Abstract
A maize histone deacetylase gene was identified as a homolog of yeast Hda1. The predicted protein corresponds to a previously purified maize deacetylase that is active as a protein monomer with a molecular weight of 48,000 and is expressed in all tissues of germinating embryos. Hda1 is synthesized as an enzymatically inactive protein with an apparent molecular weight of 84,000 that is processed to the active 48-kD form by proteolytic removal of the C-terminal part, presumably via a 65-kD intermediate. The enzymatically inactive 84-kD protein also is part of a 300-kD protein complex of unknown function. The proteolytic cleavage of ZmHda1 is regulated during maize embryo germination in vivo. Expression of the recombinant full-length protein and the 48-kD form confirmed that only the smaller enzyme form is active as a histone deacetylase. In line with this finding, we show that the 48-kD protein is able to repress transcription efficiently in a reporter gene assay, whereas the full-length protein, including the C-terminal part, lacks full repression activity. This report on the processing of Hda1-p84 to enzymatically active Hda1-p48 demonstrates that proteolytic cleavage is a mechanism to regulate the function of Rpd3/Hda1-type histone deacetylases.
INTRODUCTION
Post-translational modifications of core histones, including acetylation, methylation, phosphorylation, ubiquitination, and possibly others, represent essential elements of the epigenetic histone code (Loidl, 1988; Turner, 1993; Strahl and Allis, 2000). To decipher this code, which is recognized and interpreted by transcriptional regulators and chromatin-remodeling machines, is one of the central challenges of current chromatin research. In addition, there is a complex interrelation between DNA methylation and histone modifications (Dobosy and Selker, 2001).
Among the different types of histone modifications, acetylation is the most extensively characterized. Highly conserved Lys residues in the N-terminal extensions of core histones can be acetylated post-translationally by histone acetyltransferases; the modification is not stable and can be reversed by histone deacetylases. Both enzymes exist as multiple proteins that belong to distinct protein families.
Three main families of histone deacetylases have been identified in plants (Graessle et al., 2001; Pandey et al., 2002). Members of the HDA gene family are related to yeast Rpd3 and Hda1 (Rundlett et al., 1996; Taunton et al., 1996; Rossi et al., 1998; Lechner et al., 2000). Members of the SRT gene family are related to yeast SIR2 (Imai et al., 2000; Landry et al., 2000). In contrast to other eukaryotes, plants contain a third family of enzymes that is unrelated to the other classes, the HD2-related deacetylases, which were identified initially in maize (Lusser et al., 1997; Dangl et al., 2001).
Although many research reports have been published in recent years on the impact of histone deacetylases for gene regulation in yeast and vertebrate systems, little is known about the specific functions of these enzymes in plants. It has been shown that histone deacetylation cooperates with DNA methylation in the silencing of rRNA genes in allotetraploid Brassica (Pikaard, 1999). Both the knockdown of AtHD1 and AtHD2A expression by antisense RNA and the overexpression of Rpd3-like genes caused a variety of developmental abnormalities, suggesting a global role for these histone deacetylases in gene regulation in plants (Wu et al., 2000a, 2000b; Tian and Chen, 2001; Jang et al., 2003). There is limited knowledge of the regulation of specific genes by chromatin-dependent mechanisms (for recent reviews, see Lusser, 2002; Reyes et al., 2002). For instance, the Arabidopsis PICKLE (PKL) gene is proposed to be a chromatin-remodeling enzyme of the CHD class, based on the presence of an Snf2-related ATPase domain, a chromodomain, and PHD fingers. In pkl mutants, the expression of embryo-specific genes is disturbed, suggesting that Pkl might be a transcriptional repressor and, in analogy to the related vertebrate Mi-2 protein, function by cooperating with histone deacetylases (Ogas et al., 1999).
Maize embryos represent one of the best-characterized systems with respect to histone deacetylases in biochemical terms (Lusser et al., 2001). Four histone deacetylase activities have been characterized and purified chromatographically. These activities correspond to (1) the closely related proteins ZmRPD3I and ZmRPD3II (HD1B forms; Lechner et al., 2000); (2) HD2, as an acidic, nucleolar phosphoprotein (Lusser et al., 1997); (3) a SIR2-related enzyme (P. Loidl, unpublished data); and (4) HD1A, now designated ZmHda1. HD1A is a well-characterized enzyme. It can be purified by chromatography from maize extracts as a 48-kD protein that is subject to phosphorylation (Brosch et al., 1992, 1996). In vitro dephosphorylation of the purified enzyme is accompanied by a significant increase of histone deacetylase activity and altered substrate specificity with respect to differentially acetylated isoforms of histone H4 (Kölle et al., 1999).
Here, we identify the former HD1A as an Hda1-related enzyme. ZmHda1 encodes a 77-kD protein that is post-translationally processed by proteolysis to a catalytically active 48-kD enzyme. Protein microsequencing, mass spectrometry, in vitro protease digestion, immunological analysis, and the expression of truncated versions of ZmHda1 clearly show that the C-terminal portion of the protein has to be removed for the enzyme to gain its enzymatic activity. This is true as well for the ability of ZmHda1 to repress gene transcription, because the C-terminal part of the protein inhibits the transcription repression activity.
RESULTS
Cloning of ZmHda1
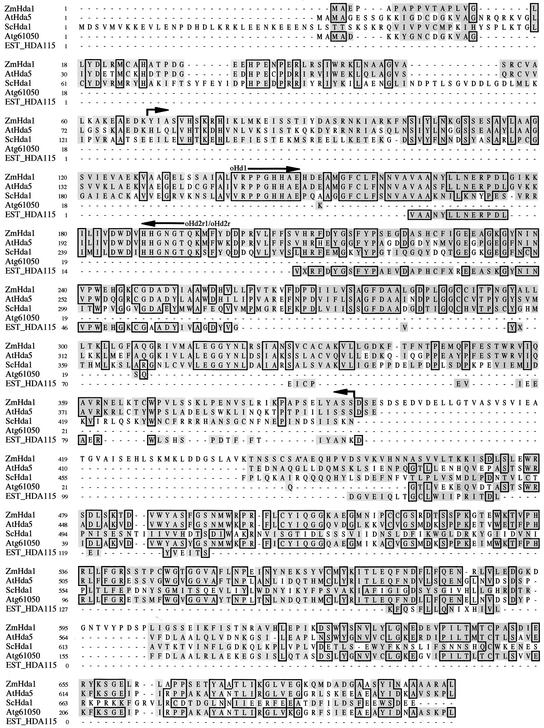
A region of the yeast Hda1 protein highly conserved among various organisms was used to design appropriate oligonucleotide primers for reverse transcriptase–mediated PCR amplification of a putative maize homolog, starting from mRNA of germinating embryos at 72 h after seed imbibition. Screening of the maize genome database (www.zmdb.iastate.edu) with the resulting product identified an EST clone of 661 bp that was used for the amplification of the 3′ and 5′ ends using the rapid amplification of cDNA ends protocol (Frohman et al., 1988). Sequencing of the final product revealed a 2103-bp open reading frame that encodes a protein of 701 amino acids with a calculated molecular weight of 76,500 (Figure 1). The deduced amino acid sequence displayed 54% sequence identity to Hda1 of Saccharomyces cerevisiae and 52% identity to Hda1 of Schizosaccharomyces pombe. The closest mammalian counterparts were the human hHDAC4 and hHDAC5, with 10.9 and 13.1% overall sequence identity and 37.4 and 36.3% sequence similarity, respectively.
Figure 1.
Comparison of the Amino Acid Sequences of ZmHda1 and Related Proteins.
ZmHda1 was aligned with the corresponding amino acid sequences of S. cerevisiae (ScHda1), two proteins of Arabidopsis (AtHda5 and Atg61050), and a maize protein (EST HDA115). Identical amino acids are boxed, and homologous amino acids are shaded. The positions of the oligonucleotide primers for the initial amplification of a ZmHda1 fragment are indicated. The area of peptides derived from protein microsequencing of ZmHda1-p48 (the former HD1A) is marked with arrows.
Arabidopsis contains a homologous gene (AtHda5) that encodes an open reading frame of 660 amino acids with 57% sequence identity and 86% sequence similarity. Another related Arabidopsis gene, which encodes a small protein of only 252 amino acids (Atg61050), contains a short amino acid stretch (17 amino acids) that is homologous with the N terminus of ZmHda1 and AtHda5; the major portion of this protein is homologous with the C-terminal part of AtHda5 (57.5% sequence identity) and the C-terminal part of ZmHda1 (47.6% sequence identity). In maize, an EST clone for a short polypeptide with 141 amino acids is available (EST HDA115) that is homologous with ZmHda1 (56% sequence identity). Neither short sequence (Atg61050 or EST HDA115) contains the sequence of the catalytic histone deacetylase domain (Figure 1).
