Abstract
Efforts to elucidate the contributions by transcription factors to plant gene expression will require increasing knowledge of their specific in vivo regulatory associations. We are systematically investigating the role of individual TGA factors in the transcriptional control of pathogenesis-related (PR) defense genes, whose expression is stimulated in leaves by salicylic acid (SA) through a stimulus pathway involving NPR1. We focused on PR-1 because its SA-induced expression in Arabidopsis is mediated by an as-1–type promoter cis element (LS7) that is recognized in vitro by TGA factors. We found that two NPR1-interacting TGA factors, TGA2 and TGA3, are the principal contributors to an LS7 binding activity of leaves that is enhanced by SA through NPR1. The relevance of these findings to PR-1 expression was investigated by the use of chromatin immunoprecipitation, which demonstrated that in vivo these TGA factors are strongly recruited in an SA- and NPR1-dependent manner to the LS7-containing PR-1 promoter. Significantly, the timing of promoter occupancy by these factors is linked to the SA-induced onset and sustained expression of PR-1. Because leaf transfection assays indicate that TGA3 activates transcription, as noted previously for TGA2, these two TGA factors are predicted to make positive contributions to the expression of this target gene. Thus, the findings presented here distinguish among different modes of regulation by these transcription factors and provide strong support for their direct role in the stimulus-activated expression of an endogenous defense gene.
INTRODUCTION
TGA factors constitute a conserved plant subfamily of basic domain/Leu zipper (bZIP) transcriptional regulators whose genomic targets are thought to include glutathione S-transferase and pathogenesis-related (PR) genes that are associated with detoxification and defense (Klinedinst et al., 2000; Niggeweg et al., 2000a; Johnson et al., 2001a, 2001b; Pontier et al., 2001). A key hallmark of this subclass of bZIP factors is their selective ability to bind as-1–type elements (Izawa et al., 1993; Ulmasov et al., 1995a) that are common to the promoters of PR and glutathione S-transferase genes and that confer transcription in response to defense hormones and xenobiotic stress cues (Liu and Lam, 1994; Ulmasov et al., 1995b; Lebel et al., 1998; Strompen et al., 1998; Niggeweg et al., 2000a; Johnson et al., 2001a; Redman et al., 2002). These observations imply that TGA factors contribute to protective gene responses that are mobilized by plants against microbial pathogens and chemical toxins.
Recent studies of a mediator protein known as NPR1 further imply a regulatory linkage between TGA factors and the expression of PR genes. This protein, which is encoded by a gene known variously as NPR1 (Cao et al., 1994), NIM1 (Ryals et al., 1997), and SAI1 (Glazebrook et al., 1996; Shah et al., 1997), functions in a signal pathway leading from salicylic acid (SA) or its analogs such as 2,6-dichloroisonicotinic acid (INA) to the induction of PR genes and the onset of a global defense program known as systemic acquired resistance (Uknes et al., 1992). Because NPR1 apparently does not bind DNA (Després et al., 2000), it presumably acts through one or more transcription factors that mediate the expression of target PR genes. Consistent with this notion, it has been shown that NPR1 binds specific TGA factors, including TGA2 (AHBP-1a) and TGA3 (Zhang et al., 1999; Després et al., 2000), through their C-terminal domains (Zhou et al., 2000; Fan and Dong, 2002). Moreover, in the absence of functional NPR1 caused by either mutations that disrupt its interaction with TGA factors (Cao et al., 1997; Ryals et al., 1997) or titration using the C-terminal domain of TGA2 (Fan and Dong, 2002), SA and INA failed to induce PR gene expression and systemic acquired resistance. NPR1 may affect the activity of endogenous TGA factors, as inferred from a recent study of a fusion protein between the yeast GAL4 binding domain and a truncated form of TGA2. When expressed in transgenic plants and recovered in nuclear extracts, GAL4-TGA2 was shown to have enhanced binding to the GAL cis element in an INA- and NPR1-dependent manner (Fan and Dong, 2002). Collectively, these findings suggest but do not prove a simple and direct regulatory role for TGA factors in the expression of PR defense genes through their promoter-specific recruitment mediated by NPR1.
However, it also is possible that TGA factors play an indirect role in regulating PR gene expression by interacting with NPR1 to upregulate the expression of other DNA binding transcription factors that are rate limiting to PR gene expression. Possible candidates include members of the WRKY transcription factor family that, like TGA factors, also are implicated in the regulation of PR gene activity (Eulgem et al., 2000). These factors bind W-box cis elements that are present in the promoters of genes that encode both PR genes (Chen et al., 2002) and genes belonging to certain WRKY transcription factors (Eulgem et al., 1999). In the latter case, this suggests the potential for autoregulation. Alternatively, because the W-box element contains within it the core as-1 motif recognized by TGA factors, it is possible that this cis-trans regulatory system mediates the observed SA-induced and NPR1-dependent activation of specific WRKY transcription factors (Yu et al., 2001; Dong et al., 2003).
To distinguish between these direct and indirect regulatory mechanisms, we investigated the contributions of specific Arabidopsis TGA factors to the expression of PR-1. This defense gene is a likely target of regulation by this family of transcription factors, because the PR-1 promoter contains two cognate as-1–type cis elements, LS5 and LS7, which mediate its basal and INA-induced activities, respectively (Lebel et al., 1998). Among the TGA factors of Arabidopsis, we specifically focused on TGA2 and TGA3, because they were shown by several groups to interact strongly with NPR1 (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000). During initial studies, we observed that both factors are present in leaf nuclear extracts and that their steady state concentrations are unaffected by either SA or the presence of NPR1. Furthermore, immunodepletion and gel-shift experiments revealed that TGA2 and TGA3 are major contributors to basal and SA-induced LS7 binding activity in vitro. To validate these findings, we subsequently used a chromatin immunoprecipitation (ChIP) assay (Johnson et al., 2001a) that was modified for use with leaves, allowing us to study the interactions of endogenous TGA2 and TGA3 with the PR-1 promoter in planta. Using ChIP, we found that both factors in leaves are recruited to this promoter in response to a stimulus induction pathway involving SA and NPR1. Moreover, as noted previously for TGA2 (Fan and Dong, 2002), we show here using leaf transfection that TGA3 is an activator of transcription. Therefore, both TGA factors are predicted to make positive contributions to PR-1 expression. Consistent with this view, the timing of promoter occupancy by these factors is linked to the SA-induced onset and sustained expression of PR-1.
RESULTS
Steady State Concentrations of Nuclear TGA2 and TGA3 Are Unaffected by SA and NPR1
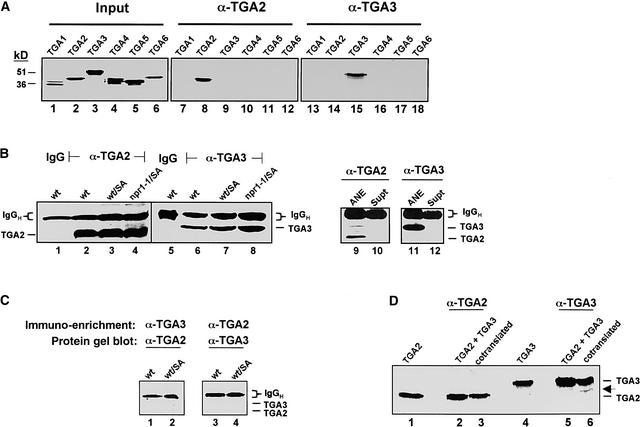
The identities and functions of TGA factors in mediating the expression of specific PR genes remain largely unresolved. To investigate these questions, we first prepared polyclonal antibodies against the most sequence-divergent domains of TGA factors (i.e., the N-terminal domains) (Lam and Lam, 1995; Xiang et al., 1997; Niggeweg et al., 2000b). After affinity purification, the specificity of these antibodies was tested in immunoprecipitation assays (Figure 1A) using in vitro–synthesized, 35S-labeled TGA factors (TGA1 through TGA6) as antigens (lanes 1 to 6). From these and other immunological assays described below, we concluded that the antibodies generated here against TGA2 and TGA3 were specific (lanes 7 to 18). Of particular importance, we observed that antibodies against TGA2 did not recognize TGA5 or TGA6; thus, we were able to distinguish among the most closely homologous members of this gene family that were tested.
