Abstract
The NS5B RNA-dependent RNA polymerase encoded by the hepatitis C virus (HCV) is a key component of the viral replicase. Reported here is the three-dimensional structure of HCV NS5B polymerase, with the highlight on its C-terminal folding, determined by X-ray crystallography at 2.1-Å resolution. Structural analysis revealed that a stretch of C-terminal residues of HCV NS5B inserted into the putative RNA binding cleft, where they formed a hydrophobic pocket and interacted with several important structural elements. This region was found to be conserved and unique to the RNA polymerases encoded by HCV and related viruses. Through biochemical analyses, we confirmed that this region interfered with the binding of HCV NS5B to RNA. Deletion of this fragment from HCV NS5B enhanced the RNA synthesis rate up to ∼50-fold. These results provide not only direct experimental insights into the role of the C-terminal tail of HCV NS5B polymerase but also a working model for the RNA synthesis mechanism employed by HCV and related viruses.
Hepatitis C virus (HCV) is a small plus-strand RNA virus responsible for a significant proportion of acute and chronic hepatitis in humans (9, 29). It is estimated that over 170 million people worldwide are potentially infected by HCV (10). Most acute HCV infections can develop into chronic hepatitis and further progress into cirrhosis and liver failure (9, 10, 29). Therefore, HCV infections represent a serious health problem globally. HCV contains a plus-strand RNA genome of ∼9.6 kb encoding a single polyprotein (24, 25). This polyprotein precursor can be processed, by both host and virally encoded proteases, to generate mature structural and nonstructural proteins that are required for virus replication and assembly (24, 25, 28). One of the nonstructural proteins, designated NS5B, is an RNA-dependent RNA polymerase (RdRp) due to the presence of the hallmark GDD motif essential for RNA polymerase function (4, 12).
The essentiality of NS5B activity to HCV replication and infection has been established in a chimpanzee model (18). Therefore, the HCV NS5B polymerase has been viewed as an important target for developing antiviral therapy. Various versions of the recombinant HCV NS5B polymerase have been produced and purified from both bacterial and insect cells (2, 11, 15, 16, 21, 22, 26, 32, 34, 35). Similar to other viral RdRps, purified HCV NS5B proteins are able to synthesize RNA by using various RNAs as templates in vitro. Recent studies suggested that HCV NS5B catalyzed two different RNA synthesis reactions, primer-dependent RNA elongation and RNA initiation, through a de novo mechanism (15, 17, 21, 22, 26, 31, 34, 36). The availability of highly purified enzyme has also facilitated the structural analysis of HCV NS5B polymerase. To date, three different groups have reported the X-ray crystal structure of the HCV NS5B varying in size and sequences (1, 5, 20).
The full-length HCV NS5B protein contains 591 amino acids, and the catalytic core domain consists of the N-terminal ∼530 amino acid (5). Previous results have shown that the last 21 amino acids at the C terminus of NS5B are hydrophobic and responsible for membrane anchorage of NS5B protein in cells (30, 34). In this report, we present both structural and biochemical evidence suggesting that the C-terminal noncatalytic region of HCV NS5B contains a regulatory motif upstream of the membrane anchor domain. This regulatory motif inhibits RNA binding and polymerase activity when tested in vitro. These findings provide a new insight into the mechanism of HCV polymerase regulation.
MATERIALS AND METHODS
Materials.
Reagents and prepacked columns used for protein purification were purchased from Pharmacia. Homopolymeric RNA templates were purchased from Pharmacia and Roche. Radiolabeled and nonlabeled nucleotides were purchased from Amersham and Gibco, respectively. Peptides used for this study were custom synthesized by Alpha Diagnostic International Inc. (San Antonio, Tex.). The RNA oligonucleotides were custom synthesized by Dharmacom Res, Inc. (Lafayette, Colo.).
Generation of HCV NS5B in bacterial cells.
For expression of a soluble form of NS5B, the cDNA encoding an HCV genotype 1b (BK) NS5B that lacks the C-terminal 21 hydrophobic amino acids (NS5B-570H) was prepared using a standard PCR method. For cloning purpose, Nhel and Xhol cleavage sites were engineered in the PCR primers so that the PCR products could be subcloned directly into the expression vector pET21b (Novagen) at these sites. The resultant expression vector would allow the expression of a tagged NS5B with six histidine residues at its C terminus to aid in protein purification. Another truncated version of the NS5B protein, deleting 51 amino acids from its C terminus (NS5B-540H), was cloned and expressed in the same way (33).
For generation of recombinant NS5B proteins, bacterial JM109-DE3 cells, transformed by one of the NS5B expression vectors described above, were first grown in TY broth containing 0.1 mg of ampicillin per ml and then induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside as described previously (16). The expressed NS5B proteins were purified through a three-step enrichment protocol. Briefly, harvested bacterial cells (∼40 g) were resuspended in 400 ml of buffer A (20 mM Tris [pH 7.5], 1 mM EDTA, 10 mM dithiothreitol [DTT], 10% glycerol, 0.5 M NaCl, protease inhibitor tablets [Roche], 200 U of DNase), and then lysed by sonication. The lysate was clarified by centrifugation at 100,000 × g for 30 min at 4°C. To remove negatively charged bacterial proteins and nucleic acids, clarified supernatant was passed through a positively charged DEAE-Sepharose column. The unbound flowthrough containing the NS5B-570H was loaded onto a 5-ml Pharmacia HiTrap heparin-Sepharose column preequilibrated with buffer B (25 mM HEPES [pH 7.5], 1 mM EDTA, 10 mM DTT, 20% glycerol). After the column was washed with 0.3 M NaCl in buffer B to the baseline absorbance, the bound proteins were eluted using a linear gradient of 0.3 to 1.0 M NaCl in buffer B. NS5B containing fractions were collected and mixed with 5 ml of Ni-nitrilotriacetic acid resin for 90 min at 4°C. The resin was then packed, washed with 30 mM imidazole, and eluted with 500 mM imidazole in buffer B. Fractions containing the histidine-tagged NS5B proteins were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the polymerase assay described below.
