Abstract
Disruption of specific components of the host cytoskeleton has been reported for several viruses and is thought to be beneficial for viral replication and spread. Our previous work demonstrated that infection of swine kidney (SK-6) cells with pseudorabies virus (PRV), a swine alphaherpesvirus, induced actin stress fiber breakdown. In the present study, using several PRV deletion mutants, we found that the US3 serine/threonine (S/T) protein kinase is involved in breakdown of actin stress fibers in different PRV-infected cell lines. Further, by transfection assays, we showed that PRV US3 itself, in the absence of other viral proteins, is able to trigger actin stress fiber breakdown when it is localized in sufficient amounts in the nucleus.
Viruses invade eukaryotic cells and complete their replication cycle by utilizing the cellular machinery of their host. Disassembly of specific components of the host cell cytoskeleton has been suggested to be important for efficient replication and spread of several viruses (19, 20, 29, 34). However, the viral proteins responsible for these phenomena remain largely unknown.
Different alphaherpesviruses can destroy cytoskeletal elements on infection of cultured cells. Almost two decades ago, Bedows et al. demonstrated disruption of actin filaments in herpes simplex virus type 1 (HSV-1)-infected Vero cells (4). More recent studies by Avitabile et al. suggested that HSV-1 infection does not induce alterations within the actin cytoskeleton of Vero or HEp-2 cells but, rather, causes disassembly of the microtubule network, which may be necessary for fragmentation and dispersal of the Golgi apparatus (2). Cultured Vero cells inoculated with equine herpesvirus 1, another alphaherpesvirus, also showed depolymerization of microtubule bundles (35).
Recently, we have shown that pseudorabies virus (PRV), a swine alphaherpesvirus, induces actin stress fiber disassembly in cultured swine kidney (SK-6) cells, from 6 h postinoculation (p.i.) onward, without altering the microtubule network (31). To date, the PRV protein(s) responsible for actin breakdown has not been identified. At least two hypotheses can be put forward on how PRV disrupts actin filaments upon infection. First, the PRV virion host shutoff (vhs) protein may be involved. PRV vhs has significant amino acid sequence similarity to its HSV-1 and bovine herpesvirus 1 (BHV-1) counterparts, and, although the HSV-1 and BHV-1 vhs proteins have not been reported to induce actin stress fiber disruption, they both are capable of downregulating several mRNAs, including actin mRNA (5, 13, 17). Second, viral serine/threonine (S/T) protein kinases may be involved in actin stress fiber breakdown. Viral S/T kinases have significant homology to cellular S/T kinases, some of which make part of actin stress fiber-controlling signaling pathways (28). The PRV genome encodes three putative S/T protein kinases: US3, UL13, and the large subunit of the ribonucleotide reductase (RR) protein, which are conserved among the alphaherpesviruses (10, 12, 32, 33, 38). Interestingly, the US3 and the RR large subunit orthologues in HSV-2 act on actin-controlling signaling pathways (Cdc42/Rac and Ras/MEK/MAPK pathway, respectively) (25, 30), and, for HSV-2 US3, transfection studies with HEp-2 cells have shown that this S/T protein kinase on its own can induce actin stress fiber breakdown (25).
To test whether the PRV vhs protein or any of the three putative PRV S/T protein kinases are involved in PRV-induced disruption of filamentous actin, we investigated the actin stress fiber organization in mock-infected and NIA3 wild-type-, NIA3 vhs null-, NIA3 US3 null-, NIA3 UL13 null-, and NIA3 RR null-infected SK-6 cells at 8 h p.i. at a multiplicity of infection (MOI) of 10. All virus strains were described previously (3, 9, 10, 11, 32).