Figure 1.
Immunodetection of TGA2 and TGA3 in Leaf Nuclear Extracts.
(A) Specificity of anti-TGA factor antibodies. Lanes 1 to 6, input fractions of in vitro–synthesized, 35S-Met–labeled Arabidopsis TGA factors (TGA1 to TGA6); lanes 7 to 18, products from immunoprecipitation reactions with normalized input fractions of the indicated TGA factors and 1 μg of affinity-purified antibodies against TGA2 (α-TGA2; lanes 7 to 12) or TGA3 (α-TGA3; lanes 13 to 18). Molecular masses (in kilodaltons) of protein markers are indicated.
(B) Immunodetection of nuclear TGA2 and TGA3 proteins. To enrich for these factors, leaf nuclear proteins (500 μg) were incubated with 1 μg of either rabbit control (IgG; lanes 1 and 5) or specific antibodies against TGA2 (α-TGA2; lanes 2 to 4) or TGA3 (α-TGA3; lanes 6 to 8). These conditions resulted in the quantitative and complete recovery of TGA factor antigens from nuclear extracts (ANE; lanes 9 and 11), because the unbound fraction after immunoprecipitation (supernatant [Supt]) lacked detectable amounts of either TGA2 (lane 10) or TGA3 (lane 12). Immunocomplexes were recovered with protein A–Sepharose, washed with RIPA buffer, and fractionated by SDS-PAGE before being examined by protein gel blot analysis with the anti-TGA factor antibodies indicated. Leaf nuclear extracts were obtained from wild-type (wt), SA-treated wild-type (wt/SA), or SA-treated npr1-1 mutant (npr1-1/SA) plants. The expected positions for TGA2 and TGA3 are indicated. IgG heavy chain (IgGH) polypeptide from the primary antibodies is present in all immunoprecipitation reactions, as expected.
(C) Coimmunoprecipitation of TGA2 and TGA3 complexes. Immunodetection of TGA2 and TGA3 was performed as described above except that the antibodies against TGA2 (α-TGA2) and TGA3 (α-TGA3) used for the initial immunoenrichment were reversed in a subsequent detection step involving protein gel blot analysis. The leaf nuclear extracts studied were from wild-type (wt) and SA-treated wild-type (wt/SA) plants. Positions corresponding to TGA2 and TGA3 polypeptides are indicated, in addition to that of the IgG heavy chain (IgGH) polypeptide from immunoprecipitation reactions.
(D) Coimmunoprecipitation assays of in vitro–synthesized TGA2 and TGA3. To test for heterodimer formation between TGA2 and TGA3, constructs that encode full-length versions of these factors were transcribed and translated in vitro in the presence of 35S-Met to label de novo proteins. Immunoprecipitation reactions were performed as described above with antibodies against either TGA2 (α-TGA2) or TGA3 (α-TGA3), with samples containing TGA2 alone (lane 1), a post-translational mixture of TGA2 and TGA3 (lanes 2 and 5), cotranslated TGA2 and TGA3 (lanes 3 and 6), or TGA3 alone (lane 4). After immunoprecipitation, immunocomplexes were fractionated by SDS-PAGE and detected by fluorography. Full-length TGA2 and TGA3 polypeptides are as shown. The arrow indicates the presence of a truncated product of TGA3.
We next determined whether TGA2 and TGA3 were present in leaf nuclear extracts and whether the relative amounts of these factors in this subcellular fraction were affected in vivo by SA and NPR1. Although recombinant forms of TGA2 and TGA3 were detected readily by protein gel blot analysis with their respective antibodies, similar efforts to detect these factors in 5-μg nuclear protein extracts of leaves were not successful. Because TGA2 and TGA3 are likely to be extremely underrepresented among total nuclear proteins, we subsequently attempted to first concentrate each of these factors by immunoprecipitation before their detection by protein gel blot analysis (Figure 1B). Compared with control reactions using rabbit IgG antibodies, which were not expected to result in the recovery of TGA factors (lanes 1 and 5), the use of antibodies against TGA2 and TGA3 resulted in the enrichment of nuclear polypeptides with apparent molecular masses that correspond to those of the factors (lanes 2 and 6). The results of immunoenrichment assays with leaf nuclear extracts from SA-treated wild-type (lanes 3 and 7) and npr1-1 (lanes 4 and 8) plants showed that steady state amounts of TGA2 and TGA3 in nuclei were unaffected by either SA or NPR1. Secondary antibodies used in protein gel blot analysis also revealed the presence of IgG heavy chain polypeptides from the immunoprecipitation reactions.
Although minor changes in the steady state amounts of TGA3 between samples were noted (lanes 6 to 8), a similar trend in the recovery of IgG polypeptide, whose input was identical between the reactions, suggested that these changes resulted from the relative efficiency of recovery of the immunocomplexes. In each case, the immunoprecipitation conditions described above resulted in the quantitative recovery of the two TGA factors (lanes 9 and 11), because the unbound fraction after immunoprecipitation lacked detectable amounts of either TGA2 (lane 10) or TGA3 (lane 12) in a subsequent round of immunorecovery and protein gel blot detection. Moreover, consistent with the notion that these two TGA factors are exclusively nuclear in the cell, we observed in experiments with a yellow fluorescent protein fused to TGA3 that it was localized exclusively to the plant cell nucleus (data not shown), as was observed recently with green fluorescent protein fused to both TGA2 and TGA3 (Pontier et al., 2002). In summary, these findings indicate that neither the rate of nuclear import/export nor the half-life of these factors is a target in their SA-responsive regulation.
TGA3 was not detected in the affinity-purified fraction containing TGA2, and vice versa (Figure 1C, lanes 1 to 4). In addition to confirming that these antibodies were specific, our data suggest that heterodimers of both factors were either absent from nuclei or at very low abundance. This view is consistent with the results of combined in vitro transcription/translation and immunoprecipitation assays, which demonstrated that TGA2 and TGA3 did not form heterodimers when cotranslated (Figure 1D, lanes 1 to 6).
TGA Factor Binding to a PR-1 Element Is SA Inducible and Mediated by NPR1
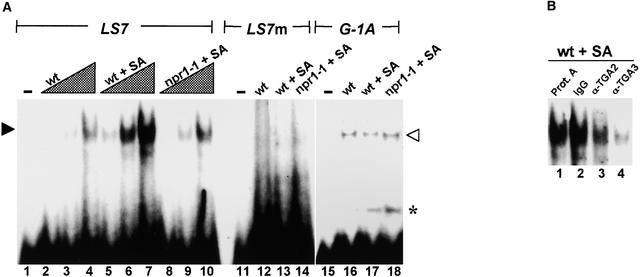
Previous gel-shift assays indicated that the as-1 binding activity of one or more endogenous TGA factors was enhanced through an SA stimulus pathway (Zhang et al., 1999; Després et al., 2000). To investigate this activity further, particularly with regard to contributions by specific TGA factors, we performed similar assays with leaf nuclear protein extracts and the as-1–type LS7 element of the PR-1 promoter (Figure 2A). This element was chosen for study because it is required for the SA-mediated activation of the PR-1 gene (Lebel et al., 1998). Increasing amounts of nuclear proteins from unstimulated leaves of wild-type plants resulted in parallel increases in binding to LS7 (lanes 2 to 4). Although a similar trend was seen in extracts from SA-treated leaves (lanes 5 to 7), the relative degree of LS7 binding activity was markedly higher, in keeping with the results cited above. To determine whether the NPR1 mediator protein affects this activity, we used leaf nuclear extracts from npr1-1 plants that had been treated with SA. The absence of stimulus-enhanced binding to LS7 under these conditions (lanes 8 to 10) indicated that NPR1 was involved in the observed SA-induced response.
Figure 2.
In Vitro Binding by Nuclear TGA Factors to the LS7 Element of PR-1.