Crystallization and structural determination of HCV NS5B.
The highly purified NS5B-570H was first concentrated in buffer B to ∼10 mg/ml using Concentrep (Amersham). Fresh DTT (final concentration, 5 mM) was added just prior to crystallization. Crystals were grown at room temperature in a hanging-drop plate using 2.0 M ammonium sulfate-to 5% 2-propanol as precipitant. Hexagonal crystals of 100 by 100 by 150 mm grew within 7 to 10 days. X-ray diffraction data were collected at −170°C with a Mar charge-coupled device detector at the IMCA beam line ID-17 at Advanced Photon Source in Argonne National Laboratories. Data were integrated and reduced using the program HKL2000. The crystals belong to space group P6522 with unit-cell dimensions of 110.82 Å and 238.57 Å. The structure was solved by molecular replacement with HCV RdRp (PDB code 1HEI) as the searching model. A truncated protein sequence amino acids (1 to 500) with side chains was used for rotation and translation searches. The initial R factor for the solution was 46.4% with a Patterson correlation coefficient of 42.5%. The structure was refined using the program CNS, and sequential model-building processes were carried out with the graphics program QUANTA. C-terminal amino acids were built with difference Fourier maps. Solvent molecules were identified with 2Fo-Fc and Fo-Fc electron density maps. The final R factor for all data is 25.4% with Rfree of 28.5%. All residues are in the allowed regions in the Ramachandran plot.
RNA polymerase filter binding assay.
A typical RNA elongation assay was performed as described previously (16), using poly(A) as the template (10 μg/ml), oligo(U)12 (1 μg/ml) as the primer, and 20 μM [α-32P]UTP as the substrate in a total volume of 25 μl of reaction mix containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 25 mM KCl, 1 mM DTT, and purified NS5B at the concentrations specified. RNA initiation assays were performed using either poly(C) or poly(U) as the template under conditions described previously in the presence of the corresponding substrate but not the primer (31). All reactions were conducted at room temperature for the time specified and then terminated by addition of 100 mM EDTA in 2× SSC solution (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Radioactive products were captured by filtering nitrocellulose membranes and quantified by liquid scintillation counting as described previously (18). Kinetic parameters were generated by fitting the collected data points into the equations specified and processed by the graphics data analysis program GraphPad Prism from GraphPad Software (San Diego, Calif.).
Gel-based RNA polymerase assay.
RNA polymerase reactions were also performed using short heteropolymeric RNAs as the template as described previously (14). A 12-mer RNA, -GCUAGGGGCCCC-, that could hybridize through the consecutive GC pairs between two molecules, was used for this study. To prepare radiolabled RNA, RNA oligonucleotides were 5′-32P-labeled using [γ-32P]ATP and T4 kinase under the conditions described previously (31). Unused [γ-32P]ATP was removed by passing the reaction mixture through Sephadex G-25 spin columns. For annealing, RNA oligonucleotides were first heated to 90°C for 1 min and then cooled to 4°C at a rate of 4°C/min. The annealed RNA primer-template complex, designated sym/sub, was stored at −20°C until used.
RNA polymerase reactions were performed in reaction mixtures containing 20 mM Tris (pH 7.5), 5 mM MgCl2, 25 mM KCl, 2 mM DTT, 0.1 to 1.5 μM sym/sub template, 1 U of RNase inhibitor per ml, 83 μM purified NS5B polymerase, and 50 to 500 μM UTP or nucleoside triphosphate (NTP) as substrate. Reactions were terminated by addition of gel loading buffer (Ambion). For product analysis, samples were heat denatured and loaded on a 7 M urea-20% polyacrylamide gel. After electrophoresis, the gel was dried and 32P-labeled products were visualized by autoradiography or by PhosphorImager analysis (Image Dynamics). Data were quantified using the ImageQuant software (Molecular Dynamics).
RNA-protein binding assay.
The radiolabeled oligo(U)12 and the preannealed sym/sub RNA duplex described above were used for RNA-binding assays. Binding reactions were performed with 100 ng of purified NS5B proteins or bovine serum albumin as the control in a total volume of 20 μl containing 25 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 25 mM KCl, and the 32P-labeled RNAs at the indicated concentrations. After a 30-min incubation at room temperature, the reaction mix was UV cross-linked for 30 s at 250 mJ/cm2. Then 5 μl of 5× SDS-PAGE sample buffer was added to the reactions mixtures. After a 5-min boiling step, the reaction products were resolved on a 4 to 20% polyacrylamide gradient SDS-PAGE gel and proteins bound to the radiolabled RNA were visualized by autoradiography.
RESULTS
Crystallization and structure determination of HCV NS5B.
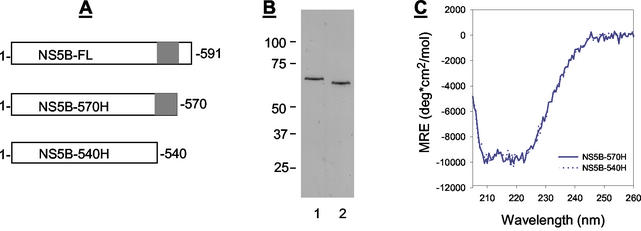
To obtain highly purified protein for structural study, we expressed the NS5B polymerase derived from HCV genotype 1b as a C-terminally histidine-tagged protein with a truncation of 21 amino acids from its C terminus because this domain is involved in membrane binding (34). This truncated protein, designated NS5B-570H (Fig. 1A), differed from the HCV NS5B protein reported previously by two or more amino acids as specified in Table 1. NS5B-570H was then enriched through a three-step purification protocol using DEAE-Sepharose, heparin-Sepharose, and Ni2+-chelating affinity columns as described in Materials and Methods. Through this approach, we could generate 5 mg of highly purified protein from 24 g of bacterial cells harvested from a 6-liter cell culture (Fig. 1B).