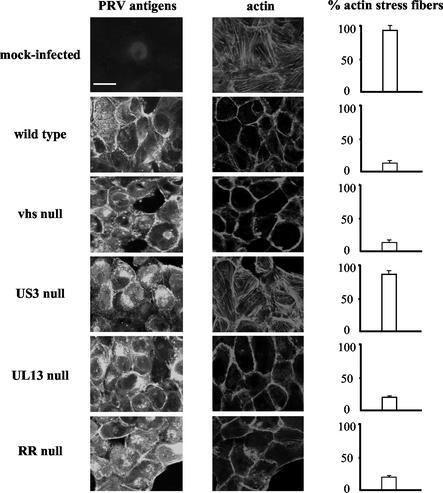
At 8 h p.i., the cells were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. Subsequently, PRV proteins and actin filaments were stained by incubating the cells for 1 h at 37°C with fluorescein isothiocyanate (FITC)-conjugated polyclonal PRV-specific antibodies (26) and phalloidin-Texas Red (Molecular Probes, Eugene, Oreg.), respectively. Finally, stained SK-6 cells were mounted and analyzed by confocal microscopy (TCS SP2 laser-scanning spectral confocal system [Leica Microsystems, Heidelberg, Germany]). Figure 1 shows that approximately 89.0% ± 7.6% of mock-infected SK-6 cells possessed intact actin stress fibers while only 13.5% ± 3.1% of NIA3 wild-type-infected SK-6 cells retained intact actin stress fibers. Furthermore, actin stress fiber disassembly observed in SK-6 cells inoculated with the NIA3 vhs null strain (14.2% ± 1.6% of the cells retained fibers), the NIA3 UL13 null strain (17.3% ± 1.5% of the cells retained fibers) or the NIA3 RR null strain (16.2% ± 2.3% of the cells retained fibers) did not differ substantially from the actin stress fiber breakdown observed in NIA3 wild-type-infected SK-6 cells. In contrast, 80.2% ± 4.8% of SK-6 cells inoculated with an NIA3 US3 null strain still possessed intact actin stress fibers at 8 h p.i. (Fig. 1). These data indicate that the PRV US3 protein, but not the vhs, UL13, or RR protein, is involved in breakdown of actin stress fibers in PRV-infected SK-6 cells.
FIG. 1.
Actin architecture of SK-6 cells infected with different PRV strains. Confluent monolayers of SK-6 cells were mock infected or infected at an MOI of 10 with an NIA3 wild-type, NIA3 vhs null, NIA3 US3 null, NIA3 UL13 null, or NIA3 RR null strain. At 8 h p.i., cells were paraformaldehyde fixed, permeabilized, and stained with FITC-conjugated polyclonal PRV-specific antibodies and phalloidin-Texas Red to visualize PRV antigens (left panels) and actin filaments (middle panels), respectively. The right panels show the percentage of cells with intact actin stress fibers (200 cells were scored). Bar, 10 μm.
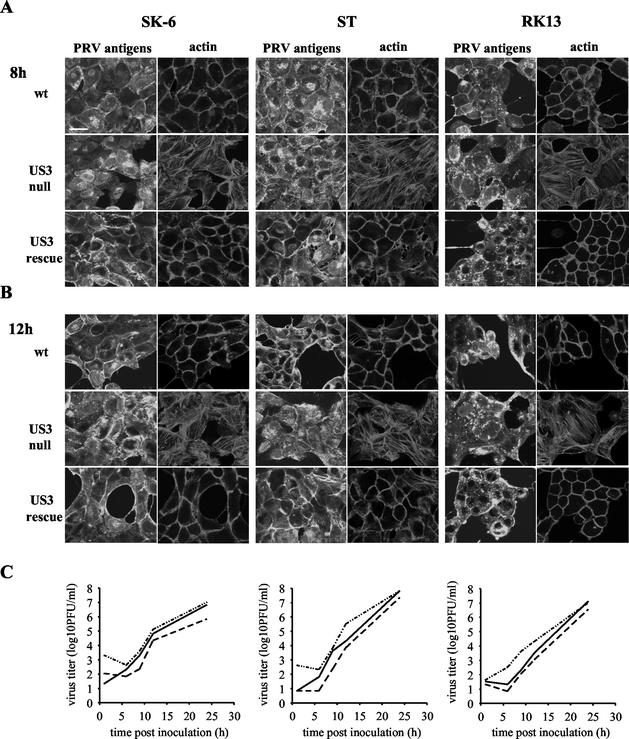
PRV US3 null virus was constructed by insertion of a 20-mer oligonucleotide, containing TAG translational stop codons in all reading frames, in the 5′ part of the US3 open reading frame (ORF) (9, 32). This NIA3 wild-type-derived mutant strain displays cell type-dependent growth retardation (22). To investigate whether the lack of actin stress fiber breakdown in PRV US3 null-infected cells may be (i) cell type dependent, and/or (ii) due to slower growth of the PRV US3 null virus, we examined the actin architecture and the extracellular virus titers in different cell types at different time points p.i. To this end, monolayers of SK-6 cells as well as monolayers of another swine cell line, swine testicle (ST) cells, and a rabbit cell line, rabbit kidney (RK13) cells, were inoculated (at an MOI of 10) with the NIA3 wild-type, NIA3 US3 null, and NIA3 US3 rescue (21) strains. Extracellular virus titers were determined at different time points (1, 6, 9, 12, and 24 h p.i.). Figure 2C shows that the PRV US3 null virus displays a 2- to 3-h delay in growth kinetics in all three cell lines tested. To determine the actin stress fiber architecture, cells were paraformaldehyde fixed at 8 or 12 h