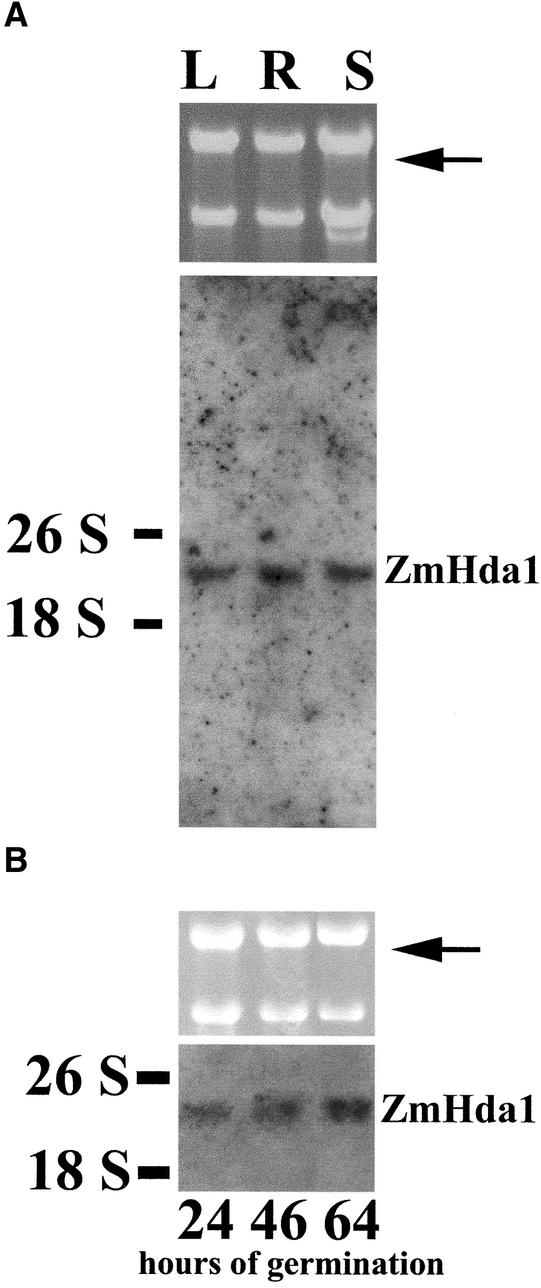
ZmHda1 mRNA is expressed in root, leaf, and shoot tissues, as demonstrated by RNA gel blot analysis (Figure 2A). The hybridization signal (at ∼4000 bp) was equally present in the maize tissues analyzed. The same uniform hybridization signal was observed at selected times during embryo germination (Figure 2B).
Figure 2.
Expression of ZmHda1 mRNA in Different Maize Organs and at Various Germination Stages of Embryos.
Total RNA was isolated from leaf (L), root (R), and shoot (S) of plant seedlings at 72 h after the start of embryo germination (A) or at 24, 46, and 64 h after the start of embryo germination (B). RNA was subjected to agarose gel electrophoresis with subsequent RNA gel blot analysis. The blot was hybridized with a ZmHda1-specific, digoxigenin-labeled, 513-bp DNA fragment. An ethidium bromide–stained gel is shown as an input control (top gels; 18 and 26 S RNA). The arrows indicate the positions of ZmHda1 mRNA.
In a parallel approach, highly purified yet unidentified maize histone deacetylase p48 (formerly designated HD1A; Brosch et al., 1996) was subjected to chymotrypsin or Lys-C endoproteinase digestion, and the resulting peptides were separated by reverse-phase HPLC. By protein microsequencing, eight peptides were identified (Table 1) with a total of 75 amino acids that matched perfectly in our ZmHda1 sequence from amino acids 68 to 391, indicating that ZmHda1 and ZmHda1-p48 (the former HD1A) originate from the same protein. Maize histone deacetylase HD1A (ZmHda1-p48) was purified originally to homogeneity as a 48-kD protein that was enzymatically active as a protein monomer and apparently not organized in a multiprotein complex (Brosch et al., 1992, 1996) (Figure 3A).
Table 1.
Amino Acid Sequences Determined by Protein Microsequencing of Peptides from Highly Purified ZmHda1-p48 (the Former HD1A)
| Peptide | Amino Acid Sequence | Sequence Position (Amino Acids; see Figure 1) |
|---|---|---|
| 1 | KYIASVHSK | 68 to 76 |
| 2 | KEISSTIYDASRNK | 83 to 96 |
| 3 | TIYDASRNKI | 88 to 97 |
| 4 | KGSSESAVL | 108 to 116 |
| 5 | AVLAAGSVI | 114 to 122 |
| 6 | GELSSAIALVRPPG | 131 to 144 |
| 7 | GEEAGKGYN | 229 to 237 |
| 8 | IKPAPSELY | 383 to 391 |
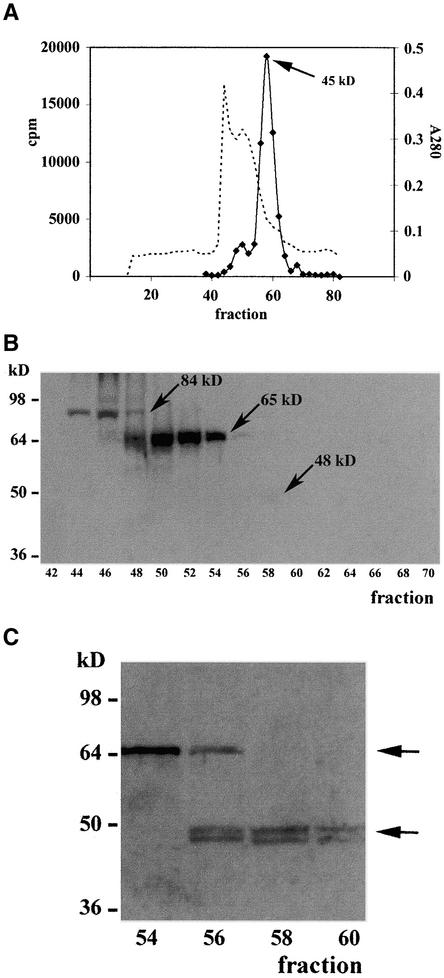
Figure 3.
Immunological Analysis of Partially Purified ZmHda1.
(A) Proteins of the soluble supernatant from chromatin preparations of maize embryos at 72 h after the start of germination were separated by Q-Sepharose Big Beads chromatography. Fractions containing histone deacetylase activity were pooled and subjected to S75 gel filtration chromatography. The bulk of enzyme activity eluted in a peak at an apparent molecular weight of ∼45,000. The dashed line depicts the A280 as a rough estimate of the protein concentration.
(B) Fractions from the S75 gel filtration chromatography were blotted onto a nitrocellulose membrane, and ZmHda1 bands were detected with anti-ZmHda1 antibodies (peptide antibodies). Prominent bands appeared at molecular weights of 84,000 and 65,000 (p84 and p65; arrows). In fraction 58, a very faint band was visible in the original wet blot at a molecular weight of ∼48,000 (p48; arrow).
(C) Chromatographic fractions (54 to 60) from S75 gel filtration chromatography were concentrated by trichloroacetic acid precipitation, subjected to SDS-PAGE with subsequent blotting onto a nitrocellulose membrane, and immunodetected with anti-ZmHda1 peptide antibodies. ZmHda1-p48 appears exactly in the histone deacetylase activity peak (see [A]). Arrows mark the positions of ZmHda1-p65 and ZmHda1-p48.
At that point, the discrepancy between the molecular weights of the putative maize Hda1 homolog and the purified ZmHda1-p48 remained unsolved. Therefore, polyclonal antibodies were raised against a synthetic peptide corresponding to ZmHda1-p48 amino acids 83 to 96 (Table 1). These antibodies were used for further studies of partially purified ZmHda1-p48. It should be noted that ZmHda1-p48 is present almost exclusively in a soluble fraction during chromatin preparation, together with cytoplasmic proteins (Brosch et al., 1996; Lusser et al., 2001). Only trace amounts of ZmRpd3 are present in this fraction; HD2 and the bulk of ZmRpd3 are tightly chromatin bound and therefore remain in the chromatin fraction (Rossi et al., 1998; Lechner et al., 2000). For this study, we used only the soluble fraction.
Biochemical and Immunological Characterization of ZmHda1
When partially purified ZmHda1-p48 (fractions after S75 gel filtration chromatography) was immunoblotted using polyclonal anti-ZmHda1 antibodies, the antibodies recognized two polypeptides of 84 and 65 kD in fractions 44 to 54. These fractions lacked significant histone deacetylase activity (Figures 3A and 3B); the very low residual activity in fractions 46 to 52 is the result of the presence of small amounts of ZmRpd3I and could be depleted efficiently by immunoprecipitation with anti-ZmRpd3I antibodies (data not shown). In the fractions containing the maximum histone deacetylase activity (56 to 62), the antibodies yielded no significant immunosignals (Figure 3B). When chromatographic fractions were concentrated >10-fold, two proteins (45 and 48 kD) became detectable that coeluted exactly with the enzyme activity (Figure 3C), suggesting that the 84- and 65-kD proteins may represent inactive pre-forms of ZmHda1-p48. The 45- and 48-kD protein bands comigrated with the phosphorylated and unphosphorylated forms of purified ZmHda1-p48 (Brosch et al., 1996; Kölle et al., 1999; G. Brosch and A. Loidl, unpublished results).
