Abstract
Membrane-bound glycerol-3-phosphate acyltransferase (GPAT; EC 2.3.1.15) mediates the initial step of glycerolipid biosynthesis in the extraplastidic compartments of plant cells. Here, we report the molecular characterization of a novel GPAT gene family from Arabidopsis, designated AtGPAT. The corresponding polypeptides possess transmembrane domains and GPAT activity when expressed heterologously in a yeast lipid mutant. The functional significance of one isoform, AtGPAT1, is the focus of the present study. Disruption of the AtGPAT1 gene causes a massive pollen development arrest, and subsequent introduction of the gene into the mutant plant rescues the phenotype, illustrating a pivotal role for AtGPAT1 in pollen development. Microscopic examinations revealed that the gene lesion results in a perturbed degeneration of the tapetum, which is associated with altered endoplasmic reticulum profiles and reduced secretion. In addition to the sporophytic effect, AtGPAT1 also exerts a gametophytic effect on pollen performance, as the competitive ability of a pollen grain to pollinate is dependent on the presence of an AtGPAT1 gene. Deficiency in AtGPAT1 correlates with several fatty acid composition changes in flower tissues and seeds. Unexpectedly, however, a loss of AtGPAT1 causes no significant change in seed oil content.
INTRODUCTION
The majority of fatty acids synthesized in a plant cell are incorporated into either membrane glycerolipids or the neutral lipid triacylglycerol (TAG). A significant body of literature demonstrates that de novo glycerolipid biosynthesis is a fundamental metabolic process that is well conserved in both prokaryotic and eukaryotic organisms (for review, see Frentzen, 1993; Wilkison and Bell, 1997; Athenstaedt and Daum, 1999; Dircks and Sul, 1999). In plant cells, there are three confirmed glycerolipid biosynthetic pathways, compartmentalized in chloroplasts, cytoplasmic membrane systems, and mitochondria (Roughan and Slack, 1982; Heinz and Roughan, 1983; Browse et al., 1986). Each of the three pathways has its own set of enzymes that essentially consist of fatty acid acyltransferases that mediate the stereospecific esterification of the glycerol backbone. However, extensive coordination and exchange of glycerolipid molecules between intracellular compartments also has been established (Browse et al., 1986; Kunst et al., 1988).
The initial and committed step of glycerolipid biosynthesis is the fatty acid acylation at the sn-1 position of glycerol-3-phosphate (G-3-P) mediated by glycerol-3-phosphate acyltransferase (GPAT). In contrast to major advancements achieved in the molecular characterization of the sn-2 acyltransferases (lysophosphatidic acid acyltransferase) (Knutzon et al., 1995; Lessner et al., 1995) and the sn-3 acyltransferases (diacylglycerol acyltransferase) (Routaboul et al., 1999; Zou et al., 1999), information regarding the membrane-bound GPATs is limited to reports concerning partially purified proteins (Fritz et al., 1986; Eccleston and Harwood, 1995; Manaf and Harwood, 2000). To date, no gene that encodes an extraplastidic membrane-bound GPAT has been reported. In the past, considerable attention was focused on the roles of acyltransferases in determining seed oil content and its fatty acid composition (Ichihara, 1984; Griffiths et al., 1985; Sun et al., 1988; Bafor et al., 1990; Frentzen, 1990; Hares and Frentzen, 1991; Katavić et al., 1995; Zou et al., 1997, 1999; Knutzon et al., 1999; Manaf and Harwood, 2000). However, because glycerolipids are the necessary building blocks of cell membranes, it also may be safely assumed that glycerolipid biosynthesis is essential for the maintenance of membrane integrity and cellular processes involving membrane biogenesis. Nonetheless, there is little information available showing the direct involvement of de novo glycerolipid biosynthesis in membrane ontogeny. Research on this topic necessarily rests on our knowledge of the fatty acid acyltransferases involved in the essential reaction steps of the biosynthetic pathway.
Pollen development is a complex process that depends on the precise coordination of developmental programs between sporophytic and gametophytic anther tissues. Both developing microspores and the surrounding tapetal cells are known to be particularly active in lipid metabolism (Ferreira et al., 1997; Piffanelli et al., 1998; Platt et al., 1998). The tapetum has been regarded as an attractive model system for studying various aspects of cellular activity because within a relatively short life span, it undergoes a remarkable maturation process that involves both structural and metabolic specialization. Tapetal differentiation also can be monitored readily by reference to the stages of microspore development. The tapetal cells of the Arabidopsis anther are of the secretory type (Murgia et al., 1991), releasing an array of proteins, lipids, and other nutrients to the anther locule. The discharge of nutrients from the tapetum involves both active secretion and, at later stages, the disintegration of the tapetal cells. The precise and temporal progression of tapetal differentiation with respect to microspore developmental stages is of major significance to the successful production of pollen (Schrauwen et al., 1996). Tapetal development also is particularly sensitive to mitochondrial dysfunction, as illustrated by a large number of cytoplasmic male sterility (CMS) mutants with abnormalities in tapetal development (Schnable and Wise, 1998). However, the molecular mechanisms that underlie most developmental defects of tapeta remain enigmatic. As prominent a metabolic feature as lipid metabolism is in tapetal development, there has been no report directly linking a de novo glycerolipid biosynthetic defect to any tapetal malfunction.
Here, we report the identification of a novel membrane-bound GPAT gene family from Arabidopsis and a functional study of one isoform, designated AtGPAT1. We provide evidence that AtGPAT1 plays a pivotal role in pollen development and male fertility through the analysis of AtGPAT1 disruption mutants. A deficiency of AtGPAT1 affects tapetal differentiation and causes most of the microspores to abort before reaching maturity.
RESULTS
Identification of the AtGPAT Gene Family in Arabidopsis
A seminal study performed by Lewin and co-workers revealed four regions of strong sequence homology, termed blocks I to IV, in various acyltransferases from species ranging from bacteria to mammals (Lewin et al., 1999). Our study with two isoforms of the yeast endoplasmic reticulum (ER)–bound GPAT confirms and further extends the view that fatty acid acyltransferases are structurally conserved across the evolutionary spectrum (Zheng and Zou, 2001). Based on enzyme structural information from bacteria, yeast, and mammals, GPATs likely are significantly larger than 1-acyl-glycerol acyltransferases (lysophosphatidic acid acyltransferase), even though the functional significance of such a difference is unknown (Wilkison and Bell, 1997). In light of the genomic information available from Arabidopsis, we made the following assumptions in search of a plant GPAT: (1) a plant membrane-bound GPAT will share certain conserved domains with other fatty acid acyltransferases, and (2) it will be significantly larger than the sn-2 acyltransferases, which generally are ∼300 amino acids long (Knutzon et al., 1995; Lessner et al., 1995).
A series of Basic Local Alignment Search Tool (BLAST) searches (Altschul et al., 1990), using a query from a partial sequence encompassing a region conserved between the yeast GPATs and other fatty acid acyltransferases, identified a gene family in Arabidopsis that we designated AtGPAT. Amino acid sequence alignment revealed that the seven members of this family (AtGPAT1 through AtGPAT7) contain the previously defined four acyltransferase domains (Figure 1). The His and Asp residues in block I, the Gly residue in block III, and the Pro residue in block IV, all of which have been shown to be catalytically important sites (Lewin et al., 1999), are absolutely conserved. Furthermore, residues critical in G-3-P binding, including the Arg in block II and the Gln and Ser in block III, also are invariant among the members of the Arabidopsis family. At the C terminus, several blocks of conserved residues extend beyond regions known to be implicated in acyltransferase enzyme function. Major sequence divergence is found at the very end of the N terminus, which in part gives rise to the variation in size of these members.
Figure 1.
Deduced Amino Acid Sequence Alignment of the Members of the AtGPAT Family in Arabidopsis.
Four conserved acyltransferase motifs are indicated above the alignment (AT I, AT II, AT III, and AT IV). Two transmembrane domains (TM I and TM II) were predicted computationally. Identical and similar amino acid residues are shaded black and gray, respectively.
Expression Patterns of the AtGPAT Genes
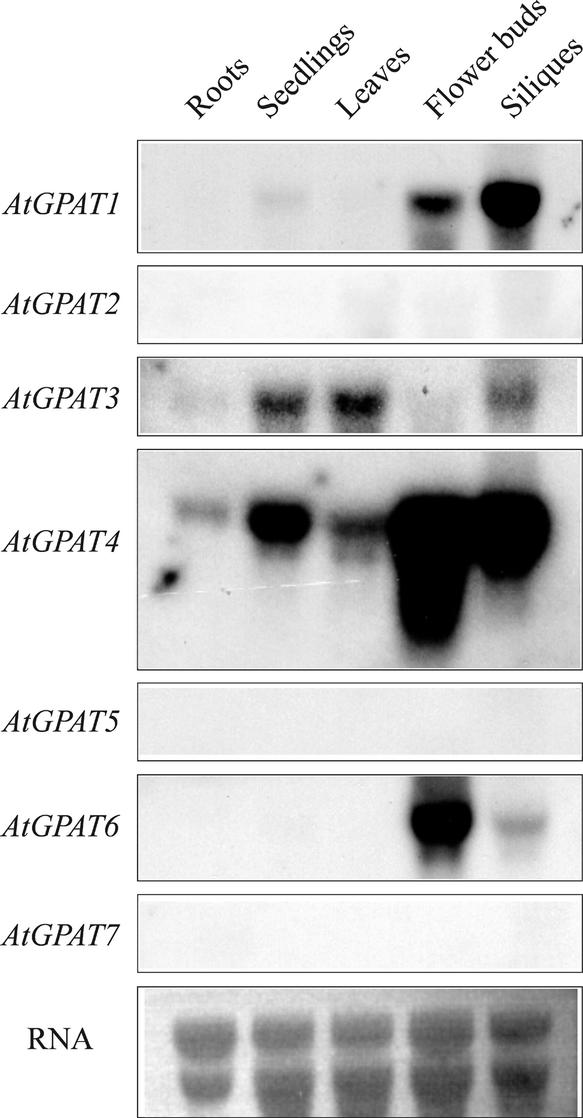
The AtGPAT cDNAs were cloned from Arabidopsis by reverse transcriptase–mediated PCR, and the expression of the corresponding genes was investigated by RNA gel blot analysis with total RNA prepared from various tissues of Arabidopsis. Hybridization was performed at high stringency. The autoradiographs shown in Figure 2 were derived from identical RNA gel blots hybridized with similar amounts of labeled probes and exposed for the same duration (24 h). AtGPAT1 transcript accumulated at a high level in developing siliques and flower buds. AtGPAT3 expression was low but present in various tissues. AtGPAT4 was the most abundantly expressed member, with transcript detectable in all tissues examined, whereas AtGPAT6 accumulated mainly in flower buds. Transcripts of AtGPAT2, AtGPAT5, and AtGPAT7 were not detected, suggesting that they were either expressed at a very low level in these particular tissues/developmental stages or induced by internal and/or external stimuli.