(A) Gel-shift binding assay with leaf nuclear extracts. Lanes 1 to 10, DNA–protein complexes between labeled LS7 probe of the PR-1 promoter and leaf nuclear proteins; lane 1, probe alone; lanes 2 to 4, 1, 3, and 9 μg of nuclear protein from leaves of untreated wild-type plants (wt); lanes 5 to 7, 1, 3, and 9 μg of nuclear protein from leaves of SA-treated wild-type plants (wt + SA); lanes 8 to 10, 1, 3, and 9 μg of nuclear protein from leaves of SA-treated npr1-1 mutant plants (npr1-1 + SA); lanes 11 to 14, labeled LS7 mutant probe (LS7m) alone (lane 11) or with 9 μg of nuclear protein from leaves of untreated wt (lane 12), wt + SA (lane 13), or npr1-1 + SA (lane 14) plants; lanes 15 to 18, labeled G-box probe (G-1A) alone (lane 15) or with 9 μg of nuclear protein from leaves of untreated wt (lane 16), wt + SA (lane 17), or npr1-1 + SA (lane 18) plants. Closed and open arrowheads indicate TGA factor and G-box factor complexes with their respective LS7 and G-1A probes. The asterisk indicates the presence of a lower mobility complex bound to the G-1A probe.
(B) Immunodepletion assay of TGA2 and TGA3. In each case, 9 μg of nuclear protein from SA-treated wild-type leaves (wt + SA) was incubated with protein A–Sepharose resin (Prot. A; lane 1), 1 μg of rabbit control (IgG; lane 2), anti-TGA2 antibody (α-TGA2; lane 3), or anti-TGA3 antibody (α-TGA3; lane 4) bound to resin. After a brief spin, supernatants from these samples were used in standard gel-shift assays with radiolabeled LS7 as a probe.
To test the specificity of these DNA–protein interactions, we used as a probe a mutant LS7 element (LS7m) that contains point mutations that block the activation of the PR-1 promoter by SA (Lebel et al., 1998). As expected, no specific DNA binding activity was observed with all extracts tested using this probe (lanes 12 to 14). To further test whether the effects of SA treatment and NPR1 were specific to LS7 binding factors, we used a G-box element (G-1A) as the gel-shift probe. Although this element shares a common ACGT core motif with as-1–type elements, it is bound selectively by G-box factors (Schindler et al., 1992). Binding of these factors to G-1A generally was similar among the nuclear extracts (lanes 16 to 18), indicating that differential changes in LS7 binding activities between the extracts were specific. Surprisingly, a lower mobility factor (asterisk) also was detected with the G-1A probe in extracts from SA-treated but not control leaves. The significance of this observation is unclear, but it suggests that binding by other nuclear factors to DNA also is stimulated by SA.
We next determined whether TGA2 and TGA3 in leaf nuclear extracts contribute to LS7 binding. To this end, we incubated leaf nuclear extracts from 16-h SA-treated leaves with specific antibodies under conditions that were shown to quantitatively recover the cognate TGA factor. These immunodepleted extracts then were incubated under standard gel-shift conditions with labeled probe to detect the remaining LS7 binding activity (Figure 2B). Control reactions included similar incubations of extracts with either protein A–Sepharose resin alone or with rabbit IgG (lanes 1 and 2). Compared with that of controls, the immunodepletion of TGA2 or TGA3 from extracts of SA-treated plants showed a marked reduction in LS7 binding (lanes 3 and 4), indicating that these two factors are the primary contributors to this activity.
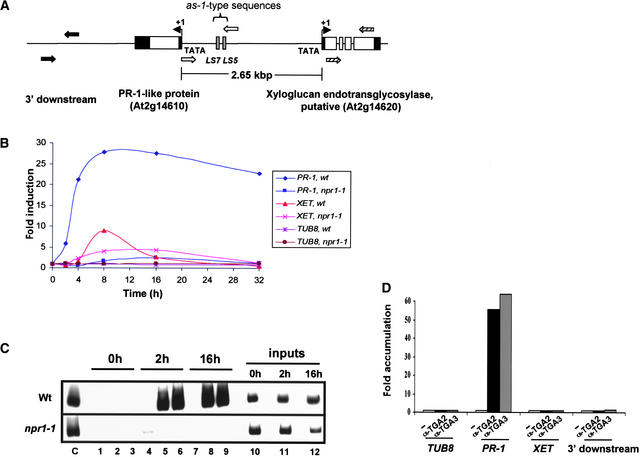
PR-1 and XET Are Transcriptionally Divergent Genes
The PR-1 promoter is embedded in an ∼2.65-kb intergenic region that lies upstream of a putative member of the xyloglucan endotransglycosylase (XET) gene family (Figure 3A). The relative orientation of PR-1 and XET to each other suggests the possibility of their divergent and coordinated regulation. To test this notion, we analyzed the relative amounts of steady state transcripts of these genes by reverse transcriptase–mediated (RT) PCR (Figure 3B). In contrast to the constitutively expressed TUB8 tubulin gene, which served as an internal control, SA treatment resulted in the maximal induction of both PR-1 and XET. Intriguingly, the timing and extent of the induction of expression of these two genes were somewhat different, as were the relative effects of NPR1 as shown in similar experiments with npr1-1 plants. Together, these data indicate that PR-1 and XET are coregulated by SA in a similar, but not identical, manner.
Figure 3.
In Vivo Recruitment of TGA2 and TGA3 to the PR-1 Genomic Promoter.
(A) Diagram of the PR-1 locus and flanking regions. The PR-1 locus on Arabidopsis chromosome 2 (T6B13 clone) contains the transcriptionally divergent genes PR-1 and a putative XET spaced ∼2.65 kb apart. (Note that “PR-1-like protein” is the designation for PR-1 in the TAIR database [http://www.tair.org].) In the shared intergenic region between PR-1 and XET, only two as-1–type elements (LS5 and LS7) are present (Lebel et al., 1998; our observations using the plantCARE software of Lescot et al. [2002]). Untranslated regions (black boxes), exons (white boxes), and the direction and start site of transcription (solid arrows at +1) are indicated. Also, primer pairs used in ChIP and RT-PCR analyses are those that amplify intergenic sequences present downstream of PR-1 (black arrows), the LS5- and LS7-containing intergenic region of the PR-1 promoter (white arrows), and coding sequences of the XET gene (hatched arrows).
(B) Effect of the SA and NPR1 signal pathway on PR-1 and XET expression in leaves. Transcripts of PR-1 and XET were measured by RT-PCR using total RNA from leaves of Arabidopsis wild-type (wt) and npr1-1 mutant (npr1-1) plants that had been treated with SA for 0 to 32 h. DNA products of these reactions were fractionated by agarose gel electrophoresis and stained with ethidium bromide for quantification using the Eagle Eye II still-video system. To facilitate comparisons between genes, changes in the amount of their corresponding transcripts were converted to fold induction by normalizing all values from SA-treated samples to those of the untreated (0-h) sample. Transcripts of a constitutively expressed β-tubulin gene (TUB8) also were analyzed as an internal control for the specificity of induction.
(C) ChIP assays of in vivo DNA binding by TGA2 and TGA3. In vivo DNA–protein complexes in chromatin were covalently cross-linked by formaldehyde and recovered from leaves of wild-type (Wt) and npr1-1 (npr1-1) Arabidopsis plants that had been treated with SA for 0 h (lanes 1 to 3), 2 h (lanes 4 to 6), or 16 h (lanes 7 to 9). Chromatin then was sonicated to yield a population of soluble fragments and incubated with BSA as a negative control (lanes 1, 4, and 7) or with specific antibodies against either TGA2 (lanes 2, 5, and 8) or TGA3 (lanes 3, 6, and 9). Target PR-1 promoter sequences were detected by PCR using specific primers and α-32P-dCTP. Reaction products were fractionated by PAGE and visualized using autoradiography. Crude chromatin (inputs) corresponding to ∼1/200th of the total sample used in the other lanes was analyzed similarly by PCR (lanes 10 to 12) to show that equal amounts of chromatin template were programmed in ChIP reactions. Lane C shows a standard PCR result using cloned PR-1 promoter as a template.
