Abstract
The import of nucleus-encoded proteins into chloroplasts is mediated by translocon complexes in the envelope membranes. A component of the translocon in the outer envelope membrane, Toc34, is encoded in Arabidopsis by two homologous genes, atTOC33 and atTOC34. Whereas atTOC34 displays relatively uniform expression throughout development, atTOC33 is strongly upregulated in rapidly growing, photosynthetic tissues. To understand the reason for the existence of these two related genes, we characterized the atTOC33 knockout mutant ppi1. Immunoblotting and proteomics revealed that components of the photosynthetic apparatus are deficient in ppi1 chloroplasts and that nonphotosynthetic chloroplast proteins are unchanged or enriched slightly. Furthermore, DNA array analysis of 3292 transcripts revealed that photosynthetic genes are moderately, but specifically, downregulated in ppi1. Proteome differences in ppi1 could be correlated with protein import rates: ppi1 chloroplasts imported the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit and 33-kD oxygen-evolving complex precursors at significantly reduced rates, but the import of a 50S ribosomal subunit precursor was largely unaffected. The ppi1 import defect occurred at the level of preprotein binding, which is consistent with a role for atToc33 during preprotein recognition. The data suggest that atToc33 is involved preferentially in the import of photosynthetic proteins and, by extension, that atToc34 is involved in the import of nonphotosynthetic chloroplast proteins.
INTRODUCTION
Like mitochondria, chloroplasts are descended from a free-living prokaryotic organism that entered the eukaryotic lineage through endosymbiosis. During the course of their evolution, chloroplasts relinquished the majority of their genes so that now, >90% of chloroplast proteins are encoded in the nucleus (Leister, 2003). Chloroplast proteins are synthesized in precursor form on cytosolic ribosomes and targeted post-translationally to chloroplasts (Keegstra and Cline, 1999). Each precursor protein (preprotein) carries an NH2-terminal targeting signal called the transit peptide, which guides the protein to the chloroplast (Bruce, 2001). Translocon complexes in the outer and inner envelope membranes of chloroplasts (Toc and Tic, respectively) mediate preprotein import in an ATP- and GTP-dependent process (Chen et al., 2000; Hiltbrunner et al., 2001a; Jarvis and Soll, 2002). Proteins are translocated across both membranes simultaneously, in extended conformation, and then processed to their mature size by a stromal processing peptidase. After translocation, newly imported proteins are folded to their final conformation by molecular chaperones or targeted to one of a number of internal compartments via separate protein targeting pathways (Keegstra and Cline, 1999).
Several components of the Toc and Tic complexes have been identified in biochemical studies using isolated pea chloroplasts. The core components of the Toc complex are called Toc159, Toc34, and Toc75, according to their molecular weights (Hirsch et al., 1994; Kessler et al., 1994; Perry and Keegstra, 1994; Schnell et al., 1994; Seedorf et al., 1995; Bölter et al., 1998). Toc159 and Toc34 are related GTPases that have been proposed to act as preprotein receptors. Both proteins are anchored in the outer envelope by COOH-terminal domains and project their GTP binding domains into the cytosol. Although their precise mode of action remains unclear (Sveshnikova et al., 2000; Hiltbrunner et al., 2001b), Toc159 and Toc34 interact with incident preproteins (Ma et al., 1996; Kouranov and Schnell, 1997) and mediate their transfer to the translocation channel, of which Toc75 is a major component. Toc75 has a β-barrel structure comprising 16 amphiphilic β-strands and forms a channel with a pore size of ∼14 to 26 Å (Hinnah et al., 2002). The Tic complex is less well characterized, and there is even some disagreement in the literature concerning the identity of its components (Jarvis and Soll, 2002). A 100-kD heat-shock protein (Hsp100) homolog, ClpC, held at the stromal face of the Tic complex has been proposed to drive chloroplast protein import, much like Hsp70 proteins drive protein translocation into mitochondria and the endoplasmic reticulum (Nielsen et al., 1997).
In Arabidopsis, several of these translocon components are encoded by multiple genes. For example, there are two Toc34-related genes in Arabidopsis, atTOC33 and atTOC34 (Jarvis et al., 1998; Gutensohn et al., 2000), and four Toc159-related genes, atTOC159, atTOC132, atTOC120, and atTOC90 (Bauer et al., 2000; Hiltbrunner et al., 2001a). There also are multiple Toc75-related sequences in Arabidopsis, but some other components are encoded by just a single gene (Jackson-Constan and Keegstra, 2001). The reason for this Toc component multiplicity in Arabidopsis is unclear, but it has been proposed that different Toc isoforms have different preprotein recognition specificities (Jarvis et al., 1998). Support for this hypothesis was obtained recently upon characterization of an atTOC159 knockout mutant called plastid protein import2 (ppi2) (Bauer et al., 2000). Homozygous ppi2 plants have a seedling-lethal, albino phenotype characterized by pronounced defects in chloroplast biogenesis. Interestingly, whereas the expression and accumulation of proteins involved directly in photosynthesis (photosynthetic proteins) are reduced strongly in ppi2, proteins involved in chloroplast functions unrelated to photosynthesis (nonphotosynthetic proteins) are expressed and accumulate normally. These observations led to the suggestion that atToc159 has recognition specificity for photosynthetic proteins and that atToc132 and atToc120 are involved preferentially in the import of nonphotosynthetic proteins (Bauer et al., 2000). The existence of such substrate-specific protein import pathways would prevent the bulk flow of highly abundant, photosynthetic proteins from outcompeting the import of less abundant but equally important nonphotosynthetic proteins.
Previously, we described the identification and preliminary characterization of an atTOC33 knockout mutant called ppi1 (Jarvis et al., 1998). Homozygous ppi1 plants are yellow-green in appearance but, unlike ppi2 plants, they are able to survive to maturity. This difference in phenotype severity may indicate that any specialization among the Toc34 homologs is less pronounced than what exists among the Toc159 homologs and that there is considerable functional redundancy between atToc33 and atToc34. Indeed, overexpression of atToc34 was shown to complement the atToc33 deficiency of ppi1 plants (Jarvis et al., 1998). Nevertheless, the possibility remains that under normal, physiological conditions, atToc33 and atToc34 operate preferentially in import pathways with different substrate specificities. Some support for this idea was provided recently by the demonstration that, in vitro, atToc33 but not atToc34 is able to inhibit the binding of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit precursor (a photosynthetic preprotein) to chloroplasts in a GTP-dependent manner (Gutensohn et al., 2000). In this study, we took an in vivo approach to further address this question by characterizing the ppi1 mutant in detail.
RESULTS
atTOC33 Expression Is Induced Strongly in Rapidly Growing Photosynthetic Tissues
We previously determined the expression levels of atTOC33 and atTOC34 over a developmental time course at the whole-plant level (Jarvis et al., 1998). We found that atTOC33 is expressed at levels several times greater than atTOC34 and that both genes are subject to developmental regulation, with the highest levels of expression occurring in young plants. These data demonstrated that both genes are expressed during periods of intense plastid biogenesis, but they provided no clues regarding the possible functional differences between the proteins, because gene expression levels in individual tissues were not investigated. Although subsequent experiments did provide spatial expression profiles for each gene (Gutensohn et al., 2000), these studies did not address the relative levels of expression of the two genes.
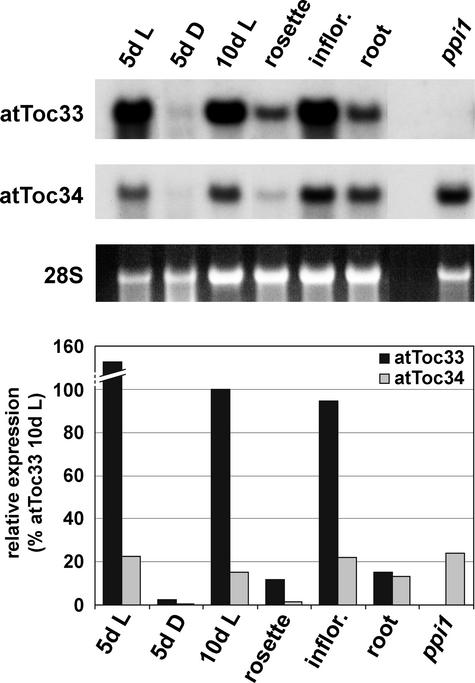
To gain insight into the reasons underlying the existence of two Toc34-related genes in Arabidopsis, we determined the relative expression levels of atTOC33 and atTOC34 at different developmental stages and in different tissue types by RNA gel blot analysis (Figure 1). Whereas atTOC34 was expressed at a relatively uniform, low level at most stages of development, atTOC33 was induced strongly in photosynthetic tissues undergoing rapid growth (in 5- and 10-day-old light-grown plants and in inflorescence tips). In fully expanded photosynthetic tissues (rosette leaves), the expression of both genes was reduced significantly, although atTOC33 continued to be expressed at a higher level than atTOC34. Most interestingly, in nonphotosynthetic root tissue, atTOC33 was downregulated specifically such that the two genes were expressed at essentially the same level. Both genes were expressed at low levels in dark-grown plants, which presumably reflects the minimal requirements for plastid biogenesis in these plants. The 10-day-old ppi1 control indicated that the hybridization probes were specific and confirmed that atTOC34 expression is not upregulated strongly in the ppi1 mutant.
