Abstract
Here, we report the identification of Karma, a LINE-type retrotransposon of plants for which continuous retrotransposition was observed in consecutive generations. The transcription of Karma is activated in cultured cells of rice upon DNA hypomethylation. However, transcription is insufficient for retrotransposition, because no increase in the copy number was observed in cultured cells or in the first generation of plants regenerated from them. Despite that finding, copy number increase was detected in the next generation of regenerated plants as well as in later generations, suggesting that the post-transcriptional regulation of Karma retrotransposition is development dependent. Our results indicate that two different mechanisms, one transcriptional and the other developmental, control the mobilization of Karma. In addition, unlike other known active plant retrotransposons, Karma is not subject to de novo methylation, and retrotransposition persists through several generations.
INTRODUCTION
The completion of the rice genome draft sequences revealed that retrotransposons account for >15% of the genome of rice (Goff et al., 2002; Yu et al., 2002). In contrast to DNA-type transposable elements, retrotransposons encode a reverse transcriptase (RT) activity and move by a replicative mechanism that involves an RNA intermediate. Thus, retrotransposons contributed greatly to the expansion and the evolution of the genome (reviewed by Kumar and Bennetzen, 1999; Prak and Kazazian, 2000; Feschotte et al., 2002).
Despite the fact that retrotransposons exist in high copy numbers in the genomes of most eukaryotes, the great majority of them are inactive or defective, and only a small portion of them retain the ability to retrotranspose (reviewed by Grandbastien, 1998; Kumar and Bennetzen, 1999). In plants, direct evidence for retrotransposition was demonstrated for only a few elements, such as Tos17 of rice (Hirochika et al., 1996) and Tto1, Tto2, and Tnt1 of tobacco (Hirochika, 1993; Lucas et al., 1995). This finding may reflect technical problems, because special techniques, such as transposon display, are required to monitor the mobility of highly repetitive elements (Melayah et al., 2001). In some cases, retrotransposition was detected only with the finding of clear-cut evidence, such as gene disruption associated with mutant phenotypes (Johns et al., 1985; Grandbastien et al., 1989; Varagona et al., 1992; Schwarz-Sommer et al., 1997).
Recently, a survey of >400,000 EST sequences of maize identified only 56 retrotransposon cDNAs, supporting the notion that most retrotransposons are inactive (Meyers et al., 2001). This search also showed that most retrotransposons found in EST libraries are low-copy elements in genomic sequences, whereas only a few of the high-copy elements are found in EST libraries. Even active retrotransposons are subject to tight regulation, so that retrotransposition occurs only rarely. A well-studied aspect of the regulation of plant retrotransposon activities is the induction of their transcription by biotic or abiotic stresses (Johns et al., 1985; Grandbastien et al., 1989; Hirochika, 1993). Epigenetic gene silencing through methylation also is an important regulatory mechanism that suppresses the activity of retrotransposons (Hirochika et al., 2000; Lindroth et al., 2001; Tompa et al., 2002). Recently, RNA silencing and RNA-directed DNA methylation of retrotransposons were demonstrated (Hamilton et al., 2002; Llave et al., 2002; Zilberman et al., 2003).
Retrotransposons can be classified as either long terminal repeat (LTR) retrotransposons or non-LTR retrotransposons, depending on the presence or absence of terminal repeats. Retrotranspositionally active elements identified from plants to date all belong to the LTR subclass, and very limited information is available concerning non-LTR retrotransposons. Plant genomes contain both the long interspersed elements (LINEs) and short interspersed elements (SINEs) types of non-LTR retrotransposons; however, these are less abundant than LTRs (Noma et al., 1999; Le et al., 2000; Turcotte et al., 2001). This is in sharp contrast to the situation in mammals, in which non-LTR retrotransposons are predominant. For example, the human L1, a LINE element, comprises 17% of the genome (Prak and Kazazian, 2000). The mechanism of mammalian L1 retrotransposition has been studied extensively (reviewed by Ostertag and Kazazian, 2001), and recently, the mobilization of SINEs by LINEs was demonstrated in eels (Kajikawa and Okada, 2002).
To fill the void of knowledge about plant non-LTR retrotransposons, isolation of active copies has been long desired. Here, the identification of Karma, a mobile LINE element in plants, is described. Karma activation in rice plants is controlled in two steps: the first occurs at transcription, and the second is post-transcriptional and development dependent. Interestingly, once the retrotransposition of Karma starts in the progeny of tissue culture–derived plants, Karma remains active for generations.