To determine whether the observed protein sizes could be the result of differentially spliced mRNAs or alternative transcription start sites, RNA gel blot analysis was performed with various RNA preparations from different tissues and various germination stages of embryos using DNA probes corresponding to the 5′ end, an internal region, and the 3′ end of the ZmHda1 cDNA. Regardless of which probe was used, hybridization under low-stringency conditions always revealed a single transcript at ∼4000 bp (data not shown), suggesting a post-translational processing of the ZmHda1 protein.
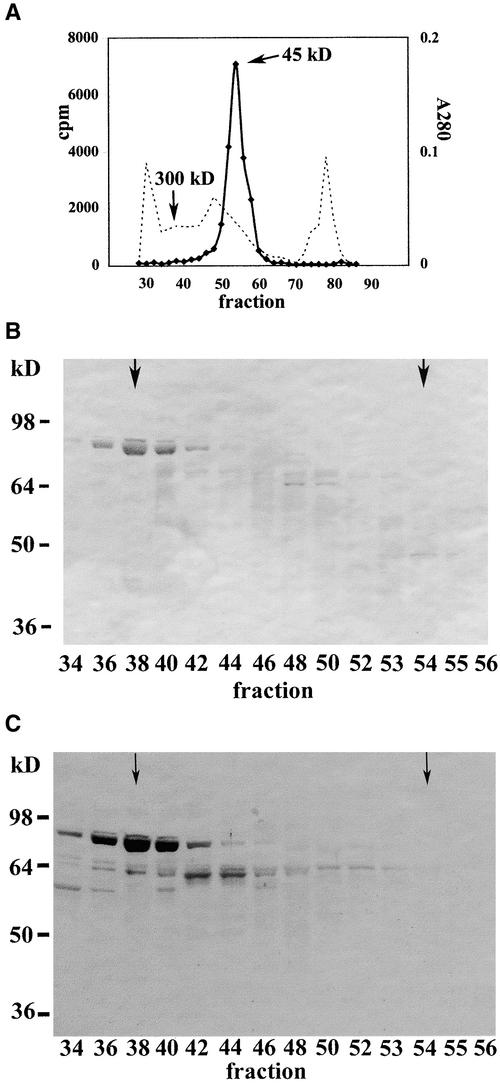
To determine whether enzymatically inactive ZmHda1-p84 is present in a higher molecular weight protein complex, we subjected the soluble fraction of 500 g of germinating maize embryos (72 h after the start of germination) to Q-Sepharose Big Beads chromatography, pooled those fractions containing ZmHda1 enzyme activity (but not ZmRpd3), and applied the concentrated pool to S200 gel filtration chromatography. Chromatographic elution revealed a unique histone deacetylase activity peak at 45 kD; no extra enzyme activity peak was detected (Figure 4A). When chromatographic fractions were immunoblotted with anti-ZmHda1 antibodies (peptide antibody), a strong immunosignal of ZmHda1-p84 was observed at a molecular weight of ∼300,000. No histone deacetylase activity was detected in the corresponding fraction (Figure 4A), indicating that ZmHda1-p84 is part of a high molecular weight protein complex that lacks enzymatic activity. Only a weak signal was observed in the peak fraction of enzyme activity (fraction 54) at 45 kD (Figure 4B). Consistent results were obtained when the immunoblot was analyzed with antibodies against the C-terminal part of ZmHda1 (amino acids 437 to 701; C-terminal antibody). The most intense immunosignal was obtained in the fractions corresponding to ∼300 kD, whereas no immunosignal was obtained in the fractions containing enzymatically active ZmHda1-p48 (fraction 54; Figure 4C).
Figure 4.
Immunological Analysis of Partially Purified ZmHda1 after S200 Gel Filtration Chromatography.
(A) The soluble supernatant from a chromatin preparation of 500 g of maize embryos at 72 h after the start of germination was subjected to Q-Sepharose Big Beads chromatography. Those fractions containing ZmHda1 activity without contamination by ZmRpd3 were pooled and subjected to S200 gel filtration chromatography. ZmHda1 enzyme activity eluted in a peak at an apparent molecular weight of ∼45,000. The dashed line depicts the A280 as a rough estimate of the protein concentration.
(B) Fractions from the S200 gel filtration chromatography were blotted onto a nitrocellulose membrane, and proteins were immunodetected with anti-ZmHda1 antibodies (peptide antibodies). Arrows in fractions 38 and 54 mark the molecular weights of 300,000 (ZmHda1-p84) and 45,000 (ZmHda1-p48), respectively.
(C) The same blot shown in (B) was immunoblotted with antibodies against the C-terminal part of ZmHda1. Arrows in fractions 38 and 54 mark the molecular weights of 300,000 (ZmHda1-p84) and 45,000 (ZmHda1-p48), respectively.
Fraction 59 from the S75 gel filtration chromatography (peak of enzyme activity; Figure 3A) was concentrated by ultrafiltration and used for immunoprecipitation with anti-ZmHda1 peptide antibodies coupled to protein A–Sepharose. The major portion of ZmHda1-p48 was precipitated into the pellet (data not shown). This result was substantiated by enzymatic activity determinations, which showed that histone deacetylase activity was precipitated by the anti-ZmHda1 antibodies, whereas a control precipitation (protein A–Sepharose alone) did not result in precipitated enzyme activity.
When crude protein extracts, which contain very low amounts of ZmHda1-p48 but large quantities of ZmHda1-p84 and ZmHda1-p65, were used for immunoprecipitation, >50% of the enzymatically inactive, high molecular weight forms of ZmHda1 (p65 and p84) were precipitated specifically into the pellet; however, no histone deacetylase enzymatic activity could be precipitated, confirming that the high molecular weight ZmHda1 forms are enzymatically inactive. The same result was obtained when those fractions from S75 gel filtration chromatography that contained only ZmHda1-p84 and ZmHda1-p65 were used for immunoprecipitation (data not shown).
Expression of Recombinant ZmHda1 in the Baculovirus Expression System
The position and alignment of the ZmHda1-p48 peptides (Table 1) derived from protein microsequencing within the ZmHda1 amino acid sequence (Figure 1) suggested that ZmHda1-p48 may represent a truncated version of ZmHda1-p84 lacking the C-terminal part of the protein. Mass spectrometric analysis of native ZmHda1-p48 confirmed this hypothesis; tandem mass spectrometry results were searched against the Hd1a sequence using SEQUEST (LCQ BioWorks; ThermoFinnigan). This analysis revealed no C-terminal peptides, indicating that the C-terminal portion (approximately amino acids 430 to 701) is missing in ZmHda1-p48. This finding was consistent with the microsequencing results of native ZmHda1-p48.
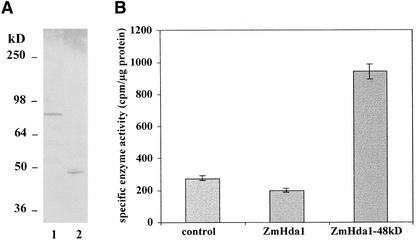
Therefore, we expressed a full-length ZmHda1 (amino acids 4 to 701) as well as a truncated version (amino acids 23 to 429), each fused to a 6xHis tag, using the baculovirus expression system (Figure 5A). When recombinant proteins were recovered on Talon affinity resin, it became evident that the full-length version of ZmHda1 was enzymatically inactive, whereas the truncated version (ZmHd1a-p48) exhibited catalytic activity (Figure 5B).
Figure 5.
Expression of ZmHda1-p84 and ZmHda1-p48 in Insect Cells Using the Baculovirus Expression System.
(A) SDS-PAGE of recombinant, His-tagged, full-length ZmHda1-p84 (amino acids 4 to 701; lane 1) and ZmHda1-p48 lacking the C-terminal 272 amino acids (amino acids 23 to 429; lane 2). Insect cell extracts were subjected to SDS-PAGE and immunoblotted using anti-ZmHda1 peptide antibodies.
(B) Histone deacetylase activity of recombinant ZmHda1-p84 and ZmHda1-p48. The control bar indicates histone deacetylase activity bound to Talon metal affinity resin of nontransfected insect cells (background activity); the ZmHda1 bar indicates histone deacetylase activity of insect cells transfected with full-length p84; the ZmHda1-48kD bar indicates histone deacetylase activity of insect cells transfected with the C-terminally truncated form.
Recombinant full-length and truncated ZmHda1 versions were subjected to mass spectrometry, and the results were consistent with the hypothesis that ZmHda1-p48 is derived from ZmHda1-p84 by proteolytic cleavage.
Protease Digestion Experiments
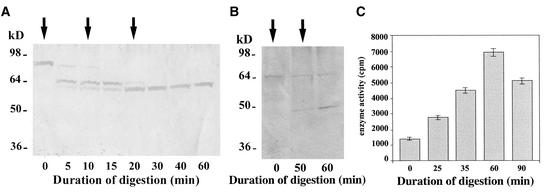
To substantiate the notion that proteolytic processing may be responsible for the conversion of ZmHda1-p84 to ZmHda1-p48, we tried to mimic this proteolysis by exoprotease (carboxypeptidase) digestion. We incubated fractions of maize extracts after S75 gel filtration chromatography (fractions 44 and 45; Figure 3) containing mainly the 84-kD form with increasing concentrations of carboxypeptidase and monitored the digestion by immunoblot analysis with the anti-ZmHda1 peptide antibody. This experiment clearly showed that the 84-kD band is converted gradually to a 65-kD intermediate (Figure 6A); further addition of carboxypeptidase did not lead to additional processing to the 48-kD form. The conversion of the 84-kD protein to p65 did not result in an increase of enzymatic activity, as measured in our standard assay (data not shown).