Figure 1.
Expression Levels of atTOC33 and atTOC34 during Seedling Development and in Different Tissues of Arabidopsis.
Fifteen-microgram samples of total RNA isolated from Arabidopsis tissues were examined by RNA gel blot analysis. RNA was isolated from wild-type seedlings grown in vitro for 5 days in the light (5d L), 5 days in the dark (5d D), and 10 days in the light (10d L), from three different tissues of 28-day-old wild-type plants grown on soil (rosette leaves, young inflorescence tips, and roots), and from ppi1 seedlings grown in vitro for 10 days in the light (ppi1). Filters were probed using 32P-labeled atTOC33 and atTOC34 cDNA probes with identical specific activities. rRNA (28S) was used as a loading control. The images shown were obtained after a 3-week exposure, but quantification was performed using a shorter exposure. Relative levels of expression of atTOC33 and atTOC34 normalized for 28S rRNA are shown in the graph at bottom; y axis values between 100 and 140 have been removed to aid visualization.
Photosynthetic Proteins Are Specifically Deficient in ppi1 Chloroplasts
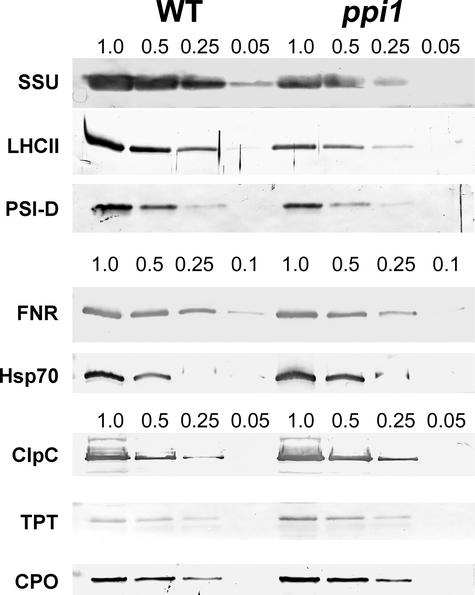
The demonstration that atTOC33 expression was induced strongly in rapidly expanding photosynthetic tissues suggests that atToc33 may be relatively more important for the assembly of the photosynthetic apparatus than atToc34. To investigate this possibility, we used immunoblot analysis to compare the abundance of a range of different photosynthetic and nonphotosynthetic proteins in chloroplasts isolated from 10-day-old wild-type and ppi1 plants (Figure 2). In this analysis, antibodies against four photosynthetic proteins (Rubisco small subunit [SSU], light-harvesting chlorophyll a/b binding protein [LHCII], photosystem I subunit D [PSI-D], and ferredoxin-NADP+ oxidoreductase [FNR]) and four nonphotosynthetic chloroplast proteins (Hsp70, the Hsp100 homolog ClpC, triose phosphate/phosphate translocator [TPT], and coproporphyrinogen oxidase [CPO]) were used. To obtain semiquantitative data, a dilution series of each chloroplast extract was examined with each antibody. By comparing the band intensities obtained, we deduced that each of the photosynthetic proteins was reduced in abundance in ppi1 chloroplasts by ∼50% or more. No evidence for preprotein accumulation in ppi1 was observed, suggesting that most nonimported preproteins are degraded rapidly or, alternatively, that gene expression is correlated closely with the efficiency of import of the encoded preprotein. By contrast, all four nonphotosynthetic proteins were present at least at wild-type levels in ppi1 chloroplasts, and some even appeared to be enriched slightly in the mutant.
Figure 2.
Immunoblot Analysis of Photosynthetic and Nonphotosynthetic Chloroplast Proteins in the ppi1 Mutant.
Dilution series of chloroplast protein preparations from the wild type (WT) and the ppi1 mutant (ppi1) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed using antibodies against photosynthetic (SSU, LHCII, PSI-D, and FNR) and nonphotosynthetic (Hsp70, ClpC, TPT, and CPO) proteins. In most cases, filters were cut in half and probed for one photosynthetic protein and one nonphotosynthetic protein (the pairings used were SSU/TPT, PSI-D/ClpC, and FNR/Hsp70); the LHCII and CPO data were derived using separate blots. The dilution steps used are given above the gels. Dilution series started with 20 μg for SSU, PSI-D, ClpC, TPT, and CPO, with 10 μg for FNR and Hsp70, and with 2 μg for LHCII. The data shown are representative of three independent experiments.
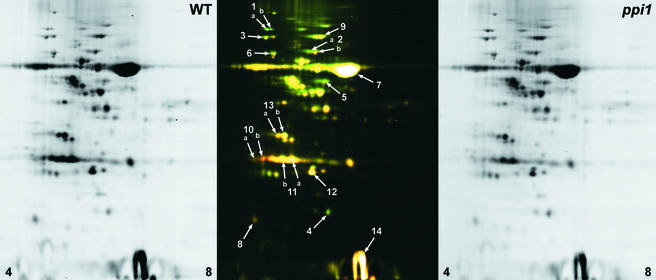
To corroborate these findings, we next adopted a proteomics approach. The chloroplast proteomes of 10-day-old wild-type and ppi1 plants were analyzed using difference gel electrophoresis (DIGE). Proteins extracted from isolated chloroplasts were labeled with nonsaturating CyDyeDIGE fluors. In a typical experiment, wild-type proteins were labeled with the Cy5 fluor and mutant proteins were labeled with Cy3. The labeled protein samples were mixed and then resolved in the first dimension by isoelectric focusing and in the second dimension by SDS-PAGE. The Cy5 and Cy3 fluorescent images of a typical gel are shown in Figure 3 (left and right, respectively). Additionally, an overlay of the Cy5 (wild type; false-colored red) and Cy3 (ppi1; false-colored green) images has been included to aid the visualization of protein abundance differences (Figure 3, middle): proteins depleted in ppi1 appear red, whereas those enriched in ppi1 appear green. The fluorescent signal associated with each protein spot was quantified and used to calculate the degree of enrichment or depletion in ppi1 chloroplasts.
Figure 3.
DIGE Analysis of the ppi1 Chloroplast Proteome.
Total chloroplast protein samples isolated from 10-day-old seedlings were analyzed using CyDyeDIGE technology. Wild-type (WT) and mutant (ppi1) protein samples (50 μg each) were labeled with Cy5 and Cy3, respectively. The samples were pooled and run on a single 24-cm, pH-3 to -10, two-dimensional gel and then imaged using parameters appropriate for each fluor. The left and right panels show corresponding sections (pH 4 to 8, as indicated) of the Cy5 and Cy3 images, respectively. The middle panel shows an overlay of the two images after inversion and the application of false color: Cy5 (wild type) is shown in red, and Cy3 (ppi1) is shown in green. Labels indicate protein spots that were identified by mass spectrometry (see Table 1).
Based on these data, spots were selected for identification by mass spectrometry, and the results obtained are summarized in Table 1. All of the proteins we identified that were depleted in ppi1 chloroplasts were components of the photosynthetic apparatus (such as SSU, OE33, and LHCII), whereas all of those showing enrichment in ppi1 were nonphotosynthetic proteins, including molecular chaperones and a component of the plastid genetic system (an EF-Tu–like translation elongation factor). Steady state levels of another genetic system component (the 50S ribosomal subunit protein, L12-C), an enzyme of carbon metabolism (transketolase), and the plastid-encoded Rubisco large subunit (LSU) were unaffected by the ppi1 mutation. Because the amounts of LSU and SSU protein normally are closely coordinated (Rodermel et al., 1996), we conclude that the moderate SSU deficiency in ppi1 is insufficient to trigger these coregulatory mechanisms.
Table 1.