Figure 2.
Expression Analysis of AtGPAT Genes.
Approximately 10 μg of total RNA in each lane was used for hybridization with the individual AtGPAT probes. Equal RNA loading was verified by ethidium bromide staining. The autoradiographs were generated by exposure for 24 h.
Members of the AtGPAT Gene Family Encode Active Acyltransferases That Specifically Acylate G-3-P
Because enzymatic characterization based on the in vitro purification and reconstitution of membrane-bound GPATs has proven very difficult (Scheideler and Bell, 1989; Lewin et al., 1999), we used an alternative strategy to establish the functionality of AtGPATs by overexpressing the AtGPAT cDNAs in a yeast mutant, gat1Δ. The gat1Δ strain harbors a mutation in a gene that encodes a major ER-bound GPAT and has a very low residual GPAT activity. The functionality of putative GPATs can be readily tested in this strain (Zheng and Zou, 2001). We assessed the GPAT activity of the total yeast lysates prepared from the gat1Δ strain expressing the Arabidopsis cDNAs and the control vector using 14C-labeled G-3-P and a mixture of stearoyl-CoA and oleoyl-CoA, respectively, in the presence of 5 mM EDTA, which, according to some studies (Eccleston and Harwood, 1992; Manaf and Harwood, 2000), increases the incorporation rate of substrates in GPAT assays. Lipid products of the reaction mixtures were separated by thin layer chromatography, and the incorporation of 14C-labeled G-3-P was measured subsequently. The calculation of enzyme activity was based on the difference in G-3-P acylation between the gat1Δ strain expressing an AtGPAT and the strain harboring the control vector. Under our assay conditions, five of the seven AtGPAT proteins demonstrated reproducible GPAT activity. The activities for AtGPAT1, AtGPAT4, AtGPAT5, AtGPAT6, and AtGPAT7 were 217.42, 58.92, 86.85, 278.56, and 58.93 pmol·min−1·mg−1 protein, respectively. Repeated assays failed to detect any activity with AtGPAT2 and AtGPAT3, which could be attributable to a lack of unidentified factors in our assay conditions.
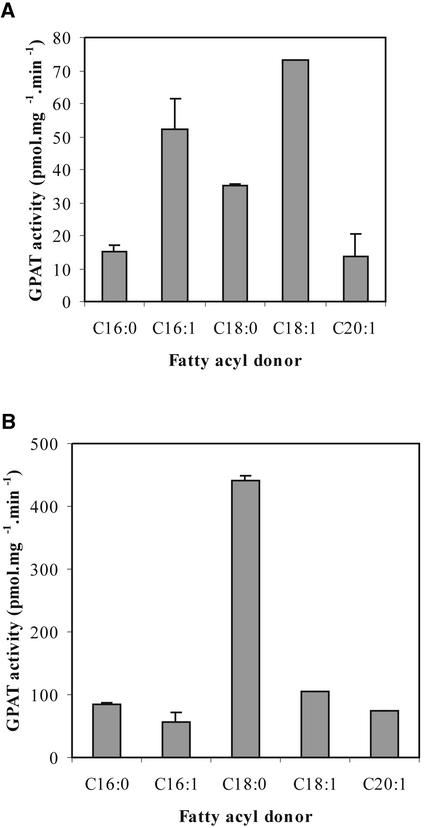
We focused on AtGPAT1 for further enzymatic characterization. To validate the notion that AtGPAT1 specifically acylates G-3-P, we performed substrate competition experiments in which potential acyl acceptors, including dihydroxyacetone phosphate, lysophosphatidic acid, and sterols, were each added at three different concentrations (25, 50, and 100 μM) to reaction mixtures containing 25 μM 14C-labeled G-3-P, after which the lipid extracts were analyzed for 14C incorporation. None of the tested metabolites affected the rate of G-3-P acylation (data not shown), thus indicating that AtGPAT1 specifically uses G-3-P as a fatty acyl acceptor. We further investigated the fatty acid substrate specificity of AtGPAT1, performing all assays in the presence and absence of EDTA because both conditions have been used in the past to evaluate membrane-bound GPAT properties (Manaf and Harwood, 2000). In the absence of EDTA, AtGPAT1 preferred 16:1 and 18:1 as fatty acyl donors (Figure 3A). However, we found that the inclusion of 5 mM EDTA in the reaction mixture, which significantly stimulated the incorporation rate of the assay, also resulted in a pronounced fatty acyl substrate preference for stearoyl-CoA (Figure 3B). It is unclear how EDTA exerts such effects, although the inhibition of lipase has been suggested (Manaf and Harwood, 2000). These results indicate that GPAT substrate specificity is difficult to evaluate due to the uncertainty of what in vitro conditions accurately reflect the in vivo situation.
Figure 3.
Fatty Acyl Specificity of AtGPAT1.
Assays of GPAT were conducted with 14C–G-3-P as described in Methods in the absence (A) and presence (B) of EDTA. C16:0, palmitoyl-CoA; C16:1, palmitoleoyl-CoA; C18:0, stearoyl-CoA; C18:1, oleoyl-CoA; C20:1, eicosenoic acyl-CoA.
Membrane Targeting of AtGPAT1 in Vitro
Analysis by two transmembrane protein prediction programs, TMpred and TopPred2, yielded results with a high degree of certainty that AtGPAT1 is a membrane-bound protein with two membrane-spanning segments located at residues 138 to 158 and 334 to 354. Similarly, regions corresponding to these two segments in other members of the AtGPAT family all were invariably predicted as transmembrane domains (Figure 1).
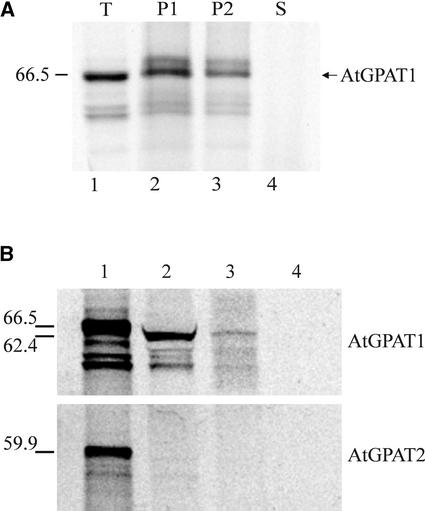
To examine the membrane localization experimentally, we performed targeting assays using canine pancreatic microsome–derived membranes and in vitro–synthesized polypeptide. The size of the in vitro translation product was consistent with the calculated molecular mass of AtGPAT1 (Figure 4A, lane 1). Polypeptide generated in the presence of microsomes was treated subsequently with a high-salt solution before spinning down the membrane fraction, and cosedimentation of AtGPAT1 with the membranes was maintained (Figure 4A, lane 2), confirming a tight association of AtGPAT1 with the membranes. Polypeptide-bound microsomes recovered after high-salt treatment were extracted further with an alkaline buffer to convert closed vesicles into open membrane sheets and release content proteins and peripheral membrane proteins into the aqueous phase (Fujiki et al., 1982). This alkaline extraction procedure entailed another round of centrifugation, and as expected, the amount of protein recovered, as shown in Figure 4A, lane 3, was less than that of the starting material in lane 2. Nonetheless, cosedimentation of the majority of the polypeptides with microsomes was clearly maintained (Figure 4A, lane 3), suggesting that the AtGPAT1 protein is integrated into the membrane systems. Accordingly, trichloroacetic acid precipitation of the supernatant revealed that the protein was present at a level too low to be detected in the alkaline-soluble fraction (Figure 4A, lane 4).
Figure 4.
Subcellular Targeting of AtGPAT1.
AtGPAT1 was synthesized in vitro using transcription/translation systems with 35S-Met.
(A) Integration of AtGPAT1 into ER-derived membranes. Lane 1, total translation products (T); lane 2, microsomal pellet (P1) of the translation mixture after the high-salt wash; lane 3, microsomal pellet (P2) derived from the translation mixture washed with the high-salt solution followed by alkaline extraction; lane 4, supernatant (S) generated from the alkaline extraction of the high-salt-washed microsomes precipitated with trichloroacetic acid. The sizes of the synthesized polypeptides are shown at left in kilodaltons.
(B) In vitro import of AtGPAT1 into pea mitochondria. Lane 1, total translation products; lane 2, labeled polypeptides imported into mitochondria; lanes 3 and 4, the imported polypeptides treated with proteinase K in the absence and presence, respectively, of 0.5% (v/v) Triton X-100. The sizes of the nonprocessed and processed proteins are indicated in kilodaltons at left. AtGPAT2 was used as a control.
Unique among AtGPATs, AtGPAT1 also possesses a long N-terminal extension that is rich in hydroxylated residues. Analyses performed with several protein-sorting prediction programs (pSORT, Predotar, and Target P V1.0; http://aramemnon.botanik.uni-koeln.de) yielded positive results for a potential mitochondrial targeting of AtGPAT1. Thus, a mitochondrial protein import experiment was performed using isolated pea mitochondria. As shown in Figure 4B, in vitro–synthesized AtGPAT1 remained tightly associated with the pea mitochondria after fractionation of the import assay mixture, indicating that AtGPAT1 is imported into the mitochondria. In addition, the mitochondrial import process resulted in a size reduction of AtGPAT1, suggesting that the protein was subjected to proteolytic cleavage of a leader sequence (Figure 4B, lane 2). To confirm that the polypeptide was not just attached to the mitochondrial surface, mitochondria purified from the import assay mixture were subjected to protease treatment and subsequently repurified. The size of the mitochondrial pellet recovered after protease treatment appeared smaller. Nevertheless, those polypeptides, once imported into the intact mitochondria, were protected from protease attack, as the polypeptides recovered after protease digestion were the same size as before the treatment (Figure 4B, lane 3). Releasing the proteins from the mitochondria by detergent treatment rendered the proteins subject to protease digestion (Figure 4B, lane 4). As a control, identical targeting experiments also were performed with AtGPAT2. Clearly, AtGPAT2 was not imported into mitochondria (Figure 4B).