p.i., permeabilized, stained for both PRV viral antigens and actin filaments, and analyzed by confocal microscopy. Figure 2 shows that US3-mediated actin stress fiber disassembly occurs in all three cell types tested and that the 2- to 3-h delay in growth kinetics of the US3 null virus is not responsible for the observed differences in actin stress fiber disassembly, since, for all three cell types, actin stress fiber breakdown was obvious at 8 h as well as 12 h p.i. in PRV wild-type- and PRV US3 rescue strain-infected cells but not in PRV US3 null strain-infected cells.
FIG. 2.
(A and B) Actin architecture of NIA3 wild-type-strain (wt)-, NIA3 US3 null strain-, or NIA3 US3 rescue strain-infected SK-6, ST, and RK13 cells at 8 h p.i. (A) and 12 h p.i. (B). At the indicated time points p.i., SK-6, ST, and RK13 cells were paraformaldehyde fixed, permeabilized, and stained with FITC-conjugated polyclonal PRV-specific antibodies and phalloidin-Texas Red to visualize PRV antigens and actin filaments, respectively. Bar, 10 μm. (C) One-step growth curves for NIA3 wild-type (solid line), NIA3 US3 null (dashed line), and NIA3 US3 rescue (dot-dash line) viruses in SK-6, ST, and RK13 cells.
Further, we used Western blot analysis to investigate whether PRV US3 affects only actin architecture or also the total amount of cellular actin. We found that the total amounts of cellular actin (42 kDa) were similar in PRV wild type- and PRV US3 null strain-infected SK-6 cells (data not shown).
In PRV-infected cells, as a result of alternative splicing and the presence of multiple translation initiation codons (ATG codons), the US3 ORF is transcribed and translated into a US3 long isoform (less than 5% of the US3 protein) of 53 kDa and an abundant US3 short isoform (greater than 95% of the US3 protein) of 41 kDa (32). The two US3 isoforms share S/T protein kinase domains but have a divergent N terminus. To investigate whether one or both of the PRV US3 S/T protein kinase isoforms on their own (in the absence of other PRV proteins) are able to trigger stress fiber breakdown, we performed transfection assays. To do this, two US3 expression vectors, one encoding the US3 long isoform (pBud/CAT/US3long) and one encoding the US3 short isoform (pBud/CAT/US3short), were constructed as follows. First, the sequence encoding the US3 long isoform was PCR amplified with Pfx-Platinum polymerase (Invitrogen, Groningen, The Netherlands) by use of the forward primer 5′CACCTGTGGGCCAGCGCGTAGTA3′ and the reverse primer 5′CACTTCATTGTTGAGCTGTGGAGAT3′, in the presence of 4% dimethyl sulfoxide. The PCR amplification consisted of an initial 10-min denaturation step at 95°C, followed by 35 cycles of denaturation (95°C for 45 s), annealing (59°C for 45 s), and extension (73°C for 1 min 30 s), and then by one final 10-min extension step. The PCR-amplified US3 long isoform-encoding sequence (1,366 bp) was cloned into pBudCE4.1/CAT/LacZ (Invitrogen) from which the lacZ reporter gene was excised, to create pBud/CAT/US3long. Subsequently, plasmid pBud/CAT/US3short was generated as follows. pBud/CAT/US3long was digested with HindIII and Eco47III, releasing a 229-bp fragment, blunt ended using T4 DNA polymerase (Invitrogen), and subsequently religated, creating an expression vector containing the ORF encoding the US3 short isoform (pBud/CAT/US3short).