Proteins Identified by DIGE and Mass Spectrometry
| Spot No. | Gene No. | Fold Change in ppi1a | Protein |
|---|---|---|---|
| 1 | At2g04030 | a, 2.41 ± 0.61; b, 1.85 ± 0.09 | Hsp90 homolog |
| 2 | At1g55490 At3g13470 |
a, 1.86 ± 0.13; b, 1.64 ± 0.05 | Chaperonin 60β |
| 3 | At4g24280 At5g49910 |
1.78 ± 0.07 | Hsp70 homolog |
| 4 | At3g62030 | 1.76 ± 0.29 | Peptidyl prolyl isomerase |
| 5 | At4g20360 | 1.72 ± 0.08 | Elongation factor, EF-Tu homolog |
| 6 | At2g28000 | 1.64 ± 0.19 | Chaperonin 60α |
| 7 | RbcL | — | Rubisco large subunit, LSU |
| 8 | At3g27850 | — | Ribosomal subunit, L12-C |
| 9 | At3g60750 | — | Transketolase-like protein |
| 10 | At3g04790 | a, 0.63 ± 0.04; b, 0.56 ± 0.03 | Ribulose-5-phosphate isomerase |
| 11a | At4g10340 | 0.61 ± 0.05 | Light-harvesting chlorophyll a/b binding protein CP26 (Lhcb5) |
| 11b | At1g29910 At1g29920 At1g29930 At2g34420 |
0.61 ± 0.05 | Light-harvesting chlorophyll a/b binding protein LHCII type 1 (Lhcb1.1, Lhcb1.2, Lhcb1.3, Lhcb1.5) |
| 12 | At1g06680 | 0.58 ± 0.03 | 23-kD subunit of oxygen-evolving complex, OE23 |
| 13 | At3g50820 At5g66570 |
a, 0.56 ± 0.09; b, 0.50 ± 0.06 | 33-kD subunit of oxygen-evolving complex, OE33 |
| 14 | At1g67090 | 0.43 ± 0.04 | Rubisco small subunit, SSU (Ats1A) |
Prefixes “a” and “b” denote protein spots that could not be distinguished upon mass spectrometric analysis. Values are means ± sd.
Expression of Photosynthetic Genes Is Downregulated Specifically in ppi1
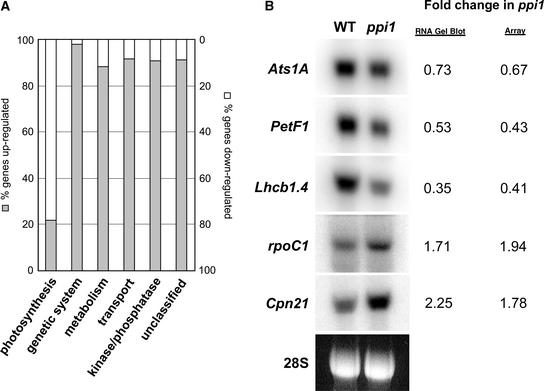
The expression of nuclear genes that encode chloroplast proteins is regulated in response to a variety of signals that emanate from plastids (Surpin et al., 2002; Jarvis, 2003). These retrograde signaling mechanisms ensure that nuclear gene expression is coupled with the needs of the cell's chloroplasts, so that the photosynthetic apparatus and other biosynthetic machineries are assembled in stoichiometric fashion. To gain further insight into the role of the atToc33 protein, we conducted a global analysis of the mRNA expression of genes of plastid-related function in ppi1 using nylon filter DNA array technology (Kurth et al., 2002; Richly et al., 2003). Of the 3292 genes analyzed, 1461 showed significantly different levels of expression from the wild type (see supplemental data online). The majority of these genes were upregulated moderately in ppi1, with only 161 genes showing reduced levels of expression. Remarkably, most of the genes of known function that were downregulated in ppi1 encode components of the photosynthetic apparatus. By contrast, genes showing increased expression levels encode components of the chloroplast's endogenous genetic system, factors involved in metabolic processes unrelated to photosynthesis, and a range of other nonphotosynthetic proteins.
We assigned all 1461 genes showing differential expression in ppi1 to six different functional categories as follows: photosynthesis, including Calvin cycle enzymes (55 genes); genetic system (152 genes); metabolism not related directly to photosynthesis (170 genes); transport of substances, including metabolites and proteins (60 genes); kinases and phosphatases (55 genes); and putative, hypothetical, or otherwise unclassifiable proteins (969 genes). The proportion of genes showing increased or decreased expression in ppi1 in each category is shown in Figure 4A. The fact that photosynthetic genes were downregulated specifically in ppi1 reinforces the hypothesis that atToc33 operates preferentially in an import pathway with specificity for photosynthetic proteins. It is conceivable that the nucleus responds to the perturbation of photosynthetic protein import, communicated via some retrograde signaling pathway, by downregulating the genes that encode the affected proteins.
Figure 4.
Analysis of the Expression of Nucleus-Encoded Chloroplast Genes in ppi1.
(A) Total RNA isolated from 10-day-old wild-type and ppi1 mutant seedlings was used to probe a nylon filter DNA array carrying 3292 gene-specific tags. All 1461 genes showing differential expression in ppi1 were placed into six different categories, as described in the text. The proportion of genes within each category showing upregulation (gray bars) and downregulation (white bars) in ppi1 is shown.
(B) Ten-microgram samples of the RNA described in (A) were analyzed further by RNA gel blot analysis. Five of the differentially regulated genes listed in Table 2 were selected for analysis. Filters were hybridized with 32P-labeled probes corresponding to the gene-specific tags described in (A). rRNA (28S) was used as a loading control. Radioactivity associated with each band shown was quantified and used to calculate the fold change in expression in ppi1 (relative to the wild type [WT]) for each gene; the corresponding data derived from the DNA array experiment ([A]; Table 2) are shown for comparison.
To further illustrate these gene expression changes, the data for 42 genes representative of four different functional categories (photosynthesis, 12 genes; genetic system, 10 genes; nonphotosynthetic metabolism, 10 genes; and protein targeting, 10 genes) as well as 8 genes belonging to other categories are shown in Table 2; the relative levels of expression of these 50 genes are illustrated further in the supplemental data online. Not only did most of the genes that show downregulated expression in ppi1 encode components of the photosynthetic apparatus, they also tended to be expressed at a higher level than genes that show upregulated expression (Table 2; see also supplemental data online). The plastome-encoded Rubisco large subunit gene, rbcL, is one obvious exception. Also of interest is the observation that all of the protein targeting–related genes were upregulated in ppi1, except for the gene that encodes Tic20-I, which is a putative channel component of the inner envelope membrane (Jackson-Constan and Keegstra, 2001). Similarly, genes that encode several different molecular chaperones, some of which have been implicated in chloroplast protein import (Schnell et al., 1994; Nielsen et al., 1997; Jackson-Constan and Keegstra, 2001), also were upregulated in ppi1 (Table 2; see also supplemental data online).
Table 2.