Identification of T-DNA Insertion Mutants
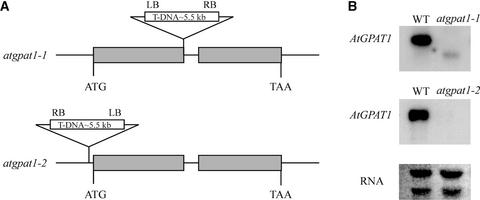
We identified two Arabidopsis mutant lines, atgpat1-1 and atgpat1-2, through PCR screening of the T-DNA–tagged Basta population (ecotype Wassilewskija) available at the University of Wisconsin (Sussman et al., 2000). Sequencing of PCR products generated from AtGPAT1-specific and T-DNA–specific primers placed the T-DNA insert at the end of exon I of the gene in atgpat1-1 and 40 bp upstream of the predicted translational start site in atgpat1-2 (Figure 5A). RNA gel blot analysis detected no AtGPAT1 mRNA in either of the two mutant lines, but a short transcript, most likely representing a truncated AtGPAT1, was detected at a low level in atgpat1-1 (Figure 5B).
Figure 5.
Genetic Analysis of the atgpat1-1 and atgpat1-2 Mutants.
(A) Genomic organization of the atgpat1-1 and atgpat1-2 loci. Gray horizontal boxes represent exons. The T-DNA insert is not drawn to scale. LB, T-DNA left border; RB, T-DNA right border.
(B) RNA gel blot analysis of AtGPAT1 transcript in the wild type (WT) and mutants. Approximately 10 μg of total RNA from siliques was loaded in each lane.
Neither of the mutant lines showed any growth or developmental defects at the vegetative stages. However, both mutant lines had severely affected seed yields, as indicated by the reduced size of their mature siliques. Heterozygous plants harboring either of the mutant alleles, on the other hand, produced siliques indistinguishable from those of wild-type plants. The effect of AtGPAT1 deficiency on seed yield was studied by systematically examining the number of seeds produced in siliques from wild-type and mutant plants grown in parallel. Compared with yields in the wild type, the average seed yield per silique in atgpat1-1 was reduced by >72%, with a similar reduction in atgpat1-2. The seed yield also varied significantly along the inflorescence axis. The siliques that formed earlier, on the lower portion of the inflorescence, had 1 or 0 seed, whereas the later-developing siliques on the upper portion of the inflorescence contained up to 29 seeds. Thus, there was a partial recovery of seed set as inflorescence development proceeded in the mutant. By contrast, the wild-type plants generally produced 40 seeds per silique.
atgpat1 Mutants Have Severely Reduced Male Fertility
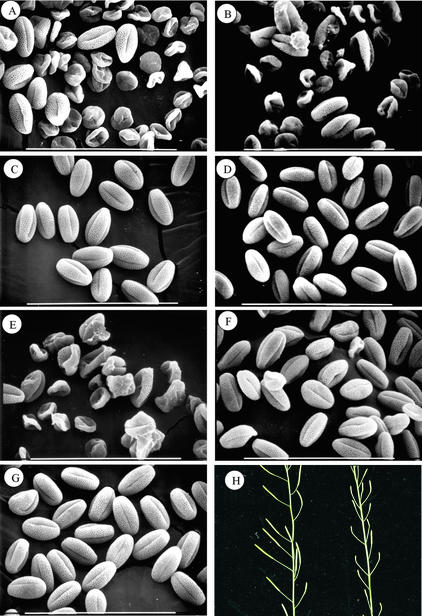
Examination of scanning electron micrographs of pollen grains produced by the atgpat1-1 and atgpat1-2 homozygous plants revealed that the majority of the pollen grains were collapsed in morphology (Figures 6A and 6B). A small portion, however, appeared normal, accounting for the ability of the mutants to produce seeds. Furthermore, pollen grains from heterozygous plants were uniform in morphology and indistinguishable from those of the wild type (Figures 6C and 6D). Thus, the collapsed nature of the pollen grains most likely was caused by a sporophytic defect in the mutants. It also was evident that the extent to which the pollen defect occurred varied among different flowers of the same mutant plant. Pollen from flowers initiated early on the inflorescence was affected most severely, whereas the proportion of normal pollen found in flowers from the upper portion of the inflorescence increased significantly (data not shown).
Figure 6.
Male Fertility in Wild-Type and Mutant Plants.
(A) to (G) Scanning electron micrographs of mature pollen grains from atgpat1-1 homozygous (A) and heterozygous (C) plants, atgpat1-2 homozygous (B) and heterozygous (D) plants, atgpat1-1 homozygous plants transformed with the control vector pRD400 alone (E) and with the vector carrying the AtGPAT1 gene (F), and a wild-type plant (G). Bars = 100 μm.
(H) Silique development in atgpat1-1 homozygous plants transformed with the AtGPAT1 gene (left) and the control vector (right).
To answer the question of whether the stigma also was affected by the mutation, we performed reciprocal crosses between atgpat1-1 and wild-type plants and found that seed yield was restored fully when atgpat1-1 plants were pollinated with wild-type pollen. By contrast, pollination of wild-type plants with the atgpat1-1 pollen generally produced only a few seeds per silique. These results strongly indicate that the function of the stigma was not compromised in the mutant.
To ascribe the impaired male fertility of the atgpat1-1 mutant to the disruption of AtGPAT1, the AtGPAT1 gene, including its promoter, and a control vector, pRD400 (Datla et al., 1992), were introduced into atgpat1-1 plants by Agrobacterium tumefaciens–mediated transformation. Examination by scanning electron microscopy of the pollen grains from transgenic plants containing AtGPAT1 showed that the pollen development defect was rescued in 12 independent transgenic lines (Figure 6F). Accordingly, seed set also was restored (Figure 6H). Pollen grains obtained from mutant plants transformed with the control vector maintained the phenotype of atgpat1-1 pollen (Figure 6E). Thus, these results provide unequivocal evidence that AtGPAT1 plays an essential role in male fertility.
The atgpat1-1 Lesion Alters Cellular Processes in the Tapetal Cells
To determine the temporal aspects of microspore development in the mutant, transverse sections of atgpat1-1 and wild-type anthers were prepared and subjected to light microscopy. The course of pollen development was analyzed according to the stages described previously by Owen and Makaroff (1995) for Arabidopsis ecotype Wassilewskija. The mutant proceeded normally with respect to the development of microspore mother cells, which are surrounded by a monolayer tapetum, middle layer, endothecium, and epidermis. Additionally, 4′,6-diamidino-2-phenylindole staining of the microsporocytes confirmed that the mutant underwent a meiotic event indistinguishable from that of wild-type plants. Tetrad release and the formation of microspore exine also ensued as normal, suggesting that microspore development before the tetrad stage was not affected.
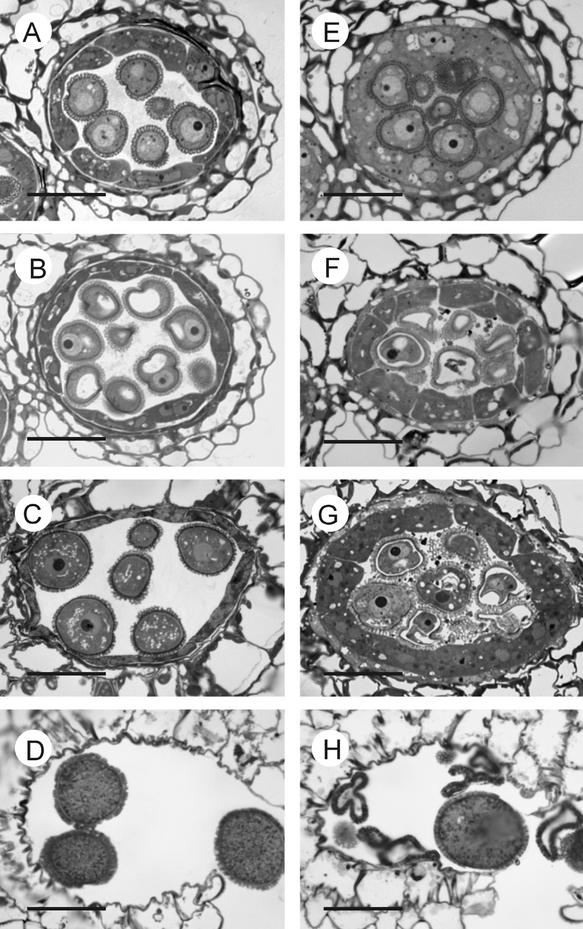
Commencing from the microspore-release stage, clear evidence of a tapetum developmental defect was detected. The size of the atgpat1-1 tapetal cells increased more conspicuously than that of the wild-type tapetal cells. The hypertrophic tapetum even invaded some space between microspores, resulting in such limited space in the atgpat1-1 locule that physical compression was imposed on the developing microspores (Figures 7A and 7E). At the ring-vacuolated stage in the wild type, a shrinkage of the tapetum layer became evident, creating excess locule space fit for the expansion of developing microspores (Figure 7B). By contrast, the atgpat1-1 tapetum continued to enlarge throughout the subsequent developmental stages (Figures 7F and 7G), disintegrating much later than the wild-type tapetum. However, the breakage of the septum and dehiscence were not affected (Figures 7D and 7H).
Figure 7.
Light Micrographs of Cross-Sections of Wild-Type and atgpat1-1 Anthers.
The left and right micrographs are from wild-type and atgpat1-1 anthers, respectively. Stages are defined according to Owen and Makaroff (1995). Bars = 20 μm.
(A) and (E) Microspore-release stage.
(B) and (F) Ring-vacuolated microspore stage.
(C) and (G) Bicellular pollen grain stage.
(D) and (H) Mature pollen grain stage.
