the stress conditions. This inducible adaptation or acclimation process has evolved throughout the plant kingdom and is critical for the survival of all plants. To understand the molecular and cellular mechanisms underlying stress acclimation and to use such mechanisms in crop production, recent studies have made significant progress in dissecting the biochemical and physiological pathways involved in stress responses. For example, a number of metabolic pathways are altered for osmotic adjustment in response to drought or hyperosmotic stress caused by high salt or low temperature (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong and Zhu, 2002). Furthermore, a large array of genes is activated by these stress conditions, so that a number of proteins are produced to join the pathways that lead to stress tolerance (Ingram and Bartel, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1997). These genes often are referred to as “stress genes.” Because dehydration represents a common stress challenge to plant cells under drought, salt, and cold conditions, a significant overlap has been identified in the repertoire of genes activated by each of these conditions. Many drought-responsive genes also are responsive to salt or cold (Shinozaki and Yamaguchi-Shinozaki, 2000; Xiong et al., 2002). The gene RD29A (also referred to as COR78 or LTI78) is an example of such a common stress-inducible gene (Shinozaki and Yamaguchi-Shinozaki, 2000). In addition, both drought and salt conditions increase the level of the stress hormone abscisic acid (ABA), which in turn also can activate RD29A (and other stress genes).
Molecular dissection of the RD29A promoter region has identified separate cis-acting elements: DRE (or CRT), which is responsive to drought, salt, and cold; and ABRE, which is responsive to ABA (Yamaguchi-Shinozaki and Shinozaki, 1994; Thomashow, 1999). Corresponding transcriptional factors that bind to each of these cis-acting elements also have been isolated (Stockinger et al., 1997; Liu et al., 1998; Choi et al., 2000; Uno et al., 2000). DREB (or CBF) proteins bind to DRE/CRT elements and AREB/ABF proteins bind to ABRE. The DREB proteins are classified into two groups, DREB1 and DREB2, which are induced specifically by cold and drought, and they are thought to play a role in cold- and drought-induced gene expression, respectively. Indeed, overexpression of DREB/CBF proteins activates the expression of many stress genes, including RD29A, and increases stress tolerance in transgenic plants (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Seki et al., 2001). These results provide a molecular basis for the regulation of RD29A gene expression by abiotic stress signals and ABA. Together with stress gene expression in ABA-deficient mutants, these studies also address the relationship between stress and ABA in stress gene expression (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Knight and Knight, 2001). In particular, drought/salt may trigger both ABA-dependent and ABA-independent pathways leading to the activation of stress genes, whereas cold signal may be transmitted by an ABA-independent pathway because cold does not appear to induce significant levels of ABA accumulation in plants (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000).
Molecular analysis of the RD29A/COR78 promoter has made a critical contribution to our understanding of the mechanism of stress gene expression. However, little is known about the molecular pathways that link stress and ABA signals to gene expression in the nucleus. A number of studies implicate Ca2+ as a second messenger in abiotic stress and ABA responses (Sanders et al., 1999, 2002; Knight and Knight, 2000; Rudd and Franklin-Tong, 2001). First, increased cellular Ca2+ is a rapid response to ABA, cold, drought, and salinity (Knight et al., 1996, 1997). Second, Ca2+ increase is required for the expression of some stress-induced genes in plants (Knight et al., 1996). Third, Ca2+ increase is sufficient for the activation of a stress gene promoter (Sheen, 1996). It is generally believed that Ca2+ transmits the stress signal downstream in the pathway by interacting with protein sensors. These calcium sensors, such as calmodulin (CaM), calmodulin domain protein kinases (CDPK), and calcineurin B–like proteins (CBLs), bind Ca2+ and change their conformation (Zielinski, 1998; Harmon et al., 2000; Luan et al., 2002; Sanders et al., 2002). Although CDPKs are themselves protein kinases, both CaM and CBL are small Ca2+ sensors that do not have apparent enzymatic activities. It is widely accepted that these small Ca2+ sensors function by interacting with and regulating the function of their target proteins (Zielinski, 1998; Luan et al., 2002). Studies have shown that CaM interacts with a number of proteins, including enzymes, transporters, and structural proteins. By contrast, CBL proteins interact with and regulate the activity of a specific group of protein kinases called CBL-interacting protein kinases (CIPKs) (Luan et al., 2002).
In the context of stress gene expression, one study has shown that activation of some CDPK members is sufficient to activate a stress gene promoter in maize protoplasts under normal conditions, implicating these CDPK members as positive regulators of stress gene expression (Sheen, 1996). A more recent study provides evidence that a CaM gene may function as a negative regulator of stress gene expression because overexpression of the CaM gene inhibits the cold-induced expression of stress gene markers (Townley and Knight, 2002). To date, however, the possible function of Ca2+ sensors in stress gene expression has not been addressed by genetic analysis of loss-of-function mutants. Using both overexpression of transgenic plants and loss-of-function mutants as models, we studied the function of a CBL-type Ca2+ sensor, CBL1, in stress gene expression and stress tolerance in plants. This study not only identifies CBL1 as a critical calcium sensor in abiotic stress responses but also provides an example of how one Ca2+ sensor can act as both a positive and a negative regulator in different signaling pathways.
RESULTS
CBL1 Is Stress Inducible
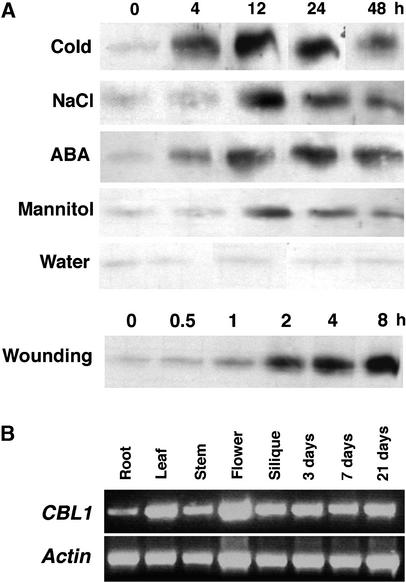
In a previous study, we showed that the CBL1 gene transcript is induced by drought, cold, and wounding (Kudla et al., 1999). Using affinity-purified CBL1 antibody, we analyzed the protein levels of CBL1 in Arabidopsis plants treated with various stress conditions and ABA. Figure 1A shows that the accumulation of CBL1, like its mRNA, was induced by hyperosmotic stress (mannitol), salt, cold, wounding, and ABA. This finding suggests that stress-induced transcription of the CBL1 gene has been translated into a protein induction that may play a role in stress responses in plants.
Figure 1.
Stress and ABA Induction of CBL1 Protein and CBL1 Gene Expression in Arabidopsis Tissues.
(A) Protein gel blot analysis of CBL1 levels under stress and ABA treatments. Plants were grown on Murashige and Skoog (1962) (MS) agar medium for 3 weeks and were treated with cold (4°C), NaCl (300 mM), ABA (100 μM), mannitol (300 mM), water (as a control), or wounding. Total protein was extracted at the indicated time points. Each lane was loaded with 30 μg of total protein for SDS-PAGE followed by protein gel blot analysis using anti-CBL1 antibody.
(B) RT-PCR analysis of CBL1 transcripts in different tissues of Arabidopsis plants. Total RNA was isolated from various tissues (root, leaf, stem, flower, or silique) of 4-week-old wild-type plants grown under long-day conditions or from germinating seeds and young seedlings (3, 7, and 21 days after sowing). RT-PCR was performed with either CBL1-specific primers (top gel) or Actin2-specific primers (bottom gel).