Selected Genes Showing Differential Expression in ppi1
| Category/Gene No. | Signal Intensity in the Wild Type |
Fold Change in ppi1 |
Gene Name/Product | Function |
|---|---|---|---|---|
| Photosynthesis | ||||
| rbcL | 68.84 | 1.26 | Plastid gene rbcL, Rubisco large subunit | Calvin cycle |
| At5g38430 | 46.48 | 0.73 | Ats1B, Rubisco small subunit | Calvin cycle |
| At4g09650 | 1.43 | 0.72 | AtpD, ATP synthase δ-subunit | ATP synthesis |
| At1g67740 | 2.55 | 0.71 | PsbY, Y-subunit of PSII | Photosystem II |
| At5g61410 | 1.55 | 0.70 | Ribulose-5-phosphate-3-epimerase | Calvin cycle |
| At1g67090 | 35.23 | 0.67 | Ats1A, Rubisco small subunit a | Calvin cycle |
| At1g76100 | 0.87 | 0.65 | PetE1, plastocyanin | Electron transport |
| At4g02770 | 6.58 | 0.63 | PsaD1, D-subunit of PSI | Photosystem I |
| At3g47470 | 8.17 | 0.60 | Lhca4, LHCI polypeptide | Light harvesting |
| At1g56190 | 1.16 | 0.43 | Phosphoglycerate kinase | Calvin cycle |
| At1g60950 | 2.75 | 0.43 | PetF1, ferredoxin | Electron transport |
| At2g34430 | 17.01 | 0.41 | Lhcb1.4, LHCII polypeptide | Light harvesting |
| Genetic system | ||||
| At1g34380 | 0.20 | 2.34 | DNA polymerase type I, putative | DNA replication |
| rpoC1 | 1.19 | 1.94 | Plastid gene rpoC1, RNA polymerase | Transcription |
| At5g26710 | 0.91 | 1.45 | Glutamyl-tRNA synthetase | Translation |
| At3g53460 | 0.78 | 1.43 | RNA binding protein, CP29 | RNA stability |
| At3g62910 | 0.47 | 1.38 | Translation releasing factor, RF-1–like | Translation |
| At4g20980 | 0.49 | 1.30 | Translation initiation factor, eIF3d-like | Translation |
| At5g24120 | 0.13 | 1.30 | σ-like factor | Transcription |
| At1g32990 | 0.91 | 1.30 | 50S plastid ribosomal protein, L11 | Translation |
| At3g27850 | 0.85 | 1.27 | 50S plastid ribosomal protein, L12-C | Translation |
| At1g74980 | 0.23 | 1.24 | 30S plastid ribosomal protein, S9 | Translation |
| Metabolism | ||||
| At1g69370 | 0.38 | 1.55 | Chorismate mutase | Amino acid biosynthesis |
| At3g48560 | 0.50 | 1.44 | Acetolactate synthase | Amino acid biosynthesis |
| At4g18240 | 0.20 | 1.40 | Starch synthase–like protein | Starch biosynthesis |
| At1g29900 | 0.78 | 1.37 | Carbamoyl phosphate synthetase, carb | Pyrimidine biosynthesis |
| At5g57030 | 0.53 | 1.34 | Lycopene ɛ-cyclase | Carotenoid biosynthesis |
| At3g02610 | 0.42 | 1.34 | Stearoyl–acyl carrier protein desaturase | Fatty acid biosynthesis |
| At4g25080 | 0.67 | 1.31 | Mg-protoporphyrin IX methyltransferase | Tetrapyrrole biosynthesis |
| At2g26670 | 0.17 | 1.31 | Heme oxygenase, HO1 | Tetrapyrrole biosynthesis |
| At1g09830 | 1.46 | 1.28 | Phosphoribosylglycineamide synthetase | Purine biosynthesis |
| At3g11670 | 0.85 | 1.26 | Digalactosyldiacylglycerol synthase | Galactolipid biosynthesis |
| Protein targeting | ||||
| At5g28750 | 0.19 | 1.85 | Tha4 | Tat pathway, thylakoids |
| At4g33350 | 13.64 | 1.77 | Tic22-IV | Protein import |
| At5g16620 | 0.23 | 1.63 | Tic40 | Protein import |
| At2g18710 | 0.33 | 1.48 | SecY | Sec pathway, thylakoids |
| At5g03940 | 0.25 | 1.46 | SRP54 | SRP pathway, thylakoids |
| At4g02510 | 0.68 | 1.38 | Toc159 | Protein import |
| At4g03320 | 0.43 | 1.31 | Tic20-IV | Protein import |
| At1g06950 | 0.30 | 1.31 | Tic110 | Protein import |
| At3g46740 | 0.78 | 1.25 | Toc75-III | Protein import |
| At1g04940 | 2.96 | 0.67 | Tic20-I | Protein import |
| Other | ||||
| At5g20720 | 0.49 | 1.78 | Cpn21 chaperonin | Molecular chaperone |
| At4g39960 | 0.70 | 1.54 | DnaJ homolog | Molecular chaperone |
| At3g13470 | 0.59 | 1.53 | Chaperonin cpn60β | Molecular chaperone |
| At2g28900 | 0.13 | 1.48 | OEP16 homolog | Outer envelope porin |
| At4g24280 | 0.69 | 1.34 | Hsp70 homolog | Molecular chaperone |
| At5g50920 | 1.14 | 1.30 | Hsp100 homolog, ClpC-V | Molecular chaperone |
| At5g24020 | 0.14 | 1.28 | MinD homolog | Chloroplast division |
| At5g55280 | 0.40 | 1.26 | FtsZ homolog | Chloroplast division |
To corroborate the findings of this DNA array experiment, we examined the expression of five genes listed in Table 2 by RNA gel blot analysis. Three photosynthetic genes (Ats1A, PetF1, and Lhcb1.4) and two nonphotosynthetic genes (rpoC1 and Cpn21) were analyzed in this way. As shown in Figure 4B, the gene expression changes observed using RNA gel blot analysis were quantitatively very similar to those observed using DNA array analysis. Therefore, these results confirm the validity of the gene expression data presented in Figure 4A and Table 2.
ppi1 Affects the Import of Different Preproteins to Different Degrees
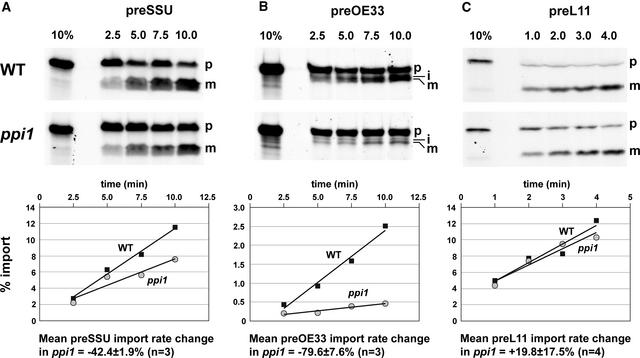
Isolated ppi1 chloroplasts were shown previously to import reduced quantities of in vitro–translated SSU, LHCII, and protochlorophyllide oxidoreductase (POR; a light-sensitive chlorophyll biosynthetic enzyme) over a fixed period of time (Jarvis et al., 1998). However, import rate comparisons for different preproteins were not conducted. To determine if the primary cause of the chloroplast proteome differences observed in ppi1 (Figures 2 and 3, Table 1) could be a differential effect of the ppi1 mutation on the import of different preproteins, we compared import rates for three different preproteins using isolated wild-type and mutant chloroplasts. Import rates were determined by measuring the accumulation of imported protein over time. For these experiments, we selected SSU (a component of the photosynthetic dark reactions that occur in the stroma), OE33 (a component of the photosynthetic light reactions that occur in the thylakoids), and L11 (a 50S ribosomal subunit protein and component of the chloroplast's endogenous genetic system) for analysis.
Initial experiments revealed that import rates were linear up to ∼10 min for SSU preprotein (preSSU) and preOE33 and up to ∼5 min for preL11 (data not shown); therefore, import rate comparisons were conducted within these periods. Resistance to thermolysin treatment confirmed that the processed forms of all three preproteins had been internalized by intact chloroplasts (see supplemental data online). As shown in Figures 5A and 5B, both preSSU and preOE33 were imported into ppi1 chloroplasts at significantly reduced rates: on average, preSSU import rates were reduced by 42.4 ± 1.9% and preOE33 import rates were reduced by 79.6 ± 7.6%. OE33 is targeted to the thylakoid lumen via a stromal intermediate (Keegstra and Cline, 1999) that could be observed between the precursor and mature OE33 bands of Figure 5B. Because this intermediate had already traversed the envelope, it was regarded as imported protein for quantification purposes. By contrast, the effect of ppi1 on the import of preL11 was slight, and import rates in the different genotypes were almost identical (Figure 5C).
Figure 5.
Comparison of Chloroplast Import Rates in the Wild Type and ppi1 for Different Preproteins.
In vitro–translated, 35S-Met–labeled preSSU (A), preOE33 (B), and preL11 (C) were imported into wild-type (WT) and mutant (ppi1) chloroplasts for 1.0 to 10.0 min, as indicated. Ten percent of the translation product added to each import reaction was loaded as a control (10%). Precursor (p), intermediate (i; OE33 only), and mature (m) protein forms are indicated at right of the gels. Quantifications for each import experiment are shown in the corresponding graph below each gel. Radioactivity associated with each mature band (intermediate and mature bands together in the case of OE33) was quantified and expressed as a percentage of the total preprotein-associated radioactivity added to each reaction. The data presented in the graphs correspond to the results shown in the gels, which are representative of several independent experiments. Mean import rate changes (±se) derived from repeated experiments are given at bottom below the graphs.