Then plasmid pBud/CAT/US3long, pBud/CAT/US3short, and control plasmid pBud/CAT/LacZ were transiently transfected in 40 to 60% confluent SK-6 cells as specified by the manufacturer (CellPhect transfection kit; Amersham Pharmacia Biotech AB). At 24 h posttransfection, the cells were paraformaldehyde fixed and permeabilized with 0.1% Triton X-100. Cells transfected with pBud/CAT/US3long or pBud/CAT/US3short were identified by subsequent incubation with anti-US3 monoclonal antibodies (kindly provided by L. Olsen and L. W. Enquist, Princeton University, Princeton, N.J.), and FITC-conjugated goat anti-mouse antibodies (Molecular Probes). Cells transfected with the pBud/CAT/LacZ control plasmid were identified by incubation with rabbit anti-chloramphenicol acetyltransferase (CAT) antiserum (Invitrogen) and FITC-labeled goat anti-rabbit secondary antibodies (Molecular Probes). For all types of transfected SK-6 cells, actin filaments were stained using phalloidin-Texas Red. Finally, stained SK-6 cells were mounted and analyzed by confocal microscopy.
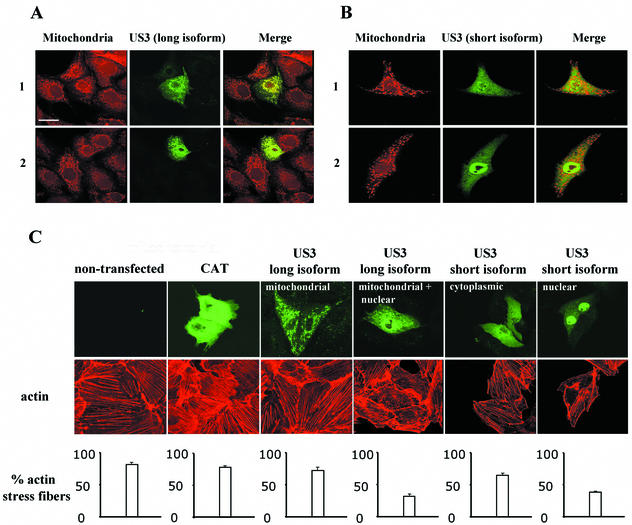
Transfection of both US3 isoforms resulted in heterogeneous localization of US3 in the cell. Since the localization of US3 appeared to be important for its effect on the actin architecture, the intracellular US3 localization is discussed in more detail before the effect of transfected US3 on the state of actin stress fibers is discussed in the next paragraph. In the majority (80%) of SK-6 cells transfected with the US3 long isoform, US3 was located exclusively in a dotted pattern in the cytoplasm, while a significantly smaller group (20%) expressed US3 in the cytoplasm as well as in the nucleus (Fig. 3C). Double immunofluorescent stainings, performed by incubating live transfected cells with the mitochondrion-specific probe MitoTracker Red CMXRos (Molecular Probes) (45 min at 37°C) and subsequently with mouse anti-US3 antibodies followed by FITC-conjugated goat anti-mouse antibodies, demonstrated that the dotted pattern of US3 long isoform localization in the cytoplasm perfectly colocalized with mitochondria (Fig. 3A). Cells transfected with the US3 short isoform also displayed a heterogeneous localization of US3, albeit different from that observed in cells transfected with the US3 long isoform. In the majority (60%) of SK-6 cells transfected with the US3 short isoform, US3 was localized predominantly in the nucleus, whereas in the remaining 40% of the cells, it was localized predominantly diffusely in the cytoplasm (Fig. 3C). Using MitoTracker Red CMXRos, we found that in contrast to the US3 long isoform, cytoplasmically localized US3 short isoform did not colocalize with mitochondria (Fig. 3B). An explanation for the intriguing finding that US3 in cells transfected with the US3 long isoform, but not in cells transfected with the US3 short isoform, shows a strong mitochondrial targeting was found by computational analysis of the amino acid sequences of the two US3 isoforms. A program specialized in the detection of mitochondrial localization signals (6) revealed the presence of a 51-amino-acid N-terminally located mitochondrial targeting signal in the US3 long isoform, which is not present in the US3 short isoform. Furthermore, this program determined that the predicted cleavage site of the mitochondrial targeting signal is 3 amino acids upstream of the US3 short-isoform ATG start codon.
