Abstract
The emergence of drug resistance-conferring mutations can severely compromise the success of chemotherapy directed against human immunodeficiency virus type 1 (HIV-1). The M184V and/or L74V mutation in the reverse transcriptase (RT) gene are frequently found in viral isolates from patients treated with the nucleoside RT inhibitors lamivudine (3TC), abacavir (ABC), and didanosine (ddI). However, the effectiveness of combination therapy with regimens containing these compounds is often not abolished in the presence of these mutations; it has been conjectured that diminished fitness of HIV-1 variants containing L74V and M184V may contribute to sustained antiviral effects in such cases. We have determined that viruses containing both L74V and M184V are more impaired in replication capacity than viruses containing either mutation alone. To understand the biochemical mechanisms responsible for this diminished fitness, we generated a series of recombinant mutated enzymes containing either or both of the L74V and M184V substitutions. These enzymes were tested for their abilities to bypass important rate-limiting steps during the complex process of reverse transcription. We studied both the initiation of minus-strand DNA synthesis with the cognate replication primer human tRNA3Lys and the initiation of plus-strand DNA synthesis, using a short RNA primer derived from the viral polypurine tract. We observed that the efficiencies of both reactions were diminished with enzymes containing either L74V or M184V and that these effects were significantly amplified with the double mutant. We also show that release from intrinsic pausing sites during reverse transcription appears to be a major obstacle that cannot be efficiently bypassed. Our data suggest that the efficiency of RNA-primed DNA synthesis represents an important consideration that can affect viral replication kinetics.
Although considerable progress has been made in the treatment of human immunodeficiency virus type 1 (HIV-1)-associated disease, the emergence of mutated variants of HIV-1 that are resistant to antiviral drugs represents a major problem. The prolonged clinical use of nucleoside reverse transcriptase (RT) analogue chain terminators, e.g., abacavir (ABC), 2′,3′-dideoxyinosine (ddI or didanosine), and (−)-2′,3′-dideoxy-3′-thiacytidine (3TC or lamivudine), gives rise to resistant viruses that contain mutations in the RT enzyme (5, 8, 11, 48, 52, 54, 56). These nucleoside RT inhibitors (NRTIs) compete with natural deoxynucleoside triphosphate (dNTP) pools after being phosphorylated by cellular kinases for incorporation into viral DNA. DNA synthesis is blocked once the chain terminator is incorporated, since these nucleoside analogues lack a 3′-OH group, which is required to continue the polymerization process (24, 44, 45).
A single amino acid substitution in RT, i.e., M184V, is sufficient to confer high-level resistance to 3TC (5, 11, 47, 54). The M184V mutation is rapidly selected both in tissue culture and in vivo. Experiments with recombinant mutant enzymes have shown that this mutation diminishes the rate of incorporation of 3TC monophosphate (MP) into DNA (27, 41). It has also been found to diminish RT processivity (4, 30) and to increase RT fidelity as measured in polymerase assays that monitor either the RNA-dependent or DNA-dependent polymerase steps in reverse transcription (20, 26, 40, 56). The M184V mutation is also known to confer low-level resistance to ddI and ABC, and susceptibility to these two drugs is further reduced in the additional presence of the L74V mutation (11, 39, 55). The L74V substitution in RT by itself is primarily associated with resistance to ddI (8, 36, 52). L74V was found to slightly increase the Ki value for incorporation of ddATP, the physiologically relevant metabolite of ddI, although this effect was not nearly as pronounced as that seen with 3TC and 3TC-resistant enzymes in the context of RTs containing the M184V mutation (35, 39).
Although M184V and L74V are located in different regions of RT (21, 46), the two mutations share some interesting features. Each can cause a diminution in viral replication in the absence of drug and can also resensitize zidovudine (ZDV)-resistant viruses that contain a number of thymidine-associated mutations (TAMs) to the drug. The M184V mutation has also been shown to increase baseline susceptibility to ZDV when present as a single mutation (30), while the L74V mutation showed a similar effect only in conjunction with mutations associated with ZDV resistance (52). A biochemical mechanism that helps to explain such resensitization effects in regard to viruses containing the M184V mutation has recently been described (13, 14). Notably, viruses and enzymes containing M184V are less able than wild types (WT) to carry out the phosphorolytic excision of incorporated nucleotide analogues, i.e., that mediated by either ATP or pyrophosphate (PPi). The ATP-dependent removal of incorporated ZDV-MP provides an important mechanism for resistance to ZDV (2, 37).