FIG. 1.
Analyses of HCV NS5B proteins. (A) Sequence comparison of the NS5B proteins used in this study. The C-terminal domain deleted from NS5B-540H is highlighted. (B) Analysis of purified HCV NS5B proteins. A 200-ng portion of purified NS5B protein was subject to separation by SDS-PAGE followed by Coomassie blue staining. Lanes: 1, NS5B-570H; 2, NS5B-540H. (C) CD spectrometric analysis of NS5B-570H (solid line) and NS5B-540H (dotted line).
TABLE 1.
Residue variation of the reported HCV NS5B crystal structures
| PDB code | Amino acid at HCV NS5B positiona:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 32 | 329 | 338 | 510 | 544 | |
| BK | S | M | R | T | V | R | R |
| 1C2P | H | H | —b | V | A | — | Q |
| 1CSJc | — | — | A | — | — | A | |
| 1QUV | — | — | — | — | — | — | Q |
| NS5B-570H | — | — | A | — | — | A | Q |
The HCV NS5B amino acid sequence, expressed in single-letter code, is numbered according to the mature protein sequence. Shown is the variation relative to the sequence of the BK strain. The first two residues listed for PDB ID 1C2P were derived from the hexahistidine tag located at the N terminus of the protein.
—, no change relative to the BK strain.
Sequence ends at amino acid 531.
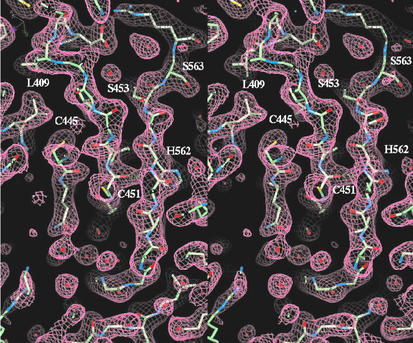
NS5B-570H was then used for a crystallization study as described in Materials and Methods. Although NS5B-570H contained similar residues to the three reported crystal structures (Table 1), the conditions we used for crystallization were different from those reported previously (1, 5, 20) and the crystals belong to a different space group. The crystal structure was refined to 2.1-Å resolution with an R factor of 25.4% and an Rfree of 28.5% (Table 2). The current refined crystallographic model contained all 570 residues derived from NS5B proteins and 458 water molecules. Figure 2 shows the 2FO-FC electron density map of the C-terminal tail that forms a β-sheet with a β-hairpin in the core structure.
TABLE 2.
Summary of crystallographic analysis
| Characteristic | Value |
|---|---|
| Data collection | |
| Resolution (Å) | 50-2.1 |
| No. of Unique reflections | 83,916 |
| Redundancy | 3.9 |
| Rsym (%)a | 4.2 |
| Completeness (%) | 95.5 |
| Space group | p6522 |
| Unit-cell parameter | a = b = 110.82 Å c = 238.57 Å |
| Refinement | |
| Rwork (%)b | 25.4 |
| Rfree (%)b | 28.5 |
| Rmsd | |
| Bonds (Å) | 0.01 |
| Angles | 1.7° |
| No. of protein atoms | 4,455 |
| No. of water molecules | 458 |
Rsym = Σ|Ih − 〈Ih〉|/ΣIh, where Ih is the intensity of reflection h.
Rwork and Rfree = Σ|FO − FC|/ΣFO, where FO and FC are the observed and calculated amplitudes, respectively. Rfree was calculated using 7% of the data excluded from refinement.
FIG. 2.
Stereo diagram of the 2FO-Fc electron density map. The map covers residues 559 to 564 from the C-terminal tail that forms a β-sheet with the β-hairpin of residues 450 to 455 and 440 to 445 from the catalytic core. The map was calculated using the native data including reflections from 20 to 2.1 Å at the 1.0 σ contour level.
Interaction of the C-terminal tail with the NS5B catalytic core structure.
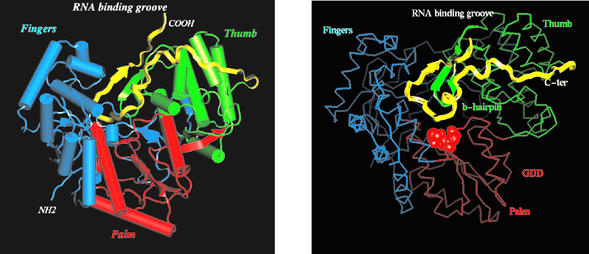
The structure of NS5B-570H is heart-shaped with approximate dimensions of 67 by 63 by 68 Å. Consistent with previously reported structures, all 21 α-helices and 18 β-sheets that are divided into three subdomains including fingers, palm, and thumb, could be seen in NS5B-570H (Fig. 3).
FIG. 3.
Overall structure of NS5B-570H. (Left) Ribbon representation of the polypeptide chain. The thumb domain is colored in green, the palm domain is in red, the fingers domain is in blue, and the C-terminal tail (residues 531 to 570) is in yellow. The putative RNA-binding groove is also indicated. (Right) Cα trace of NS5B-570H with the same color scheme as in the left panel. The C-terminal tail is highlighted in yellow ribbon. The catalytic-site -GDD- motif is shown as a red cpk ball model. The tip of the C-terminal tail reaches to the -GDD- pocket.