FIG. 3.
(A and B) Subcellular localization of US3 in SK-6 cells transfected with the gene encoding either the US3 long (A) or short (B) isoform. At 24 h posttransfection, SK-6 cells were incubated with MitoTracker Red CMXRos (left panels), paraformaldehyde fixed, permeabilized, and incubated with monoclonal anti-US3 antibodies and FITC-conjugated secondary antibodies (middle panels). Merged images are given in the right panels. Localization of the US3 long isoform is either exclusively mitochondrial (row 1) or mitochondrial and nuclear (row 2), while localization of the US3 short isoform is either predominantly cytoplasmic (row 1) or predominantly nuclear (row 2). (C) Actin architecture of US3-transfected SK-6 cells. At 24 h after mock transfection or transfection with expression vectors encoding either the CAT reporter protein or the US3 long or short isoform, cells were paraformaldehyde fixed, permeabilized, stained, and analyzed by confocal microscopy. The CAT reporter protein and US3 were stained by incubating cells with either anti-CAT or anti-US3 antibodies and subsequently with FITC-labeled secondary antibodies (top panels), and actin filaments were stained using phalloidin-Texas Red (middle panels). The bottom panels show the percentage of cells with intact actin stress fibers (100 cells were scored). Bar, 10 μm.
Both in SK-6 cells transfected with the US3 long isoform and in SK-6 cells transfected with the US3 short isoform, the state of the actin stress fiber network correlated with the localization pattern of US3. For cells transfected with the US3 long isoform, 72.3% ± 4.5% of cells with exclusive localization of US3 in the mitochondria possessed intact stress fibers, a percentage which is comparable to that for cells transfected with the control plasmid (78.1% ± 2.4%) or nontransfected cells (81.7 ± 3.8%) (Fig. 3C). However, only 31.7% ± 3.8% of the cells that showed substantial nuclear localization of US3 retained intact stress fibers (Fig. 3C). Similarly, in cells transfected with the US3 short isoform, 65.0% ± 3.6% of cells with predominant localization of US3 in the cytoplasm retained intact actin stress fibers, a percentage which is slightly lower than that for cells transfected with the control plasmid or nontransfected cells (Fig. 3C). However, only 38.3% ± 1.4% of the cells that expressed the US3 short isoform predominantly in the nucleus retained intact stress fibers (Fig. 3C). Taken together, these data indicate that in cells transfected with the US3 long or short isoform, US3 can induce actin stress fiber breakdown when it is abundantly localized in the nucleus.
Although our transfection data indicate that the US3 long isoform and especially the short isoform are efficiently transported to the nucleus in the absence of a viral chaperone, no known nuclear localization signal (NLS) was found in the amino acid sequences of the two US3 isoforms by computational analysis (7). This indicates that US3 may travel to the nucleus via a yet to be determined NLS or, alternatively, via association with a cellular chaperone or substrate protein. A putative NLS in the US3 protein is not likely to be very stringent since in the absence of a viral background, the mitochondrial localization signal in the US3 long isoform is epistatic to the putative NLS.
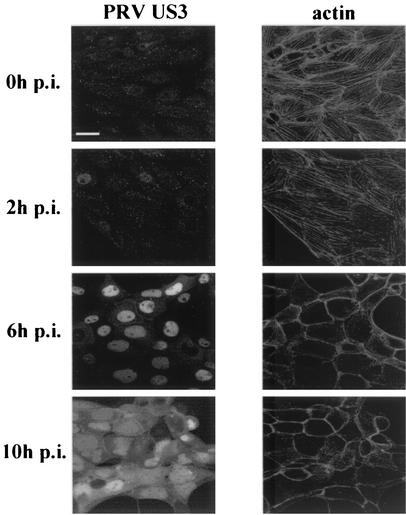
Our transfection data indicate that (i) nuclear localization of recombinant US3 is important for mediating efficient actin stress fiber breakdown and that (ii) the long isoform of US3 is localized predominantly in mitochondria. To be able to relate these transfection findings to infected cells, localization of PRV US3 and the state of actin stress fibers were examined at different stages of PRV infection. To do this, NIA3-infected cells, paraformaldehyde fixed at 0, 2, 6 and 10 h p.i., were permeabilized and stained for both PRV US3 and actin filaments. At 6 h p.i., almost all the cells had undergone stress fiber breakdown and US3 was localized predominantly in the nucleus. At 10 h p.i., US3 was localized in the nucleus as well as diffusely in the cytoplasm (Fig. 4). At none of the examined time points could colocalization between PRV US3 protein and mitochondria be observed (data not shown). Thus, in agreement with our transfection data, at the onset of actin stress fiber breakdown in PRV-infected cells, the US3 S/T kinase is localized predominantly in the nucleus.