Despite increased understanding of NRTI resistance, little is known of the biochemical basis for diminished viral replication fitness that is associated with some resistance-conferring mutations (9). Previous studies have suggested that diminished RT processivity may be one of the factors associated with the decreased fitness of viruses containing either the L74V or M184V mutation (4, 31, 51, 52). Here, we have studied the effects of these mutations on specific rate-limiting steps during the initiation of reverse transcription. It has been demonstrated that the RNA-primed initiation of plus-strand DNA synthesis, which requires relatively high concentrations of dNTPs, represents an important rate-limiting step in reverse transcription (12, 15; M. Götte, M. Kameoka, L. Cellai, and M. A. Wainberg, presented at Cold Spring Harbor Symposium on Retroviruses, Cold Spring Harbor, N.Y., 23 to 28 May 2000). A similar scenario has been described in regard to the tRNA-primed synthesis of viral minus-strand DNA [(−) ssDNA] (22, 29, 32, 33, 53). We now show that viruses containing the L74V and M184V mutations possess diminished replication capacity and that recombinant RT enzymes containing these substitutions have a diminished ability to utilize RNA primer strands.
(This work was performed by K. Diallo in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada.)
MATERIALS AND METHODS
Enzymes and nucleic acids.
WT and various mutated forms of HIV-1 RT (i.e., L74V, M184V, and L74V-M184V) were prepared and purified as previously described (31). The primer-template substrates used to study the initiation of plus-strand DNA synthesis and the incorporation of either carbovir triphosphate (CBV-TP) or ddATP into viral DNA were derived from the polypurine tract (PPT) of the HIV-1 genome (13). The following oligonucleotides were employed: 5′-TTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAGTGGCTAAGA-3′ (PPT-57), which served as a DNA template; 5′-UUAAAAGAAAAGGGGGG-3′ (17R), which served as an RNA primer; and 5′-TAAAAGAAAAGGGGGGAC-3′ (18D) and 5′-TTAAAAGAAAAGGGGGGAC-3′ (PPT-19), both of which served as DNA primers. The viral template pHIV-PBS was expressed and purified as previously described (3), and cellular tRNA3Lys was prepared and purified from human placenta (25); both of these reagents were used to study the initiation of (−) ssDNA synthesis. The RNA template and all DNA oligonucleotides were chemically synthesized by and purchased from GIBCO-BRL Inc., Montreal, Quebec, Canada.
Cells and viruses.
The human lymphoblastoid T-cell line MT-4 was used in these experiments. RT from the infectious clone pNL4-3 (1) was used to generate variants of HIV-1 containing the L74V, M184V, and L74V-M184V mutations by site-directed mutagenesis.
Drugs.
ABC and 3TC were provided by GlaxoSmithKline (Research Triangle Park, N.C.), and the active form of ABC, i.e., CBV-TP, was kindly provided by Margaret Tisdale (GlaxoSmithKline, Stevenage, Hertfordshire, United Kingdom). ddI and stavudine (d4T) were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). ddATP was ordered from MBI Fermentas Inc. (Burlington, Ontario, Canada).
Synthesis and purification of the pHIV-PBS RNA template.
RNA used as a template was synthesized in vitro, using the plasmid pHIV-PBS, which contains a 250-bp fragment of the HIV-1 5′-terminal sequence of genomic RNA, including the primer binding site (PBS). The plasmid (2.5 μM) was linearized using the restriction enzyme BssHII (3). In vitro transcription with T7 RNA polymerase was performed overnight at 37°C using the T7-MEGAshortscript in vitro transcription kit (Ambion Inc., Austin, Tex.) according to the manufacturer's instructions. The RNA product was purified using 8% polyacrylamide gels that contained 7 M urea and 50 mM Tris-borate-EDTA (TBE) and was visualized under UV light prior to elution from isolated gel slices using a solution of 0.5 M sodium acetate and 0.1% sodium dodecyl sulfate.
Initiation of (−) ssDNA synthesis.
Using a cell-free system, the efficiencies of initiation of synthesis of (−) ssDNA by WT and mutant enzymes were monitored, using the pHIV-PBS and natural tRNA3Lys primer-template system. The tRNA primer was heat annealed to the RNA template prior to initiation of DNA synthesis to ensure complete hybridization. The procedure was performed in a 150-μl reaction mixture containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, 40 nM tRNA3Lys, and 115 nM template RNA. This mixture was incubated for 2 min at 95°C, followed by 20 min at 70°C. Synthesis of (−) ssDNA was initiated by the addition of 285 nM RT in the presence of 6 mM MgCl2 and 200 μM dNTPs or dNTP-ddNTP complexes. Aliquots (2 μl) were removed at different time points, and the reactions were stopped in 8 μl of a solution containing 80% formamide and 40 mM EDTA. The reactions were monitored by using [α-32P]dCTP in the reaction mixture. The products were separated on 8% polyacrylamide-7 M urea gels containing 50 mM Tris borate (pH 8) and 1 mM EDTA and were exposed overnight. The addition of ddNTPs allowed monitoring of the efficiency of clearance of the +3 and +5 pausing sites, while the addition of dNTPs yielded synthesis of full-length DNA. The same experimental procedures were used to study DNA-primed reactions. Quantification of release from the pausing site at position +3 was preformed using Molecular Analyst software (Molecular Bioscience Group, Hercules, Calif.).