(D) Relative recoveries of chromatin sequences bound in vivo by TGA2 and TGA3. ChIP assays were performed as in (C) with chromatin from wild-type leaves treated for 16 h with SA. The resulting ChIP products then were analyzed by PCR using primers that amplify the coding sequence of a control β-tubulin gene (TUB8) that lacks identifiable TGA factor binding sites, the PR-1 promoter (PR-1), the coding sequence of the flanking XET gene (XET), and the intergenic region that is 3′ to PR-1 (3′ downstream). Histogram plots of a single experiment are shown, with PCR products corresponding to ChIP control reactions in the absence of specific antibody (white bars) or with test reactions with either anti-TGA2 antibody (α-TGA2; black bars) or anti-TGA3 antibody (α-TGA3; gray bars). Values were plotted as the relative fold accumulation of ChIP products from specific immunoprecipitation versus control (no-antibody) reactions.
SA and NPR1 Mediate the in Vivo Recruitment of TGA2 and TGA3 to the PR-1 Promoter
In vitro binding assays with the LS7 promoter element (Figure 2) suggested a regulatory role for TGA2 and TGA3 in the expression of PR-1. To validate these findings, we used ChIP to determine whether binding between these factors and the promoter of this gene occurs in vivo. ChIP involves several steps: formation of in vivo cross-links between chromatin DNA and its bound proteins by treatment with formaldehyde; recovery of cross-linked chromatin and its subsequent sonication to smaller fragments; immunoenrichment of complexes of interest; and finally, reversal of cross-links in recovered DNA to facilitate the detection of specific immunoenriched sequences by PCR (Orlando, 2000). Reactions performed without specific antibodies indicate the background noise of the system and provide a key negative control in ChIP assays. In addition, PCR of input chromatin samples serves to verify that similar amounts of starting material were used during this last step.
Three independent ChIP experiments were performed, and results from a representative assay are shown here (Figure 3C). In these experiments, cross-linked chromatin from leaves of wild-type and npr1-1 plants that had been treated with SA for 0 to 16 h were incubated with antibodies against either TGA2 or TGA3 to enrich for complexes between these TGA factors and the PR-1 promoter. Primers used to detect complexes containing the PR-1 promoter amplified an ∼1-kb fragment that contains both as-1–type elements (i.e., LS5 and LS7) that bind these factors in vitro (Després et al., 2000) (Figure 2). As expected, control reactions performed in the absence of specific antibodies did not result in the recovery of chromatin fragments containing this promoter (lanes 1, 4, and 7). In addition, leaves from wild-type plants treated for 0 h showed no evidence of recruitment of TGA2 or TGA3 (lanes 2 and 3). On the other hand, treatment with SA for 2 h (lanes 5 and 6) or 16 h (lanes 8 and 9) resulted in the marked recruitment of these TGA factors to a chromatin fragment that contains the PR-1 promoter. The amount of signal detected was ∼2% of the input fraction, which is comparable to findings from ChIP studies with yeast transcription factors (Kuras and Struhl, 1999; Li et al., 1999b) or with a tobacco TGA factor in suspension cells (Johnson et al., 2001a).
That NPR1 also is required for this recruitment was shown by negative results in analyses of npr1-1 plants. Because the amounts of PCR products from different chromatin input samples were comparable (lanes 10 to 12), the ChIP results described above are not attributable to varying amounts of starting material. PCR was performed with 28 cycles of amplification, which was within the linear range of this assay. Thus, the findings presented here demonstrate that TGA2 and TGA3 are recruited in vivo to an intergenic region containing the PR-1 promoter in response to an SA signal pathway involving NPR1.
In the process of interpreting these data, we also considered the resolving power of ChIP. Although the average size of chromatin fragments used was ∼1 kb, their range can extend to several kilobases. Thus, some TGA factor–bound chromatin fragments may contain not only the promoter of PR-1 but additional flanking regions as well. If these larger fragments also contain TGA factor binding sites, it is possible that they could contribute to the PCR signals seen here with PR-1–specific primers. To address this issue, we designed primers to proximal and adjacent sequences that flank the intergenic region under study (Figure 3A) or to the coding sequence of a distal β-tubulin gene (i.e., TUB8), which served as an additional control for the specificity of ChIP reactions. The results of PCR assays with these primers showed that neither the flanking sequences to the PR-1 intergenic region nor those of the more distal TUB8 gene were present above background in immunoenriched ChIP samples (Figure 3D). Therefore, this method is able to spatially distinguish interactions between TGA factors at the PR-1 promoter from those that might occur at flanking or distal sites. However, we note that ChIP is unable to resolve adjacent binding interactions by a common factor when these events occur within ∼150 bp of each other (Johnson et al., 2001c). Because the only known or predicted binding sites (LS5 and LS7) for TGA factors are within ∼30 bp of each other, we were unable to map specific interactions by these factors to one or the other of the sites.
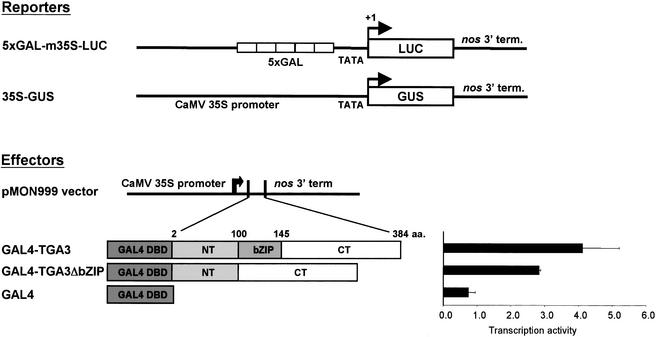
TGA3 Functions as a Transcriptional Activator
Because the SA-induced recruitment of TGA2 and TGA3 to the promoter of PR-1 was correlated strongly with an increase in the steady state expression of this gene, it follows that both TGA factors are likely to be activators of transcription. Although this function has been shown previously for TGA2 (Fan and Dong, 2002), the ability of TGA3 to mediate transcription has been unexplored to date. To address this point, the activity of this factor was tested using a heterologous transcription system in which the effectors consisted of either the yeast GAL4 DNA binding domain alone or fused to full-length TGA3 or TGA3ΔbZIP. Reporter and effector DNAs (Figure 4) were transfected into intact Arabidopsis leaves by particle bombardment. After SA treatment for 20 h, reporter gene activities were measured. Because transient transfection resulted in amounts of effector proteins that were difficult to detect by protein gel blot assay, we normalized the data by titrating the activity of each effector in a dose–response transfection assay (data not shown) and then used the amount that resulted in the maximal expression of the reporter gene.
Figure 4.
Trans-activation by Chimeric TGA3 Factors.
Effector and reporter constructs were used in transfection assays. Reporter constructs were as follows. 5xGAL-m35S-LUC, a synthetic promoter with five tandem GAL4 cis elements, was fused to the minimal TATA box–containing promoter (−46-bp region) of the 35S promoter of Cauliflower mosaic virus (CaMV) located upstream of the firefly luciferase (LUC) gene; 35S-GUS, a full-length CaMV 35S promoter was fused to the bacterial β-glucuronidase (GUS) gene (Jefferson et al., 1987). Effector constructs were as follows. The DNA binding domain (DBD) of the yeast GAL4 factor was used as a control or fused in frame to the cDNA of TGA3 or TGA3ΔbZIP. Both factors then were cloned downstream of the full-length CaMV 35S promoter in the pMON 999 vector. TGA3 is composed of an N-terminal domain (NT; amino acids [aa.] 2 to 100), a basic domain/Leu zipper domain (bZIP; amino acids 101 to 145), and a C-terminal domain (CT; amino acids 146 to 384). For the transient transfection assay, Arabidopsis rosette leaves were transfected by particle bombardment with reporter and effector DNA. The 35S-GUS reporter was used as an internal control for transfection efficiency in all experiments. All effector DNAs were used in saturating amounts that resulted in maximal expression of the 5xGAL-m35S-LUC reporter gene. After bombardment, leaves were treated with SA and incubated for 20 h. LUC activity was normalized to the activity of the GUS internal control (expressed as a ratio) and expressed as arbitrary units of activity. Means and standard errors for data from three independent experiments are shown.