We also further addressed the expression pattern of CBL1 in different Arabidopsis tissues by reverse transcriptase–mediated (RT) PCR analysis. As shown in Figure 1B, the CBL1 gene is expressed during all developmental stages of plants and in all organs examined.
Constitutive Expression of CBL1 Modifies Stress Gene Expression under Normal Conditions
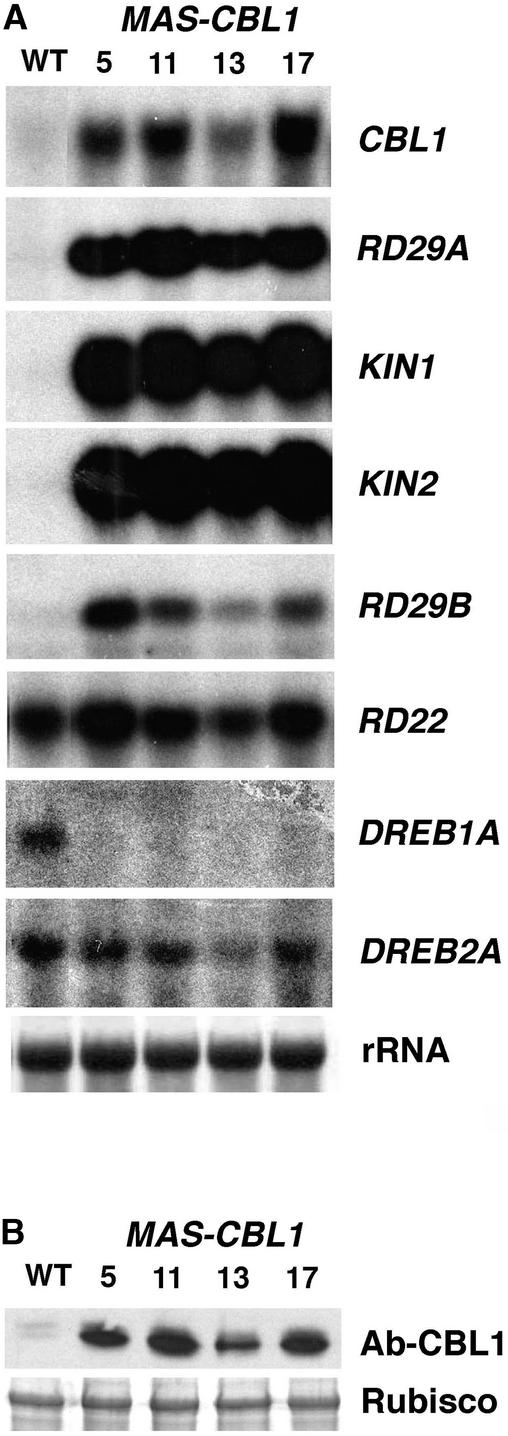
To determine whether the CBL1 level plays a role in stress response pathways, we ectopically expressed the CBL1 protein in transgenic Arabidopsis plants (MAS-CBL1) and examined the effects on the stress response in these plants. The transgenic plants harbored a construct containing CBL1 cDNA in the sense orientation under the control of a strong constitutive promoter (Ni et al., 1995). Homozygous T3 lines were obtained by selfing the 30 original T0 transformants for two generations, and four lines were selected for further analysis. We examined CBL1 mRNA and protein levels in these plants by RNA gel blot and protein gel blot analysis and found that all four independent lines showed significantly increased mRNA and protein levels compared with the wild-type control lines (Figure 2).
Figure 2.
Constitutive Expression of CBL1 in Arabidopsis Alters Stress-Responsive Gene Expression under Normal Conditions.
(A) RNA gel blot analysis in wild-type (WT) and CBL1-expressing (MAS-CBL1) plants. Total RNAs (10 μg) from 3-week-old plants grown on MS medium under normal conditions were used for RNA analysis. rRNA on the membrane was visualized by staining with methylene blue as an equal loading control. The gene markers are described in the text.
(B) CBL1 protein levels in wild-type and CBL1-overexpressing lines as revealed by protein gel blot analysis. The top gel indicates CBL1 protein level. The bottom gel shows ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) large subunit on the same blot stained with Ponceau red as an equal loading indicator.
To determine whether CBL1 overexpression has any effect on stress response pathways in transgenic plants, we chose to monitor the expression patterns of several stress gene markers, including RD29A, RD29B, KIN1/2, RD22, DREB1A, and DREB2A, in the control and CBL1-overexpressing lines. The RD29A (responsive to desiccation) gene has been shown to be responsive to ABA, drought, cold, and salinity, whereas RD29B is induced more specifically by ABA and osmotic stress (Yamaguchi-Shinozaki and Shinozaki, 1994). The KIN (cold-inducible) genes also are induced by drought, salt, cold, and ABA (Kurkela and Borg-Franck, 1992; Tahtiharju et al., 1997). DREB1A (or CBF3) is induced specifically by cold, whereas DREB2B is more specific to ABA and osmotic stress (Liu et al., 1998). As discussed previously, DREB genes often are induced earlier than RD/KIN genes, consistent with the fact that DREB proteins are transcriptional activators for RD/KIN genes. Therefore, RD/KIN and DREB gene markers can monitor the expression of both early and late stress-responsive genes. For convenience, we refer to the RD29A/COR78/LTI78 gene as RD29A and to DREB1/CBF as DREB in this report.
As shown in Figure 2A, the RD29A gene transcript was barely detectable in wild-type plants under normal conditions. By contrast, all CBL1-overexpressing plants produced high levels of mRNA for RD29A/B, KIN1/2, and RD22. The expression level of DREB2A appeared to be unchanged. Interestingly, the expression of DREB1A, a gene specifically responsive to cold, was inhibited in CBL1-overexpressing plants. Because DREB1A was expressed at a very low level under normal conditions in wild-type plants, we exposed the x-ray film for 2 weeks in this study. The result indicated that CBL1 overexpression in transgenic plants activated the expression of a number of late stress-responsive genes even under “normal” conditions. In addition, the inhibition of DREB1A suggests that CBL1 overexpression exerted negative regulation on DREB1A and possibly other cold-responsive genes.
Overexpression of CBL1 Alters the Stress Induction of Stress Gene Markers
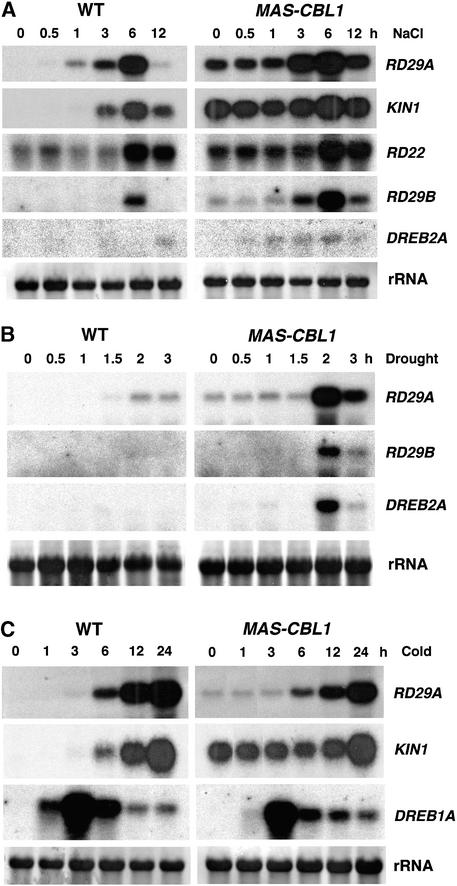
The altered expression of some stress gene markers in CBL1-overexpressing plants indicates that CBL1 might have altered stress signaling pathways. To examine this possibility further, we determined if CBL1 overexpression had altered the gene expression pattern under stress conditions (e.g., high salt, drought, and cold). Because several independent transgenic lines behaved the same way under normal conditions (Figure 2) and under stress conditions in a pilot experiment (data not shown), we used transgenic line 17 (Figure 2) for detailed analysis of the stress-responsive gene expression patterns presented in Figure 3. Consistent with previous studies (Kurkela and Borg-Franck, 1992; Yamaguchi-Shinozaki and Shinozaki, 1994; Tahtiharju et al., 1997; Liu et al., 1998), high salt treatment induced the expression of stress gene markers such as RD29A/B, KIN1/2, and RD22 in the control plants, although in our study the same conditions did not highly induce DREB2A. In CBL1-overexpressing plants, most of these genes, including RD29A/B and KIN1/2, already were active before the stress treatment, as indicated by a substantial level of mRNA accumulation, consistent with the results shown in Figure 2. On top of the high “background” level, salt treatment further induced these genes and produced higher “total” levels of mRNA than those in the control plants.