Initiation of plus-strand DNA synthesis.
The 17R primer was radiolabeled at its 5′ end. 5′-end labeling of the primer was performed with [γ-32P]ATP and T4 polynucleotide kinase (GIBCO-BRL), and the primer was purified on an 8% polyacrylamide-7 M urea gel containing 50 mM TBE. Primer 17R and template PPT-57 were prehybridized prior to incubation with RT. A mixture containing 2.72 μM template PPT-57 and 905 nM 32P-labeled primer 17R in a buffer containing 50 mM Tris-HCl (pH 7.8)-50 mM NaCl was denatured for 2 min at 95°C, followed by incubation at 72°C for 15 min and cooling for 30 min at room temperature. dNTPs (10 μM) were included in 10 μl of the preincubation mixture, together with 6 mM MgCl2. Reactions (20 μl mixtures) were initiated with 1.28 μM HIV-1 RT in a buffer containing 50 mM Tris-HCl (pH 7.8)-50 mM NaCl and were allowed to proceed for 10 min at 37°C. Aliquots (2 μl) were removed at different time points, and the reactions were stopped in 8 μl of a solution containing 80% formamide and 40 mM EDTA. The reaction products were finally analyzed on 8% polyacrylamide-7 M urea gels containing 50 mM TBE (pH 8), followed by exposure overnight. Quantification of plus pausing was performed as described above.
Incorporation of CBV-TP, ddATP, and d4T-TP.
The PPT-18 or PPT-19 primer was 5′ end labeled as previously described for the labeling of the 17R primer. The template-primer system used was PPT-57-PPT-18 for CBV-TP incorporation and PPT-57-PPT-19 for ddATP and d4T-TP incorporation. Template PPT-57 and the required primer were prehybridized prior to incubation with RT. A mixture containing 4.8 pM template PPT-57 and 1.6 pM 32P-labeled PPT-primer in a buffer containing 50 mM Tris-HCl (pH 7.8) and 50 mM NaCl was heated at 100°C for 3 min, followed by incubation at 72°C for 15 min and cooling for 30 min at room temperature. To 10 μl of the template-primer hybrid was added 4.8 pM RT in a buffer containing 50 mM Tris (pH 7.8), 50 mM NaCl, 1 μM competing dNTPs, and 10 μM other dNTPs. ddATP and CBV-TP were used at concentrations varying from 0 to 10 μM. The reaction (20-μl mixture) was initiated with 6 mM MgCl2 at 37°C and stopped by adding 100 μl of stop solution containing 100 mM ammonium acetate, 85% ethanol, and bulk tRNA. After precipitation at −80°C, the DNA was harvested by spinning it for 30 min at 13,000 rpm and was then washed with 70% ethanol. The DNA was air dried and then resuspended in 10 μl of gel loading buffer (100% formamide, bromophenol blue, and xylene); 3.5 μl of this product was loaded on an 8% polyacrylamide-7 M urea gel containing 50 mM TBE (pH 8) and visualized by autoradiography. Incorporation of d4T-TP was assessed in time course experiments; aliquots were taken at different times, and only one concentration of d4T-TP (5 μM) was used.
Site-directed mutagenesis.
Site-directed mutagenesis was performed to generate L74V, M184V, and L74V-M184V mutant viruses. The SphI-EcoRI fragment of the RT gene of the infectious clone pNL4-3 was subcloned into the cloning vector pSP72 (GIBCO-BRL), and the relevant mutation(s) was introduced using a set of primers containing the aforementioned mutations. After mutagenesis, the fragment was cloned back into the pNL4-3 plasmid, which was later extracted to yield proviral DNA containing the desired mutations.
Determinations of 50% inhibitory concentrations (IC50s) and TCID50s and viral growth experiments.
Viral stocks were prepared by transfection of proviral plasmid DNA into Cos-7 cells as described previously (31). The 50% tissue culture infective dose (TCID50) of each virus stock was determined by endpoint dilution in MT-4 cells as described previously (7). To determine the sensitivities of our constructs containing L74V, M184V, and L74V-M184V to ABC, ddI, 3TC, and d4T, 2 × 106 MT-4 cells were infected with 2 × 105 to 4 × 105 TCID50s of transfected supernatant of each virus strain, including WT virus. The cultures were incubated in 1 ml of RPMI 1640 supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics as described previously (10) for 2 h at 37°C in a CO2 incubator. At the end of incubation, the cells were washed to remove all free viral particles and plated in different concentrations of the given drugs. Samples were taken on day 4 and monitored for RT activity in culture fluids, using a poly(rA) · oligo(dT) template-primer system as described previously (6, 17, 34). To study viral growth kinetics, 2 ng of supernatant from each transfection was used to infect 106 MT-4 cells, which were incubated for 2 h at 37°C in 1 ml of medium. After incubation and removal of free viral particles, the volume was increased to 10 ml and the cultures were kept at 37°C in a CO2 incubator. Samples were taken every 2 days over a period of 10 to 12 days, and the cultures were fed twice a week. RT activities in culture fluids were determined at regular intervals and plotted.