Compared with GAL4 alone (Figure 4), the expression of the GAL4-TGA3 fusion protein resulted in an ∼4.5-fold increase in reporter gene activity. Similar results were obtained with GAL4-TGA3ΔbZIP, which lacks the bZIP domain, demonstrating that heterodimerization with native TGA factors through the Leu zipper of this domain is unlikely to explain the observed trans-activator function of TGA3. Thus, we propose that TGA3, like TGA2 (Fan and Dong, 2002), activates the transcription of in vivo target genes.
DISCUSSION
Gene expression networks in plants are governed largely by direct associations between specific transcription factors and their cognate target promoters. Here, we investigated whether specific TGA factors (i.e., TGA2 and TGA3) make direct regulatory contributions in the SA- and NPR1-dependent stimulus pathway leading to enhanced expression of the PR-1 defense gene. Both factors are inferred to be part of this transcriptional response because they interact with NPR1 and bind in vitro to an SA-responsive element (LS7) of the PR-1 promoter. Significantly, new evidence shown here demonstrates that these two trans-activating TGA factors are recruited in vivo to the PR-1 promoter in response to an SA signal pathway involving NPR1. Recruitment of these factors is correlated with the onset of PR-1 expression, thus strongly implying biological relevance. These and other studies suggest that defense- and xenobiotic stress–induced changes in sequence-specific DNA binding activity may be a common regulatory mechanism among TGA factors (Jupin and Chua, 1996; Stange et al., 1997; Johnson et al., 2001a).
Computational analysis of the intergenic sequence upstream of PR-1 showed that the only predicted binding sites for TGA factors are two as-1–type elements, LS5 and LS7, centered at nucleotides −670 and −640, respectively, from the start site of transcription. Linker-scanner mutation analysis of the PR-1 promoter indicated that these elements are functionally distinct, because each acts in concert with its cognate factor(s) to either inhibit the basal activity of PR-1 or stimulate its expression in response to SA (Lebel et al., 1998). Other distinctions between these elements include their in vivo occupancy states. For example, it was reported that the in vivo footprint at LS7 is enhanced by INA treatment, suggesting a stimulus-enhanced change in its occupancy, whereas binding to LS5 appears to be entirely constitutive (Lebel et al., 1998). Given that TGA2 and TGA3 are activators of transcription (Fan and Dong, 2002) (Figure 4) whose recruitment to the PR-1 promoter is stimulated by SA, we surmise that they selectively bind in vivo to the LS7 element. However, current limitations in the resolving power of ChIP (Johnson et al., 2001c) preclude a conclusive determination of the binding specificity of the TGA factors used here between the closely spaced LS5 and LS7 elements.
As noted above, LS5 normally represses the transcription of PR-1. Candidate binding factors may include TGA factors, which make negative contributions to PR gene expression (Pontier et al., 2001). Alternatively, another inhibitory factor that binds LS5 may confer this function. Because this as-1–type motif is part of a W-box (TTGAC) element, it may bind an inhibitory member of the WRKY family of transcription factors, which are known to specifically bind W-box elements (Eulgem et al., 2000). Future experiments to identify specific proteins that bind LS5 in vivo may distinguish between these possibilities.
Developmental and cellular expression profiles of TGA factors suggest additional levels of complexity in their regulatory interactions with the PR-1 promoter. Recent experiments show that TGA3 is expressed primarily in a subset of young leaves, whereas TGA2 is distributed more widely in leaves at different stages of development (Pontier et al., 2002). Because the leaves used here represent a spectrum of developmental stages, we surmise that the observed SA-induced interactions between these TGA factors and the PR-1 promoter represent a composite of events that occur in different leaf cells. Moreover, although TGA factors have been found to bind only their cognate cis elements as dimers (Katagiri et al., 1992; Niggeweg et al., 2000b), our evidence indicates that TGA2 and TGA3 do not form mixed dimers in vitro and in vivo. Whether they form combinatorial interactions in vivo with other proteins or TGA factors remains to be determined. Although TGA2 and TGA3 show functional redundancy in their PR-1 promoter binding and transcription activities, as demonstrated above, they may be distinguished in planta by cell type and developmentally specific roles.
Changes in nuclear trans-localization or half-life affect the DNA binding activities of some plant bZIP transcription factors (Terzaghi et al., 1997; Ang et al., 1998). Therefore, we investigated the relative amounts of nuclear TGA2 and TGA3 proteins and found that they were unaffected by either SA or NPR1. These findings are supported by the observations that green fluorescent protein and yellow fluorescent protein, when fused to TGA2 and TGA3, are localized to plant nuclei in the absence of an apparent stimulus (Pontier et al., 2002; data not shown). Thus, stimulus-induced changes in the activities of these transcription factors do not arise from their relative nuclear concentrations.
The in vivo recruitment of TGA2 or TGA3 to the PR-1 promoter precedes the SA-induced expression of a transcriptionally divergent XET gene, which encodes a putative xyloglucan endotransglycosylase. These enzymes remodel the cell wall during development and in response to environmental cues. For example, microarray studies have shown that several XTR (XET-related) genes are upregulated in response to wounding, pathogens, and abiotic stress (Cheong et al., 2002). Although the expression of the XET gene that lies upstream of PR-1 also is enhanced by SA, differences in its relative dependence on NPR1 and in the kinetics of its induction compared with PR-1 suggest that these genes are distinguished by additional levels of transcriptional control.
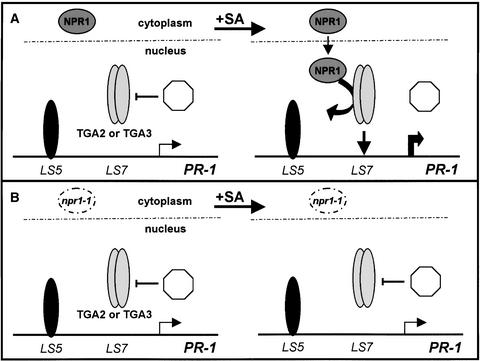
Results from this and other studies now can be integrated into the following model to account for direct contributions by TGA2 and TGA3 to the expression of PR-1 (Figure 5). In nonstimulated leaf cells, results from linker-scanner mutations and in vivo footprinting assays (Lebel et al., 1998) suggest that the LS5 element is constitutively occupied by an inhibitory protein (TGA or WRKY factor?) that keeps basal transcription to a minimum. Under these conditions, nuclear TGA2 and TGA3 factors in different cell types and developmentally staged leaves (Pontier et al., 2002) are unable to bind their target PR-1 promoter sequence. This inhibitory state is thought to result from the action of a nuclear repressor that may either alter the post-translational state of these TGA factors (Stange et al., 1997) or block their DNA binding activity through a direct physical interaction. Precedent for this mechanism of regulation among TGA factors comes from a study of TGA1a, which shows that its as-1 binding activity is reversibly inhibited by a nuclear repressor protein in vivo (Johnson et al., 2001b). In the case of TGA2 and TGA3, one plausible repressor is the protein product of SNI1, which inhibits the SA- and NPR1-mediated expression of PR genes (Li et al., 1999a). Alternatively, NIM1 (NPR1)-interacting proteins that form a ternary complex with NPR1 and TGA2 (Weigel et al., 2001) may inhibit this factor's DNA binding activity. Comparative studies of as-1 binding activities in extracts from wild-type and sni1- or nimin-defective lines may provide a critical test of these hypotheses.
Figure 5.
Model of PR-1 Transcriptional Regulation.
(A) Wild-type plants. In untreated leaves of wild-type plants, NPR1 is localized to the cytoplasm. Under these conditions, TGA2 or TGA3 in the nucleus interacts with a nuclear repressor (octagon) that inhibits its DNA binding activity, whereas an inhibitor protein (solid oval) at the LS5 element keeps basal transcription to a minimum (small arrow). After exposure to SA, the cytoplasmic form of NPR1 is mobilized to the nucleus, where it binds TGA factors and consequently displaces the repressor. The NPR1-TGA factor complex then is recruited to the target PR-1 promoter, resulting in the release of NPR1 and a net increase in the expression of this gene (large arrow).