Figure 3.
Expression of Stress-Responsive Genes in Wild-Type and CBL1-Overexpressing Arabidopsis Plants Induced by NaCl, Drought, and Cold Treatments.
rRNA on the membrane stained with methylene blue served as an equal loading control. All RNA gel blot experiments were repeated at least three times, and results from one representative experiment are shown. WT, wild type.
(A) NaCl treatment (300 mM).
(B) Drought treatment.
(C) Cold treatment.
For clarity, we can divide the stress gene mRNA in CBL1-overexpressing plants into two pools: one is “constitutive” and the other is “induced.” To assess the change in the induced pool, we subtracted the mRNA level at the 0 time point from the mRNA levels at other time points. We found that the induced mRNA pool of the RD29A and RD29B genes in CBL1-overexpressing plants was greater than that in the control plants. The induction of KIN1 and RD22 was not altered significantly (Figure 3A). The hyperinduction of stress genes was more dramatic in the drought-treated plants. As shown in Figure 3B, in the first 3 h of drought treatment, control plants showed a low level of induction of RD29A, RD29B, and DREB2A. The same genes were induced much more strongly in the CBL1-overexpressing plants.
In contrast to salt- and drought-induced gene expression, the cold-induced expression of RD29A, KIN1, and DREB1A showed the opposite pattern (Figure 3C). Consistent with the result that the background level of DREB1A expression was inhibited in CBL1-overexpressing plants, cold induction of DREB1A was reduced in these plants compared with that in control plants. More specifically, the early phase of induction (at 1 h) was reduced by 85%. An approximately twofold reduction also was observed at 3 and 6 h. Although CBL1-overexpressing plants exhibited a greater background level of RD29A and KIN1 gene expression than control plants, further cold induction of these genes was reduced in these plants compared with that in the control. Again, we subtracted the background level of mRNA from the total pool of mRNA at specific time points after cold treatment. We found that KIN1 gene expression in the CBL1-overexpressing plants remained at the “basal” level and did not increase during the first 12 h of induction, whereas control plants showed clear induction within 3 to 6 h. The induced pool of RD29A mRNA also was reduced in CBL1-overexpressing plants.
As a stress hormone that mediates a number of stress signals, ABA has been shown to activate many osmotic stress–responsive genes, including RD29A/B and KIN1/2. We surveyed the expression pattern of RD29A and KIN1 in control and CBL1-overexpressing plants after ABA treatment and found no significant changes in the induction patterns of these genes (data not shown), suggesting that CBL1 regulates stress responses without affecting ABA-regulated gene expression.
CBL1-Overexpressing Plants Display Enhanced Tolerance to Salt and Drought but Reduced Tolerance to Freezing
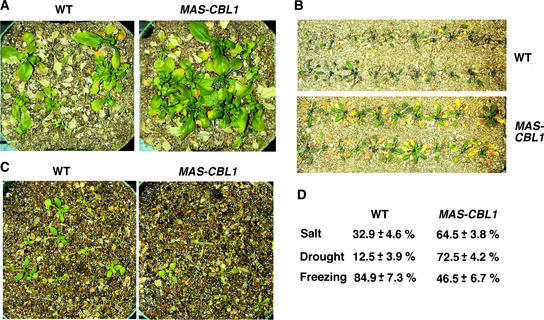
As described above, stress responses consist of many molecular and cellular pathways. Change in stress gene expression is one of the major indicators of changes in stress responses at a global level, but it does not necessarily reflect changes in the stress tolerance of whole plants. Although CBL1 overexpression modified stress gene expression, this finding may or may not be consistent with possible changes in stress tolerance in these plants. As an independent phenotypic analysis, we assayed the survival rate of CBL1-overexpressing plants compared with control plants under various stress conditions, as described previously (Jaglo-Ottosen et al., 1998; Liu et al., 1998). We performed initial assays with three independent transgenic lines, 5, 11, and 17 (Figure 2), that behaved in a similar manner (data not shown). Detailed analysis was performed on transgenic line 17, and the results are presented in Figure 4. CBL1-overexpressing plants displayed greater tolerance to salt and drought stress but showed a more sensitive phenotype to freezing. The collective results from three independent experiments are shown in Figure 4D. Under the same salt treatment, >60% of CBL1-overexpressing plants survived, compared with a 30% survival rate in control plants. Under drought conditions, ∼70% of CBL1-overexpressing plants and 10% of control plants survived. Approximately 85% of control plants and 46% of CBL1-overexpressing plants recovered from the freezing treatment. These stress assay results are consistent with those in gene marker analysis, but we should emphasize that changes in stress tolerance are not necessarily the result of altered gene expression in the CBL1-overexpressing plants. Therefore, the results from gene marker analysis and tolerance assays separately and independently support the idea that CBL1 may function in plant stress responses.
Figure 4.
Altered Stress Tolerance in CBL1-Overexpressing Plants.
(A) Increased salt tolerance. Three-week-old plants were treated with 300 mM NaCl (see Methods) and monitored for bleaching during the subsequent 2 weeks. The photographs were taken on the 10th day. WT, wild type.
(B) Increased drought tolerance. Watering was withheld from 3-week-old plants for 23 days before the photographs were taken.
(C) Reduced freezing tolerance. After freezing treatment (see Methods), plants were transferred to a growth chamber. The photographs show the difference on the 8th day after transfer to the growth chamber.
(D) Survival rate. The survival rate (%) and standard errors were calculated based on results from three independent experiments (see Methods).
Although CBL1 overexpression did not appear to change ABA-responsive gene expression in the plants, it still may alter other ABA responses, such as the inhibition of seed germination. We performed initial assays with three independent transgenic lines, 5, 11, and 17 (Figure 2), that behaved in a similar manner (data not shown). Detailed analysis was performed on transgenic lines 5 and 17, and the results are presented in Figure 5. We tested this possibility using a germination assay on medium containing ABA and found no difference between control and CBL1-overexpressing plants (Figure 5B). Interestingly, the same assay on the medium containing high concentrations of NaCl or mannitol indicated that CBL1-overexpressing seeds were more tolerant of these conditions than were control seeds (Figure 5). For example, on medium containing 200 mM NaCl, two transgenic lines overexpressing CBL1 showed 42 and 58% germination in 3 days, whereas only 10% of control seeds had germinated (Figure 5C). On medium containing 450 mM mannitol, 30% of control seeds germinated within 3 days, whereas CBL1-overexpressing seeds had achieved a 60% germination rate (Figure 5D). These results indicate that CBL1 overexpression rendered plants more resistant to high salt and hyperosmotic stress during early development (i.e., seed germination) but did not alter their ABA response.