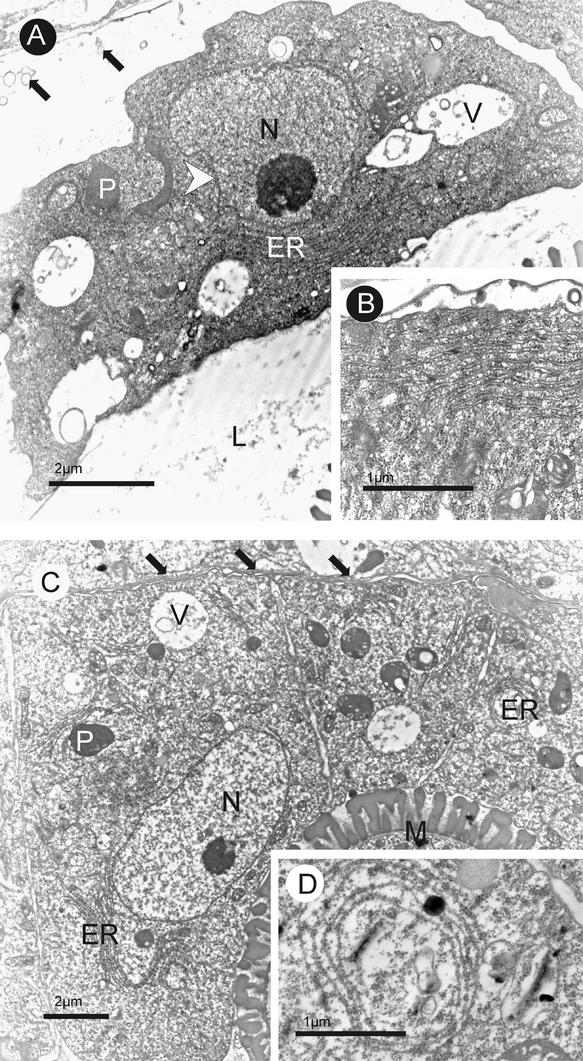
Transmission electron microscopy also detected several ultrastructural changes in the mutant tapetal cells, starting from microspore-release stage II. As described previously (Owen and Makaroff, 1995) and as observed in the wild type, standard features of tapetal cells at this stage include a complete dissolution of the cell wall (Figure 8A) and a significant dilation of ER cisternae consisting of >12 layers (Figure 8B). In some cases, the ER stacks were fused with the tapetal plasma membrane, presumably to facilitate the release of osmophilic materials into the locule (Owen and Makaroff, 1995). The cell walls of the mutant tapetal cells, on the other hand, still were intact, with one side remaining in direct contact with the middle layer (Figure 8C). In addition, ER dilation in the mutant generally was reduced, and its fusion with plasma membranes was seen only rarely (Figure 8C). Instead, unusual patterns of ER cylinders were observed occasionally (Figure 8D). The wild-type tapetal cells at this stage had distended and lobed nuclei, coalesced vacuoles, and invaginated and sometimes discontinuous plasma membranes, signifying cellular degeneration. By contrast, because these ultrastructural characteristics were not pronounced in the mutant, there was no clear indication of degeneration of the mutant tapetal cells. The cytological structural alterations were accompanied by apparent impairments in tapetal secretion in the mutant. At the ring-vacuolated stage, the wild-type anther locule was filled with fine fibrillar materials released from the tapetum (Figure 9A), whereas in atgpat1-1, such deposits were seen only occasionally (Figure 9B). Continuing into the early bicellular stage, abundant osmophilic vesicles containing inclusions were present within the wild-type locule (Figure 9C). Distinctively, vesicle deposits in the mutant locule were reduced dramatically at this stage (Figure 9D).
Figure 8.
Transmission Electron Micrographs of Cross-Sections of Wild-Type and atgpat1-1 Anthers at Microspore-Release Stage II.
(A) and (B) Wild-type anthers.
(C) and (D) atgpat1-1 anthers.
The tapetal cell wall is completely dissolved in the wild type (A) but remains intact in the mutant (C), as indicated by black arrows. The white arrowhead shows dented membranes in the nucleus of the wild-type tapetum in (A). Insets (B) and (D) show ER stack and ER ring structures in wild-type and mutant tapetal cells, respectively. ER, endoplasmic reticulum; L, locule; M, microspore; N, nucleus; P, plastid; V, vacuole.
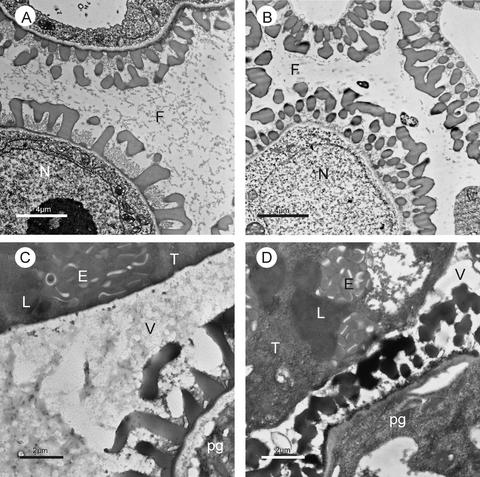
Figure 9.
Secretion Processes in Wild-Type and atgpat1-1 Tapeta.
Transmission electron micrographs of cross-sections of wild-type ([A] and [C]) and atgpat1-1 ([B] and [D]) anthers. At the ring-vacuolated microspore stage ([A] and [B]), a high concentration of fibrillar material is distributed uniformly throughout the wild-type locule (A), whereas much less material appears in the mutant locule (B). At the early bicellular stage ([C] and [D]), many osmophilic material–containing vesicles are secreted from the wild-type tapetum into the locule (C), whereas fewer vesicles are present in the mutant locule (D). E, elaioplast; F, fibrillar material; L, lipid body; N, nucleus; pg, pollen grain; T, tapetal cell; V, vesicle.
The ultrastructure of mitochondria also was examined due to the results of AtGPAT1 targeting assays. In the wild-type tapetum, the last stage at which intact mitochondria could be identified was microspore-release stage II. Although other organelles clearly were degenerating, mitochondria at this stage could be distinguished by the presence of a bilayer membrane and well-defined cristae. By contrast, at the same stage in the mutant tapetum, no intact mitochondria could be identified, despite the examination of a large number of sections. However, some spherical entities with a low electron density matrix were observed, but we could not conclude whether or not they were derived from degenerated mitochondria.
Pollen Grains Harboring the atgpat1-1 Allele Have an Altered Cytoplasmic Membrane Profile and Impaired Competitiveness in Fertilization
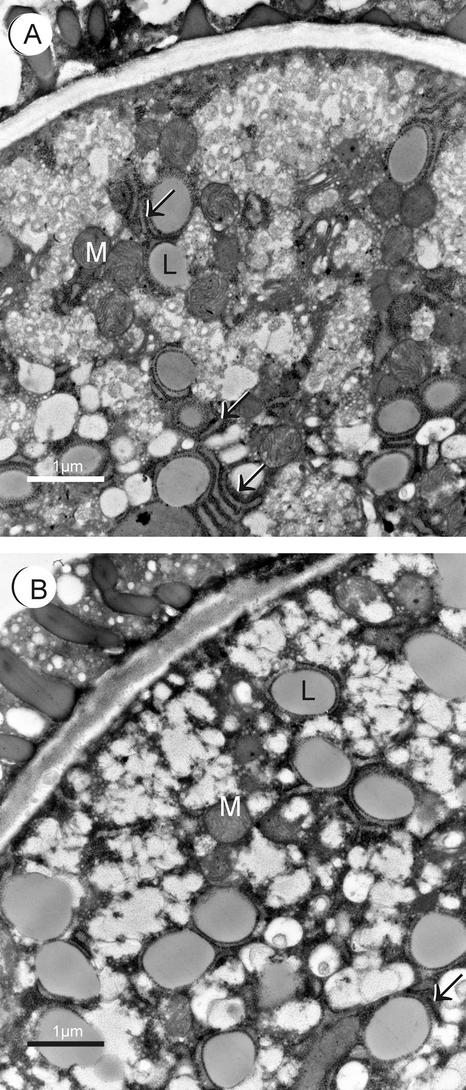
The small portion of atgpat1-1 pollen that reached maturity contained evenly distributed lipid bodies bearing ultrastructural changes, compared with the wild type, in two aspects (Figure 10B). First, it appeared that the lipid bodies in atgpat1-1 (0.867 ± 0.107 μm in diameter) were significantly larger than those of the wild type (0.635 ± 0.085 μm) (Figures 10A and 10B), as judged by the t test (P < 0.001). Second, although wild-type pollen lipid bodies often were encircled by ER cisternae (Figure 10A), these structures were associated only rarely with the lipid bodies in atgpat1-1 pollen (Figure 10B). According to Murphy and Vance (1999), unlike oil bodies in seeds, lipid bodies in pollen are not coated with a monolayer of oleosin proteins. The ER cisternae have been suggested, like the oleosins, to function as a physical barrier to prevent lipid body coalescence (Murphy and Vance, 1999). The observation of a lack of ER cisternae and hence a larger lipid body size in atgpat1-1 supports this theory. The effect of atgpat1-1 on membrane structure suggests a role for AtGPAT1 in cytoplasmic membrane biogenesis.
Figure 10.
Transmission Electron Micrographs of Mature Pollen Grains from Wild-Type and atgpat1-1 Plants.
(A) Wild-type pollen grain.
(B) atgpat1-1 pollen grain.
Arrows indicate ER cisternae. L, lipid body; M, mitochondria.
Pollen competitiveness was assessed using a protocol, as described by Mouline et al. (2002), based on the fact that the T-DNA bears a copy of the BAR gene, which confers resistance to L-phosphinothricin (L-PPT). As expected, progeny produced from a control plant heterozygous for a BAR gene that is not associated with AtGPAT1 segregated L-PPT resistance in a 3:1 ratio (325:116 L-PPT–resistant:nonresistant; χ2 = 0.40, P > 0.52). DNA gel blot analysis confirmed that atgpat1-1 has a single T-DNA insertion (data not shown). Significantly, among the progeny of self-fertilized plants heterozygous for the atgpat1-1 allele, homozygous wild-type plants accounted for 42% (425:314 L-PPT–resistant:nonresistant). Because pollination of the stigmas of atgpat1-1 homozygous plants with wild-type pollen grains resulted in full seed set, the possibility of the mutation affecting female fertility can be excluded. From the frequency of the homozygous wild-type plants (42%), the probability of fertilization by a wild-type pollen grain is inferred to be 0.84 (i.e., 0.42 × 2) in competition with an atgpat1-1 pollen grain, and the probability of fertilization by an atgpat1-1 pollen grain is only 0.16 (i.e., 1 − 0.84).