RESULTS
Karma Is a Novel LINE-Type Retrotransposon
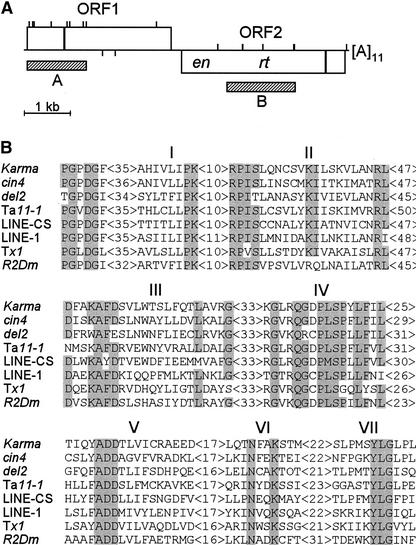
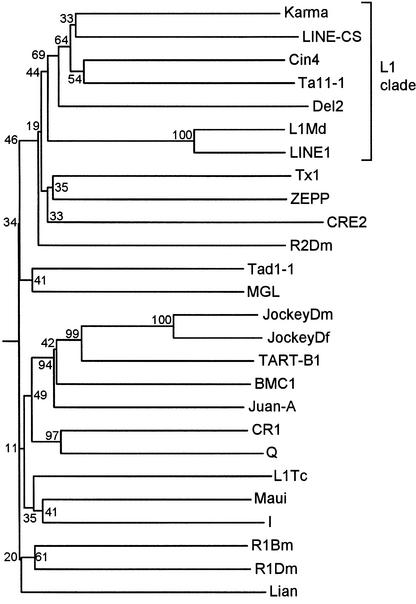
Karma was identified as an insertion in a mutant allele of the FRIZZY PANICLE2 locus (Komatsu et al., 2001, 2003). DNA sequence analysis revealed that this insertion represents a novel rice non-LTR retrotransposon of the LINE group, which was designated Karma. Karma is 7080 bp long and has a 5′ untranslated region of 39 bp, two nonoverlapping open reading frames of 3156 and 3594 bp, which are separated by an intergenic spacer of 220 bp and translated in different frames, and a 3′ untranslated region of 71 bp; it is terminated by a poly(A) tail of 11 bases (Figure 1A). Domains conserved in LINE elements are observed in Karma. Short Cys-rich motifs are present at each open reading frame, and an endonuclease and a reverse transcriptase domain are found at the second open reading frame. Karma is related more closely to other plant LINEs, in particular to the maize Cin4 element (Schwarz-Sommer et al., 1997), with which it shares 37% amino acid identity in the reverse transcriptase domain (Figure 1B). Interestingly, all known plant LINEs belong to the L1 clade (Figure 2), which includes the mouse and human LINE-1 elements (Loeb et al., 1986; Dombroski et al., 1991).
Figure 1.
Conserved Domains in Karma.
(A) Karma structure. Open boxes depict the two open reading frames of Karma (ORF1 and ORF2). ORF2 contains the endonuclease (en) and reverse transcriptase (rt) domains. Black boxes depict Cys-rich motifs. Vertical lines above the boxes depict the sites of HpaII and MspI cleavage, and vertical lines below the boxes depict the sites of EcoRV cleavage. Hatched boxes depict probes A (1317 bp) and B (1498 bp) used in DNA gel blot analysis. Bar = 1 kb.
(B) Alignment of the conserved reverse transcriptase domain. Gray boxes indicate residues conserved in at least seven of the eight sequences depicted. I to VII denote the seven domains characteristic of reverse transcriptases. Retrotransposon sequences are from rice Karma, maize cin4, lily del2, Arabidopsis Ta11-1, Cannabis LINE-CS, human LINE-1, frog Tx1, and fruit fly R2Dm.
Figure 2.
Phylogenetic Analysis of 26 LINE Reverse Transcriptase Sequences.
Sequences analyzed were from fruit fly I element, frog Tx1, fruit fly R2Dm, mouse L1Md, human LINE-1, rice Karma, lily del2, maize cin4, Arabidopsis Ta11-1, Cannabis LINE-CS, green alga ZEPP, trypanosome CRE2, mosquito Juan-A, silkworm BMC1, fruit fly TART-1, fruit fly JockeyDm, fruit fly JockeyDf, chicken CR1, mosquito Q, trypanosome L1Tc, pufferfish Maui, rice blast fungus MGL, fungi Tad1-1, mosquito Lian, silkworm R1Bm, and fruit fly R1Dm. The L1 clade is indicated. Phylogeny was constructed using the neighbor-joining algorithm from distance matrices according to Kimura's two-parameter method (Kimura, 1983). Branch lengths are proportional to genetic distance. Bootstrap values are indicated as a percentage of 1000 replicates.
Karma Distribution Varies between Rice Subspecies
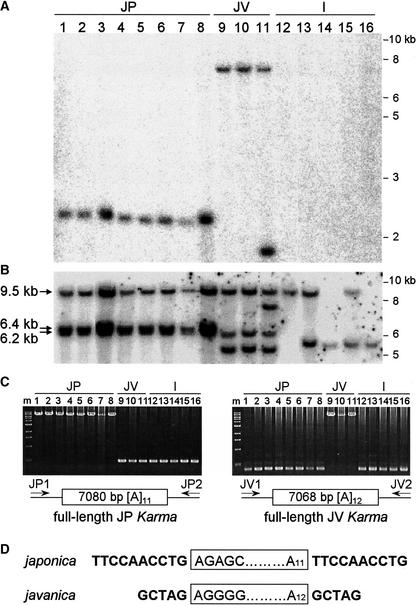
The number of Karma copies in the genome of japonica, javanica, and indica subspecies of rice was examined first by DNA gel blot analysis (Figures 3A and 3B). As indicated in Figure 1A, DNA was digested with EcoRV, and blots were hybridized with probes A and B from the 5′ and 3′ ends of Karma, respectively. In the eight japonica cultivars analyzed, only one fragment was hybridized by probe A (Figure 3A), whereas three fragments were detected by probe B (Figure 3B). All fragments were conserved in the eight cultivars, suggesting that japonica rice has one full-length and two 5′ truncated copies of Karma. In the five indica cultivars analyzed, no fragments hybridized to probe A, whereas one or two fragments were detected by probe B, suggesting that indica rice has only 5′ truncated Karma copies. Three cultivars of javanica, which also is a cultivated rice subspecies, were analyzed. One fragment detected by probe A and three fragments detected by probe B were conserved in the three javanica cultivars, suggesting that javanica rice also has one full-length and two 5′ truncated copies of Karma. An additional fragment was detected by both probes in one of the javanica cultivars.