FIG. 4.
US3 localization and actin architecture during the course of a PRV infection. SK-6 cells were infected at an MOI of 10 with an NIA3 wild-type strain. At the indicated time points (0, 2, 6, and 10 h p.i.), the cells were fixed, permeabilized, and stained for both PRV US3 and actin filaments. The left panels show the localization of US3, while the right panels show the state of the actin cytoskeleton at each of the examined time points. Bar, 10 μm.
It may be somewhat surprising that the strong mitochondrial targeting of the PRV US3 long isoform cannot be detected in infected SK-6 cells. The most obvious explanation would be that in PRV-infected cells, the US3 long isoform constitutes less than 5% of the total US3 protein content (32), possibly resulting in microscopically undetectable levels of US3 in mitochondria. Although we could not detect any US3 signal in mitochondria in infected cells, it is hard to imagine that the specific mitochondrial signal is totally irrelevant for US3 functioning. Although the role of PRV US3 in preventing apoptosis has not yet been studied, the mitochondrial localization of the US3 long isoform may have significance in this context, since several antiapoptotic proteins are located in mitochondria (15). However, this hypothesis remains speculative, especially since a similar mitochondrial targeting signal is totally absent in HSV US3, a protein known to be an effective inhibitor of apoptosis in specific cell types (14, 16, 18, 24).
Recently, Murata et al. transfected the HSV-2 orthologue of the PRV US3 ORF, which is transcribed and translated as one protein with a molecular mass of 66 kDa, in HEp-2 cells and found that HSV-2 US3 inactivates JNK, a downstream target protein of the Cdc42/Rac signaling pathway, and as a consequence blocks this pathway (25). The Cdc42/Rac pathway regulates actin stress fiber assembly and disassembly, and HSV-2 US3-mediated negative modulation of this pathway was found to result in actin stress fiber breakdown. Although it remains to be determined whether HSV-2 induces stress fiber breakdown during infection or whether HSV-2 US3 interference with the Cdc42/Rac signalling pathway occurs during infection, these data seem to be consistent with our current findings on PRV US3. A possible explanation for the importance of nuclear localization for PRV US3-mediated stress fiber breakdown may then be found when extrapolating the above-mentioned results for the HSV-2 orthologue (25). If PRV US3, like HSV-2 US3, inactivates JNK, a key component of the Cdc42/Rac pathway, and thereby induces stress fiber breakdown, nuclear localization of PRV US3 may be necessary to fulfill its function. Indeed, translocation to the nucleus of some cellular kinases, involved in actin assembly- and disassembly-regulating pathways, is vital for fulfillment of many of their activities (1, 36). Importantly, activated JNK is translocated to the nucleus, where it exerts its biological function (1). Hence, our data, which suggest that US3 may cause actin stress fiber disassembly, but only when it is localized predominantly in the nucleus, may suggest that in the nucleus, PRV US3 competes with cellular kinases that are implicated in actin stress fiber-controlling pathways.
Besides the current findings on PRV US3-mediated actin stress fiber breakdown, the PRV US3 S/T protein kinase has been indicated to play important cell type-dependent roles in virus egress from the nucleus and, recently, cell-to-cell spread (23, 27, 33). Although the exact mechanism by which PRV US3 plays its role during cell-to-cell spread is still puzzling, it has been hypothesized by Demmin et al. that certain viral proteins required for cell-to-cell spread are modified by the PRV US3 S/T protein kinase (8). Our observation of US3-mediated actin stress fiber breakdown may perhaps give an alternative explanation for the role of PRV US3 in cell-to-cell spread. Indeed, artificial disassembly of actin stress fibers in A431 cells by the addition of TPA (12-O-tetradecanoylphorbol-13-acetate) has been suggested to increase HSV-mediated cell-cell fusion (37). Further research is necessary to evaluate the possible role of PRV US3-mediated actin stress fiber breakdown in viral cell-to-cell spread.