Figure 6.
In Vitro Conversion of ZmHda1-p84 to Smaller Enzyme Forms by Carboxypeptidase Treatment.
(A) Chromatographic fractions (pool of 44 and 45; see Figure 3A) containing ZmHda1-p84 were digested with carboxypeptidase (30 units) for 60 min. The digestion process was monitored by SDS-PAGE at the indicated times. Arrows mark the addition of carboxypeptidase. The gel was blotted onto a nitrocellulose membrane, and ZmHda1 was immunodetected with anti-ZmHda1 peptide antibodies.
(B) Chromatographic fractions (pool of 52 and 53; see Figure 3A) containing ZmHda1-p65 were digested with carboxypeptidase for 60 min.
(C) Histone deacetylase activity was measured during 90 min of carboxypeptidase digestion of chromatographic fractions 52 and 53 (see Figure 3A) containing ZmHda1-p65.
When the same experiment was performed with a chromatographic fraction (S75 gel filtration chromatography) containing mainly the 65-kD intermediate form (fractions 52 and 53; Figure 3), this 65-kD protein was converted to the low molecular weight enzyme form p48 (Figure 6B). During the time course of this digestion, histone deacetylase activity increased significantly, except for during prolonged incubation periods (Figure 6C). These results are in agreement with the hypothesis that inactive ZmHda1-p84 is converted to the well-characterized p48 enzyme form via the formation of a p65 intermediate.
To exclude the possibility of nonspecific artifacts, we performed parallel digestion experiments with partially purified maize HD2 and ZmRpd3 (the former HD1B). Carboxypeptidase digestion did not stimulate enzymatic activity; rather, it caused a loss of enzymatic activity (data not shown).
ZmHda1-p84 and ZmHda1-p48 during Maize Embryo Germination
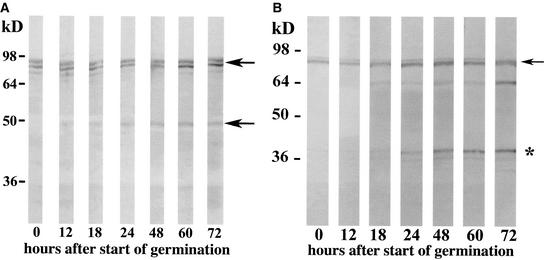
To analyze the expression of ZmHda1 and the ratio between ZmHda1-p84 and ZmHda1-p48 during embryo germination, whole embryos were harvested at selected times after the start of seed imbibition (0 to 72 h). Crude protein extracts were immunoblotted with anti-ZmHda1 peptide antibodies. Figure 7A shows that the expression of p84 increased slightly but continuously during embryo germination, although even in the dry embryos (0 h) a significant amount of p84 was present. ZmHda1-p48 was not detectable in dry embryos but appeared in very low amounts at 12 h after the start of germination and reached a maximum level at 60 h (Figure 7A); however the amount of p48 was a tiny fraction compared with the amount of p84. When the same samples were immunoblotted with the C-terminal ZmHda1 antibodies, p84 also was identified as the prominent protein band (Figure 7B); this pattern was consistent with that shown in Figure 7A. In addition to ZmHda1-p84, the C-terminal antibody should react with the putative cleavage product that is generated through the proteolytic processing of p84 but not with p48; indeed, the C-terminal antibodies reacted specifically with a protein band migrating at slightly above 36 kD. The pattern during embryo germination of this protein closely resembled the pattern seen with ZmHda1-p48 (Figure 7A): at 0 and 12 h of embryo germination, no immunosignal of the cleavage product at 36 kD was obtained. At 18 h after the start of germination, a very faint immunosignal was observed. The intensity of this signal increased continuously until 72 h after the start of germination.
Figure 7.
Immunological Characterization of ZmHda1 Forms during Maize Embryo Germination.
(A) Embryos were harvested into liquid nitrogen at various times during embryo germination (hours after the start of imbibition). Proteins were extracted, and equal amounts of protein were subjected to SDS-PAGE. The gel was blotted onto a nitrocellulose membrane, and proteins were immuno- detected with anti-ZmHda1 antibodies (peptide antibodies). Arrows mark the positions of the high molecular weight ZmHda1 form and the 48-kD form.
(B) The same blot shown in (A) was immunoblotted with antibodies against the C-terminal part of ZmHda1. The arrow marks the position of the high molecular weight ZmHda1 form, and the asterisk marks the position of the cleaved C-terminal peptide.
Transcriptional Repression Activity of ZmHda1 Constructs
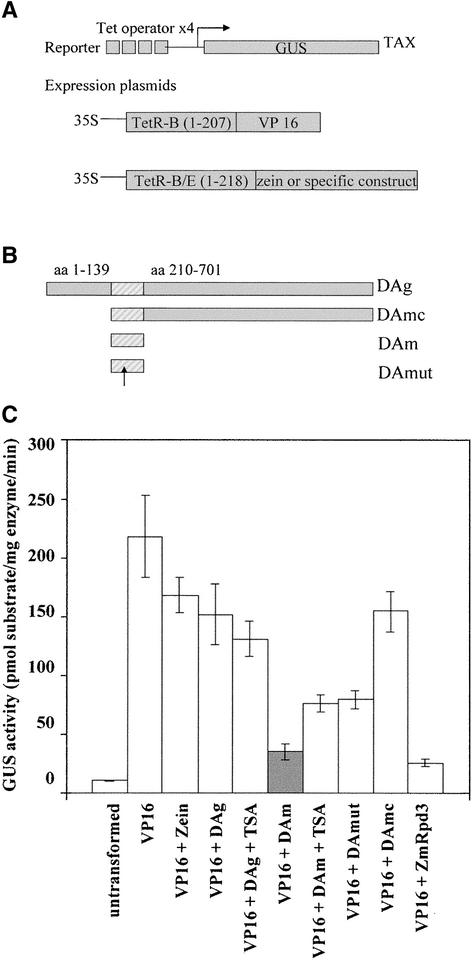
Because various histone deacetylases have been shown to be involved in transcription repression, we wished to determine whether (1) ZmHda1 could inhibit transcription when tethered to a reporter gene in vivo, (2) this repression is dependent on histone deacetylase activity, and (3) the C-terminal part affects repression activity. To this end, transcription experiments were performed using the tetracycline (TET) system (Bohner et al., 1999) (Figure 8).
Figure 8.
Effect of ZmHda1 Constructs on the Repression of Gene Transcription in Tobacco Protoplasts.
(A) Scheme of the reporter and expression plasmids used in the tobacco protoplast. The tobacco TAX line contains a reporter cassette that is integrated stably in the genome and allows for the expression of a reporter GUS controlled by a minimal promoter and four consecutive upstream TetR binding sites. Cotransfection of two plasmids with the 35S promoter driving the expression of the VP16 transcriptional activator and the protein of interest, respectively, both fused to the DNA binding domain of TetR, results in the competition of the two fusion proteins for the TET binding sites. The effect on transcription is monitored by determining GUS activity levels (Bohner et al., 1999).
(B) ZmHda1 constructs. The striped area represents the core catalytic histone deacetylase domain. The arrow depicts the position of the mutated His in the DAmut construct. aa, amino acids.
(C) Full-length ZmHda1 or ZmHda1 derivatives fused to the DNA binding domain of TetR were used together with plasmid expressing a TetR fusion of VP16 to transform protoplasts prepared from the tobacco TAX line. The effect on transcription was determined by measuring the activity of the GUS reporter gene. GUS activity is reported as pmol of 7-OH-4-methylcoumarin per milligram of protein per minute. “Untransformed” indicates protoplast samples transformed with the DNA carrier only. Error bars indicate the standard errors of assays performed on six independent transfections.
The TET system is based on the targeting of TET repressor (TetR) fusion proteins to TET binding sites upstream of a minimal promoter, which directs the transcription of the β-glucuronidase (GUS) gene (Gatz, 1997). Because the reporter construct is integrated stably into the tobacco genome (tobacco TAX transgenic line), a physiological chromatin structure can be expected (Bohner et al., 1999). Cotransfection of plasmids expressing the activator protein TetR-VP16 (Triezenberg et al., 1988) and a putative TetR-fused repressor into tobacco protoplasts results in the competition of the two proteins for the TET sites and provides a measure for transcription repression activity. To avoid heterodimerization of the TetR domains, which might interfere with VP16 transactivation efficiency, different TetR DNA binding domains unable to heterodimerize (TetR-B for VP16 fusions and TetR-B/E for the other TetR fusions; Forster et al., 1999) were used. To exclude the possibility that the observed repression is attributable solely to the competition for promoter binding sites, GUS activity values were determined for protoplasts that had been cotransformed with the TetR-VP16 protein and TetR-Zein, which is transcriptionally inert (Spena et al., 1983). These values are used as a reference for results obtained with the constructs to be tested (Rossi et al., 2003).