To provide further evidence that disruption of AtGPAT1 compromises pollen competitiveness in fertilization, we pollinated wild-type plants with pollen grains from atgpat1-2 heterozygous (atgpat1-2/AtGPAT1) plants and subsequently determined the proportion of the progeny containing the atgpat1-2 allele. PCR analysis of 112 progeny seedlings identified 37 plants bearing the mutant allele, indicating fertilization probabilities of 0.67 and 0.33 for wild-type and mutant pollen grains, respectively. This genetic pattern deviates from the 1:1 ratio (χ2 = 12.90, P < 0.001), indicating that the mutant pollen of the atgpat1-2 heterozygous line has an impaired competitiveness in fertilization. The higher fertilization probability (0.33) of the pollen grains from the atgpat1-2 heterozygote compared with the probability of 0.16 from the atgpat1-1 heterozygote may be explained by the possible existence of residual AtGPAT1 activity in the atgpat1-2 mutant, given that the insertion of T-DNA into the promoter region of AtGPAT1 may not completely abolish the presence of the gene transcript. We conclude that the ability of an individual haploid pollen grain to function effectively in the process of pollination is dependent on the presence of the AtGPAT1 gene. Thus, AtGPAT1 functions directly in the gametophytic pollen grains to modulate male fertility, most likely through its role in membrane biogenesis.
Pleiotropic Effects of AtGPAT1 Deficiency on Glycerolipid Biosynthesis
We also analyzed lipids from various tissues and organs of the mutant plants to understand the biochemical effect of a deficiency of AtGPAT1. Consistent with the AtGPAT1 expression profile, an altered glycerolipid profile was detected in the mutant flower tissues. The total lipid extract from flower buds was separated into two fractions, one containing TAGs and the other consisting of the polar lipids, which include phospholipids and glycoglycerolipids. As shown in Table 1, there was an ∼10% reduction in the content of both TAG and the polar lipids in the flower buds of the mutant compared with that in the wild type. In addition, the fatty acid composition of TAG in the mutant also changed, with a reduced proportion of palmitic acid (16:0) and increased proportions of the polyunsaturated linoleic acid (18:2) and linolenic acid (18:3).
Table 1.
Fatty Acid Composition of the Lipids from Flower Buds of Arabidopsis atgpat1-1 Mutant and Wild-Type Plants
| Lipid Fraction | Total Lipid (μmol fatty acid/g fresh weight) |
Proportion of Fatty Acid (mol %)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 14:0 | 16:0 | 16:1 | 16:3 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | ||
| TAG | ||||||||||
| Wild type | 0.86 ± 0.01 | 1.28 | 23.86 | 0.58 | 1.02 | 2.05 | 4.36 | 23.40 | 42.51 | 0.55 |
| atgpat1-1 | 0.77 ± 0.08 | 1.29 | 18.44 | 0.66 | 0.96 | 1.74 | 5.04 | 26.80 | 44.40 | 0.46 |
| Polar lipids | ||||||||||
| Wild type | 3.40 ± 0.60 | nd | 22.06 | nd | 1.30 | 2.94 | 4.83 | 25.84 | 42.65 | nd |
| atgpat1-1 | 3.05 ± 0.20 | nd | 21.97 | nd | 0.51 | 2.27 | 5.02 | 27.27 | 42.27 | nd |
nd, not detectable.
The minuscule amount of pollen grains made it impractical to quantitatively determine lipid content, so the total fatty acid composition, which is derived from membrane and storage lipids, as well as from the tryphine layer on the surface of the pollen grain, which consists mainly of neutral esters (Piffanelli et al., 1997; Hernandez-Pinzon et al., 1999), was examined. A dramatic increase in stearic acid (18:0) and a decrease in linolenic acid (18:3) in the mutant pollen grains were observed (Table 2). Previously, it was demonstrated that the majority of the stearate (18:0) of pollen grains is distributed in lipids of the pollen coat, whereas coatless pollen contains glycerolipids enriched in linolenic acid (18:3) (Hernandez-Pinzon et al., 1999). Thus, our results suggest that the surface structure on which the majority of neutral esters are distributed was established and maintained even though most of the pollen grains later collapsed in the mutant. This finding is in accordance with previous observations that the tapetum-derived exine precursors can traverse the tapetal cell wall before tapetal degeneration (Piffanelli et al., 1997). It also is consistent with the microscopic results, which indicated that early exine formation was not affected in the mutant.
Table 2.
Fatty Acid Composition of the Lipids from Pollen Grains of Arabidopsis atgpat1-1 Mutant and Wild-Type Plants
| Plant Type | Proportion of Fatty Acid (mol %)
|
|||||
|---|---|---|---|---|---|---|
| 14:0 | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | |
| Wild type | 4.75 | 27.21 | 9.64 | 7.75 | 13.69 | 36.96 |
| atgpat1-1 | 5.51 | 30.81 | 16.87 | 8.14 | 9.95 | 28.71 |
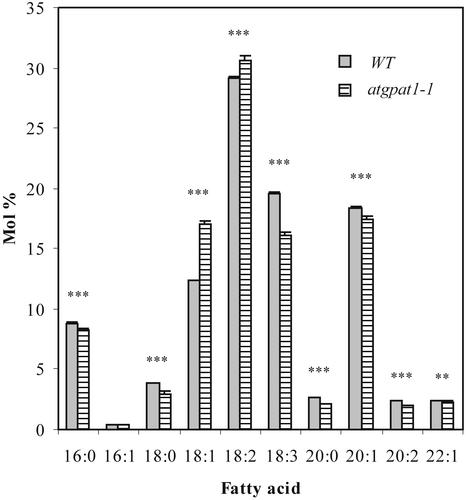
Altered fatty acid composition of seed storage lipid occurred in the mutants. The molar proportions of palmitic acid (16:0), stearic acid (18:0), linolenic acid (18:3), and the very-long-chain eicosenoic acid (20:1) decreased to varying degrees, whereas the proportions of oleic acid (18:1) and linoleic acid (18:2) increased (Figure 11). However, a <5% reduction in oil content was detected in the mutants. By contrast, repeated seed weight measurements revealed that the mutant seeds (20.55 μg/seed) were 8.7% larger than wild-type seeds (18.90 μg/seed) (t test, P < 0.01).
Figure 11.
Seed Fatty Acid Compositions of Wild-Type and Mutant Plants Grown under Identical Conditions.
Only the fatty acid composition of atgpat1-1 is shown because the two mutant lines have similar fatty acid profiles. Results shown are means ± se from five independent assays. Data were analyzed with paired t tests for individual fatty acids. Double asterisks indicate P < 0.01; triple asterisks indicate P < 0.001. WT, wild type.
DISCUSSION
Here, we report the existence of an Arabidopsis GPAT gene family that encodes membrane-bound GPATs. The AtGPAT family consists of seven members that share high sequence identity with one another, suggesting a common evolutionary origin. Distinct membrane compartments as well as the dynamics of differentiation within the same cell demand coordinated but multifaceted regulation of glycerolipid biosynthesis. Identification of the AtGPAT gene family provides a composite molecular map for future efforts to elucidate the functional significance of membrane-bound GPATs in the regulation of glycerolipid biosynthesis in plants. As illustrated by the differential expression patterns of the various isoforms, functional specialization of isoforms may exist. Discerning the functions of individual isoforms is essential to understanding the precise and coordinated developmental regulation that contributes to the diversity of glycerolipid biosynthesis in plants. Here, we focused on the study of AtGPAT1 using a reverse-genetics approach in combination with biochemical analyses.
Membrane-Targeting Properties of AtGPAT1
Membrane topology prediction programs invariably predict that members of the AtGPAT family possess two transmembrane domains, thereby distinguishing them from the soluble plastidial GPAT. Additional predictions with several protein-sorting programs (available at http://aramemnon.botanik.uni-koeln.de) suggest that AtGPAT1 may be targeted to the ER and mitochondria. To investigate the targeting properties of AtGPAT1, both in vitro microsomal and mitochondrial targeting assays were conducted. The ER membrane-targeting assay was performed according to the procedure used for the microsomal form of the 3-hydroxy-3-methylglutaryl–CoA reductase (Enjuto et al., 1994), whose membrane integration was verified through a high-salt wash plus a sodium carbonate treatment (Fujiki et al., 1982). The widely used alkaline extraction with sodium carbonate has proven effective in releasing the luminal content and peripheral membrane proteins from microsomes (Fujiki et al., 1982). This treatment did not strip the majority of AtGPAT1 off the microsomes (Figure 4A), suggesting that AtGPAT1 may be integrated into the microsomal membranes. On the other hand, the cell-free assay with purified pea mitochondria clearly demonstrated that AtGPAT1 could be imported into mitochondria. Distinguished from the microsomal targeting of AtGPAT1, the mitochondrial targeting of AtGPAT1 resulted in a size reduction of the protein (Figure 4B). Therefore, the results of the microsomal and mitochondrial targeting assays were not influenced by potential contamination of the microsomal and mitochondrial preparations with mitochondria and microsomes, respectively. It is worth noting that in a previous study, the mammalian mitochondrial GPAT also was detected immunologically in the microsomal membrane fraction, although such a result was interpreted as contamination of the ER membranes with mitochondria (Yet et al., 1993).
At present, however, the membrane-targeting properties of AtGPAT1 need to be tested vigorously, because plant mitochondria import some proteins in a promiscuous manner in vitro (Cleary et al., 2002). Thus, the precise subcellular localization of AtGPAT1 will have to be addressed in future studies using other techniques such as green fluorescent protein labeling and/or immunolocalization.
AtGPAT1 Participates in the Eukaryotic Glycerolipid Pathway
There are several lines of corroborating evidence for a role of AtGPAT1 in the eukaryotic glycerolipid pathway. Cytoplasmic membrane profile changes in the tapetum as well as in mature pollen grains harboring a defective AtGPAT1 strongly suggest that this isoform is involved in membrane lipid biosynthesis in these specialized cells. Reduced contents of phospholipid and TAG in flower tissues, both of which presumably are derived from the eukaryotic pathway, also were observed. In addition, analysis of storage lipids revealed consistent fatty acid composition changes in the mutant seeds. Within the current understanding of seed lipid biosynthesis, and as exemplified by the wrinkled seed mutant (Focks and Benning, 1998), a higher order regulatory defect would be extremely unlikely to cause fatty acid profile changes. Therefore, the significant alteration of fatty acid profiles in the mutant seeds can be attributed to the loss of function of AtGPAT1 in the synthesis of TAGs, which are generated exclusively through the eukaryotic pathway in the ER.