Figure 3.
Karma Copies in japonica, javanica, and indica Cultivars.
Lanes 1 to 8, japonica (JP) cultivars Asominori, Kinmaze, Nipponbare, Notohikari, Shiokari, Taichung65, Toride, and Kinoshitamochi; lanes 9 to 11, javanica (JV) cultivars 242, 532, and 647; lanes 12 to 16, indica (I) cultivars IR24, 108, 419, C5444, and C8005. The number of Karma copies was examined by DNA gel blot analysis of leaf DNA digested with EcoRV.
(A) Fragments hybridized by probe A as indicated in Figure 1A.
(B) Fragments hybridized by probe B as indicated in Figure 1A. Arrows indicate the fragments conserved in the eight japonica cultivars and their respective sizes.
(C) PCR using primers flanking the full-length copies of Karma in japonica and javanica rice confirms that they are localized at different positions in the genome. Left gel, PCR using primers flanking the japonica full-length Karma copy (JP1 and JP2) results in products of 7562 bp in japonica cultivars and 471 bp in javanica and indica cultivars. Right gel, PCR using primers flanking the javanica full-length Karma copy (JV1 and JV2) results in products of 7454 bp in javanica cultivars and 374 bp in japonica and indica cultivars. The molecular mass markers (m) indicate, from top to bottom, 10, 8, 6, 5, 4, 3, 2, 1.5, 1, and 0.5 kb.
(D) Direct repeats flanking the full-length Karma copies of japonica and javanica.
The full-length Karma copies conserved in japonica and javanica cultivars separated into bands of different sizes in DNA gel blot analyses. Sequencing of their flanking regions and rough mapping using recombinant inbred lines confirmed that they are localized at different positions in the genome. The japonica full-length copy is 7080 bp long and is localized on chromosome 11, whereas the javanica full-length copy is 7068 bp long and is localized on chromosome 1. PCR amplifications demonstrating the divergence in japonica and javanica full-length copy locations are shown in Figure 3C. Nondisrupted PCR products also were sequenced and showed no deletions or truncations that could indicate genome rearrangements (data not shown). Both copies were flanked by direct repeats and were terminated by poly(A) tails of different lengths (Figure 3D).
With the publication of draft sequences for the japonica and indica rice genomes (Goff et al., 2002; Yu et al., 2002) and constant updates toward their completion, it was possible to assess the structures and locations of Karma copies present in both subspecies. BLAST (Basic Local Alignment Search Tool) searches in public databases revealed that the japonica cv Nipponbare indeed has only one full-length Karma copy (Table 1). Nine 5′ truncated copies with identities ranging from 66 to >99% were found. Although not all were flanked by target site duplications, the nine elements were terminated by poly(A) repeats. 5′ truncated copies with lengths within the hybridization range of probe B also were detected by DNA gel blot analysis in lower stringency conditions (data not shown). Meanwhile, BLAST searches of the indica cv 93-11 database revealed the presence of 5′ truncated copies only, some of which were highly identical to 5′ truncated copies found in the japonica database (Table 2).
Table 1.
Results of BLAST Searches of the japonica (cv Nipponbare) Genomic Database
| Hit (PAC) | Chromosome | cM | Length | Identity (%)a | Positionb | Target Site Duplicationc | |
|---|---|---|---|---|---|---|---|
| AC134924 | 11 | 8.6 | 7080 [A]11 | 100 | 17,912 | + | TTCCAACCTG/TTCCAACCTG |
| AC135926 | 5 | 54.6 | 5764 [A]11 | 99.6 | 83,058 | − | None |
| AC138454 | 11 | 7.5 | 2126 [A]7 | 99.5 | 153,303 | + | CTTGTAAAG/CTTGTTAAG |
| AP004309 | 7 | 94.7 | 2536 [A]14 | 81 | 82,517 | + | None |
| BX000506 | 12 | 9.7 | 2822 [A]9 | 97 | 57,254 | + | None |
| AP005288 | 2 | 11.6 | 66 [A]6 | 92 | 31,725 | + | TTTATATGGGTTCT/TTTATATGGGTTCT |
| AC120539 | 11 | 19.8 | 577 [A]7 | 66 | 163,784 | + | None |
| AC135429 | 5 | 80.7 | 691 [A]10 | 69 | 106,341 | + | TTACTC/TTACTC |
| AL607005 | 4 | 52.6 | 106 [A]11 | 76 | 74,011 | + | CAAAT/CAAAT |
| AP005776 | 2 | 10.8 | 187 [A]8 | 70 | 106,179 | + | TAGG/TAGG |
cM, centimorgan; PAC, P1 artificial chromosome.
Percentage of identity to the full-length Karma copy isolated from japonica.
Position of the first nucleotide and direction within the PAC clone; +, upper strand; −, lower strand.
5′ flanking/3′ flanking duplications. Different bases within the duplications are underlined.
Table 2.
Results of BLAST Searches of the indica (cv 93-11) Genomic Database
| Hit (Contig) | Length | Identity (%)a |
Positionb | Target Site Duplication | Similar japonica Copy |
Identity (%)c |
|
|---|---|---|---|---|---|---|---|
| 1,041 | 5063 [A]12 | 99 | 6,488 | + | None | ||
| 144 | 2536 [A]13 | 81 | 13,682 | + | None | AP004309 | 99.6 |
| 23,198 | 4121 | 62 | 3,959 | − | None | ||
| 15,747 | 2336 | 68 | 2,560 | − | None | ||
| 5,471 | 2825 [A]9 | 67 | 10,621 | − | None | BX000506 | 98.8 |
| 98,735 | 52 [A]6 | 94 | 342 | − | TTTATATGGGTTCT/TTTATATGGGTTCT | AP005288 | 100 |
| 19,342 | 264 [A]7 | 70 | 4,506 | + | None | AC120539 | 99.6 |
| 9,831 | 123 [A]11 | 76 | 5,668 | − | CAAAT/CAAAT | AL607005 | 99.0 |
Percentage of identity to the full-length Karma copy isolated from japonica.