Four constructs were made to express the full-length protein (DAg), which lacks the N-terminal 139 amino acids but contains the histone deacetylase domain, and the C-terminal part (DAmc), the histone deacetylase core domain alone (DAm), or the histone deacetylase core domain containing a mutation of His-146 to Ala (DAmut), which is believed to be critical for catalytic activity (Taunton et al., 1996). Each construct was cotransfected with a TetR-VP16–expressing plasmid into tobacco protoplasts derived from the TAX line; GUS activity was assayed in protein extracts (Rossi et al., 2003).
Our results show that VP16 strongly activated GUS activity and that the activity was not reduced significantly by the presence of competing TetR-Zein (Figure 8). Conversely, cotransformation of the DAm construct of ZmHda1, representing the core catalytic domain, resulted in an approximately fourfold decrease of GUS activity compared with cotransformation of the Zein protein. Therefore, the ZmHda1 core domain is able to repress transcription in this system to almost the same extent as is maize ZmRpd3 (Rossi et al., 2003) (Figure 8). The effect was partially relieved both by treatment with the histone deacetylase inhibitor TSA and by the His-146 mutation in the histone deacetylase domain (construct DAmut), indicating that histone deacetylase activity is necessary to achieve maximum repression of reporter gene transcription. In addition, these results suggest that the domain between amino acids 139 and 210 harbors another deacetylase-independent repression activity. By contrast, protoplasts expressing full-length ZmHda1 (DAg) or the C-terminal plus the central core part (DAmc) showed very little repression compared with the Zein controls; TSA treatment in this case had no significant effect on transcription activity. These results further substantiate the notion that the presence of the C-terminal domain in ZmHda1 not only renders the protein enzymatically inactive but also corrupts its ability to repress transcription in a chromatin environment.
DISCUSSION
Histone modifications, in particular histone acetylation, play a fundamental role in chromatin structure and function. Therefore, the substrate molecules, the N-terminal extensions of the core histones, and the enzymes (histone acetyltransferases and deacetylases) are characterized by a high degree of conservation during evolution. More surprising, plants have developed enzymes and regulatory mechanisms with respect to histone deacetylation distinct from fungi and animals. The most prominent is the HD2 family of histone deacetylases (Lusser et al., 1997; Dangl et al., 2001), which seems to be plant specific. A second peculiarity of plants is the underrepresentation of the SIR2 family (sirtuins) of histone deacetylases; it was suggested that members of the HD2 family have taken over some of the functions of SIR2 in plants (Pandey et al., 2002). A further and novel mode of the regulation of ZmHda1 activity, which may be plant specific, is limited proteolysis, as shown here. A regulatory proteolytic step has not been found in yeast or animal cells for Rpd3- or Hda1-related histone deacetylases.
Limited proteolysis as a means of generating biologically active molecules from inactive pro-forms is a well-known regulatory mechanism (Seidah and Chrétien, 1997; Kageyama, 2002). In that sense, ZmHda1-p84 represents a zymogen that is converted to the active enzyme p48 via a putative 65-kD intermediate. It was reported recently that the human silent information regulator homolog hSIRT3, a nucleus-encoded protein of 44 kD, is translocated into the mitochondrial matrix; there, the protein is cleaved proteolytically to give rise to an active 28-kD deacetylase (Schwer et al., 2002). Mammalian SIR-type enzymes not only deacetylate histones but also deacetylate nonhistone proteins such as p53 and the TAFI68 subunit of the TATA-box binding protein–containing factor (Luo et al., 2001; Muth et al., 2001).
At present, we cannot determine whether a specific endoprotease is responsible for the proteolysis of the inactive p84 to the enzymatically active p48 or an autocatalytic processing mechanism might be induced by pH changes or by physiological changes during embryo development (e.g., dehydration/rehydration). However, we favor the idea of a specific endoprotease, because the conversion from p84 to p48 could not be mimicked in one step by exoproteases in vitro; proteolytic conversion of native ZmHda1-p84 led only to ZmHda1-p65 without a significant increase of enzymatic activity. Even high amounts of carboxypeptidase did not lead to further processing of p65 to p48. By contrast, when chromatographic fractions of maize extracts containing predominantly ZmHda1-p65 were used for digestion, the p65 was converted readily to p48 with a concomitant increase in histone deacetylase activity. We assume that p84 is converted to p65 by proteolysis, but most likely it has to be modified by (de)phosphorylation in vivo before it can be processed further into the enzymatically active 48-kD enzyme form. The lack of this specific phosphorylation is the most likely reason why we failed to convert p84 directly to p48 under our in vitro conditions.
This assumption is supported by earlier findings that dephosphorylation of maize extracts or purified ZmHda1-p48 results in a significant increase of histone deacetylase activity with a concomitant change of substrate specificity, in particular toward acetylated H4 isoforms (Kölle et al., 1999). In agreement with this interpretation is the fact that native ZmHda1-p48 always exists as a mixture of phosphorylated and dephosphorylated enzyme forms (Brosch et al., 1996; A. Loidl and G. Brosch, unpublished data). It should be mentioned in this context that dephosphorylation of the nucleolar maize HD2 resulted in a complete loss of enzymatic activity (Kölle et al., 1999). The regulatory role of the post-translational phosphorylation of histone deacetylases in controlling both enzymatic activity and the ability to interact with other proteins was reported recently in animal cell systems (Bertos et al., 2001; Pflum et al., 2001; Tsai and Seto, 2002; Zhang et al., 2002). It is possible that phosphorylation and limited proteolysis are interrelated in the molecular response to cellular or environmental stimuli; it also is possible that ZmHda1 is enzymatically active toward nonhistone proteins, as was shown recently for HDAC6 and tubulin (Hubbert et al., 2002; Zhang et al., 2003), and then is converted to p48 with altered substrate specificity (core histones) by phosphorylation/dephosphorylation-triggered proteolysis.
Further experimental support for the proteolytic cleavage of ZmHda1 in vivo comes from the detection of a distinct protein in extracts of maize embryos at selected times after germination by an antibody raised against the C-terminal portion of ZmHda1 (the putative cleavage product); a protein migrating at ∼37,000 on SDS-polyacrylamide gels appeared in later stages of embryo germination. The antibodies against the C-terminal part of ZmHda1 did not react with ZmHda1-p48, and the ZmHda1 peptide antibodies (raised against the peptide at amino acids 83 to 96) did not react with the putative ZmHda1 cleavage product at 37 kD (data not shown). It is interesting in this context that the enzymatically active ZmHda1-p48 was not present in significant amounts at early stages of embryo germination but appeared between 12 and 18 h after seed imbibition. The pattern of the putative 37-kD cleavage product was similar, because a faint immunosignal appeared at 18 h after the start of germination. By contrast, ZmHda1-p84 was present already in the dry embryo and increased continuously during germination. This finding indicates that the proteolytic cleavage of ZmHda1-p84 is not a constitutive process but rather is regulated during embryo germination. Moreover, it is evident that this proteolysis affects only a small proportion of the total ZmHda1-p84, because the majority of the protein remained intact and was found in a high molecular weight protein complex that lacked histone deacetylase activity.
Our transactivation experiments using different constructs of ZmHda1 also support the hypothesis that the truncated version of p48 is the biologically active protein. Full-length ZmHda1 was not effective in transcriptional repression compared with the catalytic core domain (which lacks the C-terminal portion of the protein); the same was true for the construct containing the catalytic deacetylase domain and the C-terminal part. This is a strong indication that the transcriptional repression function of ZmHda1-p48 is attributable to the enzymatic function as a deacetylase. Further evidence for this assumption was provided by analysis of the effect of the mutation of His-146 and trichostatin treatment. His-146 is a conserved residue in the catalytic domain that is essential for histone deacetylase activity. Both mutation of His-146 and trichostatin treatment strongly reduced the transcriptional repression effect. Therefore, we conclude that the C-terminal part of ZmHda1 has to be removed for full biological activity. This conclusion is in agreement with our biochemical results.
However, at present, we cannot definitively exclude an alternative explanation. One may assume that the proteolytic cleavage by an endoprotease occurs during the extraction of the enzyme, giving rise to a cleavage product with histone deacetylase activity in vitro. The intact ZmHda1-p84 organized in a 300-kD complex may be active only on nucleosomes or even on still obscure nonhistone substrates but not on free histones. It is known that the activity of different chromatin-remodeling complexes containing SWI/SNF-related ATPases can be triggered by different substrates; the ATPase activity of some SWI/SNF complexes is stimulated only by nucleosomes or (H3H4)2 tetramers, whereas the ATPase activity of others also is stimulated by free DNA (Boyer et al., 2000). A strong argument against this alternative explanation is provided by the fact that monomeric ZmHda1-p84 lacking the physiological complex partners does not exhibit histone deacetylase activity with free histones in vitro, whereas p48 does. If ZmHda1-p84 in its complex form has a distinct substrate specificity, one would assume that this substrate specificity is caused by interacting proteins present in the complex. Therefore, disruption of the complex should change the substrate specificity. However, only the cleaved p48 displays histone deacetylase activity toward free histones but not monomeric ZmHda1-p84 (Figure 3). The yeast homolog Hda1 is part of a 350-kD complex that exhibits histone deacetylase activity toward free histones in vitro (Carmen et al., 1996).