Although OE33 import was affected more strongly than SSU import in vitro (Figure 5), SSU accumulation was affected more strongly than OE33 accumulation in vivo (Figure 3, Table 1). This finding indicates that factors in addition to import efficiency influence steady state protein levels in ppi1. Nevertheless, the data clearly demonstrate that ppi1 affects the import of different preproteins to different degrees; therefore, we conclude that these differential effects are the primary cause of the selective depletion of photosynthetic proteins in ppi1 chloroplasts. Exactly how the ppi1 mutation exerts this preprotein specificity is open to interpretation. The most likely explanation is that atToc33 participates preferentially in an import pathway (which perhaps also involves atToc159) that has a degree of substrate specificity. An alternative explanation is that reduced overall levels of a functionally redundant atToc33 + atToc34 pool in ppi1 affects the import of those preproteins with the lowest affinity for the import apparatus most strongly. However, such a model does not explain why Arabidopsis has retained multiple isoforms of certain components of the import apparatus, such as Toc34 and Toc159, and only single isoforms of others, such as Tic110 and Tic40 (Jackson-Constan and Keegstra, 2001). Furthermore, this model is inconsistent with the fact that ppi1 significantly affected the import of preSSU (Figure 5A). One of the reasons that preSSU has been used in so many import studies is that it is imported into chloroplasts with high efficiency.
ppi1 Affects preSSU Import at the Level of Preprotein Binding
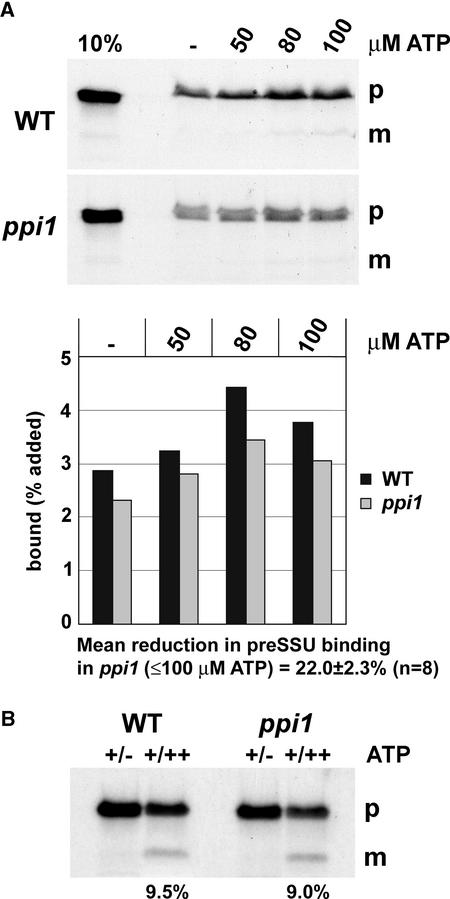
Protein import into chloroplasts can be divided into three distinct stages in vitro based on energetic requirements (Olsen et al., 1989; Olsen and Keegstra, 1992; Ma et al., 1996; Kouranov and Schnell, 1997). These stages are assumed to correspond to sequential steps in the import process that occurs in vivo. The first stage of import is energy-independent binding, and this occurs in the absence of ATP. It involves the reversible interaction of preproteins with receptor components of the Toc complex. The second stage of import is early import intermediate formation, which requires low concentrations of ATP (<100 μM) in the intermembrane space. Preproteins at this stage are inserted across the outer envelope membrane and in contact with components of the Tic complex; therefore, progression to this stage of import can be viewed as outer envelope translocation. The final stage of import is complete translocation, which requires high concentrations of ATP (>100 μM) in the stroma. Progression to this stage requires both binding and outer envelope translocation as well as inner envelope translocation and processing by the stromal processing peptidase.
To determine the stage of import affected by ppi1, we quantified the amount of preSSU bound to ATP-depleted chloroplasts in import reactions containing different, limiting concentrations of ATP (Figure 6A). All binding reactions were performed in the dark to prevent ATP synthesis, but control import reactions were performed in the light to confirm that the import competence of the chloroplasts was high (data not shown). In the absence of ATP, binding of preSSU to chloroplasts was reduced by ∼20% in ppi1 compared with the wild type (Figure 6A). This finding indicates that ppi1 affects import at the level of energy-independent binding. Increased preprotein binding as ATP is increased to micromolar concentrations is indicative of early import intermediate formation. Because the reduction in binding observed in ppi1 remained constant as ATP concentrations were increased up to 100 μM (on average, preSSU binding under ATP-limiting conditions was 22.0 ± 2.3% lower for ppi1 than for the wild type), it seems unlikely that ppi1 also affects outer envelope translocation (Figure 6A). The slight decline in the amount of bound preSSU at 100 μM ATP (seen in both genotypes) has been observed previously (Olsen et al., 1989) and presumably reflects the fact that a proportion of the preprotein has been translocated.
Figure 6.
Effect of ppi1 at Different Stages in the Import of preSSU.
(A) ATP-depleted wild-type (WT) or mutant (ppi1) chloroplasts and 35S-Met–labeled preSSU were incubated together in the absence or presence of ATP at the indicated concentrations for 10 min in darkness. Quantification of each chloroplast-bound preSSU band in the assay shown is given in the corresponding graph. The data shown are representative of four independent experiments, from which the mean reduction in preSSU binding (±se) was derived. m, mature; p, precursor.
(B) Import-chase results. Chloroplasts were incubated with preSSU under ATP-limiting conditions (50 μM). The chloroplasts then were reisolated and divided into two aliquots. Half were incubated in the absence of additional ATP (left lanes; indicated +/−), and the other half were incubated in the presence of 5 mM ATP (right lanes; indicated +/++). Values shown below the fluorograph indicate the amount of imported SSU in the lanes labeled +/++, expressed as a percentage of the amount of bound preSSU in the lanes labeled +/−. The data shown are representative of repeated experiments.
The efficiency of inner envelope translocation cannot be measured simply by quantifying the amount of protein imported in reactions containing high ATP concentrations (>100 μM), because this depends not only on the efficiency of inner envelope translocation but also on the efficiency of energy-independent binding and outer envelope translocation. To separate binding and outer envelope translocation from translocation across the inner envelope membrane, we first bound preSSU to wild-type and ppi1 chloroplasts under ATP-limiting conditions (50 μM). After the formation of import intermediates, we divided each import reaction in two. To half of each reaction, we added 5 mM ATP, whereas to the other half, no additional ATP was added. The amount of translocated protein then was expressed as a percentage of that bound initially, taking into account any differences in binding efficiency (Figure 6B). Once again, an ∼20% deficiency in binding/outer envelope translocation was observed. Although the amount of protein translocated into ppi1 chloroplasts was less in absolute terms, when normalized according to the amount of preSSU bound in each case, it was found to be essentially the same as that in the wild type (Figure 6B): 9.5% of bound protein was translocated in the wild type, and 9.0% of bound protein was translocated in ppi1. These data indicate that the effect of ppi1 is restricted to preprotein binding, which is consistent with a role for atToc33 in preprotein recognition.
DISCUSSION
We determined the relative levels of expression of atTOC33 and atTOC34 at different developmental stages and in different tissues to gain insight into the reason for the existence of these two homologous genes in Arabidopsis. The results showed that whereas atTOC34 expression is relatively uniform and low level throughout development, atTOC33 expression is upregulated strongly in new and expanding photosynthetic tissues (Figure 1). Our previous studies revealed that atTOC33 and atTOC34 both are expressed at higher levels in young plants and leaves than in older plants and leaves, but individual photosynthetic and nonphotosynthetic tissues were not investigated (Jarvis et al., 1998). The spatial expression patterns observed here are broadly in agreement with those described previously (Gutensohn et al., 2000), but this earlier study failed to address the relative expression levels of the two genes.
Our data correlate nicely with observations made using promoter–β-glucuronidase (GUS) fusions and by in situ hybridization (Gutensohn et al., 2000). The strongest expression of atTOC34-GUS was observed in roots and floral organs, whereas atTOC33-GUS expression was strongest in leaves and, in flowers, occurred only in the external, photosynthetic tissues of the sepals. In situ hybridization experiments confirmed these floral expression patterns and, furthermore, revealed that in stems, atTOC33 expression is restricted largely to the photosynthetic mesophyll tissues (Gutensohn et al., 2000). Together, these data suggest that atTOC33 is particularly important for the biogenesis of photosynthetic proteins. The observation that ppi1 affects the development of chloroplasts but not root plastids is consistent with this hypothesis (Jarvis et al., 1998; Yu and Li, 2001).
Examination of the ppi1 chloroplast proteome by immunoblot analysis and DIGE revealed that proteins involved directly in photosynthesis are specifically deficient (Figures 2 and 3, Table 1). Interestingly, we also observed a slight enrichment of several nonphotosynthetic proteins in ppi1 chloroplasts. One possible explanation for the apparent enrichment of nonphotosynthetic proteins in ppi1 is a simple concentration effect caused by the selective depletion of photosynthetic proteins. Another possibility is that these proteins are upregulated specifically in ppi1 in an attempt to compensate for perturbations in chloroplast metabolism. The upregulation of molecular chaperones could be an attempt to maximize the activity and/or half-life of those photosynthetic components that are imported successfully. Whatever the reason, the fact that ppi1 chloroplasts are able to accumulate increased quantities of these proteins suggests that the effect of ppi1 on protein import is, at least to some degree, preprotein specific.