Position of the first nucleotide and direction within the contig; +, upper strand; −, lower strand.
Percentage of identity between the indica and japonica elements.
Karma Distribution Varies in Oryza Species
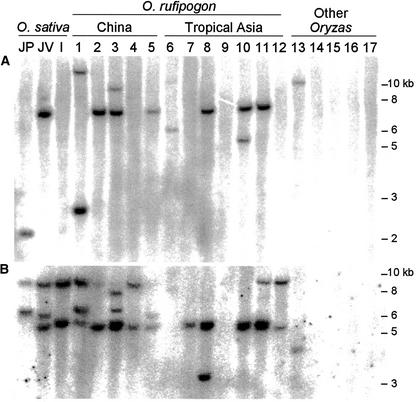
The number and distribution of Karma copies also diverged in accessions of Oryza rufipogon (Figure 4), the proposed wild ancestor of rice (Wang et al., 1992; Khush, 1997; Bautista et al., 2001), and other Oryza species. This divergence suggests that Karma transposition may have been activated occasionally during the course of rice domestication. However, the possibility that the difference in band sizes results from genome rearrangements cannot be excluded. A conserved pattern of distribution was not observed among O. rufipogon accessions from China or tropical Asia, indicating that the divergence in Karma copy distribution occurred before the domestication of rice. Accordingly, hybridization also was observed in O. longistaminata but not in other Oryza species besides O. rufipogon (Figure 4).
Figure 4.
Distribution of Karma Copies in Oryza Accessions.
The number of Karma copies was examined by DNA gel blot analysis using EcoRV. Lane JP, japonica cv Notohikari; lane JV, javanica cv 242; lane I, indica cv IR24; lanes 1 to 12, O. rufipogon accessions W1956, W1962, W1964, W1965, W1967, W108, W120, W149, W593, W630, W1972, and W1976, respectively; lanes 13 to 17, O. longistaminata, O. glumaepatula, O. meridionalis, O. glaberrima, and O. barthii, respectively. O. rufipogon accessions from China or tropical Asia are indicated.
(A) Fragments hybridized by probe A as indicated in Figure 1A.
(B) Fragments hybridized by probe B as indicated in Figure 1A.
Karma Retrotransposes in Regenerated Plants but Not in Cultured Cells
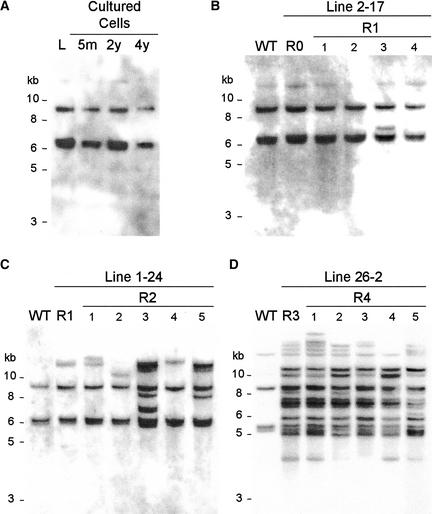
Because retrotransposons move in a “copy-and-paste” manner, an increase in copy number indicates the occurrence of retrotransposition. No increase in copy number was detected in 54 independent lineages of cells cultured for 5 months to 4 years (Table 3, Figure 5A), suggesting that Karma did not retrotranspose in cultured cells. Karma retrotransposition also was not detected in leaves of 13 independent R0 seedlings, which were regenerated from cultured cells. However, an increase in Karma copy numbers was observed in plants of R1 lineages (Table 3, Figure 5B). Most interestingly, we detected additional copy number increases in regenerated plants of consecutive generations up to R6, which was the most advanced generation analyzed (Table 3). Although we did not analyze the same lineage for more than two subsequent generations, the transmission of new insertions of the parental plant to the progeny was observed, as shown in Figures 5C and 5D. Lineages in which the copy number did not increase in the R1 generation but increased in R2 also were observed (data not shown).
Table 3.
Insertion Frequency of Active Karma Elements
| Generation | Number of Plants Testeda |
Number of New Insertions |
New Insertions per Plant |
|---|---|---|---|
| R0 (callus) | 54 | 0 | 0 |
| R0 (plants) | 13 | 0 | 0 |
| R1 | 15 | 4 | 0.27 |
| R2 | 16 | 5 | 0.31 |
| R3 | 15 | 10 | 0.67 |
| R4 | 23 | 18 | 0.78 |
| R5 | 5 | 2 | 0.40 |
| R6 | 9 | 2 | 0.22 |
Sum of plants from different lineages.
Figure 5.
Karma Transposition in Cultured Cells and Regenerated Plants.
The number of Karma copies was examined by DNA gel blot analysis using EcoRV and probe B as indicated in Figure 1A.
(A) DNA extracted from wild-type plant leaves (L) and cells cultured for 5 months (5m), 2 years (2y), and 4 years (4y).
(B) DNA extracted from leaves of a wild-type (WT) plant and line 2-17 plants, which were regenerated from cultured cells (R0) and four plants of the subsequent generation (R1).
