Abstract
Reoviruses are a leading model for understanding cellular mechanisms of virus-induced apoptosis. Reoviruses induce apoptosis in multiple cell lines in vitro, and apoptosis plays a key role in virus-induced tissue injury of the heart and brain in vivo. The activation of transcription factors NF-κB and c-Jun are key events in reovirus-induced apoptosis, indicating that new gene expression is critical to this process. We used high-density oligonucleotide microarrays to analyze cellular transcriptional alterations in HEK293 cells after infection with reovirus strain T3A (i.e., apoptosis inducing) compared to infection with reovirus strain T1L (i.e., minimally apoptosis inducing) and uninfected cells. These strains also differ dramatically in their potential to induce apoptotic injury in hearts of infected mice in vivo—T3A is myocarditic, whereas T1L is not. Using high-throughput microarray analysis of over 12,000 genes, we identified differential expression of a defined subset of genes involved in apoptosis and DNA repair after reovirus infection. This provides the first comparative analysis of altered gene expression after infection with viruses of differing apoptotic phenotypes and provides insight into pathogenic mechanisms of virus-induced disease.
The mechanisms by which viruses cause cytopathic effects in infected host cells are complex and only partially defined. Apoptosis is a direct mechanism of cellular injury and death, which can occur in the course of normal tissue development or as a pathological response to a variety of noxious stimuli. Mammalian reoviruses have served as useful models for studies of the viral and cellular mechanisms that are operative in host cell damage and death (14, 57, 80, 81). Reoviruses induce apoptosis in a multiple cell lines in vitro and in murine models of encephalitis and myocarditis in vivo (18, 58, 68). Prototype strains serotype 3 Abney (T3A) and serotype 3 Dearing (T3D) induce apoptosis more efficiently than strain serotype 1 Lang (T1L). Differences in the capacity of reoviruses to induce apoptosis map to the viral S1 gene, which encodes the viral attachment protein σ1 (15, 69, 82).
The signaling pathways by which reoviruses induce apoptosis in target cells are complex. Involvement of death receptor- and mitochondrion-mediated pathways of apoptosis as well as cysteine protease activation have been demonstrated (11, 43). Binding of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) to its cell surface death receptors—DR4 and DR5—plays a central role in reovirus-induced apoptosis in HEK293 cells and in several cancer cell lines (11, 12), and other death-inducing ligands such as FasL are equally important in neurons (68). Activation of death receptor-related apoptotic pathways results in a coordinated pattern of caspase activation (43, 44, 68). Mitochondrial apoptotic pathways act to augment death receptor-initiated apoptosis, and apoptosis can be inhibited by stable overexpression of Bcl-2 (43, 44, 69). Blockade of cysteine protease activity using selective caspase inhibitors in vitro (11, 43) and calpain inhibitors in vivo (18) results in decreased apoptosis in target cells and tissues.
Reovirus infection results in activation of cellular transcription factors, including NF-κB (16) and c-Jun (13), and this activation plays a critical role in apoptosis. In the case of c-Jun, there is an excellent correlation between the capacity of viral strains to activate the JNK/c-Jun pathway and their ability to induce apoptosis (13). Inhibition of the activation of NF-κB by stable expression of the NF-κB inhibitor IκB, whether by the use of proteosome inhibitors or by targeted disruption of the genes encoding the p65 or p55 subunits of NF-κB, results in inhibition of reovirus-induced apoptosis (16). The close correlation between transcription factor activation and reovirus-induced apoptosis strongly suggests that new gene expression is critical for this process; therefore, we investigated the cellular response to reovirus infection at the transcriptional level. This was achieved by comparing transcriptional alterations after infection with a reovirus strain that efficiently induces apoptosis (i.e., T3A) with alterations after infection with a strain that induces minimal apoptosis (i.e., T1L). These strains also differ in their potential for inducing apoptotic myocardial injury in a murine model of viral myocarditis; T3A infection causes myocarditis and apoptotic myocardial injury, whereas T1L does not.
Using high-throughput screening of over 12,000 genes by using high-density oligonucleotide microarrays, we have identified transcriptional alterations in a defined subset of genes. When grouped into functional categories, a significant proportion of altered transcripts include genes involved in apoptosis and DNA repair, and it is this subset that forms the focus of this paper. The findings described herein are the first large-scale description of virus-induced alterations in apoptotic signaling at the transcriptional level, including kinetics of these changes after infection with strains that differ in apoptosis-inducing phenotype. These findings lend important insight into specific mechanisms of viral pathogenesis, since apoptosis has previously been demonstrated to be a critical mechanism for reovirus-induced damage in vitro and in vivo.
MATERIALS AND METHODS
Cells, virus, and infection.
Human embryonic kidney 293 (HEK293) cells (ATCC CRL 1573) were plated in T75 flasks at a density of 5 × 106 cells per flask in a volume of 12 ml of Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine (Gibco-BRL), 1 mM sodium pyruvate (Gibco-BRL), and 100 U of streptomycin (Gibco-BRL) per ml. Monolayers were infected 24 h after plating, when cells were 60 to 70% confluent. Reovirus strains T3Abney (T3A) and T1Lang (T1L) (P2 stock) were used to infect monolayers at a multiplicity of infection (MOI) of 100 PFU per cell. A high MOI was used to ensure that all susceptible cells were infected and because pilot studies in our laboratory indicated that high-multiplicity infection enhanced the reproducibility of gene expression studies. Virus was adsorbed for 1 h at 37°C in a volume of 2 ml, during which time flasks were rocked every 15 min. Following adsorption, flasks were incubated at 37°C after the addition of 10 ml of fresh medium. T3A-infected cells were harvested at 6, 12, and 24 h after viral infection. T1L-infected cells were harvested at 24 h postinfection. For control infections, HEK293 monolayers were inoculated with a cell lysate suspension, which was prepared identically to viral stocks but which lacked infectious virus.
Cell harvests and RNA extraction.
Cells were harvested by gently pipetting adherent and nonadherent cells from the flasks into 50-ml centrifuge tubes. After centrifugation (DuPont Sorvall 6000) at 1,200 rpm for 5 min, cell pellets were resuspended in phosphate-buffered saline (PBS) and transferred to Eppendorf tubes for total RNA extraction (RNeasy Mini Total RNA isolation kit; QIAGEN). Total RNA was extracted from each flask independently, resulting in duplicate RNA samples for each infection condition at 6, 12 (mock, T3A), and 24 (mock, T3A and T1L) h postinfection. A total of 16 RNA samples were prepared, and the yield and purity of extracted RNA were determined by spectrophotometry.
Target preparation.
Biotinylated cRNA targets were prepared from a 10-μg aliquot of each total RNA sample by following Affymetrix instructions and protocols. Briefly, total RNA was reverse transcribed to double-stranded cDNA (Superscript Choice; Gibco-BRL) by using high-pressure liquid chromatography-purified T7-(dT)24 oligomer for first-strand cDNA synthesis. Second-strand synthesis was performed by using T4 DNA polymerase, and double-stranded cDNA was isolated by using phenol-chloroform extraction with phase-lock gels. Isolated cDNA was in vitro transcribed and labeled (by using biotin-UTP and biotin-CTP) to produce biotin-labeled cRNA (BioArray High-Yield RNA transcript labeling kit; ENZO). Labeled cRNA was isolated by using RNeasy Mini Kit spin columns (QIAGEN). Yield and purity were quantified by using spectrophotometry. Labeled cRNAs were fragmented in 100 mM potassium acetate, 30 mM magnesium acetate, and 30 mM Tris-acetate (pH 8.1) for 35 min at 94°C to produce labeled cRNA fragments of 60- to 300-bp length. For hybridization, cRNA target integrity was analyzed with Affymetrix control (Test 2) arrays to assess degradation and hybridization performance prior to hybridization to Affymetrix Human U95A high-density oligonucleotide microarrays. The U95A microarray contains cDNA oligomer that is complementary to 12,000 human genes with known function (no expressed sequence tags), which currently represents the most comprehensive coverage of the human genome represented on a single microarray. Each of the 16 prepared target cRNAs was independently hybridized to a U95A array. Eukaryotic hybridization controls bioB, bioC, bioD, and cre were also included in the hybridization cocktail. Hybridization was carried out for 16 h at 45°C with rotation at 60 rpm. Microarrays were washed and stained with streptavidin-phycoerythrin conjugate by using the Affymetrix GeneChip Fluidics Station 400, following standard Affymetrix protocols. Staining intensity was antibody amplified by using a biotinylated antistreptavidin antibody at a concentration of 3 μg per ml which was followed by a second streptavidin-phycoerythrin conjugate stain, and hybridization intensity was analyzed by scanning at 570 nM. All hybridization and scanning steps were performed at the University of Colorado Health Sciences Center Cancer Center Microarray Core Facility.
Data analysis.
Each gene on the U95A array was represented by a group of 20 different 25-base cDNA oligomers that were complementary to a cRNA target transcript (i.e., perfect-match probes). As a hybridization specificity control, each perfect match oligomer was accompanied by an oligomer differing from the perfect match sequence by a single base pair substitution (i.e., mismatch probes). The combination of perfect-match and -mismatch cDNA oligomers for each gene is termed a probe set. Affymetrix-defined mathematical analyses (metrics) were performed to assess specific versus nonspecific hybridization of experimental cRNA targets to each probe set. Data files were analyzed by using GeneChip Microarray Suite software (version 4.0).
Initially, hybridization of cRNA targets derived from each of the 16 experimental samples was analyzed independently. By using Affymetrix-defined absolute mathematical algorithms describing perfect-match and -mismatch hybridization, each gene was defined as absent or present and was assigned a raw numerical value. Next, comparisons were made between virus-infected and mock-infected chips at each of the three time points postinfection. Raw numerical values were scaled to allow comparison between arrays. Genes considered absent (excess of mismatch hybridization or no hybridization) were not excluded from analysis, since genes changing from present to absent, absent to present, or present to present (but which increased in magnitude) were all important subsets of transcriptional alteration following viral infection compared to mock infection. By using Affymetrix-defined comparison mathematical algorithms, differential hybridization (between chips) to each cDNA probe was analyzed and designated as not changed, increased, marginally increased, decreased, or marginally decreased, and a change in expression (n-fold) was calculated. Finally, a four-way comparison of both virus-infected replicates to both mock-infected replicates at a given time point was assessed, and the mean change (n-fold) was calculated and reported along with standard error of the mean.
A gene was considered upregulated following virus infection if it was present in both virus-infected samples and if its expression increased by greater than or equal to twofold in each virus-infected sample compared to both mock-infected samples. Conversely, a gene was considered downregulated if it was present in both mock-infected samples and if its expression was decreased by greater than or equal to twofold in each virus-infected sample compared to both mock-infected samples. Genes whose expression changed by less than twofold were not considered up- or downregulated. Similarly, in order to ensure the reproducibility of the data presented, genes whose expression differed from mock-infected samples in only one of the two paired viral chips were not considered up- or downregulated.