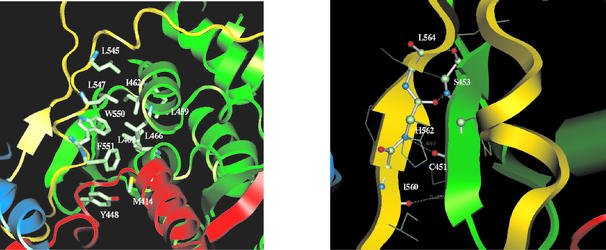
Starting from residue 530, the C-terminal tail wraps across the inner surface of the thumb subdomain (Fig. 3, left panel). After a short turn of 3 to 10 α-helices, it folds down toward the tip of the palm domain, where catalytic residues -GDD- reside (Fig. 3, right panel), and then comes off the putative RNA-binding groove to form a β-sheet with the β-hairpin structure from the thumb domain. The hydrophobic interactions between the residues in the C-terminal tail and the thumb subdomain serve as the primary force for the binding of the C-terminal tail to the catalytic core. Residues Trp550, Phe551, and Leu547 pack with Leu409, Leu459, Leu466, and Ile462 to form a well-defined hydrophobic pocket, with Leu545 and Try448 plus Met414 lying in the top and the bottom of the pocket respectively (Fig. 4, left panel). The second intensive binding site consists of a hydrogen-bonding network between the β-strand (amino acids 560 to 565) and the β-hairpin (amino acids 450 to 455). The hydrogen bond pairs are carbonyl oxygen of Ile560 with backbone amide NH of Cys451, carbonyl oxygen of Cys451 with amide NH of His562, carbonyl oxygen of His562 with amide NH of Ser453, and carbonyl oxygen of Ser453 with amide NH of Leu564 (Fig. 4, right panel). Apparently, these hydrogen bonds stabilize the β-hairpin structure. Taken together, the close proximity of the C-terminal domain to the -GDD-motif, its partial occupation at the putative RNA-binding groove, and its interaction with the β-hairpin suggest that this unique structural element might exert a regulatory role in NS5B polymerase activity.
FIG. 4.
Interactions between the C-terminal tail and the catalytic core structure. (Left) Hydrophobic interactions between conserved residues in the C-terminal tail and the thumb domain. Residues Trp550, Phe551, and Leu547 packed with Leu409, Leu459, Leu466, and Ile462 to form a well-defined hydrophobic pocket. Leu545 and Try448 plus Met414 lie in the top and bottom of this pocket. (Right) Hydrogen-bonding network between the β-strand (residues 560 to 565) and β-hairpin (residues 450 to 455). The hydrogen bond pairs are carbonyl oxygen of Ile560 with the backbone amide NH of Cys451, carbonyl oxygen of Cys451 with amide NH of His562, carbonyl oxygen of His562 with amide NH of Ser453, and carbonyl oxygen of Ser453 with amide NH of Leu564. These hydrogen bonds stabilize the β-hairpin structure.
C-terminal deletion enhances RNA binding to HCV NS5B.
Since the structural analysis indicated that the C-terminal fragment occupied the putative RNA-binding cleft, we questioned if this fragment would affect the RNA-binding efficiency of NS5B. To test this hypothesis, another histidine-tagged NS5B protein, termed NS5B-540H, was expressed, which differed from NS5B-570H by a further deletion of 30 amino acids from the C-terminus as shown in Fig. 1A. To make a better comparison, NS5B-540H was expressed, purified, and handled in the same way as for NS5B-570H. The purified NS5B-540H was shown by SDS-PAGE to be a ∼60-kDa protein (Fig. 1B), and its identity was further confirmed by amino acid sequencing.
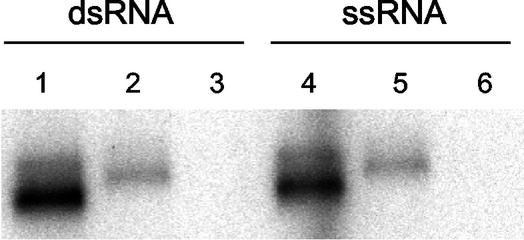
We then conducted circular dichroism (CD) spectrometry analyses to compare the overall protein folding of NS5B-540H and NS5B-570H. As shown in Fig. 1C, NS5B-540H had essentially the same CD profile as that of NS5B-570H, indicating that removal of 30 amino acids did not significantly affect the overall protein folding. Next, the two proteins were compared side by side for their ability to bind to double-stranded RNA (dsRNA) as well as single-stranded RNA (ssRNA). As seen in Fig. 5, NS5B-570H was able to bind to both ssRNA and dsRNA, but its binding efficiency was fivefold lower than that of NS5B-540H under the conditions employed. Since the only difference between NS5B-570H and NS5B-540H was the extra 30 residues at the C-terminus, these results suggested that the C-terminal motif of NS5B-570H indeed blocked the access of RNAs to the enzyme, as predicted by the structure analysis.
FIG. 5.
RNA-binding efficiency of HCV NS5B proteins. The RNA-binding efficiencies of NS5B-570H and NS5B-540H were compared using the conditions specified in Materials and Methods. Three proteins were evaluated: NS5B-540H (lanes 1 and 4), NS5B-570H (lanes 2 and 5), and bovine serum albumin as control (lanes 3 and 6). The same amounts of 32P-labeled RNAs (0.4 pmol) and protein (100 ng) were used in each experiment. Experiments were performed using dsRNA (preannealed sym/sub) and ssRNA [32P-labeled oligo(U12)].
C-terminal deletion enhances RNA elongation efficiency.
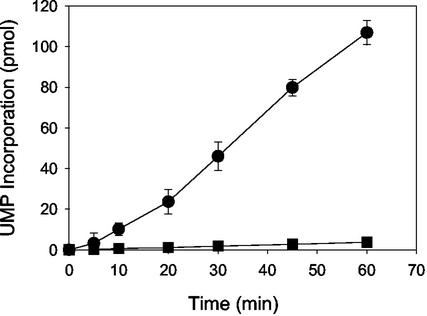
The proximity of the C-terminal domain to the -GDD- motif and its interaction with the β-hairpin structure led us to examine the effect of this fragment on RNA synthesis catalyzed by HCV NS5B. To test this hypothesis, the ability of NS5B-570H and NS5B-540H to catalyze nucleotide incorporation from a preannealed RNA template-primer was examined. The first experiment was conducted using poly(A) as the template and oligo(U12) as the primer to measure the incorporation of UTP substrate as described previously (16). As seen in Fig. 6, both NS5B-540H and NS5B-570H were able to catalyze RNA elongation in this system, although with different efficiencies. The observed steady-state rate of production formation was calculated as 0.062 ± 0.002 pmol/min for NS5B-570H and 1.33 ± 0.2 pmol/min for NS5B-540H. Apparently, the C-terminal deletion caused a ∼22-fold increase in the observed rate relative to the longer enzyme, supporting the notion that this domain had a negative impact on the primer-dependent RNA elongation catalyzed by HCV NS5B polymerase.