(C) DNA extracted from leaves of a wild-type plant, an R1 regenerated plant of line 1-24, and five plants of its progeny (R2).
(D) DNA extracted from leaves of a wild-type plant, an R3 regenerated plant of line 26-2, and five plants of its progeny (R4).
To determine if the new copies actually resulted from retrotransposition and not from DNA rearrangements, the sequences flanking five new copies were recovered using inverse PCR. As shown in Table 4, the five new copies had short or long 5′ deletions, were terminated by poly(A) tails of variable lengths, and were flanked by direct repeats of different lengths, indicating the formation of target-site duplications. These three features have been reported extensively for mammalian L1 element retrotransposition (Luan et al., 1993; Ostertag and Kazazian, 2001) and indicate that the new copies of Karma indeed resulted from retrotranspositional events.
Table 4.
Analysis of New Karma Insertions
| Designation | Insertion Sitea | Length (bp) | Target Site Duplication |
|---|---|---|---|
| Karma-1 | Chromosome 7, 105.7 to 115.5 cM (AP004570) | 7074 [A]8 | AGATGTCTAGCT/AGATGTCTAGCT |
| Karma-2 | Chromosome 2, 114 to 118.1 cM (AP004255) | 7072 [A]11 | CATCTATGGAAA/CATCTATGGAAA |
| Karma-3 | Chromosome 11, 8.6 cM (AC123526) | 4347 [A]10 | CAAACTGGACAG/CAAACTGGACAG |
| Karma-4 | Chromosome 3, 31 cM (AC104473) | 7063 [A]3 | AAAAATAATTTTT/AAAAATAATTTTT |
| Karma-5 | Chromosome 1, 143.7 cM (AC123526) | 4051 [A]15 | CAAT/CAAT |
Inferred by the position of the P1 artificial chromosome clone (in parentheses) corresponding to the region of insertion. cM, centimorgan.
Karma Transcription Is Insufficient for Transposition
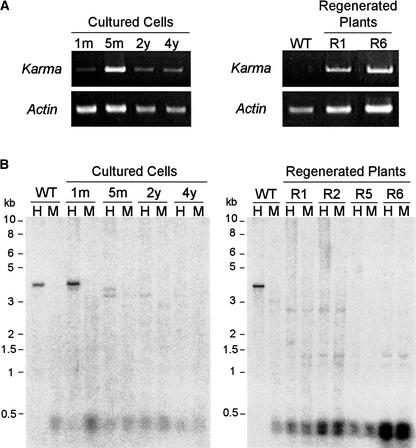
The transcription of Karma in regenerated plants and cultured cells was analyzed by RT-PCR (Figure 6A). Whereas Karma was not transcribed in leaves of normal plants, transcription was detected in leaves of regenerated plants. Transcription also was observed in cells cultured for 1 month to 4 years. The fact that Karma retrotransposition was not observed even though it was transcribed suggests that transcription is insufficient for the retrotransposition of Karma. This also was the case for R0 plants, in which transcription, but not retrotransposition, was detected (data not shown).
Figure 6.
Karma Transcription and Hypomethylation in Cultured Cells and Regenerated Plants.
(A) RT-PCR of poly(A) mRNA extracted from cells cultured for 1 month (1m), 5 months (5m), 2 years (2y), and 4 years (4y), wild-type leaves (WT), and leaves of line 2-17 R1 regenerated plants and line 26-6 R6 regenerated plants. Top gels depict Karma expression, and bottom gels depict actin expression as a loading control.
(B) DNA gel blots of cultured cells and leaf DNA treated with HpaII (H) or MspI (M) and hybridized with probe A (Figure 1A) were used to depict the methylation status of the 5′ region of the full-length Karma copy in wild-type plants, cultured cells, and regenerated plants. Cultured cells of 1 month (1m), 5 months (5m), 2 years (y), and 4 years (4y) and leaf DNA of lines 2-17 (R1 and R2) and 26-6 (R5 and R6) were used.
Karma Is Hypomethylated in Cultured Cells and Regenerated Plants
Karma methylation levels were examined by DNA gel blot analysis using the methylation-sensitive HpaII and the methylation-insensitive MspI restriction enzymes. HpaII does not cleave methylated CCGG sequences, whereas its isoschizomer, MspI, cleaves C5mCGG but not 5mCCGG sequences. Correlating with transcription, Karma methylation levels in cultured cells and regenerated plants were lower than those in wild-type plants (Figure 6B). In wild-type plants, the fragment of Karma hybridized by probe A was not digested with HpaII, indicating that Karma is methylated under normal conditions. A slight decrease in methylation levels was observed in cells cultured for 1 month. In cells cultured for 5 months, 2 years, and 4 years, methylation levels decreased considerably. Hypomethylation of Karma also was observed in lineages of regenerated plants from different generations, indicating that Karma was not subject to de novo methylation in them.
DISCUSSION
Karma Retrotransposition Continues through Several Generations
Although LINE elements are found in the genomes of several plant species, evidence of recent retrotransposition was not observed previously. We demonstrated that Karma transcription is activated by tissue culture and that Karma retrotransposes continuously in regenerated plants. Unlike plant retrotransposons reported to date, the transcription of Karma is not silenced after activation, and hypomethylated states persist through several generations. Although the precise mechanisms of the epigenetic regulation of transposons in general remain unclear, mobilization often is associated with hypomethylation and transcriptional activation (Hirochika et al., 2000; Lindroth et al., 2001; Tompa et al., 2002). Recently, it was demonstrated that short interfering RNAs derived from retroelements are found in wild-type Arabidopsis and tobacco plants and that long short interfering RNAs are correlated specifically with retroelement DNA methylation but not with mRNA degradation (Hamilton et al., 2002; Llave et al., 2002; Zilberman et al., 2003). We observed an inverse correlation between Karma methylation and mRNA levels. However, whether Karma methylation prevents transcription or RNA degradation sets Karma methylation in wild-type plants, and the specific effect of tissue culture in these processes, remain to be determined.