Figure 5.
Germination Assays of CBL1-Overexpressing Plants.
(A), (C), and (D) Germination time course (days of incubation at 23°C) on MS medium (A), MS medium containing NaCl (C), or MS medium containing mannitol (D) was recorded for wild-type (closed circles) and MAS-CBL1 (open squares [plant 5] and open triangles [plant 17]) seeds.
(B) Germination rates of wild-type and MAS-CBL1 seeds after 3 days of incubation at 23°C on MS medium containing different concentrations of ABA.
Results are presented as average values ± se from three experiments.
Disruption of the CBL1 Gene Differentially Alters Abiotic Stress Responses
The constitutive expression of CBL1 in transgenic plants resulted in significant changes in plant responses to salt, drought, and cold, suggesting that CBL1 functions in the stress signaling pathways. However, overexpression often is considered a gain-of-function approach for analyzing the function of a gene, so these results cannot be interpreted at face value. For example, a high level of CBL1 may result in altered specificity in its interaction with the target proteins, leading to an altered function in the cell. In addition, CBL1 overexpression also may have other nonspecific effects on gene expression. However, the fact that CBL1 overexpression conferred a positive effect on drought and salt response but a negative effect on cold response suggests that CBL1 function is specific even after overexpression. Nevertheless, we cannot exclude the possibility that a gain of function occurred as a result of the overproduction of CBL1.
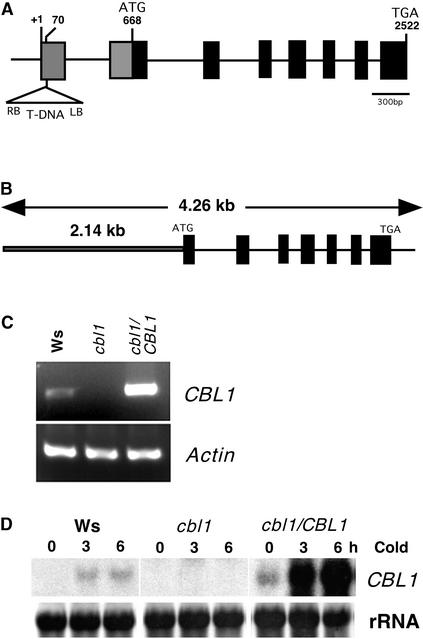
To further investigate the function of the CBL1 gene, we isolated a loss-of-function mutant in CBL1 through T-DNA insertional mutagenesis. The T-DNA insertion is located in the 5′ untranslated region of CBL1 (Figure 6A). Analysis of homozygous mutant plants using DNA gel blots indicated that the T-DNA line contained a single T-DNA insert (data not shown). The insertion should eliminate the transcription of the gene because it separated the promoter from the transcription region. Indeed, both RNA gel blot and RT-PCR analyses of CBL1 mRNA indicated that the knockout mutant plants did not produce CBL1 transcript under normal and cold stress conditions that induced the expression of the wild-type gene (Figures 6C and 6D). We also transformed homozygous mutant plants with a 4.26-kb genomic DNA fragment containing the CBL1 gene to serve as complementation lines (Figure 6B). These plants showed a higher level of CBL1 expression compared with the wild-type plants under normal conditions and were highly inducible by stress factors such as cold, a property of the endogenous CBL1 promoter (Figures 6C and 6D). A higher level of CBL1 transcript was observed in several independent complementation lines, suggesting that it was not caused by variation in the insertional position in the genome. Because we also made similar observations of other genes transformed using the same vector, the higher level of CBL1 expression in the complemented lines could be an intrinsic property related to the vector we used in this study.
Figure 6.
Isolation and Complementation of the cbl1 T-DNA Insertional Mutant.
(A) Scheme of the Arabidopsis CBL1 gene. Exons (closed boxes) and introns (lines) are indicated. The position and orientation of the T-DNA insertion are shown (not to scale). The numbers represent nucleotides, and +1 indicates the transcriptional start site for the CBL1 gene. LB, left border; RB, right border.
(B) Complementation genomic DNA fragment. A 4.26-kb DNA region containing the CBL1 gene was amplified by PCR and cloned into the pCAMBIA1300 vector for plant transformation.
(C) RT-PCR analysis of CBL1 from wild-type (Wassilewskija [Ws]), cbl1, and cbl1/CBL1 plants. RT-PCR was performed with either CBL1-specific primers (top gel) or Actin2-specific primers (bottom gel).
(D) RNA gel blot analysis of CBL1 mRNA in the wild type (Ws), the mutant (cbl1), and a complementation transgenic line (cbl1/CBL1) under cold treatment. Four independent complementation lines were analyzed with similar results, and the result from one line is shown here. Ten micrograms of total RNA from 3-week-old seedlings was probed with CBL1 cDNA. rRNA on the membrane was visualized by methylene blue as an equal loading control.
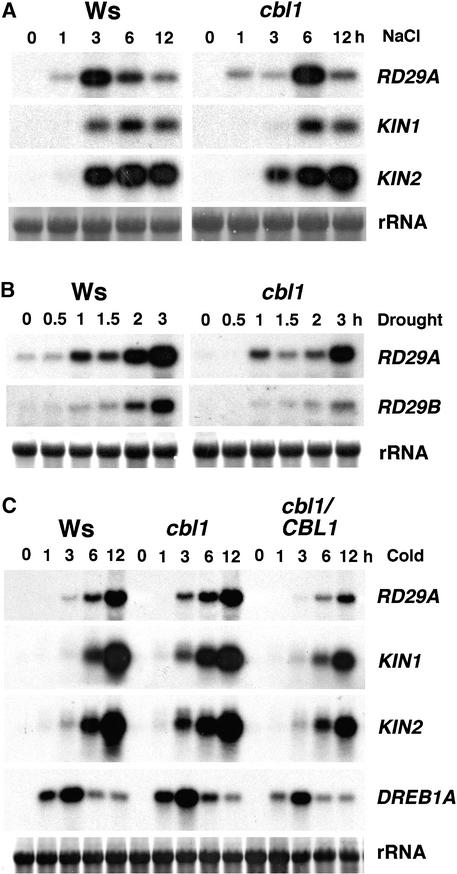
We examined mutant and wild-type plants under normal conditions during the life cycle and found no significant phenotypic changes in the mutant (data not shown). To determine if the cbl1 mutant is altered in stress signaling, we compared the stress gene expression patterns of cbl1 and the wild type by mRNA gel blot analysis (Figure 7). Under salt treatment, the expression pattern of RD29A/KIN genes showed no significant difference between the wild type and the cbl1 mutant, although the peak of RD29A mRNA accumulation appeared to be delayed in the mutant (Figure 7A). Under drought conditions (Figure 7B), the expression of RD29A and RD29B was reduced significantly in cbl1 plants compared with control plants.
Figure 7.
Expression of Stress-Responsive Genes in Wild-Type, cbl1, and cbl1/CBL1 Plants Induced by NaCl, Drought, and Cold Treatments.
rRNA on the membrane was visualized by staining with methylene blue as an equal loading control. All RNA gel blot experiments were repeated at least three times, and results from one representative experiment are shown. Ws, Wassilewskija.
(A) NaCl treatment (300 mM).
(B) Drought treatment.
(C) Cold treatment.