FIG. 6.
RNA elongation catalyzed by HCV NS5B. Reactions were performed using poly(A) as the template and [α-32P]UTP as the substrate, as described in the text. Shown is the time course for 30 nM NS5B-570H (▪) and NS5B-540H (•).
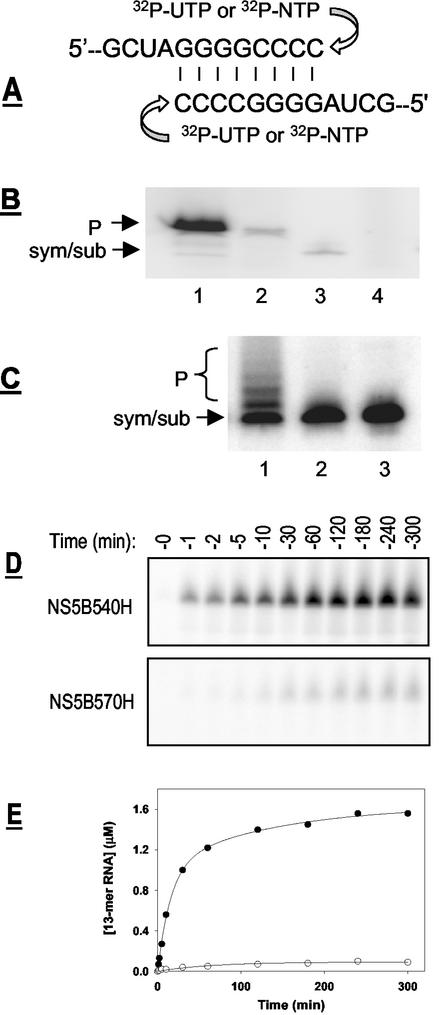
The catalytic efficiency of the two enzymes for single-nucleotide incorporation was further compared. A 12-mer RNA oligonucleotide that was able to form a symmetrical primer-template duplex was used for this study (Fig. 7A), since similar RNA templates have been used for studying the RdRps of poliovirus and HCV (3, 14). Using this preannealed template-primer, termed sym/sub, we could measure either single- or multiple-nucleotide incorporation as described in Materials and Methods. In the first experiment, either NS5B-570H or NS5B-540H was added to a reaction mixture containing nonradiolabeled sym/sub and [α-32P]UTP as the substrate. As seen in Fig. 7B, addition of a single nucleotide per sym/sub template was observed for both NS5B-570H and NS5B-540H, indicating that both enzymes could utilize this template-primer. Experiments measuring the incorporation of all four nucleotides were also performed using 32P-labeled sym/sub along with cold nucleotides (Fig. 7C). In all experiments conducted, the catalytic efficiency of NS5B-540H was found to be higher than that of NS5B-570H.
FIG. 7.
Single-nucleotide incorporation by HCV NS5B. (A) The RNA template, sym/sub, used in this study. (B) Single-nucleotide incorporation catalyzed by purified NS5B proteins. These experiments were performed using [α-32P]UTP as substrate and cold sym/sub RNA as template, as described in Materials and Methods. Lanes: 1, NS5B-540H; 2, NS5B-570H; 3, 32P-labeled sym/sub; 4, no-enzyme control. The positions for input sym/sub RNA template and 13-mer product (P) are labeled. (C) Multinucleotide incorporation assays were performed using cold NTP (ATP, GTP, CTP, and UTP) as substrates and 32P-labeled sym/sub RNA as the template, as described in Materials and Methods. Lanes: 1, NS5B-540H; 2, NS5B-570H; 3, no-enzyme control. The positions for template RNA (sym/sub) and expected 13- to 16-mer products (P) are marked. (D) Time course for single-nucleotide incorporation catalyzed by HCV NS5B. Assays were performed side by side using 0.6 μM NS5B-540H or NS5B-570H as described in Materials and Methods, with cold sym/sub RNA as the template and [α-32P]UTP as the substrate. (E) Quantitative analysis of UMP incorporation catalyzed by NS5B-540H (•) or NS5B-570H (○) as shown in panel D.
To gain a better understanding of their catalytic efficiency, time-dependent UMP incorporation into sym/sub was also investigated, as shown in Fig. 7D. A linear accumulation of product was seen in the first 30 min after addition of the enzyme, with a catalytic rate of 52.6 ± 5.0 pmol/min for NS5B-540H and 1.03 ± 0.16 pmol/min for NS5B-570H (Fig. 7E). These results revealed that NS5B-540H was approximately 51-fold more active in catalyzing UMP incorporation into sym/sub, further supporting the conclusion that the C-terminal domain inhibited the primer-dependent RNA elongation catalyzed by HCV NS5B.
C-terminal deletion affects primer-independent RNA synthesis.
Next, primer-independent RNA synthesis from the single-stranded RNA template was performed using both NS5B-570H and NS5B-540H. Since HCV NS5B could catalyze de novo RNA synthesis from homopolymeric pyrimidine RNAs (22, 31), these reactions were conducted using poly(C) and poly(U) as templates in the absence of primer, as described in Materials and Methods. This system offered no back-primed initiation due to lack of secondary structure of these homopolymeric RNA templates and thus was suitable for studying de novo RNA initiation.