(B) npr1-1 mutant plants. The absence of a functional NPR1 protein (open circle) results in a constitutive interaction between the repressor and its cognate TGA factor. Consequently, PR-1 expression no longer is activated by SA (small arrow).
The penultimate step leading to the SA-induced activation of PR-1 requires the translocation of NPR1 to the nucleus (Kinkema et al., 2000), where it interacts with the C-terminal domain of TGA2 or TGA3 (Zhou et al., 2000; Subramaniam et al., 2001; Fan and Dong, 2002). This interaction is predicted to counter the effect of the putative repressor, thus promoting the recruitment of these trans-activating TGA factors to the promoter of PR-1 with a consequent upregulation of the gene. Additional factors, such as those that bind an NF-κB–like motif of this promoter, also may contribute to this gene response (Lebel et al., 1998).
Considering that recombinant and nuclear forms of TGA2 have similar electrophoretic mobilities in gel-shift assays (Lam and Lam, 1995; Després et al., 2000; our unpublished data), it is unlikely that the DNA-bound factor continues to interact with either NPR1 or the repressor. The basis for this behavior is unknown, but it may result from a change in the conformation of the DNA-bound factor that sterically blocks the NPR1 and repressor binding site(s). Another logical assumption is that the stimulus-induced recruitment of trans-activating TGA factors overcomes the basal inhibitory effect of LS5, resulting in a net gain in transcription. By contrast, the absence of a functional NPR1 protein in npr1-1 plants blocks the SA-induced recruitment of these TGA factors and an increase in PR-1 expression.
In summary, the findings presented here distinguish among different modes of regulation by specific endogenous TGA factors and provide strong support for their direct roles in the SA- and NPR1-dependent activation of a PR defense gene. Although they provide new insights into transcriptional control in planta, the approaches described here also are relevant to efforts aimed at understanding the control of plant gene networks in a wide range of plant developmental and adaptive processes.
METHODS
Plant and Tissue Growth Conditions
Seeds of Arabidopsis thaliana ecotype Columbia (wild type) and an npr1-1 mutant line were cold shocked for 3 days, transferred to Metro-Mix 250 (Geiger Companies, Beltsville, MD), and grown to principal growth stage 5 (∼27 days after germination) under conditions described previously (Boyes et al., 2001). In some experiments, plants then were sprayed with either 1 mM salicylic acid (SA) in 0.02% Silwet L-77 (Lehle Seeds, Round Rock, TX) or with carrier solvent alone and incubated as described above.
Antibodies
Polyclonal antibodies were raised against the N-terminal (NT) ends of TGA2 and TGA3 that were expressed as glutathione S-transferase fusion proteins. TGA2-NT and TGA3-NT sequences were amplified by PCR from cDNA clones of the factors with the following primers: for TGA2, 5′-TTTGAATTCTCTGCTGATACCAGTCCGAGAAC-3′ and 5′-TTTATCGATGTCAAAGAGTCTTTTGATCCATC T-3′; for TGA3, 5′-TTTGAATTCGAGATGATGAGCTCTTCTTCTTCTACTACT-3′ and 5′-TTTATCGATGTCATTTCATCTTATCATTGATCC-3′. These sequences were subcloned initially into pBluescript KS+ vector (Stratagene), sequenced to verify the absence of mutations, and then subcloned into pGEX-4T1 (Pharmacia). Glutathione S-transferase fusion protein (∼1.5 mg) was recovered on glutathione-Sepharose 4B resin, fractionated by SDS-PAGE, and used for rabbit immunizations in a 70-day protocol (Strategic Biosolutions, Newark, DE). The antisera collected were first immunodepleted of glutathione S-transferase–specific antibodies and then affinity purified for antibodies against TGA2 and TGA3, as described (Harlow and Lane, 1988). The specificity of all antibodies was evaluated by immunoprecipitation against in vitro–synthesized, 35S-Met–labeled TGA factors.
Immunoprecipitation Assay
Leaves were harvested from ∼4-week-old Arabidopsis plants, and nuclear proteins were extracted from this material according to Lam and Lam (1995). Leaf nuclear proteins (0.5 mg) then were incubated on ice for 2 h with 1 μg of affinity-purified anti-TGA2 or anti-TGA3 antibody, or rabbit IgG (Sigma) as a negative control, and recovered with protein A–Sepharose (Pharmacia). Immunocomplexes were washed several times with RIPA buffer (Harlow and Lane, 1988), denatured in Laemmli buffer (Promega), fractionated by SDS-PAGE, and transferred to nitrocellulose. Nuclear TGA2 and TGA3 recovered by immunoprecipitation were transferred to nitrocellulose and identified using standard protein gel blot conditions with 5 μg/mL affinity-purified anti-TGA2 or anti-TGA3 as a primary antibody and donkey anti-rabbit horseradish peroxidase as a secondary antibody (Amersham). Immunocomplexes were identified using a chemiluminescent detection system (Pharmacia). Immunoprecipitation assays with in vitro–synthesized, 35S-labeled TGA factors differed only in that immunocomplexes were fractionated by SDS-PAGE and then detected by fluorography of the dried gel.
Gel-Shift Assay
DNA binding reactions (20 μL) and conditions were as described (Lam and Lam, 1995) except that samples were incubated on ice before fractionation by native PAGE. After end labeling with γ-32P-ATP (6000 Ci/mmol; Amersham) and polynucleotide kinase, sense and antisense oligonucleotides, respectively, of the following promoter sequences were used as probes in gel-shift assays with the following primers: for LS7, 5′-TTACTTACGTCATAGATGTGGC-3′ and 5′-GCCACATCTATGACGTAAGTAA-3′ (Lebel et al., 1998); for LS7m (LS7 mutated bases are underlined), 5′-TTACTTTTTCTAGATGTGTGGC-3′ and 5′-GCCACACATCTAGAAAAAGTAA-3′ (Lebel et al., 1998); for G-1A, 5′-GGGTACTTATCTTCCACGTGGCATTATTCTCAT-3′ and 5′-GGGATGAGAATAATGCCACGTGGAAGATAAGTA-3′ (Schindler et al., 1992). Radiolabeled free or bound probe was detected on dried gels by autoradiography.
Immunodepletion
Specific contributions by individual nuclear TGA factors to LS7 binding activity were identified by incubating leaf nuclear extracts with or without 1 μg of control or affinity-purified antibodies against TGA2 or TGA3 for 1 h on ice. This was followed by incubation for 30 min with 20 μL (packed volume) of protein A–Sepharose to recover immunocomplexes. After a brief centrifugation, the supernatant fraction was used in standard gel-shift assays. Relative differences in binding activity were assessed by analysis of labeled LS7-bound complexes using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Chromatin Immunoprecipitation
Cross-Linking and Isolation of Chromatin
After treatment with or without 1 mM SA, ∼50 g of both young and mature leaves was harvested from ∼4-week-old wild-type and npr1-1 mutant plants, washed briefly with water, and diced into ∼5-mm2 pieces before being placed in 400 mL of ice-cold TBS (20 mM Tris, pH 7.6, and 200 mM NaCl) plus 0.01% Silwet. To cross-link chromatin, formaldehyde was added immediately to a final concentration of 1%. Leaves were vacuum-infiltrated with fixative by two tandem vacuum-and-release cycles (25 to 30 inches of Hg), fixed for 30 to 60 min, and washed twice for 4 h with 500 mL of fresh TBS plus 0.3 M Gly at 4°C. As a control, chromatin from unfixed cells also was prepared. In all cases, leaves were frozen in liquid nitrogen and stored at −80°C until processed further. Frozen leaves were ground to a fine powder under liquid nitrogen with a mortar and pestle and resuspended in 150 mL of lysis buffer I (50 mM Hepes-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1.0% Triton X-100, and 0.1% sodium deoxycholate [DOC]) supplemented with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/mL each aprotinin, leupeptin, and bestatin, and 2 μg/mL antipain) and 10% glycerol.