Under cold stress (Figure 7C), the induction of gene markers, including RD29A, KIN1/2, and DREB1A, was altered in the cbl1 mutant. Perhaps the most significant change occurred in the level of early induction. At the 3- and 6-h time points, RD29A and KIN1/2 were induced at a higher level in cbl1 than in the wild type. By 12 h after induction, the mRNA levels were comparable between the wild type and the mutant. The change in DREB1A expression was less obvious, but mutant plants were observed reproducibly to have slightly higher levels of DREB1A mRNA than were wild-type plants. The same RNA samples also were analyzed by real-time quantitative PCR, and these small differences between the wild type and the mutant were observed as well (data not shown). Although DREB1A functions as a transcriptional activator of RD/KIN genes, we do not interpret the difference in DREB1A expression between the wild type and the mutant as the cause of changes in stress gene induction by cold in these plants. The complemented line showed lower induction of all marker genes compared with wild-type plants, consistent with the earlier observation that higher levels of CBL1 expression inhibited cold induction (Figure 3C).
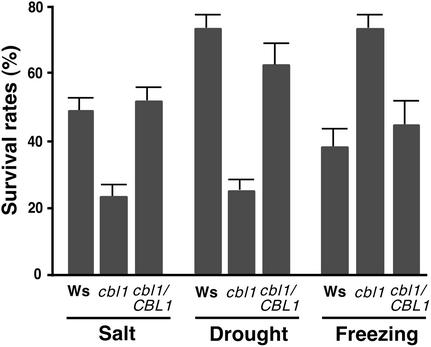
We tested stress tolerance in the cbl1 mutant together with that in wild-type and complemented lines. Figure 8 shows results from similar stress tolerance assays performed on CBL1-overexpressing plants. The cbl1 mutant plants were more tolerant of freezing treatment but less tolerant of osmotic stress (drought and high salt) than were wild-type plants. After the freezing treatment, >70% of cbl1 mutant plants recovered, whereas 40% of wild-type plants survived. By contrast, >70% of wild-type plants and only 25% of cbl1 mutant plants survived the drought treatment. Under salt conditions, the survival rate in the wild-type group was approximately twice that in the cbl1 mutant group. The results from these studies of the cbl1 mutant and from the analyses of CBL1-overexpressing plants both support the hypothesis that CBL1 functions as a positive regulator of salt and drought response but a negative regulator of cold response.
Figure 8.
Altered Stress Tolerance in cbl1 Mutant Plants.
For salt tolerance, 3-week-old plants were exposed to 300 mM NaCl solution every 3 days for three repetitions and monitored subsequently for bleaching during the next 2 weeks. For drought tolerance, watering was withheld for 23 days from 3-week-old soil-grown plants before survival rates were counted. For freezing tolerance, plants treated with freezing stress were returned to the growth chamber. Surviving plants were scored on the 8th day in the growth chamber. Survival rates and standard errors were calculated from results of three independent experiments. Ws, Wassilewskija.
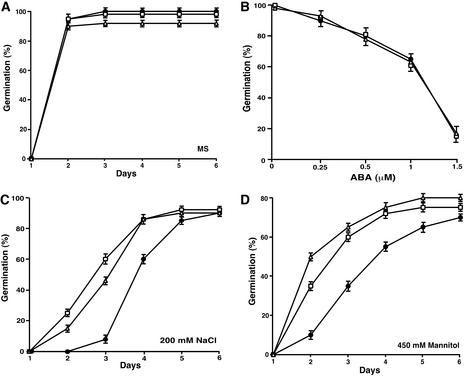
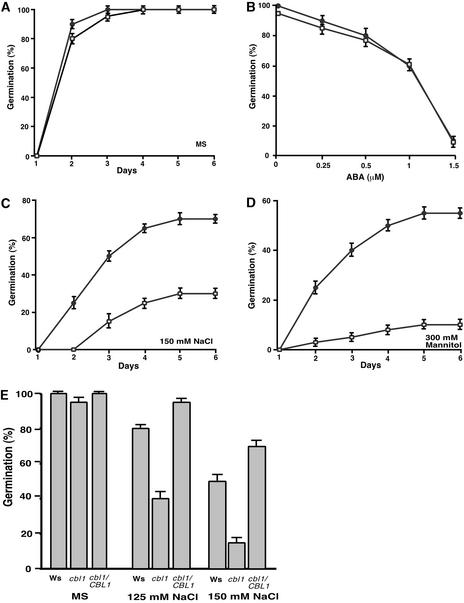
We also tested whether cbl1 mutant plants were altered in ABA response during seed germination, but again, we detected no difference between wild-type and mutant seeds (Figure 9). On the high salt and mannitol medium, however, the germination of cbl1 mutant seeds was inhibited to a greater extent than that of wild-type seeds (Figure 9). On medium with 150 mM NaCl, 50% of wild-type seeds had germinated in 3 days but only 15% of cbl1 seeds had germinated. At day 6, cbl1 mutant seeds germinated at a 25% rate, whereas 65% of wild-type seeds had germinated (Figure 9C). Likewise, in 300 mM mannitol, germination of both wild-type and cbl1 seeds was inhibited significantly. However, cbl1 seeds were much more sensitive to such hyperosmotic conditions than wild-type seeds, with a <10% germination rate in the 6-day period, whereas wild-type seeds achieved a 50% germination rate (Figure 9D). Figure 9E shows that seeds from complementation transgenic plants exhibited comparable germination rates under two different concentrations of salt, indicating that the hypersensitivity of seed germination to stress conditions in the cbl1 mutant resulted from the disruption of the CBL1 gene.
Figure 9.
Germination of cbl1 Mutant Seeds Is Hypersensitive to Osmotic Stress Conditions But Not to ABA.
(A), (C), and (D) Germination time course (days after incubation at 23°C) was recorded as described in Figure 5 on MS medium (A), MS medium containing NaCl (C), or MS medium containing mannitol (D) for wild-type (Wassilewskija) and cbl1 mutant seeds.
(B) Germination rates of wild-type and cbl1 mutant seeds at 3 days after transfer to 23°C in the presence of different concentrations of ABA.
(E) Seed germination rates in the wild type (Wassilewskija [Ws]), cbl1 mutant (cbl1), and four complementation lines (cbl1/CBL1) on MS medium (control) or MS medium containing 125 or 150 mM NaCl. Germination was scored at 3 days after incubation at 23°C. Data from one representative complementation line are shown.
Results are presented as average values ± se from three experiments. Closed circles, wild-type; open squares, cbl1 mutant.
DISCUSSION
We have identified CBL1, a calcineurin B–like calcium sensor, as a critical component in plant responses to abiotic stress signals. Importantly, CBL1 functions as a positive regulator of salt and drought responses but a negative regulator of the cold response. These findings may contribute to our understanding of several important mechanisms underlying calcium functions in stress signal transduction pathways.
CBL1 May Represent a Rate-Limiting Component in Stress Signaling Pathways
Ca2+ may serve as a second messenger in many stress signal transduction pathways, including those triggered by salt, drought, and cold (reviewed by Bush, 1995; Sanders et al., 1999, 2002; Knight and Knight, 2001). However, the molecular mechanism of Ca2+ action in plant cells is not well understood. We have shown that the CBL1 calcium sensor is highly stress responsive at both the mRNA and protein levels, suggesting that the CBL1 protein level may play a role in stress responses. Indeed, constitutive expression of CBL1 was sufficient for the activation of multiple stress genes and enhanced tolerance to both salt and drought stress in transgenic plants. This finding suggests that CBL1 is an upstream regulator of stress gene expression and serves as a rate-limiting factor in some stress response pathways. Alternatively, CBL1 overexpression may activate gene expression nonspecifically. However, results from both overexpression and loss-of-function mutant analyses suggest that CBL1 is a specific component in stress signaling processes.