Another argument for a specific endoproteolytic cleavage of ZmHda1 in vivo is based on our observation that enzymatically active p48 appears at a distinct stage of embryo germination. Again, we cannot completely exclude the possibility that the responsible protease is induced at that stage of germination, giving rise to p48 during enzyme preparation. However, this is unlikely because the conversion of ZmHda1-p84 to ZmHda1-p48 occurs irrespective of the method of enzyme extraction (salt extraction, chromatin preparation, with or without protease inhibitors; data not shown).
Interestingly, a short protein of unknown function exists in Arabidopsis (Atg61050) that has a high degree of sequence identity to the C-terminal part of ZmHda1 plus a very short N-terminal stretch; however, this protein lacks the catalytic histone deacetylase domain. A possible function of this Arabidopsis protein might be the inhibition of the transcription repression activity of histone deacetylases. In the maize sequence database, an EST clone (EST HDA115; 141 amino acids) is available that displays 56% sequence identity to ZmHda1; however, this truncated sequence lacks the sequence for the catalytic histone deacetylase domain (Figure 1). It is possible that this protein plays a specific regulatory role similar to that of ZmHda1-p84. This protein is certainly distinct from the small Atg61050 of Arabidopsis, because Atg61050 resembles a homolog of the C-terminal part of ZmHda1.
In metazoan systems, Rpd3- and Hda1-related histone deacetylases exist as large multiprotein complexes. In plants, this seems to be different. The two Rpd3-related maize deacetylases both exist as monomeric enzymes or as minimal complexes together with the retinoblastoma-related protein (Lechner et al., 2000; Rossi et al., 2003). ZmHda1 in its enzymatically active form has always been found as a monomer. However, the nonprocessed form p84 is part of a higher molecular mass protein complex (∼300 kD; Figure 4) that lacks histone deacetylase activity. p84 in this high molecular mass complex represents the majority of ZmHda1 protein, and we assume that this catalytically inactive form serves specific functions as a partner protein in regulatory complexes or as nonhistone protein deacetylases. The small Arabidopsis protein (Atg61050) probably will be the key to understanding the function of the proteolytic cleavage of ZmHda1.
It seems increasingly clear that plants have developed distinct proteins and regulatory mechanisms with respect to histone modifications, most likely as a result of the unique features of plant cells with respect to growth and development. Identification of the endoprotease responsible for the processing of ZmHda1 and its inhibition by drugs or repression of expression will provide further insight into the functional contribution of this type of plant deacetylase.
METHODS
ZmHda1 cDNA Cloning
Total RNA from maize (Zea mays) embryos was used for the generation of cDNA with SuperScript reverse transcriptase (Gibco BRL) according to the manufacturer's general information. For the design of primers, the Hda1 sequence of Saccharomyces cerevisiae was aligned with the sequence of Schizosaccharomyces pombe Clr3, mouse HDA2, human HDAC6, S. cerevisiae Rpd3, and S. cerevisiae Hos2. Consensus regions in the sequences of Hda1-like proteins from these organisms were used to design primers for PCR amplification to screen for a potential homolog of Hda1 in maize. A 158-bp DNA fragment was amplified using primers oHD1 (5′-CCNCCNGGNCAYCAYGC-3′) and oHD2r1 (5′-TGNGTNCCRTTNCCRTG-3′) for first PCR and oHD2r (5′-GTNCCRTTN- CCRTGRTG-3′) for nested PCR. Searching the Maize Genome Database (www.zmdb.iastate.edu) with the 158-bp fragment revealed an EST clone of 661 bp. Primers were designed for amplification of the 3′ and 5′ ends by the rapid amplification of cDNA ends (RACE) protocol (Frohman et al., 1988): for 3′ RACE, AW25a (5′-CAGACCTCCAGGCCACC-3′) and AW25b (5′-ACATGATGAGGCTATGGG-3′). mRNA for 5′ RACE was reverse transcribed with primer AW25cr (5′-TAACCTCCTTCAAGGGC-3′). Primers AW25dr (5′-GAAGAACAATACACGAGG-3′), AW25er (5′-TTCTGT- GTGCCATTTCCA-3′), and AW25fr (5′-ATGGTGAACATCCCAATC-3′) were used in the first and nested PCRs. The 5′ end was obtained using the Gene Racer Kit (Invitrogen, Carlsbad, CA) with the ZmHda1-specific primer Aw25hr (5′-CATCAGCTTTATATGCCTC-3′).
RNA Extraction and RNA Gel Blot Analysis
Extraction of total RNA from different tissues of germinating maize seedlings (whole embryos at selected times after the start of embryo imbibition, roots, shoots, and leaves) and RNA gel blot analysis were performed essentially as described previously (Lechner et al., 2000). A digoxigenin-labeled (Roche, Indianapolis, IN) 513-bp PCR product between ZmHda1 primers AW25b and AW25cr (see above) was used for hybridization under high-stringency conditions (Lechner et al., 2000).
For low-stringency hybridizations, additional probes were synthesized corresponding to the sequence between nucleotide positions 1305 and 2105 (3′ probe; primers Anti2 [5′-ATTGGGATCCTTGCAGTGAAGACT- AACTCAAGT-3′] and Anti12r [5′-ATTGCCCGGGTCACAAGGCTCTTGCAGCAGCAGCGTT-3′]) and the sequence between positions 73 and 627 (5′ probe; primers Akt1 [5′-ATTGCATGCAAGCGCACGCGACGCCCGAC-3′] and Aw25ABr [5′-AAGGATCCTCAGTGAACTGAGAAGAAC- AA-3′]). Membranes were incubated in roller tubes overnight at 42°C, and low-stringency washes were performed as follows: twice for 5 min in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at room temperature and twice for 20 min in 0.1× SSC and 0.1% SDS at 42°C. 2× SSC was used to rinse the membranes. Membranes were subjected to digoxigenin detection according to the manufacturer's recommendations.
Purification of ZmHda1-p48
Maize embryos (endosperm free) (strain Cuzco 251; Steirischer Landwirteverband, Graz, Austria) at 72 h after the start of germination were harvested into liquid nitrogen. The soluble cytoplasmic fraction (containing HD1A; now named ZmHda1) was prepared as described by Brosch et al. (1996). For protein microsequencing as well as mass spectrometry, ZmHda1-p48 was purified to homogeneity from 6 kg of embryos following exactly the purification scheme introduced by Brosch et al. (1996).
For biochemical and immunological experiments, ZmHda1 (p84, p65, and p48) was partially purified using Q-Sepharose Big Beads (Pharmacia) chromatography starting with the soluble fraction of 35 g of maize embryos at 72 h after the start of germination. A total of 160 mL of soluble fraction was applied to 50 mL of Q-Sepharose Big Beads. Proteins were eluted with a linear gradient of 200 mL of 0.01 to 0.4 M NaCl at a flow rate of 1 mL/min. Fractions of 4 mL were collected and assayed for histone deacetylase activity using the standard assay (Lechner et al., 2000). Fractions containing histone deacetylase activity were subjected to Superdex S75 gel filtration chromatography. When S200 gel filtration chromatography was used, the purification was increased to 500 g of maize embryos.
For distinct immunoblot experiments, chromatographic fractions were concentrated either by precipitation with trichloroacetic acid or using centrifugal ultrafiltration devices (Centricon; Amicon, Beverly, MA).
Production of Polyclonal Antibodies against ZmHda1
Three ZmHda1-specific antibodies were raised in rabbit: (1) against the peptide KEISSTIYDASRNK (amino acids 83 to 96; peptide 2 in Table 1); (2) against recombinant, full-length ZmHda1 (amino acids 1 to 701) with a 6xHis tag fused to the N terminus of the protein; and (3) against a recombinant protein comprising the C-terminal portion (amino acids 437 to 701) with a 6xHis tag fused to the N terminus.
For the production of recombinant proteins for immunization, ZmHda1 constructs were amplified by PCR using gene-specific primers (full-length ZmHda1, 5′-ATTGGGATCCGGATGTGCGCGCACGCGACGCCCGA-3′ and 5′-ATTGCCCGGGTCACAAGGCTCTTGCAGCAGCAGCGTT-3′; C-terminal part of ZmHda1, 5′-ATTGGGATCCTTGCAGTGAAGACTAAC- TCAAGT-3′ and 5′-ATTGCCCGGGTCACAAGGCTCTTGCAGCAG- CATT-3′). Restriction sites for BamHI and SmaI were used for cloning into pQE32 vector (Qiagen, Valencia, CA). Overexpression of proteins was performed in Escherichia coli (M15; pREP4). Recombinant proteins were purified with Talon affinity resin (Clontech, Palo Alto, CA). Eluted proteins were subjected to SDS-PAGE. Proteins were recovered from Coomassie Brilliant Blue R250–stained gel bands and used for the immunization of rabbits.