In summary, PRV US3 causes actin stress fiber disassembly in different PRV-infected cells, as well as in US3-transfected SK-6 cells, when localized in sufficient amounts in the nucleus. Further work is necessary to elucidate the exact mechanism of PRV US3-induced actin stress fiber breakdown as well as the role of US3-mediated stress fiber breakdown in the virus life cycle.
Acknowledgments
We thank Carine Boone and Chantal Vanmaercke for excellent technical assistance, as well as L. Olsen and L. W. Enquist (Princeton University, Princeton, N.J.) for the kind gift of monoclonal anti-US3 antibodies.
This research was supported by a cooperative research action fund of the Research Council of Ghent University.
REFERENCES
- 1.Aplin, A. E., B. P. Hogan, J. Tomeu, and R. L. Juliano. 2002. Cell adhesion differentially regulates the nucleocytoplasmic distribution of active MAP kinases. J. Cell Sci. 115:2781-2790. [DOI] [PubMed] [Google Scholar]
- 2.Avitabile, E., S. Di Gaeta, M. R. Torrisi, P. L. Ward, B. Roizman, and G. Campadelli-Fiume. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskerville, A., J. B. McFerran, and C. Dow. 1973. Aujeszky's disease in pigs. Vet. Bull. 43:465-480. [Google Scholar]
- 4.Bedows, E., K. M. Rao, and M. J. Welsh. 1983. Fate of microfilaments in Vero cells infected with measles virus and herpes simplex virus type 1. Mol. Cell. Biol. 3:712-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthomme, H., B. Jacquemont, and A. Epstein. 1993. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology 193:1028-1032. [DOI] [PubMed] [Google Scholar]
- 6.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779-786. [DOI] [PubMed] [Google Scholar]
- 7.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demmin, G. L., A. C. Clase, J. A. Randall, L. W. Enquist, and B. W. Banfield. 2001. Insertions in the gG gene of pseudorabies virus reduce expression of the upstream Us3 protein and inhibit cell-to-cell spread of virus infection. J. Virol. 75:10856-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Wind, N., A. Zijderveld, K. Glazenburg, A. Gielkens, and A. Berns. 1990. Linker insertion mutagenesis of herpesviruses: inactivation of single genes within the Us region of pseudorabies virus. J. Virol. 64:4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wind, N., J. Domen, and A. Berns. 1992. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J. Virol. 66:5200-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wind, N., A. Berns, A. Gielkens, and T. Kimman. 1993. Ribonucleotide reductase-deficient mutants of pseudorabies virus are avirulent for pigs and induce partial protective immunity. J. Gen. Virol. 74:351-359. [DOI] [PubMed] [Google Scholar]
- 12.de Wind, N., B. P. Peeters, A. Zuderveld, A. L. Gielkens, A. J. Berns, and T. G. Kimman. 1994. Mutagenesis and characterization of a 41-kilobase-pair region of the pseudorabies virus genome: transcription map, search for virulence genes, and comparison with homologs of herpes simplex virus type 1. Virology 200:784-790. [DOI] [PubMed] [Google Scholar]
- 13.Feng, X., Y. G. Thompson, J. B. Lewis, and G. B. Caughman. 1996. Expression and function of the equine herpesvirus 1 virion-associated host shutoff homolog. J. Virol. 70:8710-8718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogeneous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmacher, V. S. 2002. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84:177-185. [DOI] [PubMed] [Google Scholar]
- 16.Hata, S., A. H. Koyama, H. Shiota, A. Adachi, F. Goshima, and Y. Nishiyama. 1999. Antiapoptotic activity of herpes simplex virus type 2: the role of the US3 protein kinase gene. Microbes Infect. 1:601-607. [DOI] [PubMed] [Google Scholar]
- 17.Hinkley, S., A. P. N. Ambagala, C. J. Jones, and S. Srikumaran. 2000. A vhs-like activity of bovine herpesvirus-1. Arch. Virol. 145:2027-2046. [DOI] [PubMed] [Google Scholar]