To assess the reproducibility of hybridization results, the degree of variability in transcriptional expression among mock- and virus-infected replicate conditions was analyzed. For 99.6% of represented genes, expression was unchanged between mock-mock or virus-virus replicates. Transcriptional differences were noted in an average of 0.4% ± 0.1% of the total pool of transcripts between replicate conditions, but the genes involved represent a distinct population from the genes found to be up- or downregulated compared to virus-infected to mock-infected cells. The degree of variability in transcriptional expression as a function of time was also assessed by comparing differences in gene expression between mock infections following 6, 12, and 24 h of culture. A small proportion of transcripts were altered in response to duration of cell culture alone (1.2% ± 0.2% of the total pool). These genes were excluded from subsequent analysis.
RT-PCR.
Reverse transcriptase PCR (RT-PCR) was utilized to confirm changes in expression of selected genes as identified by analysis of oligonucleotide microarrays. For RT-PCR, RNA was extracted from infected and control HEK293 cells by using infection and extraction procedures identical to those described above. RNA was converted to cDNA by using the SuperScript preamplification system (Gibco-BRL) with the supplied oligo d(T)12-18 primer. Reverse transcription was performed at 42°C for 1 h. Semiquantitative PCR was performed by using primers generated for human DR4 (forward, 5′-CTG AGC AAC GCA GAC TCG CTG TCC AC-3′; reverse, 5′-TCC AAG GAC ACG GCA GAG CCT GTG CCA T-3′), human DR5 (forward, 5′-GCC TCA TGG ACA ATG AGA TAA AGG TGG CT-3′; reverse, 5′-CCA AAT CTC AAA GTA CGC ACA AAC GG-3′), human DCR1 (forward, 5′-GAA GAA TTT GGT GCC AAT GCC ACT G-3′; reverse, 5′-CTC TTG GAC TTG GCT GGG AGA TGT G-3′), GADD 34 (U83981) (forward, 5′-ACA CGG AGG AGG AGG AAG AT-3′; reverse, 5′-ACA GAG GAG GAA GGC AAG GT-3′), Bcl-10 (AJ006288) (forward, 5′-TCC ACA CTT CTC AGG TTG CTT-3′; reverse, 5′-AAT GGG GAA GAA GGA GAG GA-3′), caspase 3 (forward, 5′-GGT TCA TCC AGT CGC TTT GT-3′; reverse, 5′-AAC CAC CAA CCA ACC ATT C-3′; 207-bp product), Par-4 (forward, 5′-CTG AA CAT TTG CAT CCC TGT-3′; reverse, 5′ATG AAG CAG GGC AGA AAG AG-3′; 239-bp product), SMN (U80017) (forward, 5′-CCA GAG CGA TGA TGA CA-3′; reverse, 5′-TGG GTA AAT GCA ACC GTC TT-3′; 246-bp product), DNA polymerase α (L24559) (forward, 5′-TGC TTG ACC TGA TTG CTG TC-3′; reverse, 5′-ATG ACG GGA CAA AGA CAA GG-3′; 197-bp product), ParpL (AF057160) (forward, 5′-CGC AAG GTC CAG AGA GAA AC-3′; reverse, 5′-TCC CAG GTT CAC TTC TTT GG-3′; 244-bp product), SRP40 (U30826) (forward, 5′-AGA CGA AAT GCT CCA CCT GT-3′; reverse, 5′-CGA GAC CTG CTT CTT GAC CT-3′; 281-bp product), XP-C p125 (D21089) (forward, 5′-AGA GCA GGC GAA GAC AAG AG-3′; reverse, 5′-GAT GGA CAG GCC AAT AGC AT-3′; 199-bp product), and β-actin (forward, 5′-GAA ACT ACC TTC AAC TTC AAC TCC ATC-3′; reverse, 5′-CGA GGC CAG GAT GGA GCC GCC-3′ (24). PCRs were performed by using serial dilutions of each cDNA (1:5, 1:10, and 1:20) to estimate the linear range for each primer pair by interpretation of band intensity. To avoid saturation of the PCR and maximize the ability to detect relative quantitative differences between experimental samples, the highest-input cDNA dilution that produced visible PCR products was utilized for comparisons of transcript abundance, and PCR cycles were limited to 25. RT-PCR for actin was performed in parallel with each PCR of interest for each experimental sample (as a control to ensure equal input load of cDNA in each reaction). PCR cycle conditions were 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 1 min for 25 cycles. PCR products were resolved on a 2% agarose-ethidium bromide gel run at 100 V for 1 h. Products were visualized by UV illumination with Fluor-S (Bio-Rad) software imaging. Each reaction was performed at least twice in independent experiments to confirm reproducibility.
Animal infections and immunohistochemistry.
Reovirus strain 8B is an efficiently myocarditic reovirus that has been previously characterized (74) and has been shown to induce apoptotic myocardial injury in neonatal mice (18). Two-day-old Swiss-Webster (Taconic) mice were intramuscularly inoculated with 1,000 PFU of 8B reovirus (20-μl volume) in the left hind limb. Mock-infected mice received gel saline vehicle inoculation (20-μl volume; 137 mM NaCl, 0.2 mM CaCl2, 0.8 mM MgCl2, 19 mM H3BO3, 0.1 mM Na2B4O7, 0.3% gelatin). At 1 to 7 days postinfection, mice were sacrificed and hearts were immediately immersed in 10% buffered formalin solution. After mounting as transverse sections, hearts were embedded in paraffin and sectioned to 6-μm thickness. For quantification of the degree of myocardial injury, hematoxylin- and eosin-stained midcardiac sections (at least two per heart) were examined at ×125 magnification by light microscopy and evaluated for histologic evidence of myocarditis.
Immunohistochemical analysis of survivin (SMN) expression was carried out on identical sections to assess expression over the 7 days following reovirus infection. SMN antiserum was produced in New Zealand White rabbits by immunization with the amino terminus of the surviving amino peptide sequence (PTLPPAWQPFLKDHRI) linked to keyhole limpet hemocyanin by the method of Ambrosisni. Immunoglobulins from the rabbit before immunization were purified by affinity chromatography with protein A (Pierce, Rockford, Ill.). Western blot analysis against total protein extract from HeLa cells showed reactivity with a single band of protein of approximately 16.5 kDa—consistent with the expected molecular mass. Western blotting with preimmune serum showed no immunoreactivity. Slides were deparaffinized through xylene and rehydrated through a graded alcohol series. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 15 min. Antigen retrieval was performed with a 0.1 M citrate buffer for 10 min at 120°C. Primary antibody was diluted to 1.8 ng/μl in PBS (pH 7.4) with 1% bovine serum albumin applied to sections and incubated in a humidity chamber overnight at 4°C. Following three washes in PBS for 5 min each, incubation in secondary antibody labeled with polymer-linked horseradish peroxidase (Envision +; Dako, Carpinteria, Calif.) was carried out for 30 min at room temperature in a humidity chamber. Following three washes in PBS, sections were developed with 3′-diaminobenzidine (Dako) and counterstained with hematoxylin. Negative controls were performed by substitution with the preimmune immunoglobulin from the same rabbit. Positive control consisted of a colon carcinoma section that has been extensively studied in our laboratory and which shows strong staining that is consistently reproducible in this tissue.
RESULTS
Global analysis of gene expression following reovirus infection.
At 6, 12, and 24 h postinfection, transcriptional expression of each of 12,000 genes present on the HU95A microarray was compared for each pair of T3A (strongly apoptosis inducing [APO+]) virus-infected and mock-infected arrays. Similar analysis was carried out at 24 h postinfection for each pair of T1L (weakly apoptosis-inducing [APO−]) virus-infected and mock-infected arrays. The subset of genes that were transcriptionally altered following T3A (APO+) infection compared to mock infection was determined. This subset of genes was also compared to those genes that were differentially expressed following T1L (APO−) infection compared to mock infection. At 6 h post-T3A (APO+) infection, expression of 18 genes (0.2% of the total genes present on the array) was altered (all with increased expression) by twofold or greater in T3A (APO+)-infected cells compared to mock-infected cells. By 12 h post-T3A (APO+) infection, expression of 86 genes (0.7%) was altered (29 genes with increased expression and 57 genes with decreased expression) in virus-infected compared to mock-infected cells. By 24 h post-T3A (APO+) infection, expression of 309 genes (2.6%) was altered (215 with increased expression and 94 with decreased expression) in virus-infected compared to mock-infected cells. In contrast, at 24 h post-T1L (APO−) infection, expression of only 59 genes (0.4%) was altered (45 with increased expression and 14 with decreased expression) in virus-infected compared to mock-infected cells. A complete listing of all genes with twofold or greater changes in expression following T3A and T1L infection is availableonline at http://www.uchsc.edu/sm/neuro/tylerlab/personnel/completelisting.pdf. When categorized into functionally related families, a large number of differentially expressed genes following T3A (APO+) infection were noted to encode proteins involved in apoptosis (Table 1) and DNA repair (Table 2). These genes were not differentially expressed following T1L (APO−) infection [with the exception of five genes in common between T3A (APO+) and T1L (APO−)], indicating that these changes in gene expression likely correlate with differences in virus-induced pathogenicity rather than resulting from nonspecific cellular responses to viral infection.
TABLE 1.
Reovirus-induced alteration in expression of genes encoding proteins known to regulate apoptotic signaling
| Gene | GenBank accession no.a | Change in expression (n-fold)b at the indicated time (h) after infection with:
|
|||
|---|---|---|---|---|---|
| T3A
|
T1L
|
||||
| 6 | 12 | 24 | 24 | ||
| Mitochondrial signaling | |||||
| Pim-2 proto-oncogene homologue | U77735 | −2.2 ± 0.1 | |||
| MCL1 | L08246 | 2.0 ± 0.0 | 2.2 ± 0.0 | ||
| BAC 15E1-cytochrome C oxidase polypeptide | AL021546 | 2.1 ± 0.0 | |||
| Par-4 | U63809 | 2.1 ± 0.0 | |||
| HSP-70 (heat shock protein 70 testis variant) | D85730 | 2.2 ± 0.1 | |||
| BNIP-1 (BCL-2 interacting protein) | U15172 | 2.3 ± 0.2 | |||
| SMN/Btfp44/NAIP (survival motor neuron/neuronal apoptosis inhibitor protein) | U80017 | 2.5 ± 0.1 | |||
| DRAK-2 | AB011421 | 2.8 ± 0.2 | |||
| SIP-1 | AF027150 | 3.0 ± 0.2 | |||
| DP5 | D83699 | 5.5 ± 1.1 | |||
| ER stress-induced signaling | |||||
| ORP150 | U65785 | −2.4 ± 0.2 | |||
| GADD 34 | U83981 | 6.8 ± 0.2 | 3.7 ± 0.2 | 2.9 ± 0.2 | |
| GADD 45 | M60974 | 3.3 ± 0.2 | 4.9 ± 0.1 | 4.4 ± 0.1 | |
| Death receptor signaling | |||||
| Bcl-10 | AJ006288 | 5.6 ± 1.1 | |||
| PML-2 | M79463 | 3.4 ± 0.3 | |||
| Ceramide glucosyltransferase | D50840 | 4.0 ± 1.2 | |||
| Sp100 | M60618 | 6.5 ± 0.3 | |||
| 5.8 ± 0.6 | |||||
| Proteases | |||||
| Calpain (calcium-activated neutral protease) | X04366 | −2.6 ± 0.1 | |||
| Beta-4 adducin | U43959 | −2.1 ± 0.1 | |||
| Caspase 7 (lice-2 beta cysteine protease) | U67319 | 2.6 ± 0.2 | |||
| Caspase 3 (CPP32) | U13737 | 3.2 ± 0.2 | 2.8 ± 0.1 | ||
| Undefined | |||||
| Frizzled related protein | AF056087 | −2.5 ± 0.1 | −3.3 ± 0.5 | ||
| TCBP (T cluster binding protein) | D64015 | 3.3 ± 0.2 | |||
| Cug-BP/hNAb50 RNA binding protein | U63289 | 6.6 ± 1.1 | |||
GenBank accession number corresponds to sequence from which the Affymetrix U95A probe set was designed.