Although the fatty acid changes detected in the mutant appear to be inconsistent with the substrate specificity of AtGPAT1 according to in vitro assays, there are several potential explanations. Assay conditions, such as the presence or absence of the chelating agent EDTA, affect the substrate specificity of AtGPAT1. It also has been reported recently that an acyl-CoA binding protein such as BSA can alter the fatty acyl preference of the mammalian mitochondrial GPAT (Kannan et al., 2003). Likewise, the size of the fatty acyl–CoA pool is a major determinant of the fatty acid composition in glycerolipid (Dörmann et al., 2000), and in vitro assay conditions may not accurately reflect the situation in vivo. Finally, the mutant phenotype may more accurately reflect the activity of the remaining isoforms than a loss of the defective isoform.
The loss of AtGPAT1 did not cause a significant change in oil content, despite the high expression of this gene in developing siliques and its obvious involvement in seed oil biosynthesis, as reflected by the seed fatty acid profiles of the mutants. The underlying mechanism governing this phenomenon is not clear at present. One possible explanation is that plants may use an epistatic mode of interaction between molecularly homologous genes to maintain the efficient use of metabolic pathways (Chourey and Taliercio, 1994). The absence of AtGPAT1 may enhance or activate the expression of other GPAT isoforms to compensate for the loss of function normally exerted by AtGPAT1 during oil synthesis. Oil biosynthesis in the seeds of a mutant lacking AtGPAT1 also may be regulated by other hierarchical mechanisms in metabolism. As a result of the reduced seed set in the mutants, there appears to be an increased source supply being channeled to individual developing seeds. The finding that mutant seeds have a significantly greater average mass than wild-type seeds strongly supports this view. Thus, it is possible that the increased source-to-sink ratio in the mutant may result in an increased flux of fatty acids into the mutant seeds, thereby maintaining a seemingly normal rate of oil synthesis mediated by other AtGPATs.
AtGPAT1 Exerts Sporophytic and Gametophytic Effects on Pollen Development
AtGPAT1 is crucial for male fertility, as the knockout mutants exhibit a pollen development defect that can be rescued by a wild-type copy of the AtGPAT1 gene. Pollen development requires a timely differentiation of the tapetum to provide nutrients. Substances produced by the secretory tapetum, such as that of Arabidopsis, reach the developing microspores via the fluid in the locule cavity, which is generated in part by the degenerating tapetum. In angiosperms, there also exists a second type of tapetum, known as an invasive tapetum, which increases in size dramatically to invade the anther locule and relies on the close adhesion between the tapetal cytoplasm and the microspores to transfer nutrients (Pacini et al., 1985). In preparation for establishing a direct contact between an invasive tapetum and developing microspores, the walls between the tapetal cells and between the tapetum and sporogenous tissues undergo nonpolarized, complete disintegration, forming a continuous periplasmodium. Although the atgpat1-1 tapetum resembled an invasive tapetum in that it continued to enlarge, a major cytological distinction is its extended persistence of cellular integrity, particularly with regard to the delayed degeneration of cell walls. This finding suggests that the atgpat1-1 tapetum does not serve as a functional invasive tapetum. Likewise, it was evident that some of the microspores showed clear signs of developmental arrest even though they had been in direct contact with the tapetum.
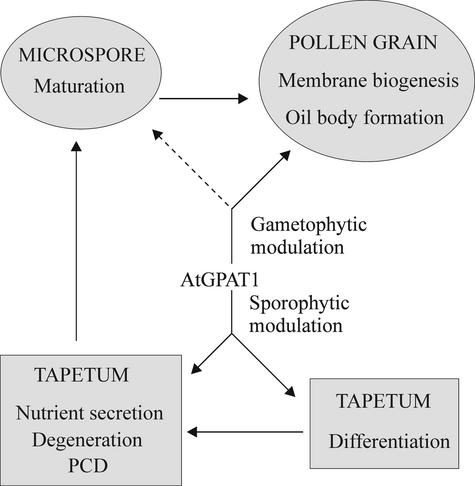
Based on the results obtained in this study, we propose a model, as depicted in Figure 12, in which AtGPAT1 plays both sporophytic and gametophytic roles in pollen development. The sporophytic effect is reflected by the involvement of AtGPAT1 in tapetal differentiation. Impairment in tapetal differentiation deprives the developing microspores of nutrients and leads to an arrest of pollen development. The gametophytic role, on the other hand, may not be essential for the microspores to reach maturity; nonetheless, it compromises the competitiveness of pollen grains to pollinate.
Figure 12.
Involvement of AtGPAT1 in Tapetal Differentiation and Pollen Development.
The placement of solid arrows is based on evidence presented in this work. The dashed arrow denotes a potential role for AtGPAT1. PCD, programmed cell death.
Here, a question arises regarding what mechanism governs the altered differentiation process of the tapetum in the atgpat1 mutants. In the atgpat1-1 tapetum, ER stacks and fusion of ER cisternae with the plasma membranes, both of which are associated with tapetal secretion, were reduced. Thus, one possibility is that a defect in AtGPAT1 reduces ER membrane biogenesis and impairs secretion, which in turn delays the onset of tapetal degeneration. Alternatively, the delayed tapetal degeneration may be caused by mitochondrial dysfunction resulting from the lack of AtGPAT1 for mitochondrial membrane biogenesis. As illustrated in a number of CMS lines, mitochondrial dysfunction has been associated with both premature and delayed degeneration of the tapetal layer (Horner, 1977; Balk and Leaver, 2001). It is worth noting that the atgpat1-1 mutant tapetum shares a remarkably similar cytological phenotype, with regard to tapetal persistence, with that of a Brassica napus CMS line (Grant et al., 1986). CMS-associated tapeta that are either normal in size and persistent or dramatically large also have been reported in carrot (Zenkteler, 1962), Cucumis, and sorghum (Overman and Warmke, 1972). In light of the general acceptance of tapetal degeneration as intrinsically governed by programmed cell death (Papini et al., 1999; Wu and Cheung, 2000), in which mitochondria play a key role, it is tempting to speculate that the role of AtGPAT1 in mitochondrial membrane lipid synthesis is important for the proper progression of tapetal differentiation. Impaired mitochondria may operate below the threshold required to instigate tapetal programmed cell death, similar to the situation in the gfa2 mutant, in which programmed cell death failed to occur in the synergid cell due to mitochondrial dysfunction (Christensen et al., 2002). As a result, the tapetum is doomed to die by necrosis (Jones, 2000), leading to a rather delayed collapse of the tapetum.
Deficiency in AtGPAT1 correlates with impaired ER membrane biogenesis and altered oil body size in the haploid pollen grains. Moreover, our results also show that pollen grains that harbor a mutation in AtGPAT1 from a heterozygous mutant plant in which a sporophytic effect may be excluded are impaired in pollination competitiveness. Although it is premature to speculate on the underlying mechanism, AtGPAT1 clearly exerts a gametophytic effect on male fertility.
METHODS
Yeast Strains and Culture Conditions
The haploid gene disruption strain gat1Δ (BY4742, Matα, his3C1, leu2C0, lys2C0, ura3C0, YKR067w::kanMX4) was purchased from Euroscarf (Frankfurt, Germany). Cells were cultured at 30°C in YPD medium containing 1% Bacto-yeast extract, 2% Bacto-peptone, and 2% glucose (Sigma).
Identification of AtGPAT Genes from Arabidopsis
Database searches using the Basic Local Alignment Search Tool (BLAST) program (Altschul et al., 1990) with sequences derived from conserved regions of the yeast glycerol-3-phosphate acyltransferases (GPATs) and other membrane-bound acyltransferases identified seven hypothetical Arabidopsis thaliana genes that encode polypeptides composed of 585, 530, 520, 503, 502, 501, and 500 amino acids. They contained segments similar to the previously recognized acyltransferase motifs. These sequences, designated AtGPAT1 to AtGPAT7, were amplified using single-stranded cDNAs transcribed from total RNA of Arabidopsis siliques and cell suspensions. The primers used for PCR were derived from Arabidopsis genomic sequences. The generated double-stranded cDNAs were cloned directly into vector pYES2.1/V5-His-TOPO (Invitrogen, Carlsbad, CA), and the resulting plasmids, AtGPAT(1-7)–pYES2.1/V5-His-TOPO, were transformed into TOP-10 cells (Invitrogen). The cDNAs were sequenced using an automated DNA sequencer (Applied Biosystems 373) and compared with the corresponding genomic sequences.
Computer Analysis of the AtGPAT Sequences
The amino acid sequences deduced from the AtGPAT cDNAs were analyzed for hydropathy profiles using the Kyte-Doolittle algorithm (DNAstar, Madison, WI). The transmembrane domains of the sequences were predicted by the TopPred2 program (von Heijne, 1992) and the TMpred program (Hofmann and Stoffel, 1993).
Heterologous Expression of the AtGPAT Genes in Yeast
The plasmids AtGPAT(1-7)–pYES2.1/V5-His-TOPO and the control vector were introduced into the yeast gat1Δ strain (Zheng and Zou, 2001). Overexpression of the AtGPAT genes in yeast was performed according to Zheng and Zou (2001) except that the induction was performed in medium containing 1% raffinose, 2% galactose, and 0.25% glycerol at 16°C for 40 h. Yeast homogenates were prepared as described previously (Zheng and Zou, 2001) except that the extraction buffer contained 50 mM Hepes, pH 7.0, 2 mM EDTA, 1 mM DTT, and 10% glycerol.
Enzyme Assays
GPAT activity was assayed at 30°C for 10 min in a 200-μL reaction mixture containing 40 mM Hepes, pH 7.0, 400 μM 14C–glycerol-3-phosphate (2.5 nCi/nmol), 67.5 μM palmitoyl-CoA and/or stearoyl-CoA or other fatty acyl donors, 1 mM DTT, 5 mM EDTA, and 2.5 mg/mL BSA unless stated otherwise. The reaction was stopped, and products were extracted as described previously (Zheng and Zou, 2001). The formed products were subjected to scintillation counting for radioactivity and thin layer chromatography analysis as described (Zheng and Zou, 2001).
Lipid Analysis
Total lipids from the tissues of wild-type and mutant plants were extracted using the procedure of Bligh and Dyer (1959). Separation of neutral lipids and polar lipids was performed using a Sep-Pak silica column (Waters, Milford, MA) according to the procedure of Uemura et al. (1995). Neutral lipids, including triacylglycerol, were resolved on thin layer chromatography plates (Si 250-PA, Baker, Phillipsburg, NJ) in a developing solvent of hexane:diethyl ether:acetic acid (80:20:2, v/v) and identified by comigration with known standards. Seed oil was isolated as described (Zou et al., 1999). The isolated lipids were transmethylated with methanolic HCl and quantified by gas chromatography as described previously (Zheng and Zou, 2001).