Immunoblot Analysis and Immunoprecipitation
Protein gel blot analysis and immunodetection of protein samples was performed as described (Lechner et al., 2000) using the three anti-ZmHda1 antibodies described above. Igs were purified using Hi-Trap protein A–Sepharose columns (Amersham). With the anti-Hda1 peptide antibodies, usually a dilution of 1:500 of a stock (4 mg/mL) was used, whereas the antibodies against the full-length ZmHda1 and those against the truncated C-terminal part of ZmHda1 were diluted 1:1500 and 1:3000, respectively (stock of 4 mg/mL).
For covalent binding of antibodies onto protein A–Sepharose, the beads (1 mL) were incubated with 2 mg of purified antibodies (1 h, room temperature, gentle rocking) and washed with 10 volumes of 0.2 M sodium borate, pH 9.0, by centrifugation at 3000g for 5 min. Beads were resuspended in 10 volumes of sodium borate. Ten microliters of beads was removed routinely for analysis by SDS-PAGE. Solid dimethylpimelimidate was added to the beads at a final concentration of 20 mM and mixed on a shaker at room temperature for 30 min. Again, 10 μL of beads was removed for SDS-PAGE analysis. The coupling reaction was stopped by washing the beads once with 0.2 M ethanolamine, pH 8.0, and subsequent incubation with 0.2 M ethanolamine with gentle mixing. Beads were centrifuged as described above and resuspended in PBS with 0.01% merthiolate for storage at 4°C. The efficiency of covalent coupling was checked by SDS-PAGE. After the coupling reaction, the beads should not liberate any Ig heavy chains. If they did, the beads were washed twice with 100 mM Gly, pH 3.0, to remove those antibodies that were only linked to protein A by noncovalent binding.
Aliquots of Superdex S75 gel filtration chromatography fractions were concentrated by ultrafiltration (Centricon-10; Amicon) from 800 to ∼160 μL. Aliquots of 40 μL of concentrated fractions were mixed with 30 μL of antibody-coupled protein A–Sepharose beads and incubated overnight with head-over-head shaking at 4°C. Beads were collected by centrifugation at 600g for 3 min. The supernatant was saved, and the beads were washed twice with 100 μL of buffer B (15 mM Tris-HCl, pH 7.9, 0.25 mM EDTA, 10% [v/v] glycerol, and 10 mM NaCl) and then resuspended in 30 μL of buffer B. Histone deacetylase activity was measured as described above in the supernatant and the resuspended pellet. Control precipitations with protein A–Sepharose beads alone (without antibodies) were performed. Analysis of immunoprecipitations was performed by SDS-PAGE and immunoblotting.
When crude protein extracts were used for immunoprecipitation experiments, they were prepared as follows. Maize seedlings at 72 h after the start of embryo germination were frozen in liquid nitrogen and ground to powder. Six mL of buffer A (0.5 M NH4Cl, 0.25 mM EDTA, 10 mM 2-mercaptoethanol, and 15 mM Tris-HCl, pH 7.9) was added to 3 g of powder. The suspension was homogenized with a potter, stirred on ice for 30 min, and centrifuged at 14,000g for 1 h at 4°C. The supernatant was used as protein extract. Protein concentration was determined according to Bradford (1976).
Carboxypeptidase Digestion
Chromatography fractions or crude protein extracts were digested with variable amounts of carboxypeptidase A (Sigma). Incubation was performed at 30°C for various time periods. The digestion process was monitored by the determination of histone deacetylase activity and immunoblotting with anti-ZmHda1 peptide antibodies.
Expression of ZmHda1 Using the Baculovirus Expression System
ZmHda1 was amplified from cDNA with primers WtXho (5′-ATACTCGAGATGGCTCCCCCGGTGACCGC-3′) and WtHis (5′-TGCGGCCGC- TAGTGATGGTGATGGTGATGGGATCCTTCTCAAGGCTCTTCAGCA- GCA-3′). ZmHda1-p48 was amplified from cDNA with primers Bac48 (5′-ATACTCGAGATGTGCGCGCACGCGACGCCCGAC-3′) and Bac48r (5′-TGCGGCCGCTAGTGATGGTGATGGTGATGCTTTGAGAGGTGTTCA- CTTATAGC-3′). A 6xHis tag was encoded within the 3′ primers to receive a recombinant protein with a C-terminal 6xHis tag. Fragments were digested with XhoI-NotI (Roche) and cloned into pBacPAK8 (Clontech). Sf21 or Sf9 insect cells were infected according to standard protocols (King and Possee, 1992). Recombinant protein was expressed in infected Sf21 or Sf9 cells for ∼48 or 72 h at 28°C. Protein content was determined using the Bradford method (Bradford, 1976).
Protoplast Transfection Experiments
Transcription experiments were performed using the tetracycline (TET) system, based on the targeting of proteins fused to the DNA binding domain of the TET repressor (TetR) to a synthetic promoter containing four TET operators upstream of a minimal promoter (Bohner et al., 1999). Four different constructs of ZmHda1 fused to TET-B/E were made to be tested for transcription repression activity in tobacco protoplasts: DAg (2100 bp) (primers AW25AE3 [5′-TAGAATTCATGGCGGAGCCGGCTCCGGCT-3′] and AW25Dr3 [5′-AAGGATCCTCACAAGGCTCTTGCAGCAG-3′] for amplification out of maize cDNA); DAm (207 bp) (primers AW25AA [5′-AAGAATTCGGCAGACCTCCAGGCCAC-3′] and AW25ABr [5′-AAGGATCCTCAGTGAACTGAGAAGAACAA-3′]); DAmc (1683 bp; primers AW25AA and AW25Dr3); and DAmut (207 bp with mutation of His-146 of ZmHda1 to Ala; corresponding to His-141 of human HDAC1) (Taunton et al., 1996). ZmHda1 fragments were digested with EcoRI and BamHI and cloned into pTET(B/E) vector in frame with the DNA binding domain of the TetR (Figure 8). The pUC-TET-VP16 construct (containing the TetR-B DNA binding domain fused to VP16) and the pTetR-B/E-Zein construct (containing the TetR-B/E DNA binding domain fused to maize zein) were described by Rossi et al. (2003). Equimolar amounts of TET-VP16 and TET-B/E fusion plasmids were used for TAX protoplast cotransformation in competition assays. The total amount of DNA used in transformation (carrier plus plasmids) was the same for each sample (100 μg). Isolation, tobacco protoplast transformation, and GUS assays were performed as described (Bohner et al., 1999). The histone deacetylase inhibitor TSA (Yoshida et al., 1990) was added to protoplasts at 12 h before GUS activity assays at a final concentration of 100 ng/μL.
Protein Microsequencing and Mass Spectrometry
HD1A (ZmHda1-p48) was excised from the dried nylon membranes and digested with chymotrypsin endoproteinase (sequencing grade; Roche). Resulting peptides after reverse-phase HPLC were subjected to Edman degradation. Peptide sequencing was performed on an Applied Biosystems model 492 Procise protein sequenator.
For mass spectrometry analysis, in-gel digestion of ZmHda1 was performed with slight modifications according to previously published methods (Wilm et al., 1996; Borchers et al., 2000). The various steps of the procedure were performed at room temperature, and all incubation steps were performed under shaking. The protein band of interest was excised from the SDS gel, cut into small pieces, transferred into a 0.25-mL polyethylene sample vial, and washed with two changes of 150 μL of 10 mM NH4HCO3, pH 8.9 (15 min at 30°C). The supernatant was discarded, and the gel pieces were shrunk by dehydration in 150 μL of 50% (v/v) acetonitrile and 10 mM NH4HCO3 (15 min at 30°C). This step was performed twice, and after removing all liquid, the gel pieces were dried in a vacuum centrifuge. The gel pieces were swollen in digestion buffer containing 10 mM NH4HCO3 and 0.05 μg/μL trypsin (sequencing grade; Roche) at 4°C for 20 min. The supernatant was removed and replaced with 20 μL of the same buffer without trypsin. Digestion proceeded overnight at 37°C. Peptides were extracted by adding 50 μL of 10 mM NH4HCO3 (37°C for 15 min) and 50 μL of acetonitrile (37°C for 15 min). After the supernatant was collected, 100 μL of 10% formic acid, 20% acetonitrile, and 20% 2-propanol was added to the gel pieces (37°C for 10 min). The supernatants were combined, dried to ∼5 μL, and stored at −20°C.
Protein digests were separated using a capillary HPLC system connected online to a mass spectrometer. The solvent delivery system consisted of a Rheos 2000 pump connected to an ERC 3215 degasser (Flux Instruments, Karlskoga, Sweden) running at a flow rate of 80 μL/min, which was reduced to a column flow of ∼500 nL/min using a precolumn T split. A fused silica microcapillary column (100 μm i.d. × 365 μm o.d.) was pulled with a model P-2000 laser puller (Sutter Instrument Co., Novato, CA) and packed with 7 cm of 5 μm of reverse-phase C18 material. The gradient (solvent A, 0.1% formic acid; solvent B, 0.1% formic acid in 85% acetonitrile) started at 0% solvent B. The concentration of solvent B was maintained at 0% for 5 min, increased linearly from 0 to 60% during 40 min, and increased from 60 to 100% during 10 min.