RESULTS
Drug susceptibility in cell culture and cell-free assays.
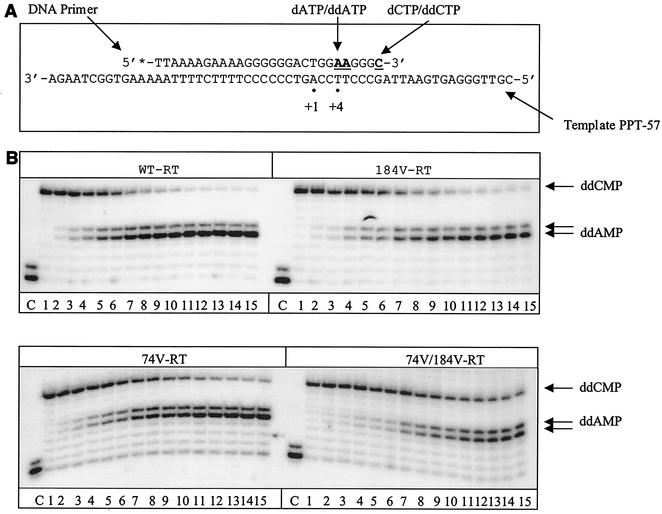
Cell culture and clinical data have shown that viruses that contain the L74V and M184V mutations display low to medium levels of resistance to ABC and ddI and that the level of resistance was increased when both mutations were present within the same RT gene (8, 16, 18, 39, 52). Here, we initially studied the efficiencies of incorporation of CBV-TP and ddATP, i.e., the active metabolites of ABC and ddI, respectively, with WT HIV-1 RT and relevant mutated enzymes in cell-free assays. Toward this end, we generated enzymes that contained the L74V and M184V mutations, either alone or together, and studied the efficiency of chain termination in dose-dependent fashion using gel-based assays. Figure 1A and C are schematic representations of the template-primer systems used in the different incorporation experiments. For each of the singly mutated enzymes, we observed a moderate decrease in chain termination for ddATP (Fig. 1B, right half of top gel and left half of bottom gel); the determination of IC50s revealed an ∼2-fold difference compared to WT RT (Table 1). This effect was amplified with the doubly mutated enzyme (Fig. 1B, right half of bottom gel); the IC50s were ∼6-fold higher than for WT RT (Table 1). In the case of CBV-TP, a significant decrease in chain termination was observed only with the double-mutant RT versus WT RT (Fig. 1D, right half of bottom gel); we measured a 10-fold difference in IC50s compared to WT RT (Table 1). The singly mutated enzymes did not show significant differences in comparison with WT RT (Fig. 1D, right half of top gel and left half of bottom gel). These effects are specific, and we did not observe differences in regard to incorporation of d4T-MP when we compared the double-mutant RT with WT RT (data not shown).
FIG. 1.
Incorporation of ddATP, CBV-TP, and d4T-TP by WT and mutant RTs. (A) Graphic representation of the cell-free system (PPT-57-PPT-19) used to monitor the incorporation of ddATP by WT and mutant RTs. The DNA template consists of 57 nucleotides designed to correspond to the PPT sequence of the HIV-1 genome. The primer was 5′ end labeled. Bold underlined letters indicate sites of chain termination. Dots define positions +1 and +4 in the reaction. (B) Dose-dependent incorporation of ddATP by WT and mutant RTs. Reactions were performed with increasing concentrations of ddATP (lanes 1 to 15). The concentration of dATP was kept at 1 μM, in order to allow incorporation of the analogue ddATP, which was employed at increasing concentrations (0, 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 μM). The other dNTPs were used at 10 μM. A control reaction was performed without RT enzyme (lanes C). (C) Graphic representation of the cell-free system (PPT-57-PPT-18) used to monitor incorporation of CBV-TP. (D) Dose-dependent incorporation of CBV-TP by WT and mutant RTs. Reactions were performed as described for panel B, except that ddATP was replaced by CBV-TP (0, 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10 μM) and the concentration of dGTP was 1 μM.
TABLE 1.
IC50s and fold resistances for incorporation of CBV-TP and ddATP
| Type of RT | ddATP
|
CBV-TP
|
||
|---|---|---|---|---|
| IC50 (μM) | Fold resistance | IC50 (μM) | Fold resistance | |
| WT | 0.5 | 0.7 | ||
| 74V | 1 | 2 | 0.7 | |
| 184V | 1 | 2 | 0.7 | |
| 74V-184V | 3 | 6 | 7 | 10 |
These biochemical data are in good agreement with data generated through the trends seen in drug susceptibility assays in cell culture (Table 2). We also infected MT-4 cells with WT HIV-1 and mutant viruses that were generated through site-directed mutagenesis and observed an ≈7- to 8-fold loss in sensitivity to both ABC and ddI on the part of each of the L74V and M184V viruses. The highest level of resistance was obtained with the L74V-M184V doubly mutated virus (i.e., ∼28-fold resistance) (Table 2). All the viruses tested retained sensitivity to d4T and were highly resistant to 3TC, except for those containing only L74V, which is known to confer only low-level resistance to the latter compound (Table 2).