Homogenization was continued by several intermittent cycles with a Polytron, and the resultant slurry was filtered through two layers of Miracloth (Calbiochem). Crude nuclei in the filtrate were collected by centrifugation at 5000g (GSA rotor; Sorvall, Newtown, CT) for 20 min at 4°C and resuspended in 10 mL of lysis buffer I plus 10% glycerol. This fraction was agitated with a vortex for 5 min for complete dispersal and collected by centrifugation as described above. This wash step was repeated two more times. The final pellet of crude nuclei was resuspended in 3 mL of lysis buffer I plus 10% glycerol and sonicated on ice, with three cycles of 15 s of continuous pulse and intermittent cooling in an ice/ethanol bath between sonication cycles. The sample then was adjusted to 8 mL with lysis buffer I plus 10% glycerol, 1.0% Triton X-100, and 0.1% DOC and clarified by centrifugation at 5000g for 15 min at 4°C. The supernatant, which contained fixed soluble chromatin, was retained. The remaining pellet was resuspended in 3 mL of lysis buffer I plus 10% glycerol and sonicated and centrifuged as described above. Combined supernatants from both steps were divided into aliquots, flash-frozen in liquid nitrogen, and stored at −80°C. Although fixed chromatin for chromatin immunoprecipitation analysis typically was stored for only a few weeks, longer term storage may be possible.
Quantification of Chromatin DNA
Aliquots of fixed chromatin were treated at 37°C with 1 μg/mL RNase A for 30 min and then with 10 μg/mL proteinase K for 30 min. Samples were incubated overnight at 65°C to reverse formaldehyde-induced cross-links and spun at the highest speed in a microcentrifuge for 5 min to clarify. The supernatant was extracted with phenol:chloroform:isoamyl alcohol (IAA) (25:24:1) followed by chloroform:IAA (95:5). DNA was collected by ethanol precipitation at −20°C for 3 h or overnight in the presence of 10 μg of linear acrylamide (Ambion, Austin, TX), after which the pellet was washed briefly with 70% ethanol and air-dried. The pellet then was resuspended in 25 μL of Tris-EDTA incubated at 37°C for several minutes and agitated vigorously to resuspend. DNA was quantified using a fluorometer and PicoGreen staining (Molecular Probes, Eugene, OR). In addition, the size range of the fragmented chromatin DNA was determined by electrophoresis with a 1% agarose gel and ethidium bromide staining.
Immunoprecipitation
Chromatin samples were thawed on ice and centrifuged at top speed in a microcentrifuge for 5 min to clarify. Aliquots corresponding to 2.5 to 5 μg of DNA were brought to a final volume of 1 mL with lysis buffer I. To these samples was added either 1 μg of acetylated BSA as a negative control or 1 μg of affinity-purified anti-TGA2 or anti-TGA3 antibody. Binding reactions were incubated overnight at 4°C with gentle mixing throughout. Then, 50 μL (packed volume) of protein A–Sepharose equilibrated 1:1 in lysis buffer I was added to samples with continued incubation for 1 h. With gentle mixing, resin was washed sequentially for 5 min at 4°C with 1 mL of the following buffers: lysis buffer I, lysis buffer II (same as lysis buffer I except with 500 mM NaCl), wash buffer (10 mM Tris, pH 8.0, 0.25 M LiCl, 0.5% Nonidet P-40 [Igepal CA-630; Sigma], 0.5% DOC, and 1 mM EDTA), and TBS (20 mM Tris, pH 7.6, and 200 mM NaCl). After the final wash, resin was resuspended in 300 μL of TES (25 mM Tris, pH 7.5, 10 mM EDTA, and 0.5% SDS). To elute immunoprecipitated material, samples were heated for 10 min at 65°C and centrifuged briefly, and the supernatant was collected (250 μL). The TES extraction and collection steps were repeated once more, after which supernatants were pooled and treated to reverse cross-links, as described above, for subsequent PCR analysis.
PCR
The general conditions used for PCR were as described previously (Johnson et al., 2001a) except that the dCTP concentration was optimized further. The primer pairs used (Figure 3A) were as follows: (1) as-1–containing sequences of PR-1 (5′-TCGGAGGGAGTATATGTTATTGCTTAGAATCA-3′ and 5′-TTGTTTCGTATCGGTAGCTTTGCCAT-3′) to show specificity; (2) PR-1 promoter flanking sequences, including the upstream XET coding sequence (same as the reverse transcriptase–mediated [RT] PCR primers) and the 3′ downstream intergenic sequences (5′-TGTTCGTGTATCGACAAACCGAAAACACTAT-3′ and 5′-TTGTTCAAATGTTATGGTACAAGAAGAATCCAGA-3′), to show the resolving power of chromatin immunoprecipitation; and (3) coding sequences of a gene distal to the PR-1 promoter, namely TUB8 (same as the RT-PCR primers), as a negative control. After amplification for 28 cycles, radiolabeled products of PCR were fractionated on 5% PAGE gels in 0.5× Tris-borate/EDTA (Sambrook et al., 1989) and detected by autoradiography.
RNA Extraction and RT-PCR
Total RNA from leaves was extracted using a hot phenol procedure (Verwoerd et al., 1989). RT-PCR was performed in a two-step method using 50 ng of RNA as a template. PCR products were fractionated by 1% agarose gel and ethidium bromide staining and quantified using the Eagle Eye II still-video system (Stratagene). Primers that span portions of the coding sequences of the following genes were used for RT-PCR analysis: for PR-1, 5′-CTTTGTAGCTCTTGTAGGTGCTCTTGTTC-3′ and 5′-TCCTGCATATGATGCTCCTTATTGAAATACTGAT-3′; for XET, 5′-AAATCGCTCAAAACCATTTGTACTTCTCG-3′ and 5′-TATTTAGGAAGAGAACATTCCTTCGGTAGC-3′; for TUB8, 5′-CTCCTGCACTTCCACTTCGTCTTC-3′ and 5′-CGTGGATCACAGCAATACAGAGCC-3′. The linear range of PCR product synthesis was established for each primer pair, and based on these findings, the conditions used in RT-PCR assays were chosen to reflect the midpoint of this range.
Transient Transfection
Reporter and Effector Constructs
Reporter constructs included 5xGAL-m35S-LUC (Padidam and Cap, 2001) and 35S-GUS (Pascuzzi et al., 1998). Effector constructs were made as described previously (Pascuzzi et al., 1998). Using TGA3 cDNA as a template, full-length TGA3 was amplified by PCR using the following primers: TGA3 (4), 5′-TTTGAATTCGAGATGATGAGCTCTTCTTCTTCTACTACT-3′; and TGA3 (1152), 5′-TTTATCGATGTCAAGTGTGTTCTCGTGGACGAG-3′. TGA3ΔbZIP required a more complex approach of first amplifying the N-terminal domain with TGA3 (4) and TGA3-NT (a fusion of amino acids 433 and 294; 5′-ATTGCGTACGCATAATCCCTTATCATTGATCCGGTC-3′) primers and amplifying the C-terminal (CT) domain with TGA3-CT (a fusion of amino acids 294 and 433; 5′-GGATTATGCGTACGCAAT-3′) and TGA3 (1152). To make the TGA3ΔbZIP construct, the products from TGA3 (4) + NT and CT + TGA3 (1152) in the first amplification step were mixed and used as templates in a second round of amplification using the TGA3 (4) and TGA3 (1152) primers. All products were subcloned into pBluescript KS+ vector containing a FLAG epitope sequence and sequenced. The yeast GAL4 DNA binding domain (amino acids 1 to 114) was obtained by EcoRI digestion of GAL4-TGA1a (Pascuzzi et al., 1998) and ligated into the cognate EcoRI site at the C-terminal end of the FLAG epitope. Finally, the DNAs that coded for translational fusion proteins of FLAG-GAL4, FLAG-GAL4-TGA3, and FLAG-GAL4-TGA3ΔbZIP were subcloned into the pMON 999 expression vector.