To gain a more global view of the effects of the ppi1 mutation, we used DNA array technology to survey the expression of 3292 different genes: 2661 genes with predicted or actual chloroplast transit peptides and 631 others. Remarkably, the ppi1 mutation resulted in the specific downregulation of highly expressed, photosynthetic genes. By contrast, genes related to the plastid genetic system, nonphotosynthetic metabolism, and various transport processes were upregulated moderately in ppi1 (Figure 4, Table 2; see also supplemental data online). Comparison of these data with those obtained previously (Kurth et al., 2002; Strand et al., 2003) indicates that the gene expression changes observed in ppi1 are not a general response to the perturbation of chloroplast biogenesis. An Arabidopsis knockout for the L11 protein of the 50S ribosomal subunit (Pesaresi et al., 2001)—a mutant that has a visible phenotype similar to that of ppi1—was found to exhibit upregulated expression of both photosynthetic and plastid genetic system genes (Kurth et al., 2002). On the other hand, more severe defects in chloroplast biogenesis caused by the growth of plants on the herbicide norflurazon resulted in the downregulation of both photosynthetic and plastid genetic system genes (Strand et al., 2003). Therefore, it seems plausible that the specific downregulation of photosynthetic genes observed in ppi1 is an adaptive response to the reduction in import efficiency of the corresponding proteins.
When we analyzed the import kinetics of three different preproteins in ppi1, we found that the effect of the mutation varied according to the preprotein in question (Figure 5). Interestingly, the two preproteins that were affected most strongly (preSSU and preOE33) are components of the photosynthetic apparatus, whereas the preprotein that was affected least (preL11) is a component of the chloroplast's endogenous genetic system. These data indicate that the selective depletion of photosynthetic proteins in ppi1 most likely is caused by a differential effect of the mutation on the import of different preproteins. Consequent deficiencies in photosynthetic proteins would, in turn, be predicted to cause downregulated expression of the corresponding nuclear genes through retrograde signaling (Surpin et al., 2002; Jarvis, 2003). If we extend this argument to its logical conclusion, one might suppose that all of the genes that are downregulated in ppi1 encode preproteins that require atToc33 for their import into chloroplasts. However, it also is possible that ppi1 affects the import of just a few proteins and that it is the consequent deficiencies in these proteins that perturb, indirectly, the expression and/or accumulation of a range of other (functionally related) proteins. Thus, although it seems likely that many of the proteins listed in Tables 1 and 2 use atToc33 preferentially during their import into chloroplasts, it is not necessarily the case that they all do.
Although studies on protein import kinetics in ppi1 have not been conducted previously, ppi1 chloroplasts were shown to import reduced quantities of SSU, LHCII, and two different isoforms of POR over a fixed period of time (Jarvis et al., 1998). Although POR is not involved directly in photosynthesis, it is a light-dependent enzyme that catalyzes the committed step in chlorophyll biosynthesis. These import data are therefore consistent with the results presented here and with the idea that atToc33 is involved preferentially in the import of abundant proteins that play roles closely related to photosynthesis.
By manipulating the ATP concentration in import assays, we found that the effect of ppi1 on preSSU import is restricted to preprotein binding (Figure 6). This finding is consistent with a role for atToc33 in preprotein recognition. The precise role played by atToc33 (and other Toc34-related proteins) during preprotein binding remains uncertain, however, because current data suggest two different models for Toc receptor function. In the first model, Toc34 and Toc159 remain stably associated with the outer envelope membrane and interact with incident preproteins directly, much like the mitochondrial protein import receptors Tom20 and Tom22 (Pfanner and Geissler, 2001). In support of this model, preproteins can be cross-linked to pea Toc34 (psToc34) and/or psToc159 very early during import into pea chloroplasts (Perry and Keegstra, 1994; Ma et al., 1996; Kouranov and Schnell, 1997), and a direct and specific interaction between preproteins and psToc34 has been observed in vitro (Sveshnikova et al., 2000; Schleiff et al., 2002). However, not all cross-linking studies resulted in the identification of psToc34 (Perry and Keegstra, 1994; Ma et al., 1996), and in those that did, psToc34 appeared to interact with the mature region of the preprotein rather than with the transit peptide, as would be expected of a receptor (Kouranov and Schnell, 1997).
In the second model, soluble Toc159 binds to preproteins in the cytosol and then mediates their targeting to the chloroplast surface by docking at membrane-bound Toc34. In support of this model, an abundant cytosolic form of atToc159 has been observed (Hiltbrunner et al., 2001b), and the crystal structure of psToc34 suggests that heterodimerization between Toc34 and Toc159 may be possible (Sun et al., 2002). This model is reminiscent of signal recognition particle (SRP)–dependent protein targeting, in which the GTP binding protein SRP54 initiates preprotein translocation by docking at receptors that are themselves GTP binding proteins (Keenan et al., 2001).
Thus, preprotein recognition specificity by atToc33 may be mediated by its preferential interaction with a particular subset of preproteins, or alternatively by its preferential interaction with one or more of the different Toc159 isoforms. Bearing these possibilities in mind, it is interesting that atToc33 and atToc34 have been shown to exhibit differential competitor properties in an in vitro preSSU chloroplast binding assay: heterologously expressed atToc33 inhibited the binding of preSSU (translated using a rabbit reticulocyte system) to chloroplasts in a GTP-dependent manner, whereas atToc34 did not (Gutensohn et al., 2000). These data support the former possibility and suggest that the specific effect of ppi1 on photosynthetic protein import is a reflection of differential preprotein binding properties of atToc33 and atToc34. Interestingly, during our analyses of protein import in the ppi1 mutant, we consistently observed increased binding of unprocessed preL11 to mutant chloroplasts (Figure 5C). Increased abundance of a receptor with specificity for nonphotosynthetic preproteins (not necessarily associated with Toc complexes) might account for this observation. Although atTOC34 transcript levels were not increased strongly in ppi1 (Figure 1), it is possible that atToc34 accumulates to a higher level in ppi1 as a result of post-transcriptional processes.
Whatever the mode of action of the Toc GTPases, it seems likely that the proposed substrate specificity of the import apparatus is dependent on differences between the transit peptides of different preproteins. Transit peptides share a few general characteristics (e.g., they are rich in hydroxylated residues and deficient in acidic residues), but they differ widely in size and primary sequence and, unlike mitochondrial presequences, they do not share a common secondary structure (Bruce, 2001). This variability, together with the absence of precise information regarding the recognition process, makes it very difficult to predict which preproteins might be imported through a particular Toc complex or import pathway. Nevertheless, examination of the transit peptide sequences of the three preproteins used in this study revealed one property that correlated with import efficiency in the ppi1 mutant: at pH 7.0, the preSSU, preOE33, and preL11 transit peptides are predicted to have net charges of 2.877, 3.046, and 6.206, respectively. It is conceivable that the higher charge of the preL11 transit peptide favored interaction with atToc34 (or atToc132- and atToc120-related proteins in the wheat germ translation system) and that the less highly charged preSSU and preOE33 transit peptides interact more favorably with atToc33 (or atToc159-related wheat germ proteins).
We have demonstrated that the ppi1 mutation affects, with remarkable specificity, the expression, chloroplast import, and accumulation of photosynthetic proteins. Furthermore, the ppi1 import defect appears to be restricted to the preprotein binding step, which is consistent with a role for atToc33 in preprotein recognition. Together, these data suggest that atToc33 is involved preferentially in the recognition of highly abundant, photosynthetic preproteins and that atToc34 may have preference for less abundant, nonphotosynthetic preproteins.
METHODS
Isolation of Arabidopsis Chloroplasts
Chloroplasts were isolated from 10-day-old wild-type and ppi1 Arabidopsis thaliana plants (both Columbia-0 ecotype) grown in vitro as described previously (Aronsson and Jarvis, 2002). Plant material was homogenized for 3 to 4 s (wild type) or 2 to 3 s (ppi1) using a Polytron. After the first centrifugation, the homogenate (in 0.5 to 2 mL of isolation buffer) was loaded onto a linear Percoll gradient and centrifuged in a swing-out rotor at 7800g for 10 min. Intact chloroplasts were recovered and washed once with HS buffer (50 mM Hepes and 0.3 M sorbitol). The yield and intactness of the chloroplasts were determined as described previously (Aronsson and Jarvis, 2002).