The persistence of Karma hypomethylation through generations may result from the low activity of plant de novo methyltransferases (Vongs et al., 1993; Kakutani et al., 1999). Alternatively, it may be a consequence of the small number of additional Karma copies present in early generations of regenerated plants. The increase in copy number correlates positively with both the restoration of DNA methylation levels and the inactivation of the Tto1 LTR retrotransposon already in the R0 generation (Hirochika et al., 2000). Also, a decrease in transcription of the Drosophila I LINE element was associated with increasing copy numbers, indicating the occurrence of repeat-induced gene silencing (Chaboissier et al., 1998). Therefore, the possibility that the greater accumulation of new insertions through the generations will eventually lead to the inactivation of Karma cannot be excluded.
Regulation of Karma Retrotransposition
The general steps of retrotransposition include transcription, RNA processing, mRNA export, translation, post-translational modifications, entry into the nucleus, reverse transcription, and integration (reviewed by Grandbastien, 1998; Ostertag and Kazazian, 2001). Any of these steps can limit the activity of retrotransposons. The developmental and environmental regulation of the transcription of plant LTR retrotransposons has been studied extensively and involves the alternating use of different cis-regulatory sequences present in the LTRs (Suoniemi et al., 1996; Casacuberta et al., 1997; Grandbastien et al., 1997; Takeda et al., 1999). Although the mechanisms of LINE retrotransposition in plants are unknown, studies in mammals demonstrated that an internal promoter localized in the 5′ untranslated region is required for the in vitro transcription of human L1 elements (Swergold, 1990) and suggested that the expression of mouse L1 elements is germ line specific (Branciforte and Martin, 1994; Trelogan and Martin, 1995).
In the case of Karma, we showed that transcripts were not detected in leaves of wild-type plants but were observed in cultured cells and leaves of regenerated plants. Therefore, although we did not analyze the transcription of Karma in other tissues of wild-type plants, the suppression of transcription or the degradation of transcripts might be the first stage in the regulation of Karma retrotransposition. However, transcription is required but is insufficient for the retrotransposition of Karma, because new copies were not detected in cultured cells or in the R0 generation of regenerated plants despite the fact that transcripts were observed. Therefore, post-transcriptional regulation may prevent additional steps of Karma retrotransposition. Evidence of transcription but no recent integration has been reported for some LTR and SINE retrotransposons (Bi and Laten, 1996; Deragon et al., 1996; Turcich et al., 1996; Meyers et al., 2001), suggesting that the regulation of post-transcriptional steps prevents the complete retrotransposition of these elements. The fact that an increase in Karma copy number was observed in regenerated plants of the R1 and later generations indicates that Karma retrotransposition also is regulated developmentally. Tight control of retrotransposition is necessary to prevent an indiscriminate increase in copy number that could be deleterious to the host. The restriction of post-transcriptional steps to certain stages of the host development could be such a control. Although the precise mechanisms for the development-dependent regulation of Karma remain elusive, the two-step regulation may have evolved to minimize defects to hosts and to ensure the effective transmission of retrotransposed copies to offspring.
METHODS
Plant Materials and Cell Culture
Regenerated rice (Oryza sativa) plants were obtained from 18 independent transgenic callus lineages that were transformed and cultivated as described previously (Izawa et al., 1991; Hiei et al., 1994; Nakagawa et al., 2002).
DNA Extraction and DNA Gel Blot Analysis
Extraction of total DNA from calli and mature leaves, blotting, preparation of probes, and hybridization were performed as described previously (Enoki et al., 1999). Probes were amplified by PCR using the following primers: for probe A, 5′-TTGGTGAATAGGGAAACGTGG-3′ and 5′-CGCATTCTCACTAACCTCCATG-3′; for probe B, 5′-CAGGCCATCCAAAGCAAGG-3′ and 5′-CGCATTCTCACTAACCTCCATG-3′.
Characterization of Karma Copies by Inverse PCR
Two micrograms of genomic DNA extracted from leaves was digested with EcoRV and self-ligated using the DNA Ligation Kit Version 2 (Takara Bio, Otsu, Japan).
Ligated DNA was subjected to PCR using the following primer pairs: 5′-CGCATTCTCACTAACCTCCATG-3′ and 5′-GATGTGATTGCCATGTTGGAG-3′ (full-length 5′ flanks); 5′-AGGAGATTGTCAGCGAGAAGTG-3′ and 5′-GATGTGATTGCCATGTTGGAG-3′ (5′ truncated 5′ flanks); and 5′-CGGACAGCATAGTGTTGTGTTG-3′ and 5′-GAATGTTGTTGTGGTTTGCAATG-3′ (3′ flanks). The full-length Karma copies of japonica and javanica cultivars were amplified using primers JP1 and JP2 (5′-TAGCTCCGAAAGCAACTACAGAG-3′ and 5′-CAGGCAGGAACTGAGGAAAG-3′, respectively) and JV1 and JV2 (5′-TGAGAAGGCCTTCTTCCTTTG-3′ and 5′-TACGAGTATGCAGATGGCCC-3′, respectively). PCR samples of 20 μL contained 0.5 units of ExTaq DNA polymerase (Takara Bio), 1× PCR reaction buffer (Takara Bio), 0.2 mM of each deoxynucleotide triphosphate, 10 μM of each primer, 4% DMSO, and ∼30 ng of genomic DNA. PCR conditions included an initial step at 94°C for 2 min, 30 cycles of amplification (30 s at 94°C, 30 s at 60°C, and 2 min at 72°C), and a final step at 72°C for 5 min.