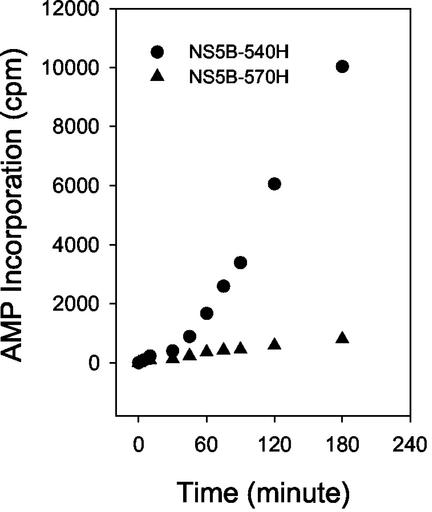
De novo RNA synthesis from a poly(U) template was performed in the absence of primer. Reactions were started by addition of either NS5B-540H or NS5B-570H, and ATP incorporation was monitored at different time points. The final products, generated through de novo RNA initiation, were plotted as a function of time. As shown in Fig. 8, both NS5B540H and NS5B 570H showed a lag phase at the early stage of the reaction (0 to 30 min), perhaps because they were involved in assembly of the replication complex. However, product formation quickly increased as the reaction progressed. The observed rate for product generation through the de novo mechanism was calculated to be 72.08 ± 3.23 min−1 for NS5B-540H and 1.82 ± 0.20 min−1 for NS5B-570H, which represents about a 40-fold difference. These data suggested that the presence of the C-terminal tail negatively affects the primer-independent RNA synthesis catalyzed by the NS5B enzyme. We also performed primer-independent RNA synthesis using poly(C) as the template and GTP as the substrate and found that similar results were obtained, with NS5B-570H demonstrating much lower activity in catalyzing primer-independent RNA synthesis (data not shown).
FIG. 8.
RNA initiation catalyzed by HCV NS5B. Primer-independent RNA synthesis reactions were performed using NS5B-540H or NS5B-570H with poly(U) as the template and [α-32P]ATP as the substrate as described in Materials and Methods. At the time point specified, an aliquot of the reaction mix was taken out and subjected to quantitative filter binding and scintillation counting.
Inhibition of HCV NS5B polymerase activity by peptides derived from the C-terminal domain.
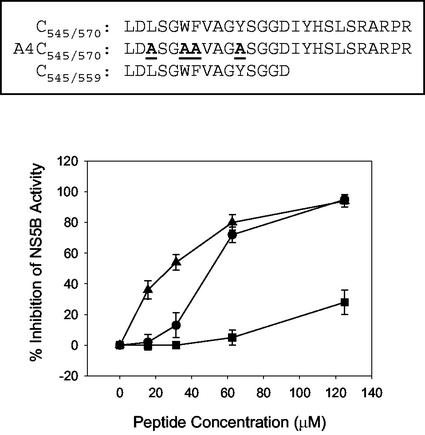
Encouraged by the results obtained from the assays described above, we designed and synthesized a peptide representing the 26 amino acids from residues 545 to 570 deleted from the C terminus of NS5B-570H (Fig. 9) to see if it could mimic the original C-terminal tail and then affect NS5B-540H polymerase activity. When this peptide, termed C545/570, was included in the primer-dependent polymerase assay using poly(A)-poly(U12) as the template-primer pair, dose-dependent inhibition of NS5B-540H was identified, as shown in Fig. 9. As a control, another peptide termed A4C545/570, in which the four key residues involved in the formation of the hydrophobic pocket in NS5B were all mutated to alanine (Fig. 4, left panel), was also evaluated. Compared to the native peptide C545/570, the mutant peptide A4C545/570 did not inhibit NS5B-540H significantly at the concentrations tested (Fig. 9). Similarly, a shorter peptide, C545/559, representing residues 545 to 559, also showed inhibitory activity against NS5B-540H (Fig. 9). In contrast, inhibition of NS5B-570H by these peptides was not observed at the same doses (data not shown). It is plausible that the native peptides, C545/570 and C545/559, but not the mutant one, were able to bind to NS5B-540H at the RNA-binding site occupied by the C-terminal tail and then exerted a similar impact on the enzyme catalytic efficiency. Taken together, these data supported the notion of the C-terminal domain as a regulatory element to HCV NS5B polymerase activity.
FIG. 9.
Effect of synthetic C-terminal peptides on HCV polymerase activity. Amino acid sequences of the peptides used for this study are shown at the top. Below are shown the results of the primer-dependent RNA synthesis reactions of NS5B-540H that were performed using poly(A) as the template and [α-32P]UTP as the substrate in the presence of different peptides. Shown is the percent inhibition of NS5B-540H activity at different concentrations of individual peptides: C545/570 (•), A4C545/570 (▪), and C545/559 (▴).
Key residues of the C-terminal domain are highly conserved.
To gain a better understanding of the significance of the C-terminal domain of NS5B, we performed a sequence comparison of NS5B proteins encoded by different HCV genotypes and subtypes to assess the degree of conservation of this motif. Analysis of 142 HCV isolates found in the database revealed several highly conserved amino acids in this fragment (Table 3). It is interesting that the four residues Leu545, Leu547, Trp550, and Phe551, forming part of the hydrophobic pocket (Fig. 4, left panel), are completely conserved among all 142 HCV isolates. Residues implicated in hydrogen bonding with the β-hairpin are less highly conserved; this may reflect the fact that these hydrogen bonds are formed through backbone atoms rather than side chains of these residues, as illustrated in Fig. 4 (right panel).
TABLE 3.