Data are means ± standard errors of the means.
TABLE 2.
Reovirus-induced alteration in expression of genes encoding proteins known to be involved in DNA repair
| Gene | GenBank accession no.a | Change in expression (n-fold)b at the indicated time (h) after infection with:
|
|||
|---|---|---|---|---|---|
| T3A
|
T1L | ||||
| 6 | 12 | 24 | 24 | ||
| DNA ligase 1 | M36067 | −8.2 ± 1.1 | |||
| PARPL | AF057160 | −6.3 ± 0.7 | |||
| XP-C repair complementing protein (p125) | D21089 | −3.4 ± 0.1 | |||
| DNA polymerase gamma | U60325 | −1.9 ± 0.1 | −2.9 ± 0.1 | ||
| ERCC5 | L20046 | −2.7 ± 0.1 | |||
| DNA polymerase alpha | L24559 | −2.5 ± 0.2 | |||
| HLP (helicase-like protein) | U09877 | −2.4 ± 0.1 | |||
| GTBP | U28946 | −2.0 ± 0.1 | −2.1 ± 0.0 | ||
| DDB2 (p48 subunit) | U18300 | −2.0 ± 0.0 | |||
| RAD 54 homologue | X97795 | −2.0 ± 0.1 | |||
| Mi2 autoantigen | X86691 | −1.3 ± 0.1 | −2.0 ± 0.1 | ||
| MMS2 | AF049140 | 2.1 ± 0.0 | |||
| Rad-51-interacting protein | AF006259 | 2.6 ± 0.2 | |||
| Rec-1 | AF084513 | 2.4 ± 0.4 | |||
GenBank accession number corresponds to sequence from which the Affymetrix U95A probe set was designed.
Data are means ± standard errors of the means.
Reovirus-induced alteration in expression of genes involved in apoptosis.
Expression of 24 genes related to the regulation of apoptosis was altered in T3A (APO+) reovirus-infected cells. These genes encode proteins that participate in apoptotic signaling involving death receptors, endoplasmic reticulum (ER) stress, mitochondrial signaling, and cysteine proteases (Table 1). For 22 of these 24 genes, significant alteration (>2-fold) in expression was not apparent until 24 h postinfection. Only five of these genes were also differentially expressed following T1L (APO−) infection.
Altered expression of genes involved in death receptor signaling pathways.
We have previously shown that members of the TNF receptor superfamily of cell surface death receptors, including DR4, DR5, and their apoptosis-inducing ligand, TRAIL, play a critical role in reovirus-induced apoptosis (11, 12). We wished to determine whether alterations in expression levels of genes encoding these proteins were altered following reoviral infection. Using oligonucleotide microarrays, we did not detect significant changes in gene expression of TRAIL or the death receptors DR5, decoy receptor 1 (DcR-1), or DcR-2 at 6, 12, and 24 h postinfection in reovirus-infected cells compared to mock-infected controls (Table 3). DR4 was not represented on the U95A microarray. Expression of genes encoding other important members of the TNF receptor superfamily and their ligands was also unchanged in reovirus-infected cells including Fas, Fas ligand, TNF-α, TNF-β, and TNF receptor and TNF receptor-related protein (Table 3).
TABLE 3.
Genes encoding proteins known to be involved in apoptotic signaling which were not differentially expressed following reovirus T3A or T1L infection
| Gene | GenBank accession no.a |
|---|---|
| Death receptor signaling | |
| TRAIL | U37518 |
| DR5 (TRAIL-R2) | AF014794 |
| Decoy receptor 1 (TRAIL-R3) | AF014794 |
| Decoy receptor 2 | AF029761 |
| TNF | M16441 |
| TNF-α | X02910 |
| TNF-β | D12614 |
| TNF receptor | M58286 |
| TNF receptor 2-related protein | L04270 |
| Fas (Apo-1/CD95) | X83490, X83492, Z70519, X82279, X63717 |
| Fas ligand (FasL) | U11821, D38122 |
| Mitochondrial signaling | |
| Bcl-2 | M13994, M14745b, M13995b |
| BAX alpha | L22473 |
| BAX beta | L22474 |
| BAX gamma | L22475b |
| BAX delta | U19599 |
| Proteases | |
| Caspase 2 (Ich-1) | U13021,bU13022b |
| Caspase 9 (Mch-6) | U60521b |
| Caspase 4 (ICErel-II) | U28014 |
| Caspase 5 (ICErel-III) | U28015 |
| Caspase 6 (Mch2)-isoform alpha | U20536 |
| Caspase 8 (MACH-alpha 1, MACH-beta 1, MACH-beta 2) | X98172, X98176, X98175 |
| Caspase 10 (Mch4) | U60519 |
GenBank accession number corresponds to sequence from which the Affymetrix U95A probe set was designed. Multiple accession numbers are noted for multiple representations of a particular gene (unique probe sets) on the U95A microarray.
Indicates multiple representations of a particular gene derived from the same GenBank sequence.
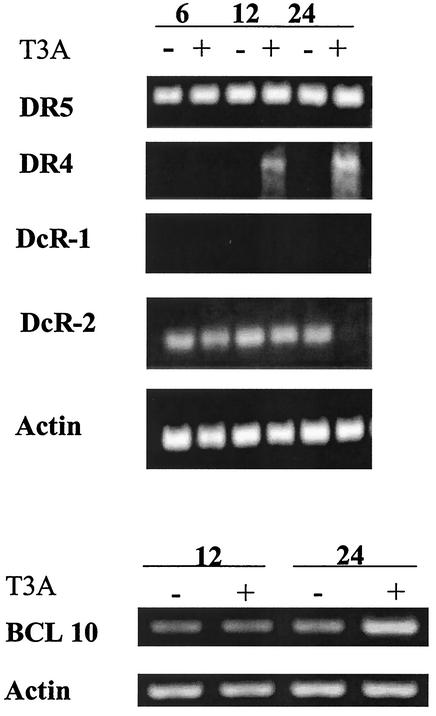
We performed additional analysis of death receptor-related gene expression, including DR4, DR5, DcR-1, and DcR-2 following T3A (APO+) infection by RT-PCR (Fig. 1). DR4 expression was increased at 12 and 24 h post-T3A infection compared to mock infection. In contrast, expression of DR5 was unchanged following T3A infection, thus confirming results obtained from microarray analysis. DcR-1 transcripts were not detectable in either mock- or T3A-infected cells. DcR-2 expression appeared to be decreased at 24 h post-T3A infection compared to mock-infected cells in this RT-PCR analysis. However, consistent with the microarray results, this decrease was not seen in RNase protection assays (data not shown).
FIG. 1.
Expression of genes related to death receptor-mediated apoptotic signaling is altered following reovirus T3A infection. HEK293 cells were either mock (−) or T3A (+) infected at an MOI of 100 PFU per cell. mRNA was collected at 6, 12, and 24 h postinfection and analyzed by semiquantitative RT-PCR for expression of selected transcripts encoding proteins involved in death receptor-mediated apoptotic signaling.
Microarray analysis also revealed differential expression of four genes that encode proteins that may be involved in modulation of death receptor-associated signaling cascades in T3A (APO+)-infected cells at 24 h postinfection: PML-2, ceramide glucosyltransferase, Bcl-10, and Sp100 were increased 3.4 ± 0.3-, 4.0 ± 1.2-, 5.6 ± 1.1-, and 6.1 ± 0.5-fold, respectively. We used RT-PCR to confirm the upregulated expression of BCL-10 at 24 h post-T3A (APO+) infection (Fig. 1). Taken together, these results suggest that, with the exception of DR4 and DcR-2, changes in death receptor and ligand gene expression are unlikely to play a critical role in reovirus-induced apoptosis. However, transcriptional alterations in genes encoding proteins that modulate death receptor signaling may play a role in reovirus-induced apoptosis.
Altered expression of genes involved in mitochondrial signaling pathways.
Mitochondrial pathways play an important role in reovirus-induced apoptosis in HEK293 cells (43, 44). Following reovirus infection, both cytochrome c and Smac/DIABLO are released from mitochondria and trigger the activation of caspase 9 as well as the degradation of inhibitors of apoptosis (43, 44). Reovirus-induced apoptosis in MDCK and HEK293 cells is inhibited by stable overexpression of Bcl-2 (43, 44, 69), which is consistent with a significant role for the mitochondrial apoptosis pathway during reovirus infection.
Among genes whose expression was altered in T3A (APO+)-infected cells were 10 genes encoding proteins involved in mitochondrial signaling. This group included genes encoding a number of proteins known to interact with Bcl-2, including SMN, whose expression was increased 2.5 ± 0.1-fold, and SMN-interacting protein 1 (SIP-1), whose expression was increased 3.0 ± 0.2-fold. Expression of genes encoding several other Bcl-2-interacting proteins included the Bcl-2 family member MCL-1, prostate apoptosis response 4 (Par-4), DAP kinase-related apoptosis-inducing protein kinase 2 (DRAK-2), neuronal death protein 5 (DP-5), and Bcl-2-interacting protein 1 (BNIP-1), each of which were upregulated two- to sixfold following T3A (APO+) infection (Table 1). With the exception of MCL-1, expression of these genes was unaltered following T1L (APO−) infection. Bcl-2 itself was not differentially expressed in virus-infected cells, nor was the proapoptotic Bcl-2 family member BAX (Table 3).