DNA Sequence Analysis
PCR products were sequenced directly with the Big Dye Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems, Foster City, CA) and analyzed with an Applied Biosystems Prism 3100 Genetic Analyzer.
Mapping and Basic Local Alignment Search Tool Analysis
Rough mapping of the japonica and javanica full-length Karma loci was performed using cleaved amplified polymorphic sequence markers, which were constructed based on sequence information provided by the National Institute of Agrobiological Sciences (Tsukuba, Japan). BLAST (Basic Local Alignment Search Tool) searches (Altschul et al., 1990) were performed on National Institute of Agrobiological Sciences and University of Tokyo servers using the National Center for Biotechnology Information, Rice Genome Research Program, and Beijing Genomics Institute BLAST services. Chromosomal locations of the P1 artificial chromosome clones were obtained from the servers of International Rice Genome Sequencing Project members. Phylogenetic analyses were performed using CLUSTAL W (Thompson et al., 1994) from the Pasteur Institute suite of online programs. Pairwise alignment was performed using the LFasta local alignment tool from the Pole Bio-Informatique Lyonnais World Wide Web server.
RNA Isolation and Reverse Transcription PCR Analysis
Total RNA was isolated using the guanidinium method (Chomczynski and Sacchi, 1987). Poly(A)+ mRNA was obtained using the Oligotex-Mag mRNA Purification Kit (Takara Bio) according to the manufacturer's instructions. For reverse transcription PCR analysis, 1 μg of poly(A)+ mRNA was reverse transcribed using RAV2 reverse transcriptase (Amersham Pharmacia Biotech, Buckinghamshire, UK) according to the manufacturer's instructions. The product of the first-strand cDNA synthesis reaction was amplified by PCR using the same primers that were used for probe B of the DNA blots. Primers 5′-CAATCGTGAGAAGATGACCC-3′ and 5′-GTCCATCAGGAAGCTCGTAGC-3′ were used to amplify actin cDNA as a loading control.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact J. Kyozuka, akyozuka@mail.ecc.u-tokyo.ac.jp.
Accession Numbers
The accession numbers for the sequences mentioned in this article are as follows: rice Karma, AB81316; maize cin4, Y00086; lily del2, Z17425; Arabidopsis Ta11-1, L47193; Cannabis LINE-CS, AB013908; human LINE-1, M80340; frog Tx1, M26915; fruit fly R2Dm, X519667; fruit fly I, M14954; mouse L1Md, AAA66024; green alga ZEPP, D899938; trypanosome CRE2, U19151; mosquito Juan-A, U87543; silkworm BMC1, AB018558; fruit fly TART-1, U14101; fruit fly JockeyDm, M22874; fruit fly JockeyDf, M38437; chicken CR1, U88211; mosquito Q, U03849; trypanosome L1Tc, X83098; pufferfish Maui, AF086712; rice blast fungus MGL, AF018033; fungi Tad1-1, L25662; mosquito Lan, U87543; silkworm R1Bm, M19755; and fruit fly R1Dm, X51968.
Acknowledgments
We are grateful to T. Ishii for providing DNA samples and seeds of rice cultivars and accessions and to N. Okada for assistance with the phylogenetic analysis. We thank J. Finnegan for critical reading of the manuscript and M. Nobuhara and Y. Satake for technical assistance. M.K. was supported by a Japan Society for the Promotion of Science Research Fellowship for Young Scientists.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.011809.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bautista, N.S., Solis, R., Kamijima, O., and Ishii, T. (2001). RAPD, RFLP and SSLP analyses of phylogenetic relationships between cultivated and wild species of rice. Genes Genet. Syst. 76, 71–79. [DOI] [PubMed] [Google Scholar]
- Bi, Y.-A., and Laten, H.M. (1996). Sequence analysis of a cDNA containing the gag and prot regions of the retrovirus-like element, SIRE-1. Plant Mol. Biol. 30, 1315–1319. [DOI] [PubMed] [Google Scholar]
- Branciforte, D., and Martin, S.L. (1994). Developmental and cell type specificity of LINE-1 expression in mouse testis: Implications for transposition. Mol. Cell. Biol. 14, 2584–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta, J.M., Vernhettes, S., Audeon, C., and Grandbastien, M.A. (1997). Quasispecies in retrotransposons: A role for sequence variability in Tnt1 evolution. Genetica 100, 109–117. [PubMed] [Google Scholar]
- Chaboissier, M.C., Bucheton, A., and Finnegan, D.J. (1998). Copy number control of a transposable element, the I factor, a LINE-like element in Drosophila. Proc. Natl. Acad. Sci. USA 95, 11781–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Deragon, J.M., Gilbert, N., Rouquet, L., Lenoir, A., Arnaud, P., and Picard, G. (1996). A transcriptional analysis of the S1Bn (Brassica napus) family of SINE retroposons. Plant Mol. Biol. 32, 868–878. [DOI] [PubMed] [Google Scholar]
- Dombroski, B.A., Mathias, S.L., Nanthakumar, E., Scott, A.F., and Kazazian, H.H., Jr. (1991). Isolation of an active human transposable element. Science 254, 1805–1808. [DOI] [PubMed] [Google Scholar]
- Enoki, H., Izawa, T., Kawahara, M., Komatsu, M., Koh, S., Kyozuka, J., and Shimamoto, K. (1999). Ac as a tool for the functional genomics of rice. Plant J. 19, 605–613. [DOI] [PubMed] [Google Scholar]
- Feschotte, C., Jiang, N., and Wessler, S.R. (2002). Plant transposable elements: Where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. [DOI] [PubMed] [Google Scholar]
- Grandbastien, M.A. (1998). Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3, 181–187. [Google Scholar]
- Grandbastien, M.A., Lucas, H., Morel, J.-B., Mhiri, C., Vernhettes, S., and Casacuberta, J.M. (1997). The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100, 241–252. [PubMed] [Google Scholar]