Immunoblot Analysis
Standard methods were used for SDS-PAGE, protein gel blot analysis, and detection. Wild-type and ppi1 chloroplast preparations were quantified using Bradford reagent (Bio-Rad) and solubilized in 2× SDS-PAGE sample buffer (20% [v/v] glycerol, 10% [v/v] 2-mercaptoethanol, 4% [w/v] SDS, 0.125 M Tris/HCI, pH 6.8, and 0.04% [w/v] bromophenol blue). Amounts of protein loaded per lane ranged from 20 to 0.1 μg. To confirm equal loading of the samples, control gels were stained with Coomassie Brilliant Blue R 250 (Fisher), and five different bands, selected at random, were quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). On average, differences between the samples were ∼6%. Where possible, filters were cut across the lanes so that one half could be probed with a “photosynthetic” antibody and the other half could be probed with a “nonphotosynthetic” antibody. Primary antibodies were raised in rabbits and were kindly provided by Kenneth Cline (University of Florida, Gainesville) (Hsp70; raised against pea stromal Hsp70), Ulf-Ingo Flügge (University of Cologne, Germany) (TPT; raised against the spinach protein), Bernhard Grimm (Humboldt University, Germany) (CPO; raised against the tobacco protein), Neil Hoffman (National Science Foundation, Arlington, VA) (LHCII; raised against the pea protein), Kenneth Keegstra (Michigan State University, East Lansing) (ClpC; raised against pea stromal ClpC), Kenton Ko (Queen's University, Kingston, Ontario, Canada) (SSU; raised against the pea protein), and Henrik Scheller (The Royal Veterinary and Agricultural University, Copenhagen, Denmark) (PSI-D and FNR; both raised against barley proteins). The secondary antibody was an anti-rabbit IgG alkaline phosphatase conjugate (Sigma), and the detection reagent was 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium alkaline phosphatase substrate (Sigma).
Chloroplast Proteomics
Isolated chloroplasts were precipitated using 0.1 M ammonium acetate in 100% methanol and resuspended in 10 mM Tris/HCl, pH 8.5, 8 M urea, and 2% (w/v) amidosulfobetaine-14 (Calbiochem). Protein concentrations were determined using a detergent-compatible protein quantification kit (Bio-Rad). Protein samples from wild-type and mutant chloroplasts (50 μg each) were labeled with complementary CyDyeDIGE fluors (Cy3 and Cy5; Amersham Biosciences) (Unlu et al., 1997), pooled, and run on either 13- or 24-cm, pH-3 to -10 or pH-4 to -7 immobilized pH gradient strips (Amersham Biosciences). Resolution in the second dimension was performed using Hoeffer SE 600 or Ettan DALT SDS-PAGE systems (Amersham Biosciences). Gels were scanned using a 2920-2D MasterImager (Amersham Biosciences), and images were exported as 16-bit TIFF files. Image analysis was performed using DeCyder software (Amersham Biosciences). Protein spots showing >1.5-fold change in volume ratio were excised manually after colloidal Coomassie blue staining and digested to peptides using trypsin on a MassPrepStation (Micromass, Manchester, UK). The resulting peptides were examined by liquid chromatography/tandem mass spectrometry (Q-Tof, CapLC, Micromass/Waters, www.waters.com). Fragmentation data were used to search the National Centre for Biotechnology Information database using the MASCOT search engine. Multiple chloroplast preparations from each genotype were analyzed. Each combination of samples was run in triplicate for each pH range, including one experiment in which the CyDyeDIGE fluor labeling was reversed. Spots excised from each gel were treated individually so that multiple identifications were performed for each protein.
Protein Import into Arabidopsis Chloroplasts
Template DNA for the in vitro transcription/translation of preproteins was amplified by PCR from cDNA clones using M13 primers. The preSSU and preL11 cDNA clones were described previously (Aronsson and Jarvis, 2002), and the preOE33 cDNA clone was obtained from the ABRC (Columbus, OH) as clone 119E10T7. Transcription/translation was performed using a wheat germ system (Promega) containing 35S-Met and T7 RNA polymerase according to the manufacturer's instructions.
Import reactions were performed in HMS buffer (50 mM HEPES, 3 mM MgSO4, and 0.3 M sorbitol) containing 20 mM gluconic acid (potassium salt), 10 mM NaHCO3, and 0.2% (w/v) BSA (Aronsson and Jarvis, 2002). Each 150-μL import assay contained 107 chloroplasts, 5 mM MgATP, 10 mM Met, and translation product not exceeding 10% of the total volume. Import, thermolysin treatment, and quantification were performed according to Aronsson and Jarvis (2002).
To deplete endogenous ATP, chloroplasts were kept in the dark at room temperature for 20 min. Small molecules, including ATP, were removed from the translation products by Sephadex G 25 filtration (Pharmacia). MgATP was added to import reactions containing dark-adapted chloroplasts and ATP-depleted preprotein at different concentrations (50 to 5000 μM) or was omitted completely. Reactions were incubated in the dark at 25°C for 10 min and then analyzed as usual.
Import-chase experiments used similarly prepared, ATP-depleted chloroplasts and translation products and were performed in the dark. First, 2 × 107 chloroplasts were incubated with translation product in the presence of 50 μM ATP, in a total volume of 300 μL, for 10 min at 25°C. The chloroplasts then were reisolated by spinning through a 35% Percoll cushion in HS buffer at 2000g for 6 min and resuspended (with a cut 0.2-mL pipette tip) in 300 μL of buffer containing all of the components of an import reaction except chloroplasts, ATP, and translation product. The reaction then was divided into two equal aliquots of 150 μL. To one aliquot, ATP was added to a final concentration of 5 mM, and to the other aliquot, an equal volume of water was added. The import reactions were allowed to proceed for 10 min at 25°C and then analyzed as described previously.
RNA Isolation
RNA was isolated from plant material grown in vitro (5 and 10 days old) or on soil (28 days old). Plant material (up to 4 g) was ground to a powder in liquid nitrogen, and nucleic acids were extracted with equal volumes (∼5 mL each) of 100 mM Tris/HCl, pH 9.0, and phenol in 15-mL tubes. Samples were centrifuged at 3000g for 30 min at 4°C and then purified by phenol:chloroform:isoamyl alcohol (25:24:1) extraction. Nucleic acid was precipitated by adding 1.5 mL of 3.0 M NaOAc, pH 5.2, and 2 volumes of 100% ethanol and incubating overnight at −20°C. After centrifugation at 3000g for 45 min at 4°C, samples were dried, resuspended in 500 μL of water, and transferred to 1.5-mL tubes. RNA was precipitated by adding 200 μL of 10 M LiCl and incubating on ice overnight. Samples were centrifuged at 20,000g for 15 min at 4°C, and the pellets were washed with 70% (v/v) ethanol, dried, and resuspended in sterile water. Heating to 65°C was necessary to dissolve the RNA.
RNA Gel Blot Analysis
Total RNA was fractionated by electrophoresis on 1.3% (w/v) agarose gels containing 1.85% (v/v) formaldehyde and Mops buffer [20 mM 3-(N-morpholino)-propanesulfonic acid, 1 mM EDTA, pH 8.0, and 50 mM NaOAc, pH 7.0] and transferred to Hybond NX membranes (Amersham Pharmacia Biotech) in 10× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). Membranes were probed with α-32P-dCTP–labeled DNA fragments (Feinberg and Vogelstein, 1983) in 0.3 M sodium phosphate buffer, pH 7.2, containing 7% (w/v) SDS, 1 mM EDTA, pH 8.0, and 2% (w/v) BSA at 65°C overnight. Filters were washed at 65°C twice with 2× SSC and 0.1% (w/v) SDS for 30 min, once with 1× SSC and 0.1% (w/v) SDS for 20 min, and once with 0.5× SSC and 0.1% (w/v) SDS for 10 min. Band quantification was performed using ImageQuant software (Molecular Dynamics).
Identical filters were probed simultaneously with atTOC33 and atTOC34 probes. The atTOC33 and atTOC34 probes corresponded to full-length cDNAs and were almost identical in length (1132 and 1158 bp, respectively) and GC content (40.99 and 41.28%, respectively). The probes were labeled simultaneously under identical conditions using the same isotope and were shown to have identical specific activities by scintillation counting. Hybridization, washing, and exposure steps were performed simultaneously under identical conditions. Cross-hybridization of the probes between atTOC33 and atTOC34 (sequence homology of 50.2%) can be excluded, because the wash stringency used allowed only probe/target combinations with >83% identity to remain hybridized.