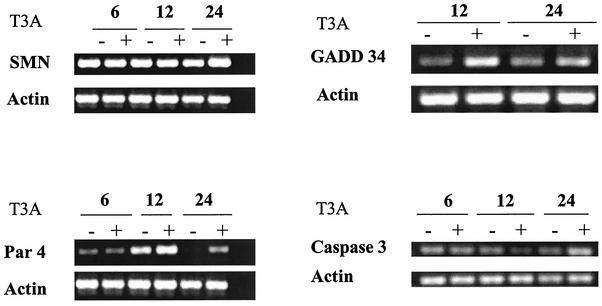
We used RT-PCR to confirm changes in the expression of selected genes involved in mitochondrial signaling and interaction with Bcl-2 (Fig. 2). We found that the expression of SMN was increased at 24 h post-T3A (APO+) infection but not at earlier time points. Expression of Par-4 was increased at 12 and 24 h post-T3A (APO+) infection, thus confirming results obtained with oligonucleotide arrays.
FIG. 2.
Expression of genes related to mitochondrion-, ER stress-, and protease-mediated apoptotic signaling is altered following reovirus T3A infection. HEK293 cells were either mock (−) or T3A (+) infected at an MOI of 100 PFU per cell. mRNA was collected at 6, 12, and 24 h postinfection and analyzed by semiquantitative RT-PCR for expression of selected transcripts encoding proteins involved in mitochondrial (SMN and Par-4) and ER stress (GADD 34)-mediated apoptotic signaling as well as for caspase 3, a cysteine protease involved in apoptosis.
These results add to previous data demonstrating that Bcl-2 plays an important regulatory role in reovirus-induced apoptosis by revealing a complex interplay of Bcl-2 regulatory factors at the transcriptional level in T3A (APO+)-infected cells.
Altered expression of genes involved in ER stress pathways.
In addition to death receptor and mitochondrial pathways of apoptosis, stress signals originating in the Golgi apparatus and ER can also trigger apoptosis (79, 86). Viral proteins are potent inducers of ER stress responses (7, 22, 64). Three genes encoding proteins that are potentially involved in ER stress-related responses were differentially expressed following both T3A (APO+) and T1L (APO−) infection (Table 1). Two of these genes encode growth arrest and DNA damage (GADD)-inducible proteins GADD 34 and GADD 45. Alterations in the expression of these genes were among the earliest changes in gene expression detected in T3A (APO+)-infected cells. The increased expression of GADD 45 following reovirus infection has been discussed previously in terms of its role in reovirus-induced cell cycle regulation (65). Expression of GADD 34 was increased 6.8 ± 0.2-fold as early as 6 h post-T3A (APO+) infection. This was the largest increase in expression found for any apoptosis-related gene at any time following infection. Expression remained increased at 24 h postinfection, although the magnitude of the increase declined (3.7 ± 0.2-fold). Expression of GADD 34 and GADD 45 was also upregulated at 24 h post-T1L (APO−) infection—by 2.9 ± 0.2-fold and 4.4 + 0.1-fold, respectively. In order to confirm the increased expression of GADD 34 detected by using oligonucleotide arrays, we performed RT-PCR on reovirus-infected HEK293 cells by using GADD 34-specific primers. GADD 34 transcripts were increased at 12 and 24 h post-T3A (APO+) infection compared to mock infection (Fig. 2), thus confirming microarray results. In addition to transcriptional upregulation of genes encoding GADD proteins, transcripts for ORP150—an ER resident protein involved in the misfolded protein rescue response—were downregulated by 2.4 ± 0.2-fold following T3A (APO+)—not T1L (APO−)—infection.
These results suggest that ER stress-induced apoptotic signaling may play a role in reovirus infection. However, the fact that altered expression in GADD genes occurred following both T1L (APO−) and T3A (APO+) infection (although at lower levels in T1L [APO−] infection) suggests that these pathways may play a less-critical role in determining virus-induced apoptotic injury and rather are activated as a nonspecific cellular response to reoviral infection.
Altered expression of genes encoding cysteine proteases.
Initiation of apoptosis through death receptor, mitochondrial, or ER and Golgi pathways results in the activation of specific initiator caspases. These caspases in turn activate additional caspases, culminating in the activation of effector caspases. Effector caspases, exemplified by caspases 3 and 7, act on substrates, including laminins and the caspase-activated DNase responsible for inducing the morphological features of apoptosis in target cells (28). Caspases 3, 8, and 9 are activated in reovirus-infected cells, and inhibition of caspase activation inhibits apoptosis (43).
Expression of genes encoding the effector caspases 3 and 7 were increased at 24 h following T3A (APO+) reovirus infection but not at earlier time points, consistent with their role as downstream effector caspases that are activated at the terminus of caspase cascades. Caspase 7 expression was increased 2.6 ± 0.2-fold, and that of caspase 3 was increased 3.2 ± 0.2-fold (Table 1). Caspase 3 expression was also increased at 24 h post-T1L (APO−) infection at lower levels (2.8 + 0.1-fold). Expression of genes encoding other caspases was not significantly altered following reovirus infection, including that of caspases 2, 4, 5, 6, 8, 9, and 10 (Table 3). Caspases 11, 12, 13, and 14 were not represented on the U95A microarray.
Because of the importance and central role of caspase 3 as a common effector in death receptor, mitochondrial, and ER and Golgi apoptotic signaling pathways, we wished to confirm the increased expression of this gene by using RT-PCR. Caspase 3 expression was increased over expression levels in mock-infected cells at 24 h post-T3A (APO+) infection (Fig. 2), thus confirming results obtained via oligonucleotide microarrays. Although not detected by microarray analysis, caspase 3 expression was noted to be decreased at 12 h post-T3A (APO+) infection, preceding the increase seen at 24 h postinfection.
These results suggest that although caspase activity is clearly modulated at the protein level following reovirus infection, transcriptional upregulation of genes encoding effector caspases may also play a role in effecting reovirus-induced apoptotic injury. The fact that caspase 3 transcripts were noted to initially decrease at 12 h postinfection and then increase at 24 h postinfection indicates that transcriptional regulation is likely a complex and dynamic process that is tightly linked to rapid changes in caspase protein levels and states within infected cells.
Reovirus-induced alteration in expression of genes related to DNA repair.
DNA damage is one of the basic stimuli that induces apoptosis. Cells have evolved complex mechanisms for sensing both single-strand and double-strand DNA breaks and initiating their repair (67). Expression of 14 genes encoding multiple classes of DNA repair enzymes was altered in T3A (APO+)-infected cells (Table 2). For 9 of these 14 genes, significant alteration (>2-fold) in expression was not apparent until 24 h postinfection, and 11 of these 14 alterations represented downregulation of expression. Transcription of DNA repair genes was not altered following T1L (APO−) infection. This global transcriptional downregulation of multiple classes of DNA repair enzymes following T3A (APO+) infection, which ranged from two- to eightfold, has not previously been appreciated.
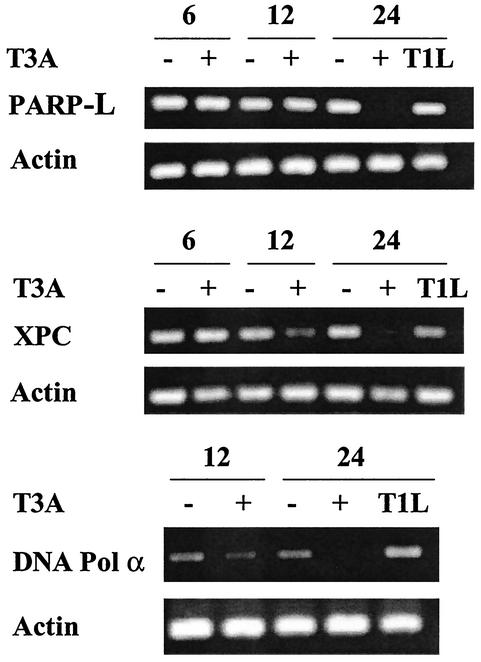
Among downregulated DNA repair enzymes, expression of poly(ADP-ribosyl)-transferase (PARPL) was decreased 6.3 ± 0.7-fold, XP-C repair complementing protein 125 was decreased 3.4 ± 0.1-fold, and DNA polymerase α was decreased 2.5 ± 0.2-fold compared to expression levels in mock- or T1L-infected cells. We used RT-PCR to confirm transcriptional alterations following T3A (APO+) infection compared to mock and T1L (APO−) infection in these three DNA-repair enzymes (Fig. 3). Transcripts for XP-C and DNA polymerase α were decreased at 12 and 24 h post-T3A (APO+) infection, and PARPL was downregulated at 24 h post-T3A (APO+) infection, thus confirming the decreases in expression detected by microarray analysis. In contrast, in T1L (APO−)-infected cells, transcripts for PARPL were unchanged, transcripts for XPC were only minimally decreased (much less so than the dramatic reductions seen in T3A [APO+]-infected cells), and transcripts for DNA polymerase α were increased.
FIG. 3.
Genes encoding DNA repair proteins are differentially expressed following reovirus infection. HEK293 cells were either mock (−) or T3A (+) infected at an MOI of 100 PFU per cell, and mRNA was collected at 6, 12, and 24 h postinfection. HEK293 cells were also infected with T1L at an MOI of 100 PFU per cell, with mRNA collected at 24 h postinfection. Samples were analyzed by semiquantitative RT-PCR for expression of selected transcripts encoding proteins important for DNA repair.
These results suggest that, as well as directly stimulating proapoptotic signaling pathways, T3A (APO+) reovirus infection may also facilitate apoptosis by downregulating the host cell's transcription of genes encoding proteins that have the capacity to repair DNA damage.
Translation of microarray data into the in vivo model of reovirus-induced myocarditis.
Although changes in mRNA levels do not necessarily represent changes in protein expression, we next investigated whether previously unrecognized changes in gene expression identified by microarray analysis of reovirus-infected cells in vitro could be directly translated into delineation of pathogenic mechanisms of reovirus-induced apoptosis in vivo. Specifically, we wished to determine if an observed alteration in gene expression would be predictive of changes in expression of the relevant protein in a model of reovirus-induced tissue injury characterized by apoptosis. To this end, we analyzed murine myocardial tissue following reovirus strain 8B infection, since 8B is efficiently myocarditic in neonatal mice (74), and we have previously shown that apoptosis is an important component of myocardial tissue injury following 8B infection (18).