- Grandbastien, M.A., Spielmann, A., and Caboche, M. (1989). Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337, 376–380. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Okamoto, H., and Kakutani, T. (2000). Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika, H., Sugimoto, K., Otsuki, Y., Tsugawa, H., and Kanda, M. (1996). Retrotransposons of rice involved in mutations induced by tissue culture. Proc. Natl. Acad. Sci. USA 93, 7783–7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Miyazaki, C., Yamamoto, M., Terada, R., Iida, S., and Shimamoto, K. (1991). Introduction and transposition of the maize transposable element Ac in rice (Oryza sativa L.). Mol. Gen. Genet. 227, 391–396. [DOI] [PubMed] [Google Scholar]
- Johns, M.A., Mottinger, J., and Freeling, M. (1985). A low copy number, copia-like transposon in maize. EMBO J. 4, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa, M., and Okada, N. (2002). LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111, 433–444. [DOI] [PubMed] [Google Scholar]
- Kakutani, T., Munakata, K., Richards, E.J., and Hirochika, H. (1999). Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush, G.S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. [PubMed] [Google Scholar]
- Kimura, H. (1983). The Neural Theory of Molecular Evolution. (Cambridge, UK: Cambridge University Press).
- Komatsu, M., Chujo, A., Shimamoto, K., and Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development, in press. [DOI] [PubMed]
- Komatsu, M., Maekawa, M., Shimamoto, K., and Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. [DOI] [PubMed] [Google Scholar]
- Kumar, A., and Bennetzen, J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33, 479–532. [DOI] [PubMed] [Google Scholar]
- Le, Q.H., Wright, S., Yu, Z., and Bureau, T. (2000). Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 7376–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., and Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, D.D., Padgett, R.W., Hardies, S.C., Shehee, W.R., Comer, M.B., Edgell, M.H., and Hutchison, C.A., III (1986). The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol. Cell. Biol. 6, 168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, D.D., Korman, M.H., Jakubczak, J.L., and Eickbush, T.H. (1993). Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 72, 595–605. [DOI] [PubMed] [Google Scholar]
- Lucas, H., Feuerbach, F., Kunert, K., Grandbastien, M.A., and Caboche, M. (1995). RNA-mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana. EMBO J. 14, 2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melayah, D., Bonnivard, E., Chalhoub, B., Audeon, C., and Grandbastien, M.A. (2001). The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 28, 159–168. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, M., Shimamoto, K., and Kyozuka, J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. [DOI] [PubMed] [Google Scholar]
- Noma, K., Ohtsubo, E., and Ohtsubo, H. (1999). Non-LTR retrotransposons (LINEs) as ubiquitous components of plant genomes. Mol. Gen. Genet. 261, 71–79. [DOI] [PubMed] [Google Scholar]
- Ostertag, E.M., and Kazazian, H.H., Jr. (2001). Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538. [DOI] [PubMed] [Google Scholar]
- Prak, E.T., and Kazazian, H.H., Jr. (2000). Mobile elements and the human genome. Nat. Rev. Genet. 1, 134–144. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Leclercq, L., Gobel, E., and Saedler, H. (1997). Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 9, 3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suoniemi, A., Narvanto, A., and Schulman, A.H. (1996). The BARE-1 retrotransposon is transcribed in barley from an LTR promoter active in transient assays. Plant Mol. Biol. 31, 295–306. [DOI] [PubMed] [Google Scholar]
- Swergold, G.D. (1990). Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10, 6718–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Sugimoto, K., Otsuki, H., and Hirochika, H. (1999). A 13-bp cis-regulatory element in the LTR-promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18, 383–393. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, R., McCallum, C.M., Delrow, J., Henikoff, J.G., van Steensel, B., and Henikoff, S. (2002). Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 12, 65–68. [DOI] [PubMed] [Google Scholar]
- Trelogan, S., and Martin, S.L. (1995). Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 92, 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcich, M.P., Bokhari-Riza, A., Hamilton, D.A., He, C., Messier, W., Stewart, C.-B., and Mascarenhas, J.P. (1996). PREM-2, a copia-type retroelement in maize is expressed preferentially in early microspores. Sex. Plant Reprod. 9, 65–74. [Google Scholar]
- Turcotte, K., Srinivasan, S., and Bureau, T. (2001). Survey of transposable elements from rice genomic sequences. Plant J. 25, 169–179. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J., Purugganan, M., and Wessler, S.R. (1992). Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs, A., Kakutani, T., Martienssen, R.A., and Richards, E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Second, G., and Tanksley, S.D. (1992). Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor. Appl. Genet. 83, 565–581. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., Cao, X., and Jacobsen, S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719. [DOI] [PubMed] [Google Scholar]