TABLE 2.
IC50s of various recombinant viruses for antiviral nucleosidesa
| Virus | TCID50/ml | ABC
|
ddI
|
d4T
|
3TC
|
||||
|---|---|---|---|---|---|---|---|---|---|
| IC50 (μM) | Fold resistance | IC50 (μM) | Fold resistance | IC50 (μM) | Fold resistance | IC50 (μM) | Fold resistance | ||
| WT | 1.23 × 108 | 0.15 ± 0.1 | 0.72 ± 0.1 | 1.64 ± 0.4 | 0.6 ± 0.3 | ||||
| 74V | 3.24 × 106 | 1.18 ± 0.2 | 7.8 | 4.91 ± 0.6 | 6.8 | 2.82 ± 0.4 | 1.7 | 2.43 ± 0.3 | 4.1 |
| 184V | 3.05 × 106 | 1.22 ± 0.2 | 8 | 6.79 ± 0.1 | 9.4 | 1.29 ± 0.2 | >100 | >150 | |
| 74V-184V | 5.94 × 105 | 4.27 ± 0.1 | 28 | 17.16 ± 0.8 | 24 | 2.14 ± 0.3 | 1.3 | >100 | >150 |
Values are means from at least three independent experiments ± standard errors.
Replication characteristics of mutant viruses.
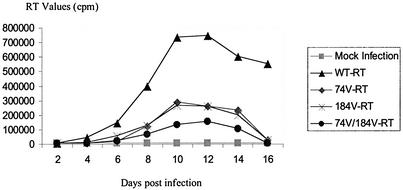
In order to assess and compare the replication capacities of mutant viruses and WT HIV-1, we determined the TCID50s of our stock viruses, as well as the kinetics of viral growth. Our data show that TCID50s were 3 log units lower for the double mutant and 2 log units lower for each of the single mutants than for WT HIV-1 in our assay (Table 2). Growth curves also showed delays for the L74V/M184V-containing viruses, which were associated with lower levels of RT activity (Fig. 2). In agreement with previous studies, we found decreases in RT activity for each of the singly mutated viruses; however, each of the singly mutated viruses was able to produce higher levels of RT than the doubly mutated virus. The cultures were also tested for levels of capsid (CA) (p24) antigen, and the results were consistent with those obtained by RT assay; notably, the double mutant had the lowest levels of p24 compared with WT virus and all the other mutant viruses that were tested (data not shown).
FIG. 2.
Growth curve of HIV-1 replication after infection of MT-4 cells by viruses harvested from transfected Cos-7 cells. One million MT-4 cells were infected with either the pNL4-3-WT, pNL4-3-L74V, pNL4-3-M184V, or pNL4-3-L74V-M184V virus, using 2 ng of p24 antigen in each case. Virus production was monitored by RT assay of culture fluids.
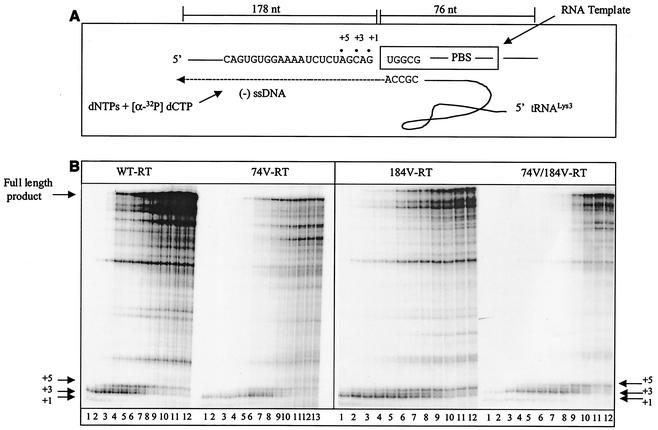
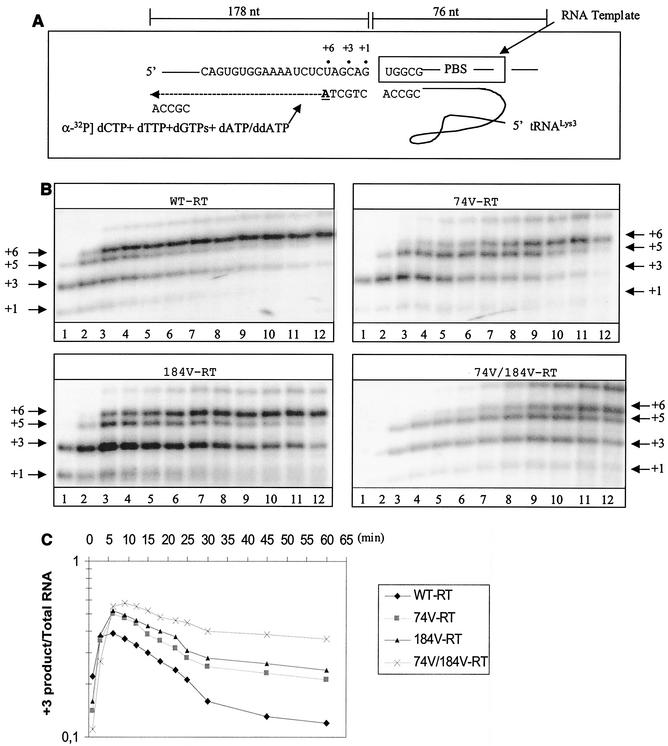
Initiation of synthesis of (−) ssDNA.