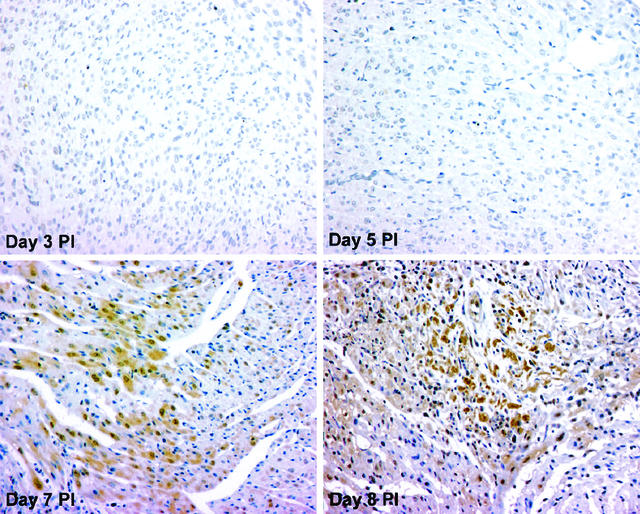
Altered expression of genes encoding Bcl-2 regulatory proteins (which have an impact on mitochondrial apoptotic signaling) were among the most abundant changes detected by microarray analysis. We selected one of these Bcl-2 regulatory proteins, SMN, for analysis in vivo, since transcripts for SMN were noted to be selectively increased at 24 h following infection with the apoptosis-inducing strain in the microarray experiment, and this result was confirmed by RT-PCR. We therefore analyzed myocardial tissues from reovirus 8B-infected mice by immunohistochemistry on days 1 to 7 postinfection for expression of SMN in relation to histologic evidence of virus-induced apoptotic tissue damage. SMN was maximally expressed within myocardial lesions in temporal and spatial concordance with histologically detectable apoptotic myocardial injury on days 7 and 8 postinfection (Fig. 4). SMN was not detected in myocardial tissue without evidence of apoptotic myocardial injury on days 1, 3, and 5 postinfection, nor was it detected in tissues from mock-infected animals. The significance of SMN upregulation within injured myocardial tissue is being investigated further. However, these results illustrate that microarray analysis of transcriptional changes following reovirus infection may provide a useful springboard toward the delineation of critical virus-induced pathogenic signaling pathways that are operative at the protein level.
FIG. 4.
SMN expression is increased in reovirus 8B-infected myocardial tissues, coincident with myocardial apoptotic injury. Neonatal Swiss-Webster mice were intramuscularly infected with 1,000 PFU of 8B virus. Myocardial tissues were analyzed on days 1 to 7 postinfection for histologic evidence of myocarditis as well as for expression of SMN by immunohistochemistry, since we have previously shown that apoptotic myocardial injury is detected at 7 days postinfection in this model. SMN protein was detected (brown stain) in infected myocardial tissue beginning on day 7 postinfection (at the time that histologic evidence of myocarditis was detected, within discrete myocardial lesions). Neither SMN nor evidence of myocardial injury was detected at earlier time points postinfection, as demonstrated on days 3 and 5 postinfection. Original magnification, ×40.
DISCUSSION
Transcriptional changes related to apoptosis.
Using high-throughput microarray analysis, we now demonstrate that reovirus infection is associated with differential expression of genes encoding proteins that participate in apoptotic signaling, including death receptor-, mitochondrion-, and ER stress-mediated pathways as well as DNA repair. These results represent the first large-scale description of virus-induced alterations in apoptotic signaling at the transcriptional level, including the kinetics of these changes following infection with viral strains that differ in apoptosis-inducing phenotype. The fact that the majority of these alterations occurred preferentially in T3A (APO+)- and not T1L (APO−)-infected cells suggests that interpretation of these alterations may provide important insight into critical mechanisms of reovirus-induced pathogenesis.
Microarray analysis has been increasingly utilized to investigate global transcriptional alterations following viral infection of many types, including human immunodeficiency virus (20, 83), herpesviruses (i.e., herpes simplex virus, varicella-zoster virus, Epstein-Barr virus, cytomegalovirus, and Kaposi's sarcoma-associated herpesvirus) (10, 29, 36, 39, 90, 93), rotavirus (17), Sindbis virus (38), hepatitis B (32) and C (5) viruses, measles virus (8), influenza virus (23), enterovirus (63), and papillomavirus (55). Although several groups have reported isolated alterations in transcription of genes related to apoptotic signaling following viral infection, none of these studies was specifically designed to understand the specific pathogenic mechanisms by which apoptosis-inducing viruses inflict damage on infected cells. In a comparison of two strains of Sindbis virus that differed in neurovirulence, differential transcriptional alteration of several genes related to mitochondrial apoptotic signaling was noted, including Bcl-2 family members mcl-1, bfl-1, and PBR (38). Transcriptional alteration of genes related to mitochondrial apoptotic signaling, including cytochrome c and inhibitors of apoptosis was also reported following rotavirus infection of caco-2 cells (17). Altered transcripts for several members of death receptor-mediated signaling pathways were reported following hepatitis C infection of hepatocytes (5), including TRAIL, TNF-R, and Fas. Likewise, TRAIL and Fas transcripts were altered in a study of papillomavirus infection of cervical keratinocytes (55). Altered transcripts for caspase 8 and TRAF4, known to be involved in death receptor signaling, have also been reported following varicella-zoster virus infection of skin fibroblasts (39). In addition to these alterations, several groups have noted transcriptional alteration in genes encoding serpins, which are known to inhibit caspases (17, 39). Transcripts for NF-κB and c-Jun—which have been linked to apoptotic signaling pathways—have also been altered following several types of viral infection.
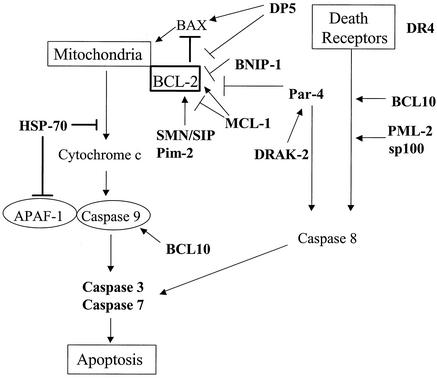
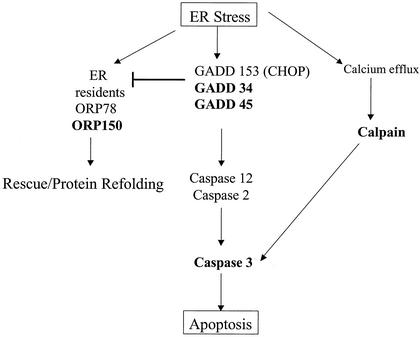
Our study is the first that was specifically designed to dissect virus-induced alteration in apoptosis-related transcription and the first to report alteration in coordinated groups of genes with relevance to several major apoptotic signaling pathways, as well as being the first to mention global (or even isolated) downregulation of DNA repair gene transcripts following viral infection. The potential implications of identified transcriptional alterations are discussed in further detail below. Schematics that summarize the apoptosis-related transcriptional changes identified following reovirus infection are depicted in Fig. 5 (mitochondrial and death receptor signaling) and Fig. 6 (ER stress and death receptor signaling) for reference in this discussion.
FIG. 5.
Schematic of mitochondrion- and death receptor-related transcriptional alterations detected by microarray analysis following reovirus infection. Transcripts that were differentially expressed in HEK293 cells following reovirus infection compared to mock infection are indicated in boldface type. Please see Discussion for details of each indicated transcript.
FIG. 6.
Schematic of ER stress-related transcriptional alterations detected by microarray analysis following reovirus infection. Transcripts that were differentially expressed in HEK293 cells following reovirus infection compared to mock infection are indicated in boldface type. Please see Discussion for details of each indicated transcript.
Death receptor pathways.
Members of the TNF receptor superfamily of cell surface death receptors—specifically DR4, DR5 and their apoptosis-inducing ligand, TRAIL—play a critical role in reovirus-induced apoptosis in HEK293 cells (11). We did not detect significant alteration in the expression of any TNF receptor superfamily members or their ligands by microarray analysis, but we did detect upregulation of transcripts for DR4 and downregulation of the decoy receptor DcR-2 by RT-PCR. This suggests, with the possible exception of DR4 and DcR-2, that reovirus activation of death receptor pathways does not involve changes in gene expression but rather occurs predominantly at the protein level. However, microarray analysis did detect alteration in the expression of genes encoding BCL-10, PML-2, and ceramide glucosyltransferase. The proteins encoded by these genes can modulate death receptor signaling, suggesting that reovirus-induced alterations in expression of these genes might influence death receptor signaling cascades.
Bcl-10 (derived from B-cell malt lymphomas) binds to TRAF2, a key accessory mediator of TNF-R signaling (78, 91). This binding can perturb TRAF-related activation of mitogen-activated protein kinases (MAPK), including JNK, and can induce the activation of the transcription factor NF-κB (25, 76, 78, 87, 91, 92). Bcl-10 contains a caspase activation and recruitment domain and can bind to procaspase-9, thereby promoting its autoproteolytic activation (45, 76, 92). Overexpression of Bcl-10 induces apoptosis in a variety of cells, including HEK293 cells (92), and it may provide a potential link between the capacity of reoviruses to activate JNK cascades, activate the transcription factor NF-κB, and induce apoptosis. PML-2 (encoded by the acute promyelocytic leukemia gene) has been shown to enhance activation of death receptor-mediated pathways involving Fas-Fas ligand and TNF and TNF-R (84, 85), suggesting the possibility that it could also potentiate signaling through DR4 and DR5. The mechanism of action of PML is unclear but appears to involve the activation of effector caspases, including caspase 3 (85). Ceramide has been implicated as an important signaling intermediary involved in both Fas-Fas ligand-mediated apoptosis and activation of MAPK cascades (27, 56, 62) Although expression of genes encoding ceramide synthesis were not altered in infected cells, the gene encoding ceramide glucosyltransferase was upregulated. This enzyme catalyzes the initial glycosylation step in glycosphingolipid synthesis to produce glucosylceramide, and its upregulation could potentially enhance ceramide signaling.
Bcl-2 and mitochondrial signaling pathways.
Pro- and antiapoptotic members of the Bcl-2 family interact at the surface of the mitochondria, where complex homo- and heterodimeric interactions regulate release of proapoptotic molecules including cytochrome c, Smac/DIABLO, and apoptosis-inducing factor (28). Reovirus infection is associated with release of cytochrome c from the mitochondria into the cytoplasm and with activation of caspase 9 (43). Stable overexpression of the antiapoptotic molecule Bcl-2 inhibits reovirus-induced apoptosis in both MDCK (69) and HEK293 cells (43, 44). These results suggest that, in order to induce apoptosis, reovirus must overcome the antiapoptotic effects of Bcl-2 and related family members in order to activate mitochondrial apoptotic pathways.
Genes encoding several proteins that inhibit the activity of Bcl-2 and therefore facilitate apoptosis were found to be up- regulated in reovirus-infected cells. These included Par-4, DRAK-2, DP5, and BNIP-1. Par-4 was initially identified in prostate tumor cells undergoing apoptosis but is widely expressed in human tissues (50). Although the exact mechanism of action of Par-4 is not known, it can facilitate apoptosis by suppression of Bcl-2 expression, inhibition of NF-κB activation, and activation of caspase 8 (2, 9, 21). DRAK-2 is a member of a novel family of nuclear serine-threonine kinases that can induce apoptosis (72). These kinases are known to associate with Par-4, and coexpression of Par-4 and the Zip kinase (42) (related to DRAK-2) enhances apoptosis (60). DP-5 (a death-promoting gene) contains a BH3 domain that allows it to interact with Bcl-2 family proteins. Overexpression of DP5 induces apoptosis in a variety of cells (33, 34). DP5 activation is linked with calcium release from ER stores, suggesting that DP-5 may play a role as a link between ER stress-induced and mitochondrial apoptotic pathways (33). BNIP-1 is a member of a novel BH3 domain-containing protein family that interacts with both Bcl-2 and Bcl-xL to inhibit their antiapoptotic actions, thereby enhancing apoptosis induction (49).