Protein digests were analyzed using a LCQ ion-trap instrument (ThermoFinnigan, San Jose, CA) equipped with a nanospray interface. The nanospray voltage was set at 1.6 kV, and the heated capillary was held at 170°C. Tandem mass spectrometry spectra were searched against the Hda1 sequence using SEQUEST (LCQ BioWorks; ThermoFinnigan).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact A. Pipal, Alexandra.Pipal@uibk.ac.at.
Accession Numbers
The GenBank accession number for ZmHda1 is AF322918. Accession numbers for other sequences mentioned are as follows: Saccharomyces cerevisiae Hda1, P53973; Schizosaccharomyces pombe Clr3, P56523; mouse HDA2, T13964; human HDAC6, AAD29048; S. cerevisiae Rpd3, NP014069; S. cerevisiae Hos2, NP011321; and a 661-bp EST clone, AW256098.
Acknowledgments
We thank Gerald Brosch and Hubertus Haas for technical help and numerous discussions. A. Lusser is the recipient of an Austrian Programme for Advanced Research and Technology fellowship from the Austrian Academy of Sciences. This work was supported in part by grants from the Austrian Science Foundation (FWF-P-14528) and the Austrian National Bank (P-7415 and P-9000) to P.L.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.013995.
References
- Bertos, N.R., Wang, A.H., and Yang, Y.J. (2001). Class II histone deacetylases: Structure, functions, and regulations. Biochem. Cell Biol. 79, 243–252. [PubMed] [Google Scholar]
- Bohner, S., Lenk, I.I., Rieping, M., Herold, M., and Gatz, C. (1999). Technical advances: Transcriptional activator TGV mediates dexamethasone-inducible and tetracycline-inactivable gene expression. Plant J. 19, 87–95. [DOI] [PubMed] [Google Scholar]
- Borchers, C., Peter, J.F., Hall, M.C., Kunkel, T.A., and Tomer, K.B. (2000). Identification of in-gel digested proteins by complementary peptide mass fingerprinting and tandem mass spectrometry data obtained on an electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem. 72, 1163–1168. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., Logie, C., Bonte, E., Becker, P.B., Wade, P.A., Wolffe, A.P., Wu, C., Imbalzano, A.N., and Peterson, C.L. (2000). Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275, 18864–18870. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brosch, G., Georgieva, E., López-Rodas, G., Lindner, H., and Loidl, P. (1992). Specificity of Zea mays histone deacetylase is regulated by phosphorylation. J. Biol. Chem. 267, 20561–20564. [PubMed] [Google Scholar]
- Brosch, G., Goralik-Schramel, M., and Loidl, P. (1996). Purification of histone deacetylase HD1-A of germinating maize embryos. FEBS Lett. 393, 287–296. [DOI] [PubMed] [Google Scholar]
- Carmen, A.A., Rundlett, S.E., and Grunstein, M. (1996). HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J. Biol. Chem. 272, 15837–15844. [DOI] [PubMed] [Google Scholar]
- Dangl, M., Brosch, G., Haas, H., Loidl, P., and Lusser, A. (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213, 280–285. [DOI] [PubMed] [Google Scholar]
- Dobosy, J.R., and Selker, E.U. (2001). Emerging connections between DNA methylation and histone acetylation. Cell. Mol. Life Sci. 58, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, K., Helbl, V., Lederer, T., Urlinger, S., Wittemburg, N., and Hillen, W. (1999). Tetracycline-inducible expression systems with reduced basal activity in mammalian cells. Nucleic Acids Res. 27, 708–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz, C. (1997). Chemical control of gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 89–108. [DOI] [PubMed] [Google Scholar]
- Graessle, S., Loidl, P., and Brosch, G. (2001). Histone acetylation: Plants and fungi as model systems for the investigation of histone deacetylases. Cell. Mol. Life Sci. 58, 704–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert, C., Guardiola, A., Shao, R., Kawaguchi, Y., Ito, A., Nixon, A., Yoshida, M., Wang, X.F., and Yao, T.P. (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458. [DOI] [PubMed] [Google Scholar]
- Imai, S., Armstrong, C.M., Kaeberlein, M., and Guarente, L. (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Pahk, Y.M., Song, S.I., Kwon, H.J., Nahm, B.H., and Kim, J.K. (2003). Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1-3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. Plant J. 33, 531–541. [DOI] [PubMed] [Google Scholar]
- Kageyama, T. (2002). Pepsinogens, progastricsins, and prochymosins: Structure, function, evolution, and development. Cell. Mol. Life Sci. 59, 288–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, L.A., and Possee, R.D. (1992). The Baculovirus Expression System: A Laboratory Guide. (London: Chapman & Hall).
- Kölle, D., Brosch, G., Lechner, T., Pipal, A., Helliger, W., Taplick, J., and Loidl, P. (1999). Different types of maize histone deacetylases are distinguished by a highly complex substrate and site specificity. Biochemistry 38, 6769–6773. [DOI] [PubMed] [Google Scholar]
- Landry, J., Sutton, A., Tafrov, S.T., Heller, R.C., Stebbins, J., Pillus, L., and Sternglanz, R. (2000). The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, T., Lusser, A., Pipal, A., Brosch, G., Loidl, A., Goralik-Schramel, M., Sendra, R., Wegener, S., Walton, J., and Loidl, P. (2000). RPD3-type histone deacetylases in maize embryos. Biochemistry 39, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Loidl, P. (1988). Towards an understanding of the biological function of histone acetylation. FEBS Lett. 227, 91–95. [DOI] [PubMed] [Google Scholar]
- Luo, J., Nikolaev, A.Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001). Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148. [DOI] [PubMed] [Google Scholar]
- Lusser, A. (2002). Acetylated, methylated, remodeled: Chromatin states for gene regulation. Curr. Opin. Plant Biol. 5, 437–443. [DOI] [PubMed] [Google Scholar]
- Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. [DOI] [PubMed] [Google Scholar]
- Lusser, A., Kölle, D., and Loidl, P. (2001). Histone acetylation: Lessons from the plant kingdom. Trends Plant Sci. 6, 59–65. [DOI] [PubMed] [Google Scholar]
- Muth, V., Nadaud, S., Grummt, I., and Voit, R. (2001). Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Sommerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R., Müller, A., Napoli, C.A., Slinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflum, M.K., Tong, J.K., Lane, W.S., and Schreiber, S.L. (2001). Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276, 47733–47741. [DOI] [PubMed] [Google Scholar]
- Pikaard, C.S. (1999). Nucleolar dominance and silencing of transcription. Trends Plant Sci. 4, 478–483. [DOI] [PubMed] [Google Scholar]
- Reyes, J.C., Hennig, L., and Gruissem, W. (2002). Chromatin-remodeling and memory factors: New regulators of plant development. Plant Physiol. 130, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, V., Hartings, H., and Motto, M. (1998). Identification and characterization of an RPD3 homologue from maize (Zea mays L.) that is able to complement an rpd3 null mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 258, 288–296. [DOI] [PubMed] [Google Scholar]
- Rossi, V., Locatelli, S., Lanzanova, C., Boniotti, B., Varotto, S., Pipal, A., Goralik-Schramel, M., Lusser, A., Glatz, C., Gutierrez, C., and Motto, M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51, 401–413. [DOI] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Kobayashi, R., Bavykin, S., Turner, B.M., and Grunstein, M. (1996). HDA1 and RPD3 are members of distinct histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., North, B.J., Frye, R.A., Ott, M., and Verdin, E. (2002). The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah, N.G., and Chrétien, M. (1997). Eukaryotic protein processing: Endoproteolysis of precursor proteins. Curr. Opin. Biotechnol. 8, 602–607. [DOI] [PubMed] [Google Scholar]
- Spena, A., Viotti, A., and Pirrotta, V. (1983). Two adjacent genomic zein sequences: Structure, organization and tissue-specific restriction pattern. J. Mol. Biol. 169, 799–811. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and Allis, C.D. (2000). The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Taunton, J., Hassig, C.A., and Schreiber, S.L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3. Science 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg, S.J., Kingsbury, R.C., and McKnight, S.L. (1988). Functional dissection of VP16, the transactivator of herpes simplex virus immediate early gene expression. Genes Dev. 2, 718–729. [DOI] [PubMed] [Google Scholar]
- Tsai, S.C., and Seto, E. (2002). Regulation of histone deacetylase 2 by protein kinase CK2. J. Biol. Chem. 277, 31826–31833. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. (1993). Decoding the nucleosome. Cell 8, 5–8. [PubMed] [Google Scholar]
- Wilm, M., Shevchenko, A., Houthaeve, T., Breit, S., Schweigerer, L., Fotsis, T., and Mann, M. (1996). Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379, 466–469. [DOI] [PubMed] [Google Scholar]
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000. a). Functional analysis of a RPD3 histone deacetylase homolog in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000. b). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990). Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]
- Zhang, C.L., McKinsey, T.A., Chang, S., Antos, C.L., Hill, J.A., and Olson, E.N. (2002). Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Li, N., Caron, C., Matthias, G., Hess, D., Khochbin, S., and Matthias, P. (2003). HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 22, 1168–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]