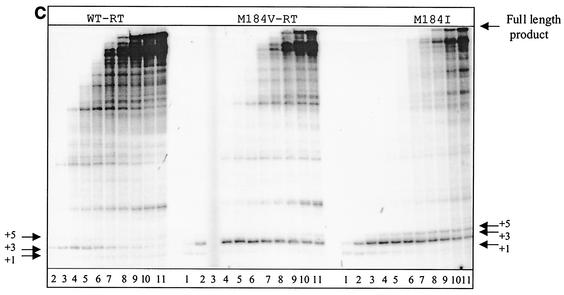
To study mechanisms potentially associated with the low replication capacities of the mutated viruses, we evaluated the efficiency of initiation of (−) ssDNA synthesis using a gel-based assay system as described previously (57). First, we heat annealed natural tRNA3Lys to an in vitro synthesized RNA template containing the HIV-1 PBS to form a binary complex that could be used as a substrate for RT (Fig. 3A). The preformed tRNA-RNA complexes were incubated with either WT or mutant RT enzymes to initiate the RT reaction in the presence of dNTPs. We then performed time course experiments, and these pointed to a decrease in the total amount of tRNA primer extension catalyzed by the mutants, as well as decreased release of pausing at positions +3 and +5, when both of the singly mutated RTs, i.e., L74V and M184V, were assessed (Fig. 3B, right half of left gel and left half of right gel). However, the decrease in product formation was most pronounced in the case of the doubly mutated enzyme, i.e., RT-L74V-M184V (Fig. 3B, right half of right gel). Thus, a correlation may exist between diminished rates of initiation of (−) ssDNA synthesis and diminished viral replication competence. In both cases, the order of efficiency was WT > M184V > L74V > L74V-M184V.
FIG. 3.
Synthesis of full-length DNA using WT and mutant enzymes. (A) Graphic representation of the cell-free system used for the synthesis of full-length (−) ssDNA. The RNA template (pHIV-PBS) used in this system consists of 258 nucleotides (nt) at the 5′ end of the HIV-1 genome, which contains the R, U5, and PBS regions. (B) Synthesis of (−) ssDNA from the pHIV-PBS RNA template by each of the WT and mutant RTs. A time course experiment was performed by heat annealing tRNA3Lys to the RNA template to yield a complex which was incubated together with each RT studied at 37°C for 5 min. Reactions were initiated by the addition of dNTPs and monitored by incorporation of [α-32P]dCTP into the growing DNA chain. The reactions were stopped at different times during a period of 60 min (lanes 1 to 12). Lane 13 exceptionally shows one of these reactions after 90 min. (C) Synthesis of (−) ssDNA from the pHIV-PBS RNA template by WT, M184V, and M184I RTs. Reaction conditions were as described for panel B.
To further substantiate the relationship between diminished viral replication and the initiation of reverse transcription, we also studied the synthesis of (−) ssDNA in the context of other mutations that are known to be associated with diminished viral replication capacity. For example, the M184I mutation appears early after initiation of 3TC monotherapy and is later replaced by the M184V mutated virus because the latter replicates more efficiently (4, 30). The results in Fig. 3C show the same trend in regard to efficiency of initiation of (−) ssDNA synthesis, i.e., WT > M184V > M184I. Again, the decreased formation of the full-length (−) ssDNA product was correlated with an accumulation of paused products at early stages of the reaction.