It is generally believed that cellular Ca2+ levels hold the key to the activation of stress response pathways. Normally, cellular Ca2+ fluctuates below the “threshold value.” Stress signals typically boost the Ca2+ levels over the threshold, leading to the activation of calcium sensors and the downstream components in the pathway. Consistent with this view, an increase of cellular Ca2+ by ionophores is sufficient to activate a stress- and ABA-induced promoter in the absence of stress or ABA application (Sheen, 1996). Based on our findings in this report, we propose that levels of some calcium sensors also may serve as a threshold (rate-limiting) factor for the responsiveness of plants to stress signals. In the case of CBL1, its basal level is low under normal conditions and is induced by stress signals. The stress-responsive increase in CBL1 level may serve as a feedback mechanism for increasing the sensitivity to stress signals. In CBL1-overexpressing transgenic plants, CBL1 levels are constantly high, so that some stress response pathways are activated to a certain degree even under normal conditions, when calcium level is below the threshold. Stress conditions such as drought and high salt further activate the signaling pathways by increasing the level of calcium, another threshold factor.
This hypothesis also can explain the results from another study in which the overexpression of pathogen-inducible CaM isoforms constitutively activated pathogen-related gene expression, leading to enhanced pathogen resistance in the plants (Heo et al., 1999). In a similar manner, pathogen signaling pathways also involve calcium fluctuations (Rudd and Franklin-Tong, 2001). Both this study and the study on CaM function in pathogen signaling (Heo et al., 1999) identified functional specificities of different isoforms of the calcium sensor studied. In the case of CaM function in pathogen resistance (Heo et al., 1999), only the pathogen-inducible isoforms, but not other isoforms, conferred resistance when overexpressed in plants. In our study, CBL1, but not other noninducible isoforms, conferred stress tolerance to plants (data not shown). Again, this isoform specificity makes it unlikely that these gene products exert nonspecific effects on gene expression in plant cells. In both cases, the basal level of calcium sensors (CBL and CaM) is low under normal conditions and highly inducible by corresponding stress signals. We hypothesize that the levels of both calcium and its sensor proteins are important threshold parameters in calcium-mediated signal transduction in plants. Sufficient increase in either calcium or its sensors will lead to a certain degree of activation of the downstream pathway. An increase in both will lead to a hyperactive response. This may represent a general mechanism in many calcium signaling pathways in eukaryotic systems, although to our knowledge such a phenomenon has not been reported in animal systems. Our study provides an opportunity to further address how CBL1 serves as a threshold factor in stress signaling pathways.
CBL1 Mediates Calcium Function in Specific Stress Signaling Pathways
One of the earliest responses to stress signals is an increase in cellular calcium in plants (Bush, 1995; Knight et al., 1997; Sanders et al., 2002). Later responses include changes in the expression profiles of a large number of stress genes (Ingram and Bartel, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1997; Thomashow, 1999). From calcium to gene expression, a number of intermediate components may play a role in the signaling process. These include calcium sensors such as CaM, CDPK, and CBL, which are immediate downstream components after calcium changes (Zielinski, 1998; Harmon et al., 2000; Luan et al., 2002; Sanders et al., 2002). To understand the function of calcium sensors in stress signaling pathways, a critical approach involves performing loss-of-function mutant analysis of each calcium sensor and observation of the consequences to the stress response pathways. Our study showed that disruption of CBL1 function in the cbl1 mutant resulted in a reduced level under drought conditions but a more rapid gene induction under cold conditions. By contrast, increased expression of CBL1 activated the drought- and salt-induced expression but reduced the cold-induced expression of stress gene markers such as RD29A/B and KIN1/2. Furthermore, the whole-plant phenotype in stress tolerance also was altered by overexpression or disruption of the CBL1 gene. Together, these results suggest that CBL1 serves as a positive regulator of the salt and drought signaling pathways and as a negative regulator of the cold response pathway. This study not only places a calcium sensor between stress signals and gene expression and stress tolerance but also reveals the specificity and complexity of CBL1 function in different stress signaling pathways.
Salt, drought, and cold often activate the expression of the same genes. One explanation for this fact is that these signals may share a common pathway leading to the activation of the genes. Recent studies have identified separate cis-acting elements (often present in the same gene promoter) and transcription factors that bind to each of them (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). These studies immediately suggest that different signals may activate the same gene by distinct mechanisms. Nevertheless, studies also suggest that these pathways may share common components that serve as crosstalk nodes between the different pathways (Knight and Knight, 2001). Our study provides evidence for both the specificity and overlap of abiotic stress and the ABA response pathways. For example, drought and cold have been shown to activate RD29A gene expression by activating the same cis-acting element, DRE/CRT. However, evidence suggests that different transcription factors, DREB1 and DREB2, may be involved in drought and cold responses, implicating separate pathways linking drought and cold to RD29A expression (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000).
Here, we show that CBL1 regulates the expression of the same gene differently depending on which stress signal is applied to the plants. Under drought conditions, RD29A was upregulated by a higher level of CBL1 and downregulated by the disruption of CBL1 expression. Conversely, the cold induction of RD29A gene expression was inhibited by a higher level of CBL1 protein and enhanced in the cbl1 null mutant plants. This result clearly shows that distinct pathways exist for cold- and drought-induced gene expression. Most of the previous studies have been directed to the transcriptional control of stress genes (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). If CBL1 regulates stress genes at the transcription level, it may interact with its target kinases, which in turn regulate the activity of different transcription factors, such as DREB1A or DREB2A, leading to the activation or inactivation of their target genes. There is little evidence that stress signals regulate gene expression by a post-transcriptional mechanism. However, we cannot exclude the possibility that such post-transcriptional regulation exists and that CBL1 may be part of this mechanism instead of or in addition to its function in transcriptional regulation. Further studies are needed to address the mechanism underlying CBL1 action in the regulation of stress gene expression.
Another result further defines specificity in drought/salt and ABA signal transduction. It is known that drought/salt triggers an increase in ABA levels that partly mediates drought/salt responses, including gene induction. As a result, drought/salt signal is transmitted by both ABA-dependent and ABA-independent pathways. Here, we show that CBL1 positively regulates the drought/salt response but does not alter the ABA response. This would happen if CBL1 were involved in the regulation of an ABA-independent branch of the drought/salt signaling pathway. Alternatively, CBL1 may function specifically in the early steps of abiotic stress signal transduction upstream of ABA production.
CBL1 and the Complex CBL-CIPK Network in Stress Signaling Pathways
How can one calcium sensor serve as both a positive and a negative regulator to closely related signal transduction pathways triggered by cold and drought that often activate the same genes? Possible interpretations are related to the mode of action of CBL proteins.
In Arabidopsis, the CBL family consists of at least 10 members that share significant sequence homology. Each of the 10 members interacts with one or more partner protein kinases (CIPKs) that constitute a unique kinase family of at least 25 members (Luan et al., 2002). Some closely related CBL members may share the same partner CIPKs, although the interaction affinity may differ (Shi et al., 1999; Kim et al., 2000; Albrecht et al., 2001). Some more distantly related CBL members interact with completely different CIPKs. Thus, CBL–CIPK interactions form a complex network to mediate Ca2+ signaling functions (reviewed by Luan et al., 2002). Upon calcium increase by stress signals, CBL1 may interact with multiple CIPKs. Indeed, CBL1 has been shown to interact with a number of CIPKs in the yeast two-hybrid system (Kim et al., 2000; Albrecht et al., 2001). Some CBL1-interacting CIPKs may positively regulate the salt and drought signaling pathways and some may negatively regulate the cold pathway. Alternatively, the same CIPK may have a positive effect on the drought pathway and a negative effect on the cold pathway. Upon overexpression of CBL1, both positive and negative regulation would be enhanced. By contrast, disruption of CBL1 will lead to downregulation of the positive and negative effects on the drought and cold responses, respectively. This is consistent with the finding that drought-responsive RD29A gene expression is reduced but cold induction is enhanced in cbl1 mutant plants.