C-terminal consensus sequence of HCV NS5B polymerasea
| AA number | 545 | 550 | 555 | |||||||||||
| Consensus residue | S | Q | L | D | L | S | G | W | F | V | A | G | Y | S |
| % Conservation | 70 | 54 | 100 | 100 | 100 | 100 | 67 | 100 | 100 | 65 | 81 | 99 | 83 | 71 |
| AA number | 560 | 565 | 570 | |||||||||||
| Consensus residue | G | G | D | I | Y | H | S | L | S | R | A | R | P | R |
| % Conservation | 100 | 95 | 99 | 97 | 98 | 99 | 100 | 60 | 100 | 78 | 98 | 100 | 95 | 100 |
Sequence alignment of HCV NS5B proteins from 142 different genotypes and subtypes was performed using CLUSTAL W. (v. 1.81). Shown is the fragment representing the C-terminal amino acids 543 to 570 upstream of the membrane anchor domain of HCV polymerase (the amino acid position is labeled “AA number” at the top). The revealed consensus residues (middle line) are expressed in single-letter code. Percent conservation represents the percentage of conservation calculated from sequence comparison. Residues involved in the formation of the hydrophobic pocket and hydrogen-bonding network are underlined and in bold, respectively.
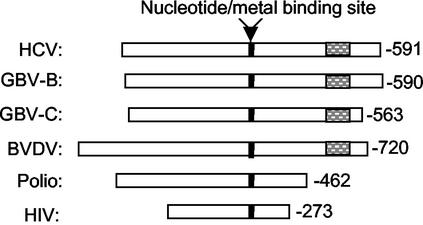
We also performed sequence alignment among the known viral RNA polymerases to see if this structural feature is also present in other viral polymerases. As reported previously, the key residues and motifs A to E, originally identified in poliovirus 3D polymerase and human immunodeficiency virus RNA polymerase (13), are contained in the N-terminal section of HCV NS5B (1, 5, 20). On the other hand, the NS5B polymerases encoded by flaviviruses, including HCV, bovine viral diarrhea virus, and GB-virus B, all contain a much longer C-terminal tail compared to the poliovirus 3D polymerase and human immunodeficiency virus reverse transcriptase (Fig. 10). These data suggest that the C-terminal structural features, including the membrane anchor domain and the above-mentioned regulatory motif, might be specific to the viruses belonging to the flavivirus family.
FIG. 10.
Sequence alignment of viral RNA polymerases. Amino acid residues of the listed viral polymerase were aligned to the center of the nucleotide/metal-binding site (solid box). For most polymerases listed, the nucleotide/metal-binding site is referred as the -GDD- motif. The C-terminal regulatory motif found in HCV NS5B and related viruses is highlighted (hatched box).
DISCUSSION
HCV NS5B polymerase, composed of 591 amino acids, is ∼130 residues longer than poliovirus 3D polymerase, the best-characterized viral RNA polymerase. Sequence alignment has indicated that these additional residues are located at the C-terminal extremity of the NS5B protein (5) (Fig. 10). Previous results have suggested that these extra residues, particularly the last 21 amino acids of the C terminus, define a membrane-anchor domain that is responsible for endoplasmic reticulum-membrane association during viral replication process (30, 34). With the catalytic core domain consisting of ∼530 amino acids (5), that still leaves ∼100 residues with unknown utility but documented sequence conservation. In this report, we present both structural and biochemical evidence suggesting that another structural element, located upstream of the membrane-anchor domain, plays a regulatory role in HCV NS5B polymerase activity and RNA binding.
The X-ray crystal structure of HCV NS5B has been reported by three different groups (1, 5, 20). These studies revealed several interesting structural features for NS5B, including a specific β-hairpin that has recently been shown through biochemical analysis to be implicated in guiding RNA initiation (14). Of the three NS5B crystal structures disclosed, one was solved using an NS5B protein containing the first 531 amino acids, which did not cover the C-terminal ∼40 amino acids upstream of the membrane-anchor domain (5). The other two groups obtained the structure of NS5B containing ∼570 residues; however, detailed structural information regarding the C-terminal tail was not discussed (1, 20). Ago et al. indeed described the folding of the C-terminal region; unfortunately, the crystal structure beyond residue 556 was disordered (1). On the basis of this limited information, they proposed that C-terminal residues 532 to 570 might serve as an autoinhibitory sequence, but no biochemical data was presented to support the hypothesis (1).
Our crystal structure for HCV NS5B, solved at a resolution of 2.1 Å, showed that the C-terminal region, together with the β-hairpin defined previously (14), protruded into the RNA-binding cavity and would overlap with the modeled template-primer duplex. In addition, we found that several residues of this segment are involved in the formation of a hydrophobic pocket that is very close to the -GDD- motif responsible for nucleotide-metal binding (Fig. 3B). Thus, this study provides additional information that was not included in previous reports. More importantly, the high-resolution structure of HCV NS5B provided an excellent background for biochemical studies of this fragment. On the basis of the structural information, we designed a mutant NS5B protein lacking this fragment to study its potential role in regulating NS5B polymerase activity.
Our biochemical data confirmed that the C-terminal region, comprising residues 545 to 564, interfered with RNA binding and consequently affected the observed rate of product formation as shown in several experiments designed. Deletion of this domain not only enhanced polymerase-RNA binding but also caused a 20- to 51-fold increase in RNA synthesis efficiency, depending on the assay conditions. Most likely, due to its specific folding and position at the RNA-binding site, this C-terminal domain would be a blockade for RNA template entry and/or would lead to an inefficient assembly of the elongation complex consisting of the annealed template-primer and the nucleotide. It is also possible that this domain would block the incoming nucleotide since it is very close to the -GDD- motif (Fig. 4, right panel). In either case, it would exert an inhibitory effect on overall product formation when tested in vitro. Further supporting evidence for this hypothesis came from the peptide studies (Fig. 9). On the other hand, the enhanced catalytic efficiency may result not only from the observed better access of the RNA-template to the truncated enzyme (Fig. 5) but also from improved processivity as well as release of the newly synthesized RNA products. Unfortunately, we were unable to separate these steps from each other because of the current experimental design. To address these questions, additional experiments must be performed.