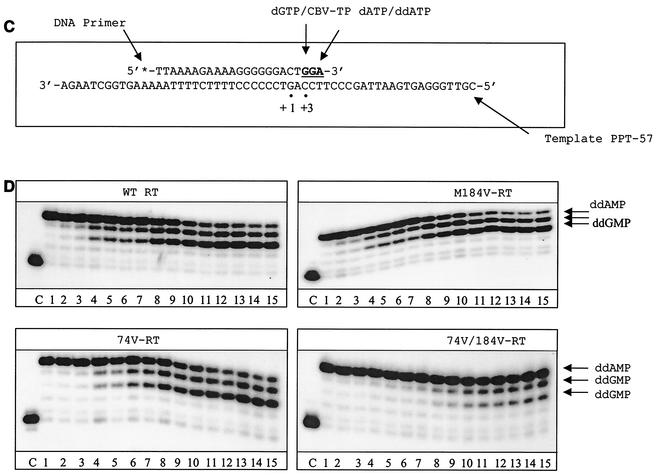
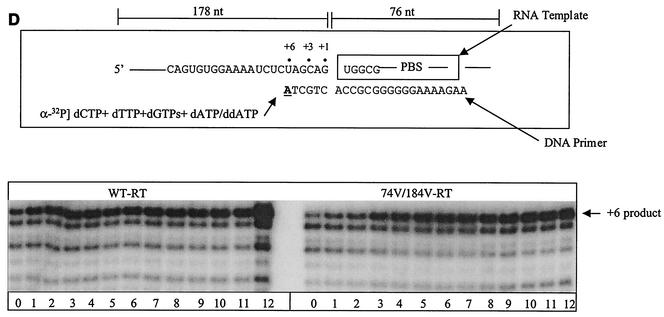
To further analyze these changes in pausing patterns, we modified the assay described above and restricted DNA synthesis to the initiation stage by the addition of a ddNTP at position +6 (Fig. 4A). These data are in good agreement with the results shown in Fig. 3B. Furthermore, release from the pausing sites at positions +3 and +5 was severely compromised when the L74V and M184V mutations were present (Fig. 4B). Graphic representation of these data revealed that the rate of escape from pausing at position +3 reached a plateau before this process was completed, and again, the strongest effect was seen with the L74V-M184V double mutant (Fig. 4C). In contrast, we did not detect major differences between the WT RT and the double-mutant enzyme when these reactions were initiated with a DNA primer, although the levels of synthesis were modest in both cases (Fig. 4D). These results suggest that mutated enzymes containing the L74V and/or M184V mutation may not be able to effectively utilize RNA primers. To further test this hypothesis, we next studied the efficiencies of the RNA-primed initiation of plus-strand DNA synthesis using WT RT and mutated enzymes.
FIG. 4.
Initiation of (−) ssDNA synthesis by WT and mutant enzymes. The same system and conditions were used as for Fig. 3, except that ddATP was used instead of dATP in the dNTP mixture to restrict synthesis during initiation, with the addition of a stop nucleotide (nt) (ddATP) at position +6. A mixture of ddATP, dGTP, and dTTP was used at 10 μM, and dCTP was employed at 1 μM to allow incorporation of [α-32P]dCTP, which was used as a tracer during the reaction. (A) Schematic representation of the reaction. Underlined letters indicate sites of chain termination. Dots define specific positions in the reaction, as indicated. (B) Data obtained with WT versus mutant RTs. (C) Quantification of the data shown in panel B. The graphs show the time-dependent release from pausing at position +3. (D) Limited DNA synthesis in reactions performed with a corresponding DNA primer.
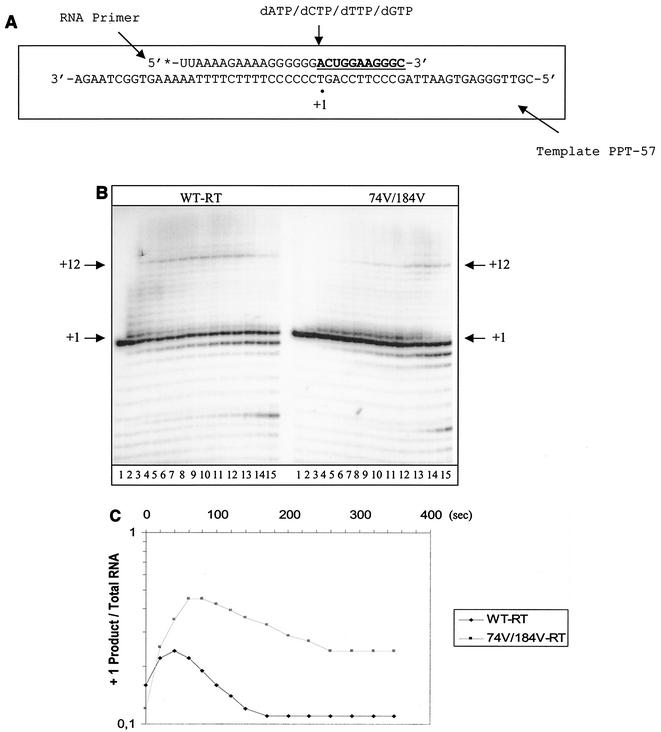
Initiation of synthesis of plus-strand DNA.