Another possibility is that CBL1 may interact with some CIPKs that also interact with other CBLs. Such an interaction network may cause both synergistic and antagonistic effects on the function of each CBL member. For example, CIPK member CIPKx may interact with CBL1 and CBLy (“x” can be any member of the CIPK family and “y” can be any other member in the CBL family). If both interactions positively regulate the drought response, these two CBL members may have a synergistic effect on the drought response. If the CBL1-CIPKx complex functions as a positive regulator of the drought response and the CBLy-CIPKx complex serves as a positive regulator of the cold response, CBL1 and CBLy may have an antagonistic relationship resulting from competition for the same pool of CIPKx. Normally, this competition will reach equilibrium with CIPKx partitioned to serve both roles. When CBL1 is mutated, however, the CBLy member will be dominant and the equilibrium will be tipped to the CBLy function. In the cbl1 mutant, CIPKx will be dedicated solely to the function of CBLy, which serves as a positive regulator of the cold response, resulting in an increased response to cold. In CBL1-overexpressing plants, the opposite tipping of the equilibrium will occur: more CIPKx will be allocated to CBL1-dependent function (i.e., upregulation of the drought response) and less CIPKx will be dedicated to the cold response in the CBLy-dependent pathway. These interpretations may not be mutually exclusive, because they can occur simultaneously in plants.
Because cold and drought both have a dehydration component, some of the cellular responses to these two factors are similar (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). In this context, why do plant cells need to have an antagonistic mechanism between the cold and drought response pathways? This question is difficult to answer, but it certainly reminds us of the differences between the two pathways and our limited knowledge of these differences. This study raises the possibility that calcium sensors provide crosstalk points for the cell to “coordinate” such differences. A recent study (Kim et al., 2003) shows that CIPK3, a member of the CIPK family, functions as a positive regulator of the cold response but has no effect on the drought response, consistent with the hypothesis that the cold and drought responses involve different regulators. Because CIPK3 does not interact with CBL1 (Kim et al., 2000), other CIPKs may mediate CBL1 function in plant cells. It is expected that many other components are involved in the crosstalk between the different pathways, weaving a complex network. Further functional dissection of the calcium sensors, their targets, and their interplay in signaling pathways will contribute to our understanding of this signaling network.
METHODS
Plant Materials, Stress Treatments, and RNA Analysis
Plants (Arabidopsis thaliana ecotype Wassilewskija [Ws] or Columbia) were grown in a greenhouse under long-day conditions (16-h-light/8-h-dark cycle) to flowering stage for plant transformation and RNA analysis. For treatment under different stress conditions, 3-week-old seedlings grown on Murashige and Skoog (1962) (MS) medium were used. Sterilized seeds were plated on MS medium solidified with 0.8% agar. For abscisic acid (ABA) treatment, 100 μM (±)-cis,trans–ABA was sprayed on the seedlings to ensure total coverage of the foliage area. The plants were incubated at room temperature under white light. For NaCl or mannitol treatment, 300 mM NaCl or mannitol was added to the seedlings on MS plates and incubated at room temperature under white light. For cold treatment, seedlings were transferred to a 4°C cold room under white light. For drought treatments, 4-week-old plants grown in potting soil were carefully removed and dehydrated on filter paper as described by Yamaguchi-Shinozaki and Shinozaki (1994). Wounding was performed by puncturing leaves with a hemostat, as described by Kudla et al. (1999). For RNA gel blot analysis, total RNA (10 μg) isolated using a Tripure isolation reagent (Roche Diagnostics, Indianapolis, IN) was separated by electrophoresis on a 1.2% agarose gel, transferred to GeneScreen Plus nylon membranes, and hybridized with 32P-labeled specific probes as described in Results. The membranes were autoradiographed with Kodak XAR film. Membrane-bound 23S rRNA was stained with methylene blue and used as a loading control. The images were scanned with a high-resolution scanner and processed using Adobe Photoshop (Mountain View, CA). All RNA gel blot experiments were repeated at least three times, and results from one representative experiment are shown in the figures.
Generation of CBL1-Overexpressing Transgenic Plants
To make CBL1 overexpression construct (MAS-CBL1), the full-length CBL1 cDNA (Kudla et al., 1999) was digested with NotI and cloned into pATC940 vector under the control of super promoter (Ni et al., 1995). The orientation and the junction of the cloned fragment were confirmed by DNA sequencing. This construct was transferred into Agrobacterium tumefaciens GV3101 and used for Arabidopsis transformation (see below). Seeds were harvested from these plants and screened on selection medium (half-strength MS medium, 1× Gamborg's vitamins, and 50 μg/mL kanamycin) for transformants. The putative transformants (T1) were rescued from plates and grown in a greenhouse under long-day conditions. T2 plants were raised to produce seeds that were germinated to produce T3 plants. Those T2 plants that produced 100% kanamycin-resistant plants in the T3 generation were considered homozygous transformants and were used for further experiments.
For plant transformation, Arabidopsis plants were grown in a greenhouse under long-day conditions (16-h-light/8-h-dark cycle) for 4 weeks before a floral-dip procedure (Clough and Bent, 1998). Briefly, Agrobacterium cells was grown in Luria-Bertani broth for 24 h at 30°C. The cells were collected by centrifugation and resuspended in infiltration medium (half-strength MS medium, 5% sucrose, 1× Gamborg's vitamins, 0.044 μM benzylamino purine, and 0.04% Silwet L77) to an OD600 of 1.5 to 2.0. Plants were dipped into this suspension for 30 s and transferred to a greenhouse.
Isolation and Complementation of the cbl1 T-DNA Insertional Mutant
The CBL1 insertion allele was isolated by screening 60,480 T-DNA–tagged Arabidopsis lines (ecotype Ws) at the University of Wisconsin Arabidopsis Knockout Facility. The primer JL-202 (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) annealing to the T-DNA left border was used together with CBL1-specific primers (forward, 5′-AGGTACAATTAGGTTCGGTCTAACTGAAA-3′; reverse, 5′-ATTACACCGATTAGCTCAGACTTGTTCTT-3′) for the identification of putative mutant lines. The T-DNA insertion in the mutant (cbl1) was confirmed by DNA gel blot analysis, and its exact position was determined by sequencing. Plants homozygous for the cbl1 mutant were used for further analysis.
For complementation of the cbl1 mutant, a 4.26-kb fragment including the CBL1 coding region and 2.14 kb of the 5′ flanking DNA upstream the ATG codon was amplified by PCR from Arabidopsis genomic DNA (Ws ecotype) with forward (5′-ATAAGCTTATTAGTACAGAGGGAAGTG-3′) and reverse (5′-TATCTGCAGCGTATGATGTAATATGTACA-3′) primers. The PCR product was cloned into the binary vector pCAMBIA1300 (CAMBIA GPO, Canberra, Australia) using HindIII and PstI restriction sites (underlined in the primer sequences). The constructs were transformed into Agrobacterium strain GV3101 and introduced into Arabidopsis plants by the floral-dip method (Clough and Bent, 1998). Transgenic seeds were plated on half-strength MS medium containing 0.8% (w/v) agarose, 112 mg/L Gamborg's B5 vitamin mixture, 50 μg/mL kanamycin, and 15 μg/mL hygromycin. The resistant seedlings were transplanted to soil and grown in a greenhouse to produce seeds. Homozygous complemented lines (denoted cbl1/CBL1) were used for further analysis. All of the PCR procedures were performed using Pfu DNA polymerase (Stratagene, La Jolla, CA) to enhance fidelity. All constructs were verified by DNA sequencing.