In addition to the upregulation of transcripts encoding Bcl-2-inhibitory proteins, downregulation of transcripts encoding proteins that promote Bcl-2 activity could contribute to promotion of apoptosis following reovirus infection. Pim 2 proto-oncogene homologue is a serine-threonine kinase that is highly expressed in a variety of tissues that may play an antiapoptotic role by enhancing Bcl-2 expression (3, 75). It is one of the few apoptosis regulatory genes that was downregulated following reovirus infection. Since the Pim 2 gene product enhances Bcl-2 expression, its downregulation might reduce levels of Bcl-2 in infected cells and thereby enhance apoptosis.
In parallel with these transcriptional alterations with expected anti-Bcl-2 implications, alterations were also noted in expression of transcripts encoding proteins expected to promote the action of Bcl-2 and block apoptosis. Genes encoding two proteins that may act as positive modulators of Bcl-2—SMN and SIP—were found to be upregulated following reovirus infection. SMN interacts with Bcl-2 to enhance its antiapoptotic activity (35). SIP-1 interacts with SMN to form a heterodimeric complex (22a). Coexpression of SMN and Bcl-2 provides a synergistic protective effect against Bax-induced or Fas-mediated apoptosis (35, 73). Expression of the gene encoding the Bcl-2 family member MCL-1 was also upregulated following reovirus infection. MCL-1 may exert either pro- or antiapoptotic activity by modulation of the activity of Bcl-2 or by acting independently (6).
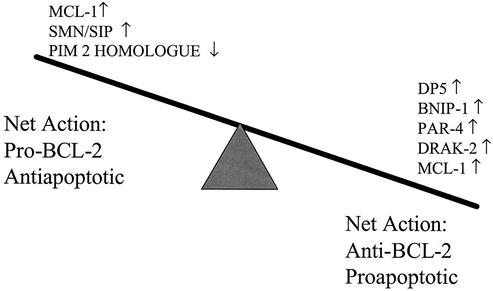
Taken together, these results suggest a model in which reovirus infection is associated with the altered expression of multiple modulators of Bcl-2 in infected host cells, with the balance tipped toward genes encoding proteins that would be predicted to inhibit Bcl-2 activity and thereby promote apoptosis (Fig. 7). Although changes in mRNA levels do not necessarily predict changes in protein expression, we demonstrated that at least one of these proteins, SMN, is altered in vivo in a biologically relevant model of reovirus infection, in close temporal and spatial association with evidence of virus-induced apoptotic myocardial tissue injury.
FIG. 7.
Transcriptional regulation of Bcl-2 modulatory proteins is a central theme in reovirus-induced apoptosis. Expression of eight transcripts encoding proteins known to influence Bcl-2 activity was altered following reovirus infection. The predicted result of the observed transcriptional alterations would be net inhibition of Bcl-2, and thus promotion of apoptosis, in reovirus-infected cells. Please see Discussion for details of each indicated transcript.
In addition to the modulatory struggle at the Bcl-2 level, alteration of a transcript encoding a protein with a separate role in mitochondrial apoptotic signaling was detected. HSP-70 homologue expression was increased following reovirus infection. HSP-70 is an antiapoptotic chaperone protein (52) that inhibits mitochondrial release of cytochrome c and blocks the recruitment of procaspase 9 to the apoptosome complex (4, 47, 71). Although the cellular actions of HSP-70 are predominantly antiapoptotic, the protein also plays a role during reovirus replication in facilitating the trimerization of the reovirus σ1 protein (46). Since this protein is a critical determinant of reovirus apoptosis and reovirus-induced activation of specific MAPK signaling cascades (13), HSP-70 homologue may also facilitate apoptosis through its actions during virion assembly.
ER stress pathways.
The accumulation of abnormal quantities of protein or of malfolded proteins in the Golgi apparatus or ER may trigger kinase cascades that result in the activation of caspase 12 (7, 53, 54). This initiator caspase triggers activation of effector caspases, such as caspase 3. One of the cellular markers of Golgi and ER stress is an increase in the quantities of specific GADD proteins such as GADD 135/CHOP (41, 94). Expression of genes encoding two of the five currently described members of the GADD family, GADD 34 and GADD 45, were increased following reoviral infection. These genes were the earliest ones found to be significantly upregulated following reovirus infection. The upregulation of ER stress apoptosis-inducing transcripts was complemented by downregulated expression of the gene for ORP 150, which encodes a protein involved in refolding malfolded protein transcripts within the ER (32a). In addition to a potential role for GADD 34 for ER stress-mediated pathways, GADD 34 induction parallels that of BAX in other models of apoptosis (30), thus suggesting a possible link to mitochondrion-regulated apoptosis signaling pathways, which are known to play an important role in reoviral infection. These results suggests that ER stress pathways may be activated as an early event following reoviral infection and that ER stress-induced apoptotic signaling may contribute to reovirus-induced apoptosis. However, the fact that altered gene expression occurred following both T1L (APO−) and T3A (APO+) infection [although at lower levels in T1L (APO−) infection] suggests the possibility that this pathway may play a less-critical role in determining virus-induced apoptotic injury and rather is activated as a nonspecific cellular response to reoviral infection.
Cysteine proteases.
Death receptor, mitochondrial, and Golgi and ER pathways of apoptosis all initiate the activation of specific initiator caspases, which in turn trigger the activation of a limited set of downstream effector caspases, including caspase 3. Caspases 3, 8, and 9 are all activated during reovirus infection (43), and inhibition of this activation inhibits reovirus-induced apoptosis. Expression of the genes encoding caspases 3 and 7 were increased following reovirus infection. This suggests that, as well as inducing the activation of specific caspases at the posttranslational level, reovirus infection also results in upregulation of caspase gene expression that would be predicted to increase the levels of key effector caspases in infected cells.
We have previously shown that the cysteine protease calpain is also activated in reovirus-infected fibroblasts and myocardiocytes in vitro (19) and in the heart in vivo (18). This activation appears to be an extremely early event that can be detected as early as 30 min following infection of cells with purified virus. Inhibition of calpain activation inhibits apoptosis and reduces reovirus-induced cytopathic effects in vitro and prevents reovirus-induced apoptotic myocardial injury in vivo (18). The mechanism for the proapoptotic actions of calpains is not fully understood but may involve activation of and activation by several caspases (40, 51, 53, 66, 70, 88). Calpain can also facilitate activation of NF-κB by degrading its cytoplasmic inhibitor, IκB (1).
Surprisingly, rather than being upregulated, the expression of the gene encoding calpain was downregulated in reovirus-infected cells. This downregulation was apparent only at 24 h postinfection and could potentially represent a negative feedback response to calpain activation at the protein level: initial increases in calpain activity in infected cells could potentially be countered by downregulation of calpain expression at the transcriptional level at later time points.
Transcriptional changes related to DNA repair.
Expression of transcripts encoding multiple classes of DNA repair enzymes was decreased following T3A (APO+)—but not T1L (APO−)—reovirus infection. Most DNA repair mechanisms involve a recognition step in which single- or double-stranded DNA breaks are identified, followed by the sequential action of helices that unwind damaged segments, nucleases that incise the damaged region, polymerase that resynthesizes DNA, and ligases that repair DNA strand breaks. Failure of any of these steps can result in accumulation of DNA damage and the subsequent induction of apoptosis (67).
DNA nucleotide mismatches or mutations are recognized by a mismatch-binding factor that consists of two distinct proteins—hMSH2 and G/T mismatch binding protein (GTBP)—both of which are required for mismatch-specific binding (61). The gene encoding GTBP was downregulated >2-fold at both 12 and 24 h post-T3A (APO+) infection. Downregulation of GTBP would be expected to impede recognition of single-strand DNA breaks or mutations that distort the structure of the DNA helix.
Once damage has been sensed, helicases unwind damaged DNA as a precursor to excision of the damaged segments. RAD54 homologue, Mi2 autoantigen, and helicase-like protein are three helicases (26) whose transcripts were downregulated following reovirus infection. Genes encoding two nucleotide excision repair enzymes, ERCC5 and XP-C repair-complementing protein p125, were both downregulated following T3A (APO+) reovirus infection. These enzymes are involved in repairing single-strand DNA breaks or in repairing nucleotide mutations that distort the structure of the DNA helix (67). Damaged DNA binding protein 2 (DDB2) may also play a role in nucleotide excision repair (95)—and like ERCC5 and XP-C repair-complementing protein p125, expression of genes encoding DDB2 was also downregulated in T3A (APO+)-infected cells.
Once damage is recognized, the helix is unwound, the damaged area is excised, and new DNA synthesis is required to replace the damaged nucleotides. At least nine DNA polymerases involved in various aspects of DNA replication and repair have been identified in eukaryotes (31). DNA polymerase α and DNA polymerase γ transcripts were both downregulated following T3A (APO+) reovirus infection. DNA polymerase α is primarily required for DNA replication but also interacts with and coordinates other DNA polymerases and cellular factors required for DNA repair. DNA polymerase γ is the sole polymerase required for mitochondrial replication and plays an important role in the efficient repair of mismatched DNA in vitro as well as in the repair of damaged DNA (31, 89).
Following DNA synthesis, the repaired segment must be religated with the rest of the helix. DNA ligases promote the rejoining of both double- and single-stranded DNA breaks (37, 77). Expression of the gene encoding DNA ligase 1 showed the most change in expression of any DNA repair-related gene analyzed (downregulated 8.2 ± 1.1-fold at 24 h post-T3A infection). The second-largest change in gene expression following T3A reovirus infection was that of PARPL. PARPL attaches poly(ADP-ribose) chains to damaged DNA, a process termed ribosylation. Ribosylation is a key step during DNA repair and transcription that prevents binding of transcription factors to regions of damaged DNA (48). Thus, PARPL may serve as a molecular switch between transcription and repair of DNA to avoid expression of damaged genes (59).
The downregulation of genes encoding DNA damage and repair has not previously been appreciated as a consequence of reoviral infection. The fact this global downregulation occurred following T3A (APO+) and not T1L (APO−) reovirus infection suggests that, as well as directly stimulating proapoptotic signaling pathways, T3A reovirus infection may result in signaling pathways that facilitate apoptosis by hampering the capacity of the host cell to repair DNA damage.