- 18.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H. Y. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 73:8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joachims, M., and D. Etchison. 1992. Poliovirus infection results in structural alteration of a microtubule-associated protein. J. Virol. 66:5797-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joachims, M., K. C. Harris, and D. Etchison. 1995. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology 211:451-461. [DOI] [PubMed] [Google Scholar]
- 21.Kimman, T. G., N. De Wind, N. Oei-Lie, J. M. A. Pol, A. J. M. Berns, and A. L. J. Gielkens. 1992. Contribution of single genes within the unique short region of Aujeszky's disease virus (suid herpesvirus type 1) to virulence, pathogenesis and immungenicity. J. Gen. Virol. 73:243-251. [DOI] [PubMed] [Google Scholar]
- 22.Kimman, T. G., N. De Wind, T. De Bruin, Y. De Visser, and J. Voermans. 1994. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology 205:511-518. [DOI] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 24.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata, T., F. Goshima, T. Daikoku, H. Takakuwa, and Y. Nishiyama. 2000. Expression of herpes simplex virus type 2 US3 affects the Cdc42/Rac pathway and attenuates c-Jun N-terminal kinase activation. Genes Cells 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 26.Nauwynck, H. J., and M. B. Pensaert. 1995. Effect of specific antibodies on the cell-associated spread of pseudorabies virus in monolayers of different cell types. Arch. Virol. 140:1137-1146. [DOI] [PubMed] [Google Scholar]
- 27.Pol, J. M., F. Wagenaar, and A. L. Gielkens. 1991. Morphogenesis of three pseudorabies virus strains in porcine nasal mucosa. Intervirology 32:327-337. [DOI] [PubMed] [Google Scholar]
- 28.Ridley, A. J. 2001. Rho family proteins: coordinating cell responses. Trends Cell Biol. 11:471-477. [DOI] [PubMed] [Google Scholar]
- 29.Smith, G. A., and L. W. Enquist. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18:135-161. [DOI] [PubMed] [Google Scholar]
- 30.Smith, C. C., J. Nelson, L. Aurelian, M. Gober, and B. B. Goswami. 2000. Ras-GAP binding and phosphorylation by herpes simplex virus type 2 RR1 PK (ICP10) and activation of the Ras/MEK/MAPK mitogenic pathway are required for timely onset of virus growth. J. Virol. 74:10417-10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Minnebruggen, G., G. R. Van de Walle, H. W. Favoreel, H. J. Nauwynck, and M. B. Pensaert. 2002. Temporary disturbance of actin stress fibers in swine kidney cells during pseudorabies virus infection. Vet. Microbiol. 86:89-94. [DOI] [PubMed] [Google Scholar]
- 32.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus: comparision with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747-1755. [DOI] [PubMed] [Google Scholar]
- 33.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 34.Wallin, M., J. Deinum, L. Goobar, and U. H. Danielson. 1990. Proteolytic cleavage of microtubule-associated proteins by retroviral proteinases. J. Gen. Virol. 71:1985-1991. [DOI] [PubMed] [Google Scholar]
- 35.Walter, I., and N. Nowotny. 1999. Equine herpes virus type 1 (EHV-1) infection induces alterations in the cytoskeleton of vero cells but not apoptosis. Arch. Virol. 144:1827-1836. [DOI] [PubMed] [Google Scholar]
- 36.Yoneda, Y. 2000. Nucleocytoplasmic protein traffic and its significance to cell function. Genes Cells 5:777-787. [DOI] [PubMed] [Google Scholar]
- 37.Yura, Y., J. Kusaka, T. Bando, S. Yamamoto, H. Yoshida, and M. Sato. 2000. Enhancement of herpes simplex virus-induced polykaryocyte formation by 12-6-tetradecanoyl formol 13-acetate: association with the reorganization of actin filament and cell motility. Intervirology 43:129-138. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, G., R. Stevens, and D. P. Leader. 1990. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and in infected cells. J. Gen. Virol. 71:1757-1765. [DOI] [PubMed] [Google Scholar]