DNA Array Analysis
The 1827 gene-specific tags (GSTs) described previously (Kurth et al., 2002) were combined with an additional 1465 GSTs from genes that encode proteins featuring a chloroplast transit peptide predicted by TargetP (Emanuelsson et al., 2000), resulting in a 3292-GST array (Richly et al., 2003). Amplification, quantification, verification, and spotting (in duplicate) of PCR products on nylon filters were performed as described (Varotto et al., 2001; Kurth et al., 2002). At least three experiments with different filters and independent cDNA probes from plant pools were performed for each condition, thus minimizing variation between individual plants, filters, or probes. cDNA probe synthesis was primed by a mixture of oligonucleotides matching the 3292 genes in the antisense orientation and hybridized to the GST array as described (Kurth et al., 2002). Images were read using a Storm PhosphorImager (Molecular Dynamics) imported into the ArrayVision program (version 6.0; Imaging Research, St. Catharines, Ontario, Canada), in which data were normalized with reference to all spots on the array (Kurth et al., 2002). After import of expression data into the ArrayStat program (version 1.0; Imaging Research), a z test (nominal α set to 0.05) was performed to identify statistically significant differential expression values. The data obtained for all significantly differentially regulated genes are given in the supplemental data online.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Paul Jarvis, rpj3@le.ac.uk.
Supplementary Material
Acknowledgments
We thank Svenja Hester, Julie Howard, Christine Jackson, Adrian Shirlin, and Anthony Wardle for technical assistance and Jocelyn Bédard, Jonathan Combe, Sabina Kovacheva, Enrique López-Juez, and Simon Geir Møller for their insightful comments on the manuscript. We are grateful to Kenneth Cline, Ulf-Ingo Flügge, Bernhard Grimm, Neil Hoffman, Kenneth Keegstra, Kenton Ko, and Henrik Scheller for generously providing antibodies. This work was supported by the Royal Society Rosenheim Research Fellowship (to P.J.), by Biotechnology and Biological Sciences Research Council (BBSRC) Grants 91/C12976 and 91/P12928 (to P.J.), by the Deutsche Forschungsgemeinschaft (to D.L.), and by the Bundesministerium für Bildung und Forschung (to J.K. and D.L.). The GARNet proteomics facility at the University of Cambridge was funded by the BBSRC Investigating Gene Function Initiative.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012955.
Footnotes
Online version contains Web-only data.
References
- Aronsson, H., and Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529, 215–220. [DOI] [PubMed] [Google Scholar]
- Bauer, J., Chen, K., Hiltbrunner, A., Wehrli, E., Eugster, M., Schnell, D., and Kessler, F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Bölter, B., May, T., and Soll, J. (1998). A protein import receptor in pea chloroplasts, Toc86, is only a proteolytic fragment of a larger polypeptide. FEBS Lett. 441, 59–62. [DOI] [PubMed] [Google Scholar]
- Bruce, B.D. (2001). The paradox of plastid transit peptides: Conservation of function despite divergence in primary structure. Biochim. Biophys. Acta 1541, 2–21. [DOI] [PubMed] [Google Scholar]
- Chen, K., Chen, X., and Schnell, D.J. (2000). Mechanism of protein import across the chloroplast envelope. Biochem. Soc. Trans. 28, 485–491. [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Gutensohn, M., Schulz, B., Nicolay, P., and Flugge, U.I. (2000). Functional analysis of the two Arabidopsis homologues of Toc34, a component of the chloroplast protein import apparatus. Plant J. 23, 771–783. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Bauer, J., Alvarez-Huerta, M., and Kessler, F. (2001. a). Protein translocon at the Arabidopsis outer chloroplast membrane. Biochem. Cell Biol. 79, 629–635. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Bauer, J., Vidi, P.A., Infanger, S., Weibel, P., Hohwy, M., and Kessler, F. (2001. b). Targeting of an abundant cytosolic form of the protein import receptor at Toc159 to the outer chloroplast membrane. J. Cell Biol. 154, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnah, S.C., Wagner, R., Sveshnikova, N., Harrer, R., and Soll, J. (2002). The chloroplast protein import channel Toc75: Pore properties and interaction with transit peptides. Biophys. J. 83, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, S., Muckel, E., Heemeyer, F., von Heijne, G., and Soll, J. (1994). A receptor component of the chloroplast protein translocation machinery. Science 266, 1989–1992. [DOI] [PubMed] [Google Scholar]
- Jackson-Constan, D., and Keegstra, K. (2001). Arabidopsis genes encoding components of the chloroplastic protein import apparatus. Plant Physiol. 125, 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, P. (2003). Intracellular signalling: The language of the chloroplast. Curr. Biol. 13, R314–R316. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., Chen, L.J., Li, H., Peto, C.A., Fankhauser, C., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., and Soll, J. (2002). Toc, Tic, and chloroplast protein import. Biochim. Biophys. Acta 1590, 177–189. [DOI] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, R.J., Freymann, D.M., Stroud, R.M., and Walter, P. (2001). The signal recognition particle. Annu. Rev. Biochem. 70, 755–775. [DOI] [PubMed] [Google Scholar]
- Kessler, F., Blobel, G., Patel, H.A., and Schnell, D.J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Kouranov, A., and Schnell, D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139, 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth, J., Varotto, C., Pesaresi, P., Biehl, A., Richly, E., Salamini, F., and Leister, D. (2002). Gene-sequence-tag expression analyses of 1,800 genes related to chloroplast functions. Planta 215, 101–109. [DOI] [PubMed] [Google Scholar]
- Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19, 47–56. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Kouranov, A., LaSala, S.E., and Schnell, D.J. (1996). Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Akita, M., Davila-Aponte, J., and Keegstra, K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, L.J., and Keegstra, K. (1992). The binding of precursor proteins to chloroplasts requires nucleoside triphosphates in the intermembrane space. J. Biol. Chem. 267, 433–439. [PubMed] [Google Scholar]
- Olsen, L.J., Theg, S.M., Selman, B.R., and Keegstra, K. (1989). ATP is required for the binding of precursor proteins to chloroplasts. J. Biol. Chem. 264, 6724–6729. [PubMed] [Google Scholar]
- Perry, S.E., and Keegstra, K. (1994). Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell 6, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesaresi, P., Varotto, C., Meurer, J., Jahns, P., Salamini, F., and Leister, D. (2001). Knock-out of the plastid ribosomal protein L11 in Arabidopsis: Effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., and Geissler, A. (2001). Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349. [DOI] [PubMed] [Google Scholar]
- Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Co-variations in the nuclear chloroplast transcriptome reveal a regulatory master switch. EMBO Rep. 4, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel, S., Haley, J., Jiang, C.Z., Tsai, C.H., and Bogorad, L. (1996). A mechanism for intergenomic integration: Abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl. Acad. Sci. USA 93, 3881–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff, E., Soll, J., Sveshnikova, N., Tien, R., Wright, S., Dabney-Smith, C., Subramanian, C., and Bruce, B.D. (2002). Structural and guanosine triphosphate/diphosphate requirements for transit peptide recognition by the cytosolic domain of the chloroplast outer envelope receptor, Toc34. Biochemistry 41, 1934–1946. [DOI] [PubMed] [Google Scholar]
- Schnell, D.J., Kessler, F., and Blobel, G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., Waegemann, K., and Soll, J. (1995). A constituent of the chloroplast import complex represents a new type of GTP-binding protein. Plant J. 7, 401–411. [DOI] [PubMed] [Google Scholar]
- Strand, A., Asami, T., Alonso, J., Ecker, J.R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. [DOI] [PubMed] [Google Scholar]
- Sun, Y.J., Forouhar, F., Li Hm, H.M., Tu, S.L., Yeh, Y.H., Kao, S., Shr, H.L., Chou, C.C., Chen, C., and Hsiao, C.D. (2002). Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat. Struct. Biol. 9, 95–100. [DOI] [PubMed] [Google Scholar]
- Surpin, M., Larkin, R.M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14 (suppl.), S327.–S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveshnikova, N., Soll, J., and Schleiff, E. (2000). Toc34 is a preprotein receptor regulated by GTP and phosphorylation. Proc. Natl. Acad. Sci. USA 97, 4973–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unlu, M., Morgan, M.E., and Minden, J.S. (1997). Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077. [DOI] [PubMed] [Google Scholar]
- Varotto, C., Richly, E., Salamini, F., and Leister, D. (2001). GST-PRIME: A genome-wide primer design software for the generation of gene sequence tags. Nucleic Acids Res. 29, 4373–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T.S., and Li, H. (2001). Chloroplast protein translocon components atToc159 and atToc33 are not essential for chloroplast biogenesis in guard cells and root cells. Plant Physiol. 127, 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.