Regulatory mechanisms involved in apoptosis, DNA repair, and cell cycle regulation are highly integrated and involve a number of overlapping and intersecting signaling pathways and proteins. Expression analysis performed by using high-density oligonucleotide arrays provides a unique opportunity to investigate the complex mechanisms responsible for pathogenic effects in reovirus-infected cells and tissues. Transcriptional analysis may not only be directly translatable into in vivo models of myocarditis and encephalitis, but more importantly, it may provide testable hypotheses that may not have been explored in the absence of a large-scale analysis of multiple concurrent signaling networks. Work is in progress to further investigate the models suggested in this report by manipulating genes with potentially central themes and observing effects on apoptosis and cell cycle arrest as well as effects on signaling cascades known to be operative in the reovirus model.
Acknowledgments
We thank Samuel Pan for his contribution to RT-PCR validation of microarray data and Vicki VanPutten for technical assistance in analysis of microarray data.
REFERENCES
- 1.Baghdiguian, S., M. Martin, I. Richard, F. Pons, C. Astier, N. Bourg, R. T. Hay, R. Chemaly, G. Halaby, J. Loiselet, L. V. Anderson, D. M. Lopez, M. Fardeau, P. Mangeat, J. S. Beckmann, and G. Lefranc. 1999. Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IκB α/NF-κB pathway in limb-girdle muscular dystrophy type 2A. Nat. Med. 5:503-511. [DOI] [PubMed] [Google Scholar]
- 2.Barradas, M., A. Monjas, M. T. Diaz-Meco, M. Serrano, and J. Moscat. 1999. The downregulation of the pro-apoptotic protein Par-4 is critical for Ras-induced survival and tumor progression. EMBO J. 18:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baytel, D., S. Shalom, I. Madgar, R. Weissenberg, and J. Don. 1998. The human Pim-2 proto-oncogene and its testicular expression. Biochim. Biophys. Acta 1442:274-285. (Erratum, 1444:312-313, 1999.) [DOI] [PubMed]
- 4.Beere, H. M., B. B. Wolf, K. Cain, D. D. Mosser, A. Mahboubi, T. Kuwana, P. Tailor, R. I. Morimoto, G. M. Cohen, and D. R. Green. 2000. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2:469-475. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingle, C. D., R. W. Craig, B. M. Swales, V. Singleton, P. Zhou, and M. K. Whyte. 2000. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J. Biol. Chem. 275:22136-22146. [DOI] [PubMed] [Google Scholar]
- 7.Bitko, V., and S. Barik. 2001. An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J. Cell Biochem. 80:441-454. [DOI] [PubMed] [Google Scholar]
- 8.Bolt, G., K. Berg, and M. Blixenkrone-Moller. 2002. Measles virus-induced modulation of host-cell gene expression. J. Gen. Virol. 83:1157-1165. [DOI] [PubMed] [Google Scholar]
- 9.Camandola, S., and M. P. Mattson. 2000. Pro-apoptotic action of PAR-4 involves inhibition of NF-κB activity and suppression of BCL-2 expression. J. Neurosci. Res. 61:134-139. [DOI] [PubMed] [Google Scholar]
- 10.Carter, K. L., E. Cahir-McFarland, and E. Kieff. 2002. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76:10427-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, P., S. M. Meintzer, S. Gibson, C. Widmann, T. P. Garrington, G. L. Johnson, and K. L. Tyler. 2000. Reovirus-induced apoptosis is mediated by TRAIL. J. Virol. 74:8135-8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, P., S. M. Meintzer, A. C. Spalding, G. L. Johnson, and K. L. Tyler. 2001. Caspase 8-dependent sensitization of cancer cells to TRAIL-induced apoptosis following reovirus-infection. Oncogene 20:6910-6919. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, P., S. M. Meintzer, C. Widmann, G. L. Johnson, and K. L. Tyler. 2001. Reovirus infection activates JNK and the JNK-dependent transcription factor c-Jun. J. Virol. 75:11275-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke, P., and K. L. Tyler. 2003. Reovirus-induced apoptosis: a minireview. Apoptosis 8:141-150. [DOI] [PubMed] [Google Scholar]
- 15.Connolly, J. L., E. S. Barton, and T. S. Dermody. 2001. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J. Virol. 75:4029-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connolly, J. L., S. E. Rodgers, P. Clarke, D. W. Ballard, L. D. Kerr, K. L. Tyler, and T. S. Dermody. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J. Virol. 74:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 76:4467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeBiasi, R. L., C. L. Edelstein, B. Sherry, and K. L. Tyler. 2001. Calpain inhibition protects against virus-induced apoptotic myocardial injury. J. Virol. 75:351-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeBiasi, R. L., M. K. Squier, B. Pike, M. Wynes, T. S. Dermody, J. J. Cohen, and K. L. Tyler. 1999. Reovirus-induced apoptosis is preceded by increased cellular calpain activity and is blocked by calpain inhibitors. J. Virol. 73:695-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Fuente, C., F. Santiago, L. Deng, C. Eadie, I. Zilberman, K. Kehn, A. Maddukuri, S. Baylor, K. Wu, C. G. Lee, A. Pumfery, and F. Kashanchi. 2002. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 3:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Meco, M. T., M. J. Lallena, A. Monjas, S. Frutos, and J. Moscat. 1999. Inactivation of the inhibitory κB protein kinase/nuclear factor κB pathway by Par-4 expression potentiates tumor necrosis factor α-induced apoptosis. J. Biol. Chem. 274:19606-19612. [DOI] [PubMed] [Google Scholar]
- 22.Everett, H., and G. McFadden. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160-165. [DOI] [PubMed] [Google Scholar]
- 22a.Fischer, U., Q. Liu, and G. Dreyfuss. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90:1023-1029. [DOI] [PubMed] [Google Scholar]
- 23.Geiss, G. K., M. C. An, R. E. Bumgarner, E. Hammersmark, D. Cunningham, and M. G. Katze. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith, T. S., S. R. Wiley, M. Z. Kubin, L. M. Sedger, C. R. Maliszewski, and N. A. Fanger. 1999. Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J. Exp. Med. 189:1343-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guiet, C., and P. Vito. 2000. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-κB activation. J. Cell Biol. 148:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammermann, R., U. Warskulat, and D. Haussinger. 1998. Anisoosmotic regulation of the Mi-2 autoantigen mRNA in H4IIE rat hepatoma cells and primary hepatocytes. FEBS Lett. 435:21-24. [DOI] [PubMed] [Google Scholar]
- 27.Hannun, Y. A. 1996. Functions of ceramide in coordinating cellular responses to stress. Science 274:1855-1859. [DOI] [PubMed] [Google Scholar]
- 28.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 29.Hobbs, W. E., and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollander, M. C., Q. Zhan, I. Bae, and A. J. Fornace, Jr. 1997. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 272:13731-13737. [DOI] [PubMed] [Google Scholar]
- 31.Hubscher, U., H. P. Nasheuer, and J. E. Syvaoja. 2000. Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25:143-147. [DOI] [PubMed] [Google Scholar]
- 32.Iizuka, N., M. Oka, H. Yamada-Okabe, N. Mori, T. Tamesa, T. Okada, N. Takemoto, A. Tangoku, K. Hamada, H. Nakayama, T. Miyamoto, S. Uchimura, and Y. Hamamoto. 2002. Comparison of gene expression profiles between hepatitis B virus- and hepatitis C virus-infected hepatocellular carcinoma by oligonucleotide microarray data on the basis of a supervised learning method. Cancer Res. 62:3939-3944. [PubMed] [Google Scholar]
- 32a.Ikeda, J., S. Kaneda, K. Kuwabara, S. Ogawa, T. Kobayashi, M. Matsumoto, T. Yura, and H. Yunagi. 1997. Cloning and expression of cDNA encoding the human 150 kDa oxygen-regulated protein, ORP150. Biochem. Biophys. Res. Commun. 230:94-99. [DOI] [PubMed] [Google Scholar]
- 33.Imaizumi, K., T. Morihara, Y. Mori, T. Katayama, M. Tsuda, T. Furuyama, A. Wanaka, M. Takeda, and M. Tohyama. 1999. The cell death-promoting gene DP5, which interacts with the BCL2 family, is induced during neuronal apoptosis following exposure to amyloid beta protein. J. Biol. Chem. 274:7975-7981. [DOI] [PubMed] [Google Scholar]
- 34.Imaizumi, K., M. Tsuda, Y. Imai, A. Wanaka, T. Takagi, and M. Tohyama. 1997. Molecular cloning of a novel polypeptide, DP5, induced during programmed neuronal death. J. Biol. Chem. 272:18842-18848. [DOI] [PubMed] [Google Scholar]
- 35.Iwahashi, H., Y. Eguchi, N. Yasuhara, T. Hanafusa, Y. Matsuzawa, and Y. Tsujimoto. 1997. Synergistic anti-apoptotic activity between Bcl-2 and SMN implicated in spinal muscular atrophy. Nature 390:413-417. [DOI] [PubMed] [Google Scholar]
- 36.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, A. P., and M. P. Fairman. 1997. The identification and purification of a novel mammalian DNA ligase. Mutat. Res. 383:205-212. [DOI] [PubMed] [Google Scholar]
- 38.Johnston, C., W. Jiang, T. Chu, and B. Levine. 2001. Identification of genes involved in the host response to neurovirulent alphavirus infection. J. Virol. 75:10431-10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato, M., T. Nonaka, M. Maki, H. Kikuchi, and S. Imajoh-Ohmi. 2000. Caspases cleave the amino-terminal calpain inhibitory unit of calpastatin during apoptosis in human Jurkat T cells. J. Biochem. (Tokyo) 127:297-305. [DOI] [PubMed] [Google Scholar]
- 41.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 42.Kogel, D., H. Bierbaum, U. Preuss, and K. H. Scheidtmann. 1999. C-terminal truncation of Dlk/ZIP kinase leads to abrogation of nuclear transport and high apoptotic activity. Oncogene 18:7212-7218. [DOI] [PubMed] [Google Scholar]
- 43.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ. 9:926-933. [DOI] [PubMed] [Google Scholar]
- 44.Kominsky, D. J., R. J. Bickel, and K. L. Tyler. 2002. Reovirus-induced apoptosis requires mitochondrial release of Smac/DIABLO and involves reduction of cellular inhibitor of apoptosis protein levels. J. Virol. 76:11414-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koseki, T., N. Inohara, S. Chen, R. Carrio, J. Merino, M. O. Hottiger, G. J. Nabel, and G. Nunez. 1999. CIPER, a novel NF κB-activating protein containing a caspase recruitment domain with homology to herpesvirus-2 protein E10. J. Biol. Chem. 274:9955-9961. [DOI] [PubMed] [Google Scholar]