RNA Analysis
RNA extractions and RNA gel blot analysis were performed as described (Zheng et al., 2001). The cDNA fragments of the AtGPAT genes were used as the probe templates for hybridization.
Microsomal Targeting of in Vitro–Synthesized Polypeptide
AtGPAT1 cDNA was cloned into the pYES2.1/V5-His-TOPO vector bearing the T7 promoter. The in vitro synthesis of the polypeptide was conducted using TNT Quick Coupled Transcription/Translation systems (Promega) in the presence of canine pancreatic microsomes (Promega) labeled with 35S-Met. To test for the insertion of the polypeptide into microsomal membranes, the procedure followed was that described by Enjuto et al. (1994) for microsomal targeting of 3-hydroxy-3-methylglutaryl–CoA reductase. The 35S-labeled translation products were diluted to yield a solution with a final concentration of 50 mM Hepes-KOH, pH 7.6, 120 mM sucrose, 500 mM potassium acetate, 3 mM magnesium acetate, and 5 mM DTT (buffer 1) to eliminate nonspecific binding of the peptide onto the surface of the microsomal membranes. Samples treated with this high-salt solution were layered onto cushion 1 (0.5 M sucrose in buffer 1) and centrifuged at 100,000g for 30 min at 4°C. The microsomal pellets containing the cosedimented peptides were resuspended in 0.1 M Na2CO3, pH 11, vortexed briefly, and incubated on ice for 30 min (Fujiki et al., 1982). The resulting open membrane sheets were repelleted by centrifugation though cushion 2 (250 mM sucrose in 0.1 M Na2CO3, pH 11) at 120,000g for 1 h at 4°C. The supernatant was mixed with an equal volume of ice-cold 20% trichloroacetic acid and incubated at 4°C for 1 h, and the protein precipitate collected by centrifugation was washed twice with ice-cold acetone. Both the membrane pellet and the protein extract were dissolved in the electrophoresis buffer. Labeled proteins were analyzed using 10 to 20% SDS-polyacrylamide gels (Bio-Rad) and a BenchMark prestained protein ladder (GIBCO) as a molecular weight marker and visualized by autoradiography.
Isolation of Mitochondria and Import Studies
The in vitro synthesis of the polypeptide was performed in the absence of microsomes as described above. Isolation of pea mitochondria and import of the in vitro–synthesized polypeptide into the mitochondria were performed as described by Shen et al. (2003). The import reaction containing freshly prepared mitochondria, equivalent to 100 μg of protein, 20 mM Tes-NaOH, pH 7.2, 0.3 M mannitol, 80 mM KCl, 10 mM MgCl2, 2 mM MnCl2, 4 mM sodium phosphate, 2 mM DTT, 5 mM Met, 5 mM Cys, 10 mM malate, 5 mM ATP, 50 μM ADP, 4% (w/v) fatty acid–free BSA, and 15 μL of the translation product (without the microsomes) in a total volume of 200 μL, was incubated at 25°C for 30 min. After incubation, three of the reaction mixtures were mixed together and then divided into three equal portions. One portion was kept on ice without treatment; one portion was treated with 50 μg/mL proteinase K on ice for 40 min followed by termination with 3 mM phenylmethylsulfonyl fluoride; and one portion was digested with protease in the presence of 0.5% (v/v) Triton X-100. Intact mitochondria were isolated by centrifugation though a 20% sucrose cushion (20% [w/v] sucrose and 20 mM Tes-NaOH, pH 7.2) at 14,000g for 8 min at 4°C. Samples were analyzed by SDS-PAGE and visualized by autoradiography as described above.
Screening of T-DNA–Tagged Mutants
A total of 72,960 T-DNA–tagged, Basta-resistant Arabidopsis (ecotype Wassilewskija) plants generated at the Arabidopsis Knockout Facility of the University of Wisconsin were screened by PCR. The T-DNA left border primer (5′-CATTTTATAATAACGCTGCGGACATCTAC-3′) was selected for screening in combination with each of two primers (5′-CTTCTCTCTCTACGCCATAGCTATGGTTT-3′ and 5′-GCACTAAAGCGACAGGTTGAGATTATGGA-3′) designed according to sequences corresponding to the 5′ and 3′ ends of the AtGPAT1 gene. The generated PCR products were subjected to DNA gel blot analysis, and those that tested positive and were of the appropriate size were cloned into the pCR2.1TOPO vector (Invitrogen) and sequenced to confirm the insertion of T-DNA into the AtGPAT1 gene. To determine the T-DNA insertion number, DNA gel blot analysis was conducted using the BAR gene of the T-DNA cassette as a probe of the genomic DNA isolated from the T2 homozygotes.
Genetic Complementation
The AtGPAT1 genomic sequence comprising 2029 bp of promoter region and 2491 bp of coding region was inserted into the binary vector pRD400 carrying a kanamycin resistance marker. The self-pollinated progeny of plants homozygous for atgpat1-1 were used to transform the empty vector and the recombinant vector using standard Agrobacterium tumefaciens–mediated infiltration transformation. The transformants were selected with 50 μg/mL kanamycin, confirmed by PCR, and used to examine pollen and silique development.
Microscopy
To visualize the morphological differences between the mature pollen of wild-type and mutant plants, pollen grains were mounted on stubs and coated with gold particles from a sputter coater (S150B; Edwards, Crawley, UK). Specimens then were examined with a scanning electron microscope (SEM 505; Philips, Eindhoven, The Netherlands) at an accelerating voltage of 30 kV. To conduct sectioning experiments, Arabidopsis flower buds were fixed with 3% glutaraldehyde (Electron Microscopy Sciences, Fort Washington, PA) in 25 mM phosphate buffer, pH 6.8, overnight at 4°C and postfixed in 1% osmium tetroxide (Electron Microscopy Sciences) for 8 h on ice. The samples were dehydrated in a graded ethanol series and embedded in acrylic resin (London Resin Company, London, UK). Sections were made on a Reichert Jung Ultracut E microtome (Leica, Vienna, Austria). For light microscopy, semithick (1 to 2 μm) sections were stained with 1% toluidine blue O in 1% borax. For transmission electron microscopy, ultrathin sections (50 to 70 nm) were double stained with 2% (w/v) uranyl acetate and 2.6% (w/v) lead citrate aqueous solution. Observations and micrographs were made using a Philips CM-10 transmission electron microscope at an accelerating voltage of 60 kV, and the images were photographed on Kodak electron image film (Electron Microscopy Sciences).
Determination of the Competitiveness of Pollen Grains to Pollinate
Pollen competitiveness in pollination was assessed essentially as described by Mouline et al. (2002). The T2 seeds derived from T1 plants heterozygous for the atgpat1-1 mutation were subjected to segregation analysis based on L-phosphinothricin (L-PPT) resistance. The seeds were soaked in 14 mg/mL L-PPT solution at 4°C for 3 days and then grown at 23°C for 9 days under 24 h of light. Both L-PPT–sensitive and L-PPT–resistant plants were scored for their segregation ratios. This ratio depends on the probability of fertilization by the atgpat1-1 pollen and the wild-type pollen.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact J. Zou, jitao.zou@nrc-cnrc.gc.ca.
Accession Numbers
The accession numbers for AtGPAT1 to AtGPAT7 are At1g06520, At1g02390, At4g01950, At1g01610, At3g11430, At2g38110, and At5g06090, respectively.
Acknowledgments
We are grateful to R. Datla and V. Katavić for critical reading of the manuscript. We also acknowledge the Arabidopsis Knockout Facility at the University of Wisconsin for assisting us in atgpat1 mutant screening. This work was supported by the National Research Council–Plant Biotechnology Institute core program. This is National Research Council of Canada publication 45234.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.012427.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Athenstaedt, K., and Daum, G. (1999). Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266, 1–16. [DOI] [PubMed] [Google Scholar]
- Bafor, M., Stobart, A.K., and Stymne, S. (1990). Properties of the glycerol acylating enzyme in microsomal preparations from the developing seeds of safflower (Carthamus tinctorius) and turnip rape (Brassica campestris) and their ability to assemble cocoa-butter type fats. J. Am. Oil Chem. Soc. 67, 217–225. [Google Scholar]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh, E.G., and Dyer, W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Browse, J., Warwick, N., Somerville, C.R., and Slack, C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the 16:3 plant Arabidopsis thaliana. Biochem. J. 235, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourey, P.S., and Taliercio, E.W. (1994). Epistatic interaction and functional compensation between the two tissue- and cell-specific sucrose synthase genes in maize. Proc. Natl. Acad. Sci. USA 91, 7917–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, C.A., Gorsich, S.W., Brown, R.H., Jones, L.G., Brown, J., Shaw, J.M., and Drews, G.N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14, 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, S.P., Tan, F-C., Nakrieko, K-A., Thompson, S.J., Mullineaux, P.M., Creissen, G.P., von Stedingk, E., Glaser, E., Smith, A.G., and Robinson, C. (2002). Isolated plant mitochondria import chloroplast precursor proteins in vitro with the same efficiency as chloroplasts. J. Biol. Chem. 277, 5562–5569. [DOI] [PubMed] [Google Scholar]
- Datla, R.S., Hammerlindl, J.K., Panchuk, B., Pelcher, L.E., and Keller, W. (1992). Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene 122, 383.-384. [DOI] [PubMed] [Google Scholar]
- Dircks, L., and Sul, H.S. (1999). Acyltransferases of de novo glycerophospholipid biosynthesis. Prog. Lipid Res. 38, 461–479. [DOI] [PubMed] [Google Scholar]
- Dörmann, P., Voelker, T.A., and Ohlrogge, J.B. (2000). Accumulation of palmitate in Arabidopsis mediated by the acyl-acyl carrier protein thioesterase FATB1. Plant Physiol. 123, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston, V., and Harwood, J.L. (1992). Glycerol 3-phosphate acylation by microsomal fractions from avocado mesocarp. Biochem. Soc. Trans. 20, 169S. [DOI] [PubMed] [Google Scholar]
- Eccleston, V., and Harwood, J.L. (1995). Solubilization, partial purification and properties of acyl-CoA:glycerol-3-phosphate acyltransferase from avocado (Persea americana) fruit mesocarp. Biochim. Biophys. Acta 1257, 1–10. [DOI] [PubMed] [Google Scholar]
- Enjuto, M., Balcells, L., Campos, N., Caelles, C., Arro, M., and Boronat, A. (1994). Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc. Natl. Acad. Sci. USA 91, 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M.A., de Almeida Engler, J., Miguens, F.C., van Montagu, M., Engler, G., and de Oliveira, D.E. (1997). Oleosin gene expression in Arabidopsis thaliana tapetum coincides with accumulation of lipids in plastids and cytoplasmic bodies. Plant Physiol. Biochem. 35, 729–739. [Google Scholar]
- Focks, N., and Benning, C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen, M. (1990). Comparison of certain properties of membrane bound and solubilized acyltransferase activities of plant microsomes. Plant Sci. 69, 39–48. [Google Scholar]
- Frentzen, M. (1993). Acyltransferases and triacylglycerols. In Lipid Metabolism in Plants, T.S. Moore, Jr., ed (Boca Raton, FL: CRC Press), pp. 195–220.