Leaf Transfection
Particle bombardment was performed with the PDS-1000/He Biolistic Delivery System (Bio-Rad). The distance parameters were as follows: from the rupture disk to the macrocarrier was 2.2 cm; from the macrocarrier to the stopping screen was 1.2 cm; and from the macrocarrier to the leaf sample was 7 cm. The leaf sample consisted of two to three rosette leaves from 4- to 5-week postgermination plants placed abaxial side up in the center of 0.5× Murashige and Skoog (1962) plates (0.7% phytagel agar). Each plate was shot twice using for each shot 0.5 mg of 1.6-μm gold particles covered with 1.5 μg of DNA in a ratio of 4:1:1 (reporter:effector:internal control); the amounts of each effector used (250 ng) corresponded to activities at saturating levels as determined by dose–response assays (data not shown). The particles were prepared according to the manufacturer's instructions except that a single sonication pulse was applied to the DNA-covered gold using a Fisher 500 sonicator and 15% output. The other settings include the use of 1350-p.s.i. rupture disks and vacuum pressure of 27 inches of Hg. After bombardment, leaf samples were treated with or without 1 mM SA and 0.02% Silwet L-77 for 20 h in an incubator (Percival Scientific, Perry, IA) set at 22°C under an 18-h-light/6-h-dark cycle.
Enzymatic Assays
Whole-cell extracts were first prepared by collecting transfected leaves and grinding them under liquid nitrogen using minimal ground glass, followed by resuspension in 250 μL of 1× CCLR buffer (Promega). Samples were vortexed for 30 s and spun at top speed in a microcentrifuge for 20 min at 4°C to recover supernatants. Luciferase enzyme assays were performed immediately according to the manufacturer's instructions (Promega) using a TD-20/20 Dual-Luciferase Ready Luminometer (Turner Designs, Sunnyvale, CA). For methyl-umbelliferylglucuronidase enzyme assays, whole-cell extracts and 2 mM methyl-umbelliferylglucuronidase (prepared in 1× CCLR) were mixed in equal volumes of 100 μL each and incubated at 37°C for 8 h. Reaction samples of 10 μL were quenched in 190 μL of 0.2 M Na2CO3 stopping buffer, and the fluorescence of each sample (100 μL) was measured on a PE LS55 Luminescence Spectrometer (Perkin-Elmer).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact J. Arias, arias@umbi.umd.edu.
Acknowledgments
We thank the following individuals for generously providing the reagents listed: Xinnian Dong (npr1-1 mutant line), Eric Lam (pET23a constructs of TGA2 through TGA6), and Malla Padidam (5xGAL-m35S-LUC reporter). We also thank Christiane Gatz, David Straney, and Marisela Morales for insightful comments on the manuscript. This work was supported by National Science Foundation Grants MCB-9817820 and MCB-0209697 to J.A. and by a Howard Hughes Medical Institute Undergraduate Research Fellowship to E.B. We thank Susan Kim for her help in growing plants and doing ChIP assays.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012211.
References
- Ang, L., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Boyes, D., Zayed, A., Ascenzi, R., McCaskill, A., Hoffman, N., Davis, K., and Gorlach, J. (2001). Growth stage–based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S., Gordon, A., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clark, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–64. [DOI] [PubMed] [Google Scholar]
- Chen, W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y., Chang, H.-S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P., Robatzek, S., and Somssich, I. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P., Schmelzer, E., Hahlbrock, K., and Somssich, I. (1999). Early nuclear events in plant defence signaling: Rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E., and Ausubel, F. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 179, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D., eds (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Izawa, T., Foster, R., and Chua, N.-H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230, 1131–1144. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., Desai, M., Pascuzzi, P., and Arias, J. (2001. a). In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 28, 237–243. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Glover, G., and Arias, J. (2001. b). Regulation of DNA binding and trans-activation by a xenobiotic stress-activated plant transcription factor. J. Biol. Chem. 276, 172–178. [DOI] [PubMed] [Google Scholar]
- Johnson, T., Wilson, H., and Roesler, W. (2001. c). Improvement of the chromatin immunoprecipitation (ChIP) assay by DNA fragment size fractionation. Biotechniques 31, 740–742. [DOI] [PubMed] [Google Scholar]
- Jupin, I., and Chua, N.-H. (1996). Activation of the CaMV as-1 cis-element by salicylic acid: Differential DNA-binding of a factor related to TGA1a. EMBO J. 15, 5679–5689. [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Seipel, K., and Chua, N.-H. (1992). Identification of a novel dimer stabilization region in a plant bZIP transcription activator. Mol. Cell. Biol. 12, 4809–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinedinst, S., Pascuzzi, P., Redman, J., Desai, M., and Arias, J. (2000). A xenobiotic-stress activated transcription factor and its cognate target genes are preferentially expressed in root tip meristem. Plant Mol. Biol. 42, 679–688. [DOI] [PubMed] [Google Scholar]
- Kuras, L., and Struhl, K. (1999). Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399, 609–613. [DOI] [PubMed] [Google Scholar]
- Lam, E., and Lam, Y. (1995). Binding site requirements and differential representation of TGA factors in nuclear ASF-1 activity. Nucleic Acids Res. 23, 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., Rouzé, P., and Rombauts, S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999. a). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Li, X.Y., Virbasius, A., Zhu, X., and Green, M. (1999. b). Enhancement of TBP binding by activators and general transcription factors. Nature 399, 605–609. [DOI] [PubMed] [Google Scholar]
- Liu, X., and Lam, E. (1994). Two binding sites for the plant transcription factor ASF-1 can respond to auxin treatments in transgenic tobacco. J. Biol. Chem. 269, 668–675. [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000. a). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000. b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked chromatin immunoprecipitation. Trends Biochem. Sci. 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Padidam, M., and Cap, Y. (2001). Elimination of transcriptional interference between tandem genes in plant cells. Biotechniques 31, 328–334. [DOI] [PubMed] [Google Scholar]
- Pascuzzi, P., Hamilton, D., Bodily, K., and Arias, J. (1998). Auxin-induced stress potentiates trans-activation by a conserved plant basic/leucine-zipper factor. J. Biol. Chem. 273, 26631–26637. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Miao, Z.-H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Privat, I., Trifa, Y., Zhou, J.-M., Klessig, D., and Lam, E. (2002). Differential regulation of TGA transcription factors by post-transcriptional control. Plant J. 32, 641–653. [DOI] [PubMed] [Google Scholar]
- Redman, J., Whitcraft, J., Johnson, C., and Arias, J. (2002). Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis. Plant Cell Rep. 21, 180–185. [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.-Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schindler, U., Terzaghi, W., Beckmann, H., Kadesch, T., and Cashmore, A. (1992). DNA binding site preferences and transcriptional activation properties of the Arabidopsis transcription factor GBF1. EMBO J. 11, 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessing, D. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Stange, C., Ramirez, I., Gomez, I., Jordana, X., and Holuigue, L. (1997). Phosphorylation of nuclear proteins directs binding to salicylic acid-responsive elements. Plant J. 11, 1315–1324. [DOI] [PubMed] [Google Scholar]
- Strompen, G., Gruner, R., and Pfitzner, U. (1998). An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19, 769–772. [DOI] [PubMed] [Google Scholar]
- Terzaghi, W., Bertekap, R., Jr., and Cashmore, A. (1997). Intracellular localization of GBF proteins and blue light-induced import of GBF2 fusion proteins into the nucleus of cultured Arabidopsis and soybean cells. Plant J. 11, 967–982. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Liu, Z., Hagen, G., and Guilfoyle, T. (1995. a). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Ohmiya, A., Hagen, G., and Guilfoyle, T. (1995. b). The soybean GH2/4 gene that encodes a glutathione S-transferase has a promoter that is activated by a wide range of chemical agents. Plant Physiol. 108, 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T., Dekker, B., and Hoekema, A. (1989). A small-scale procedure for rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, R., Bauscher, C., Pfitzner, A., and Pfitzner, U. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46, 143–160. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415. [DOI] [PubMed] [Google Scholar]
- Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, Z., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.-M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D. (2000). NPR1 differentially interacts with members of the TGA-OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–200. [DOI] [PubMed] [Google Scholar]