Reverse Transcriptase–Mediated PCR Analysis
To analyze the expression of CBL1 by reverse transcriptase–mediated PCR, total RNA (0.2 μg) from wild-type (Ws), cbl1, and cbl1/CBL1 plants was heated to 65°C for 7 min and then subjected to reverse transcription reaction using SuperScript II RNase H− reverse transcriptase (200 units per reaction; Invitrogen, Carlsbad, CA) with oligo(dT) primer for 50 min at 42°C. PCR amplification was performed with initial denaturation at 94°C for 2 min followed by 25 cycles of incubations at 94°C for 20 s, 55°C for 40 s, and 72°C for 1.0 min, with a final extension at 72°C for 10 min using the CBL1-specific forward (5′-CATCTGTAAATGGGCTGCTTCCAC-3′) and reverse (5′-CAGTTTGTTTCTTCATGTGGCAATC-3′) primers with AmpliTaq polymerase (2 units per reaction; Perkin-Elmer). Actin expression level was used as a quantitative control. Aliquots of individual PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under UV light.
Protein Isolation and Protein Gel Blot Analysis
Leaves of 3-week-old Arabidopsis plants were ground to a fine powder in liquid nitrogen with a pestle and mortar and transferred to a centrifuge tube containing extraction buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 2 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, and 5 μg/mL leupeptin, aprotinin, and pepstatin). The suspension was mixed thoroughly for 30 s and centrifuged at 14,000g for 30 min at 4°C. The supernatant was used as a total protein extract and examined by SDS-PAGE and protein gel blot analysis. Recombinant CBL1 was expressed and antiserum was generated as described previously (Kudla et al., 1999; Shi et al., 1999). For protein gel blot analysis, proteins in the gel were blotted electrically to nitrocellulose membranes (Nitrobind, Osmonics, Minnetonka, MN). Membranes were blocked with TBST (25 mM Tris-HCl, pH 7.5, 137 mM NaCl, and 0.2% Tween 20) containing 5% nonfat dry milk for at least 4 h and incubated with affinity-purified polyclonal CBL1 antibody (1:1000) for 2 h in TBST containing 1% nonfat dried milk. After multiple washes with TBST, bound antibodies were detected with peroxidase-conjugated secondary antibodies and a chemiluminescence kit (Amersham Pharmacia).
Drought, Salt, and Freezing Tolerance Analyses
For the drought assays, watering was withheld from 3-week-old soil-grown plants for specified times. To minimize experimental variations, the same number of plants in each comparison group was grown on the same tray. Watering was withheld from the plants for 23 days before photographs were taken and survival rates were counted. For the salt tolerance assays, 3-week-old plants were exposed to 300 mM NaCl solution every 3 days for three repetitions and subsequently monitored for bleaching for the next 2 weeks. Photographs were taken and survival rates were counted on the 10th day. For the freezing assays, 3-week-old plants grown in soil in a growth chamber (22 ± 2°C, 16 h of light and 8 h of dark) were treated at 4°C under light conditions for 12 h for acclimation. After that, the plants were subjected to freezing at −7°C for 6 h. After this treatment, plants were transferred immediately to 4°C under white light and incubated for 12 h. The plants then were transferred to a growth chamber. Surviving plants were scored on the 8th day in the growth chamber. All experiments were repeated at least three times, and results from one representative experiment are shown in Figure 4. Survival rates and standard errors (Figures 4D and 8) were calculated from the results of three independent experiments.
Germination Assay
Approximately 100 seeds each from the wild type (Ws), the cbl1 mutant, and the cbl1/CBL1 complemented line were planted in triplicate on MS medium with different concentrations of ABA, NaCl, or mannitol and incubated at 4°C for 4 days before being placed at 23°C under long-day conditions. Germination (emergence of radicals) was scored daily for 6 days.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact S. Luan, sluan@nature.berkeley.edu.
Acknowledgments
We are grateful to the ABRC (Ohio State University, Columbus, OH) for Arabidopsis seeds and DNA clones and to the Arabidopsis Gene Knockout Facility (University of Wisconsin, Madison, WI) for screening the cbl1 mutant. This work was supported by the National Science Foundation and by Syngenta Research and Technology.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012393.
References
- Albrecht, V., Ritz, O., Linder, S., Harter, K., and Kudla, J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Bush, D.S. (1995). Calcium regulation in plant cells and its role in signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 95–122. [Google Scholar]
- Choi, H.I., Hong, J.H., Ha, J.O., Kang, J.Y., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Harmon, A.C., Gribskov, M., and Harper, J.F. (2000). CDPKs: A kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159. [DOI] [PubMed] [Google Scholar]
- Heo, W.D., Lee, S.H., Kim, M.C., Kim, J.C., Chung, W.S., Chun, H.J., Lee, K.J., Park, C.Y., Park, H.C., Choi, J.Y., and Cho, M.J. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 96, 766–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram, J., and Bartel, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Kim, K.-N., Cheong, Y.H., Grant, J.J., Pandey, G.K., and Luan, S. (2003). CIPK3, a calcium sensor–associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.-N., Cheong, Y.H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124, 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2000). Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J. Exp. Bot. 51, 1679–1686. [DOI] [PubMed] [Google Scholar]
- Knight, H., and Knight, M.R. (2001). Abiotic stress signalling pathways: Specificity and cross-talk. Trends Plant Sci. 6, 262–267. [DOI] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1996). Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8, 489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, H., Trewavas, A.J., and Knight, M.R. (1997). Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 96, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela, S., and Borg-Franck, M. (1992). Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 19, 689–692. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B–like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 (suppl.), S389.–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Ni, M., Cui, D., Einstein, J., Narasimhulu, S., Vergara, C.E., and Gelvin, S.B. (1995). Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7, 661–676. [Google Scholar]
- Rudd, J.J., and Franklin-Tong, V.E. (2001). Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol. 151, 7–33. [DOI] [PubMed] [Google Scholar]
- Sanders, D., Brownlee, C., and Harper, J.F. (1999). Communicating with calcium. Plant Cell 11, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, D., Pelloux, J., Brownlee, C., and Harper, J.F. (2002). Calcium at the crossroads of signaling. Plant Cell 14 (suppl.), S401.–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y., and Shinozaki, K. (2001). Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (1996). Ca2+-dependent protein kinases and stress signal transduction in plants. Science 274, 1900–1902. [DOI] [PubMed] [Google Scholar]
- Shi, J., Kim, K.-N., Ritz, O., Albrecht, V., Gupta, R., Harter, K., Luan, S., and Kudla, J. (1999). Novel protein kinases associated with calcineurin B–like calcium sensors in Arabidopsis. Plant Cell 11, 2393–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju, S., Sangwan, V., Monroy, A.F., Dhindsa, R.S., and Borg, M. (1997). The induction of kin genes in cold-acclimating Arabidopsis thaliana: Evidence of a role for calcium. Planta 203, 442–447. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Townley, H.E., and Knight, M.R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128, 1169–1172. [DOI] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.), S165.–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., and Zhu, J.K. (2002). Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ. 25, 131–139. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, R.E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725. [DOI] [PubMed] [Google Scholar]