- Fritz, P.J., Kauffman, J.M., Robertson, C.A., and Wilson, M.R. (1986). Cocoa butter biosynthesis, purification and characterization of a soluble sn-glycerol-3-phosphate acyltransferase from cocoa seeds. J. Biol. Chem. 261, 194–199. [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: Application to endoplasmic reticulum. J. Cell Biol. 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, I., Beversdorf, W.D., and Peterson, R.L. (1986). A comparative light and electron microscopic study of microspore and tapetal development in male fertile and cytoplasmic sterile oilseed rape (Brassica napus). Can. J. Bot. 64, 1055–1068. [Google Scholar]
- Griffiths, G., Stobart, A.K., and Stymne, S. (1985). The acylation of sn-glycerol-3-phosphate and the metabolism of phosphatidate in microsomal preparations from the developing cotyledons of safflower (Carthamus tinctorius L.) seed. Biochem. J. 230, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hares, W., and Frentzen, M. (1991). Substrate specificities of the membrane-bound and partially purified microsomal acyl-CoA:1-acylglycerol-3-phosphate acyltransferase from etiolated shoots of Pisum sativum (L.). Planta 185, 124–131. [DOI] [PubMed] [Google Scholar]
- Heinz, E., and Roughan, P.G. (1983). Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 72, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Pinzon, I., Ross, J.H.E., Barnes, K.A., Damant, A.P., and Murphy, D.J. (1999). Composition and role of tapetal lipid bodies in the biogenesis of the pollen coat of Brassica napus. Planta 208, 588–598. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). Tmbase: A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374, 166. [Google Scholar]
- Horner, H.T. (1977). A comparative light- and electron-microscopic study of microsporogenesis in male-fertile and cytoplasmic male-sterile sunflower (Helianthus annuus). Am. J. Bot. 64, 745–759. [Google Scholar]
- Ichihara, K. (1984). sn-Glycerol-3-phosphate acyltransferase in a particulate fraction from maturing safflower seeds. Arch. Biochem. Biophys. 232, 685–698. [DOI] [PubMed] [Google Scholar]
- Jones, A.M. (2000). Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci. 5, 225–230. [DOI] [PubMed] [Google Scholar]
- Kannan, L., Knudsen, J., and Jolly, C.A. (2003). Aging and acyl-CoA binding protein alter mitochondrial glycerol-3-phosphate acyltransferase activity. Biochim. Biophys. Acta 1631, 12–16. [DOI] [PubMed] [Google Scholar]
- Katavić, V., Reed, D.W., Taylor, D.C., Giblin, E.M., Barton, D.L., Zou, J., MacKenzie, S.L., Covello, P.S., and Kunst, L. (1995). Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon, D.S., Hayes, T.R., Wyrick, A., Xiong, H., Davis, H.M., and Voelker, T.A. (1999). Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiol. 120, 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon, D.S., Lardizabal, K.D., Nelsen, J.S., Bleibaum, J.L., Davis, H.M., and Metz, J.G. (1995). Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol. 109, 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst, L., Browse, J., and Somerville, C. (1988). Altered regulation of lipid biosynthesis in a mutant Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85, 4143–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner, M.W., Levering, C.K., Davids, H.M., and Knutzon, D.S. (1995). Lysophosphatidic acid acyltransferase from meadowform mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic rapeseed oil. Plant Physiol. 109, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin, T.M., Wang, P., and Coleman, R.A. (1999). Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38, 5764–5771. [DOI] [PubMed] [Google Scholar]
- Manaf, A.M., and Harwood, J.L. (2000). Purification and characterization of acyl-CoA:glycerol 3-phosphate acyltransferase from oil palm (Elaeis guineensis) tissues. Planta 210, 318–328. [DOI] [PubMed] [Google Scholar]
- Mouline, K., Very, A.A., Gaymard, F., Boucherez, J., Pilot, G., Devic, M., Bouchez, D., Thibaud, J.B., and Sentenac, H. (2002). Pollen tube development and competitive ability are impaired by disruption of a Shaker K(+) channel in Arabidopsis. Genes Dev. 16, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia, M., Charzynska, M., Rougier, M., and Cresti, M. (1991). Secretory tapetum of Brassica oleracea L.: Polarity and ultrastructural features. Sex. Plant Reprod. 4, 28–35. [Google Scholar]
- Murphy, D.J., and Vance, J. (1999). Mechanisms of lipid-body formation. Trends Biochem. Sci. 24, 109–115. [DOI] [PubMed] [Google Scholar]
- Overman, M.A., and Warmke, H.E. (1972). Cytoplasmic male sterility in sorghum. II. Tapetal behaviour in fertile and sterile anthers. J. Hered. 63, 227–234. [Google Scholar]
- Owen, H.A., and Makaroff, C.A. (1995). Ultrastructure of microsporogenesis and microgametogenesis in Arabidopsis thaliana (L.) Heynh. ecotype Wassilewskija (Brassicaceae). Protoplasma 185, 7–21. [Google Scholar]
- Pacini, E., Franchi, G., and Hesse, M. (1985). The tapetum: Its form, function, and possible phylogeny in Embryophyta. Plant Syst. Evol. 149, 155–185. [Google Scholar]
- Papini, A., Mosti, S., and Brighigna, L. (1999). Programmed-cell-death events during tapetum development of angiosperms. Protoplasma 207, 213–221. [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1997). Intra- and extracellular lipid composition and associated gene expression patterns during pollen development in Brassica napus. Plant J. 11, 549–562. [DOI] [PubMed] [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11, 65–80. [Google Scholar]
- Platt, K.A., Huang, A.H.C., and Thomson, W.W. (1998). Ultrastructural study of lipid accumulation in tapetal cells of Brassica napus L. cv. Westar during microsporogenesis. Int. J. Plant Sci. 159, 724–737. [Google Scholar]
- Roughan, P.G., and Slack, C.R. (1982). Cellular organization of glycerolipid metabolism. Annu. Rev. Plant Physiol. 33, 97–132. [Google Scholar]
- Routaboul, J.M., Benning, C., Bechtold, N., Caboche, M., and Lepiniec, L. (1999). The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol. Biochem. 37, 831–840. [DOI] [PubMed] [Google Scholar]
- Scheideler, M.A., and Bell, R.M. (1989). Phospholipid dependence of homogeneous, reconstituted sn-glycerol-3-phosphate acyltransferase of Escherichia coli. J. Biol. Chem. 264, 12455–12461. [PubMed] [Google Scholar]
- Schnable, P.S., and Wise, R.P. (1998). The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3, 175–210. [Google Scholar]
- Schrauwen, J.A.M., Mettenmeyer, T., Croes, A.F., and Wullems, G.J. (1996). Tapetum-specific genes: What role do they play in male gametophyte development? Acta Bot. Neerl. 45, 1–15. [Google Scholar]
- Shen, W., Wei, Y., Dauk, M., Zheng, Z., and Zou, J. (2003). Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: Evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Lett. 536, 92–96. [DOI] [PubMed] [Google Scholar]
- Sun, C., Cao, Y.Z., and Huang, A.H.C. (1988). Acyl coenzyme A preference of the glycerol phosphate pathway in the microsomes from the maturing seeds of palm, maize and rapeseed. Plant Physiol. 88, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis Knockout Facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, M., Joseph, R.A., and Steponkus, P.L. (1995). Cold acclimation of Arabidopsis thaliana: Effect on plasma membrane lipid composition and freeze-induced lesions. Plant Physiol. 109, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1992). Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494. [DOI] [PubMed] [Google Scholar]
- Wilkison, W.O., and Bell, R.M. (1997). sn-Glycerol-3-phosphate acyltransferase from Escherichia coli. Biochim. Biophys. Acta 1348, 3–9. [DOI] [PubMed] [Google Scholar]
- Wu, H.M., and Cheung, A.Y. (2000). Programmed cell death in plant reproduction. Plant Mol. Biol. 44, 267–281. [DOI] [PubMed] [Google Scholar]
- Yet, S.-H., Lee, S., Hahm, Y.T., and Sul, H.S. (1993). Expression and identification of p90 as the murine mitochondrial glycerol-3-phosphate acyltransferase. Biochemistry 32, 9486–9491. [DOI] [PubMed] [Google Scholar]
- Zenkteler, M. (1962). Microsporogenesis and tapetal development in normal and male sterile carrots (Daucus carota). Am. J. Bot. 49, 341–348. [Google Scholar]
- Zheng, Z., Uchacz, T.M., and Taylor, J.L. (2001). Isolation and characterization of novel defence-related genes induced by copper, salicylic acid, methyl jasmonate, abscisic acid and pathogen infection in Brassica carinata. Mol. Plant Pathol. 2, 159–169. [DOI] [PubMed] [Google Scholar]
- Zheng, Z., and Zou, J. (2001). The initial step of the glycerolipid pathway: Identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae. J. Biol. Chem. 276, 41710–41716. [DOI] [PubMed] [Google Scholar]
- Zou, J., Katavić, V., Giblin, E.M., Barton, D.L., Mackenzie, S.L., Keller, W.A., Hu, X., and Taylor, D.C. (1997). Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9, 909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J., Wei, Y., Jako, C., Kumar, A., Selvaraj, G., and Taylor, D.C. (1999). The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 19, 645–653. [DOI] [PubMed] [Google Scholar]