As discussed above, poliovirus 3D polymerase is shorter than HCV polymerase at its C-terminal extremity. Consequently, the β-hairpin and the C-terminal domain are not present in the 3D polymerase, which coincides with the fact that poliovirus replicates its genome through protein priming catalyzed by the 3D polymerase (27). Unlike the 3D polymerase, HCV NS5B is thought to replicate the viral genome through primer-independent de novo initiation (17, 22, 26, 31, 36). Such a structural difference, accompanied by their unique replication mechanisms, suggests that the β-hairpin and the C-terminal domain may play a role in controlling HCV replication. Recently, the β-hairpin structure of HCV NS5B has been proposed to be important in permitting an appropriate initiation of replication. It should be noted that the RNA polymerase encoded by the double-stranded RNA bacteriophage φ6 also contains a β-hairpin-like structure (6). More importantly, this structural element was shown to be important for forming an initiation platform and stabilizing the first two incoming nucleotides in φ6 polymerase (19). Interestingly, deletion of the β-hairpin from HCV NS5B or mutation of the β-hairpin-like loop in φ6 RNA polymerase also causes enhancement of primer-dependent elongation (19, 23), similar to the data reported here.
In summary, this report provides direct structural and experimental insights into the role of the C-terminal tail of HCV NS5B polymerase. On the basis of the data presented in this report, together with the results published previously, we propose a working model highlighting the role of the C-terminal domain in regulating HCV genome replication. According to this model, the C-terminal domain, together with the β-hairpin, forms a rigid and bulky loop at the active site, which could prevent abnormal RNA replication, such as RNA synthesis from the middle of the viral genome or back-primed initiation. In addition, the C-terminal domain might help with the β-hairpin to form an initiation platform interacting with and stabilizing the first two incoming nucleotides during the initiation process. Supporting evidence is that C-terminal residues form a well-defined hydrogen bond network with the β-hairpin (Fig. 4, right panel). In this role, the C-terminal domain stabilizes the initiation complex by interacting with the β-hairpin structure. Furthermore, the C-terminal domain might play a role in recognizing the key secondary structure at the 3′-untranslated region of the HCV genome to ensure correct initiation. Although there are no data to support this hypothesis to date, HCV NS5B has been reported to bind to the 3′ untranslated region of the HCV genome, and the binding site on the 3′ untranslated region, but not on NS5B, has been mapped recently (8). The unique hydrophobic pocket, formed among several completely conserved residues in the C-terminal domain together with Tyr448 of the β-hairpin structure (Fig. 4, left panel), might be critical in this regard. It has been shown recently that the β-hairpin is required for HCV genome replication in cells (7); it remains to be determined if the C-terminal regulatory motif is also essential for HCV replication. Further experiments should help to address these specific issues and allow us to gain a better understanding of the roles of these unique structural elements in regulating HCV NS5B polymerase function.
ADDENDUM
During the preparation of the manuscript, we noticed that Adachi et al. had published similar results (1a).
Acknowledgments
V. Lévêque and R. Johnson contributed equally to this work.
We thank Mel Johnson, Kati Case, and Jirong Lu for technical assistance and Eric Kool (Stanford University) for great suggestions about this work.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 1a.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.AI, R. H., Y. Xie, Y. Wang, and C. H. Hagedorn. 1998. Expression of recombinant hepatitis C virus non-structural protein 5B in Escherichia coli. Virus Res. 53:141-149. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): assembly of stable, elongation-competent complexes by using a symmetrical primer/template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 7.Cheney, I. W., S. Naim, V. C. H. Lai, S. Dempsey, D. Bellows, M. P. Walker, J. H. Shim, N. Horscroft, Z. Hong, and W. Zhong. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298-306. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, J. C., M. F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo, Q.-L., G. Kuo, A. J. Weiner, R. L. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. 1999. The scientific challenge of hepatitis C. Science 285:26-30. [DOI] [PubMed] [Google Scholar]
- 11.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagedorn, C. H., E. H. van Beers, and C. De Staercke. 2000. Hepatitis C virus RNA-dependent RNA polymerase. Curr. Top. Microbiol. Immunol. 242:225-260. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 14.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. B., X. L. Sun, M. A. Hockman, E. C. Villarreal, M. Wakulchik, and Q. M. Wang. 2000. Specificity and mechanism analysis of hepatitis C virus RNA-dependent RNA polymerase. Arch. Biochem. Biophys. 377:129-134. [DOI] [PubMed] [Google Scholar]
- 17.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirement for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurila, M. R. L., E. V. Makeyev, and D. H. Bamford. 2002. Bacteriophage φ6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277:17117-17124. [DOI] [PubMed] [Google Scholar]
- 20.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully enriched active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 24.Major, M. E., and S. M. Feinstone. 1997. The molecular virology of hepatitis C. Hepatology 25:1527-1538. [DOI] [PubMed] [Google Scholar]
- 25.Miller, R. H., and R. H. Purcell. 1990. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus subgroups. Proc. Natl. Acad. Sci. USA 87:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 28.Reed, K. E., and C. M. Rice. 1998. Molecular characterization of hepatitis C virus. Curr. Stud. Hematol. Blood Transfus. 62:1-37. [DOI] [PubMed] [Google Scholar]
- 29.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q.-L. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 31.Sun, X. L., R. B. Johnson, M. A. Hockman, and Q. M. Wang. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem. Biophys. Res. Commun. 268:798-803. [DOI] [PubMed] [Google Scholar]
- 32.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, C. A. Case, J. Lu, S. Parsons, F. Zhang, R. Radhakrishnan, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 35.Yuan, Z. H., U. Kumar, H. C. Thomas, Y. M. Wen, and J. Monjardino. 1997. Expression, purification, and partial characterization of HCV RNA polymerase. Biochem. Biophys. Res. Commun. 232:231-235. [DOI] [PubMed] [Google Scholar]
- 36.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]