- 46.Leone, G., M. C. Coffey, R. Gilmore, R. Duncan, L. Maybaum, and P. W. Lee. 1996. C-terminal trimerization, but not N-terminal trimerization, of the reovirus cell attachment protein is a posttranslational and Hsp70/ATP-dependent process. J. Biol. Chem. 271:8466-8471. [DOI] [PubMed] [Google Scholar]
- 47.Li, C. Y., J. S. Lee, Y. G. Ko, J. I. Kim, and J. S. Seo. 2000. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase 3 activation. J. Biol. Chem. 275:25665-25671. [DOI] [PubMed] [Google Scholar]
- 48.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 49.Low, B. C., Y. P. Lim, J. Lim, E. S. Wong, and G. R. Guy. 1999. Tyrosine phosphorylation of the Bcl-2-associated protein BNIP-2 by fibroblast growth factor receptor-1 prevents its binding to Cdc42GAP and Cdc42. J. Biol. Chem. 274:33123-33130. [DOI] [PubMed] [Google Scholar]
- 50.Mattson, M. P., W. Duan, S. L. Chan, and S. Camandola. 1999. Par-4: an emerging pivotal player in neuronal apoptosis and neurodegenerative disorders. J. Mol. Neurosci. 13:17-30. [DOI] [PubMed] [Google Scholar]
- 51.McGinnis, K. M., M. E. Gnegy, Y. H. Park, N. Mukerjee, and K. K. Wang. 1999. Procaspase-3 and poly(ADP)ribose polymerase (PARP) are calpain substrates. Biochem. Biophys. Res. Commun. 263:94-99. [DOI] [PubMed] [Google Scholar]
- 52.Mosser, D. D., A. W. Caron, L. Bourget, A. B. Meriin, M. Y. Sherman, R. I. Morimoto, and B. Massie. 2000. The chaperone function of hsp70 is required for protection against stress-induced apoptosis. Mol. Cell. Biol. 20:7146-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa, T., and J. Yuan. 2000. Cross-talk between two cysteine protease families—activation of caspase-12 by calpain in apoptosis. J. Cell Biol. 150:887-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakagawa, T., H. Zhu, N. Morishima, E. Li, J. Xu, B. A. Yankner, and J. Yuan. 2000. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403:98-103. [DOI] [PubMed] [Google Scholar]
- 55.Nees, M., J. M. Geoghegan, T. Hyman, S. Frank, L. Miller, and C. D. Woodworth. 2001. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J. Virol. 75:4283-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obeid, L. M., C. M. Linardic, L. A. Karolak, and Y. A. Hannun. 1993. Programmed cell death induced by ceramide. Science 259:1769-1771. [DOI] [PubMed] [Google Scholar]
- 57.Oberhaus, S. M., T. S. Dermody, and K. L. Tyler. 1998. Apoptosis and the cytopathic effects of reovirus. Curr. Top. Microbiol. Immunol. 233(Reovir.ii):23-49. [DOI] [PubMed] [Google Scholar]
- 58.Oberhaus, S. M., R. L. Smith, G. H. Clayton, T. S. Dermody, and K. L. Tyler. 1997. Reovirus infection and tissue injury in the mouse central nervous system are associated with apoptosis. J. Virol. 71:2100-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oei, S. L., J. Griesenbeck, M. Schweiger, and M. Ziegler. 1998. Regulation of RNA polymerase II-dependent transcription by poly(ADP-ribosyl)ation of transcription factors. J. Biol. Chem. 273:31644-31647. [DOI] [PubMed] [Google Scholar]
- 60.Page, G., D. Kogel, V. Rangnekar, and K. H. Scheidtmann. 1999. Interaction partners of Dlk/ZIP kinase: co-expression of Dlk/ZIP kinase and Par-4 results in cytoplasmic retention and apoptosis. Oncogene 18:7265-7273. [DOI] [PubMed] [Google Scholar]
- 61.Palombo, F., P. Gallinari, I. Iaccarino, T. Lettieri, M. Hughes, A. D'Arrigo, O. Truong, J. J. Hsuan, and J. Jiricny. 1995. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science 268:1912-1914. [DOI] [PubMed] [Google Scholar]
- 62.Perry, D. K. 1999. Ceramide and apoptosis. Biochem. Soc. Trans. 27:399-404. [DOI] [PubMed] [Google Scholar]
- 63.Pietiainen, V., P. Huttunen, and T. Hyypia. 2000. Effects of echovirus 1 infection on cellular gene expression. Virology 276:243-250. [DOI] [PubMed] [Google Scholar]
- 64.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 65.Poggioli, G. J., R. L. DeBiasi, R. Bickel, R. Jotte, A. Spalding, G. L. Johnson, and K. L. Tyler. 2002. Reovirus-induced alterations in gene expression related to cell cycle regulation. J. Virol. 76:2585-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rami, A., R. Agarwal, G. Botez, and J. Winckler. 2000. μ-Calpain activation, DNA fragmentation, and synergistic effects of caspase and calpain inhibitors in protecting hippocampal neurons from ischemic damage. Brain Res. 866:299-312. [DOI] [PubMed] [Google Scholar]
- 67.Rich, T., R. L. Allen, and A. H. Wyllie. 2000. Defying death after DNA damage. Nature 407:777-783. [DOI] [PubMed] [Google Scholar]
- 68.Richardson-Burns, S. M., D. J. Kominsky, and K. L. Tyler. 2002. Reovirus-induced neuronal apoptosis is mediated by caspase 3 and is associated with the activation of death receptors. J. Neurovirol. 8:365-380. [DOI] [PubMed] [Google Scholar]
- 69.Rodgers, S. E., E. S. Barton, S. M. Oberhaus, B. Pike, C. A. Gibson, K. L. Tyler, and T. S. Dermody. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J. Virol. 71:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruiz-Vela, A., D. B. Gonzalez, and A. Martinez. 1999. Implication of calpain in caspase activation during B cell clonal deletion. EMBO J. 18:4988-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saleh, A., S. M. Srinivasula, L. Balkir, P. D. Robbins, and E. S. Alnemri. 2000. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2:476-483. [DOI] [PubMed] [Google Scholar]
- 72.Sanjo, H., T. Kawai, and S. Akira. 1998. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J. Biol. Chem. 273:29066-29071. [DOI] [PubMed] [Google Scholar]
- 73.Sato, K., Y. Eguchi, T. S. Kodama, and Y. Tsujimoto. 2000. Regions essential for the interaction between Bcl-2 and SMN, the spinal muscular atrophy disease gene product. Cell Death Differ. 7:374-383. [DOI] [PubMed] [Google Scholar]
- 74.Sherry, B., F. J. Schoen, E. Wenske, and B. N. Fields. 1989. Derivation and characterization of an efficiently myocarditic reovirus variant. J. Virol. 63:4840-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shirogane, T., T. Fukada, J. M. Muller, D. T. Shima, M. Hibi, and T. Hirano. 1999. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11:709-719. [DOI] [PubMed] [Google Scholar]
- 76.Srinivasula, S. M., M. Ahmad, J. H. Lin, J. L. Poyet, T. Fernandes-Alnemri, P. N. Tsichlis, and E. S. Alnemri. 1999. CLAP, a novel caspase recruitment domain-containing protein in the tumor necrosis factor receptor pathway, regulates NF-κB activation and apoptosis. J. Biol. Chem. 274:17946-17954. [DOI] [PubMed] [Google Scholar]
- 77.Teo, S. H., and S. P. Jackson. 1997. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 16:4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thome, M., F. Martinon, K. Hofmann, V. Rubio, V. Steiner, P. Schneider, C. Mattmann, and J. Tschopp. 1999. Equine herpesvirus-2 E10 gene product, but not its cellular homologue, activates NF-κB transcription factor and c-Jun N-terminal kinase. J. Biol. Chem. 274:9962-9968. [DOI] [PubMed] [Google Scholar]
- 79.Tirasophon, W., K. Lee, B. Callaghan, A. Welihinda, and R. J. Kaufman. 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14:2725-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tyler, K. L. 1998. Pathogenesis of reovirus infections of the central nervous system. Curr. Top. Microbiol. Immunol. 233 (Reovir.ii):93-124. [DOI] [PubMed] [Google Scholar]
- 81.Tyler, K. L., P. Clarke, R. L. DeBiasi, D. Kominsky, and G. J. Poggioli. 2001. Reoviruses and the host cell. Trends Microbiol. 9:560-564. [DOI] [PubMed] [Google Scholar]
- 82.Tyler, K. L., M. K. Squier, S. E. Rodgers, B. E. Schneider, S. M. Oberhaus, T. A. Grdina, J. J. Cohen, and T. S. Dermody. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein σ1. J. Virol. 69:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 85.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 86.Welihinda, A. A., W. Tirasophon, and R. J. Kaufman. 1999. The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 7:293-300. [PMC free article] [PubMed] [Google Scholar]
- 87.Willis, T. G., D. M. Jadayel, M. Q. Du, H. Peng, A. R. Perry, M. Abdul-Rauf, H. Price, L. Karran, O. Majekodunmi, I. Wlodarska, L. Pan, T. Crook, R. Hamoudi, P. G. Isaacson, and M. J. Dyer. 1999. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 96:35-45. [DOI] [PubMed] [Google Scholar]
- 88.Wood, D. E., and E. W. Newcomb. 1999. Caspase-dependent activation of calpain during drug-induced apoptosis. J. Biol. Chem. 274:8309-8315. [DOI] [PubMed] [Google Scholar]
- 89.Wood, R. D., and M. K. Shivji. 1997. Which DNA polymerases are used for DNA repair in eukaryotes? Carcinogenesis 18:605-610. [DOI] [PubMed] [Google Scholar]
- 90.Yang, W. C., G. V. Devi-Rao, P. Ghazal, E. K. Wagner, and S. J. Triezenberg. 2002. General and specific alterations in programming of global viral gene expression during infection by VP16 activation-deficient mutants of herpes simplex virus type 1. J. Virol. 76:12758-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoneda, T., K. Imaizumi, M. Maeda, D. Yui, T. Manabe, T. Katayama, N. Sato, F. Gomi, T. Morihara, Y. Mori, K. Miyoshi, J. Hitomi, S. Ugawa, S. Yamada, M. Okabe, and M. Tohyama. 2000. Regulatory mechanisms of TRAF2-mediated signal transduction by Bcl10, a MALT lymphoma-associated protein. J. Biol. Chem. 275:11114-11120. [DOI] [PubMed] [Google Scholar]
- 92.Zhang, Q., R. Siebert, M. Yan, B. Hinzmann, X. Cui, L. Xue, K. M. Rakestraw, C. W. Naeve, G. Beckmann, D. D. Weisenburger, W. G. Sanger, H. Nowotny, M. Vesely, E. Callet-Bauchu, G. Salles, V. M. Dixit, A. Rosenthal, B. Schlegelberger, and S. W. Morris. 1999. Inactivating mutations and overexpression of BCL10, a caspase recruitment domain-containing gene, in MALT lymphoma with t(1;14)(p22;q32). Nat. Genet. 22:63-68. [DOI] [PubMed] [Google Scholar]
- 93.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zinszner, H., M. Kuroda, X. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zolezzi, F., and S. Linn. 2000. Studies of the murine DDB1 and DDB2 genes. Gene 245:151-159. [DOI] [PubMed] [Google Scholar]