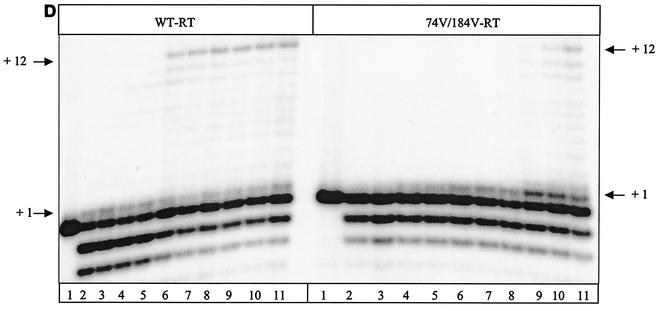
It has been shown that the initiation of plus-strand DNA synthesis is an important rate-limiting step in reverse transcription (15, 16). Low concentrations of dNTPs caused dramatic reductions in rates of DNA synthesis, which is primarily a result of significantly increased Km values during the first two nucleotide incorporation events. Here, we studied the efficiency of the reaction with the doubly mutated RT, which showed the strongest effect on the tRNA-primed synthesis of (−) ssDNA. Figure 5A is a schematic representation of the reaction that was employed. Time course experiments revealed that the pause site at position +1 was rapidly released by the WT RT, while this reaction was slow and incomplete with the L74V-M184V double mutant (Fig. 5B and C). This resulted in a significant reduction in synthesis of plus-strand DNA, as monitored by the second pause site at position +12, at which WT RT was able to clear pausing in 120 s. In contrast, 200 s was necessary in order for the L74V-M184V RT to accomplish this task. In these reactions, the detection of larger products is impeded by the RNase H activity that cleaves at the newly formed DNA-RNA junction, and the results in Fig. 5D show that relatively high concentrations of dNTPs were required to overcome rate-limiting steps during the initiation reaction.
FIG. 5.
Initiation of synthesis of full-length plus-strand DNA by WT and L74V-M184V RTs. (A) Graphic representation of the system used to monitor the initiation of plus-strand DNA synthesis. Underlined letters indicate sites of chain termination. The dot defines the +1 position in the reaction. (B) Time course determination of the initiation of plus-strand DNA synthesis. The dNTP concentrations were 10 μM, and samples were removed every 20 s for 5 min (lanes 1 to 15). (C) Quantification of the data shown in panel B. The graphs show the time-dependent release from pausing at position +1. (D) Efficiency of initiation of plus-strand DNA synthesis in the presence of increasing concentrations of dNTPs. Lanes 1 to 11 show 5-min reactions in the presence of 0, 1.5, 3, 6, 12, 25, 50, 100, 200, and 400 μM (each) concentrations of the four dNTPs.
Taken together, our biochemical data show that the L74V and M184V mutations cause significant reductions in rates of initiation of both minus- and plus-strand viral DNA synthesis. Clearance from pausing appears to be a major obstacle that is not effectively bypassed by enzymes containing these mutations.
DISCUSSION
A combination of the L74V and M184V mutations in HIV-1 RT is the most frequent pattern associated with resistance to both ABC and ddI (18, 38, 39, 55, 58). These mutations constitute important cornerstones of resistance to these drugs, since prolonged therapy is often associated with the emergence of additional mutations that increase the level of nonresponsiveness. Here, we generated singly and doubly mutated enzymes containing these substitutions, as well as relevant recombinant viruses, to study enzyme activities and viral phenotypic resistance.
We have shown that viruses that contain mutations associated with low levels of resistance to ABC and ddI can exert additive effects in regard to phenotypic sensitivity to these compounds. Although mutated enzymes containing either L74V or M184V showed only slight reductions in incorporation of CBV-MP and ddAMP, the doubly mutated RT discriminated more efficiently between the two nucleoside inhibitors and their natural counterparts. Levels of resistance to ABC and ddI were higher in the phenotypic assay than in biochemical experiments. This is consistent with earlier findings (39, 42, 43) and may reflect the fact that chain-terminating nucleotides can be incorporated at multiple sites during reverse transcription of the entire RNA genome, while the use of a model primer-template system offers only a limited number of incorporation sites. Thus, small effects in biochemical assays may be amplified either in vivo or in tissue culture systems. Alternatively, one may invoke the possibility of increased rates of excision of incorporated CBV-MP and ddAMP associated with L74V and M184V. However, we did not detect any significant differences with respect to PPi- and ATP-dependent primer unblocking with these two chain terminators when comparing reactions conducted with the WT or mutated RTs. Conceivably the additional acquisition of TAMs and/or other mutations is required to increase rates of primer unblocking and to further augment levels of resistance. Indeed, the M41L, D67N, and T215Y mutations are known to facilitate the excision of many NRTIs, including ZDV-MP, d4T-MP, and CBV-MP. Combinations of TAMs and M184V and/or L74V are associated clinically with higher levels of resistance to both ABC and ddI.
It should be noted that additional mutations may also compensate for the diminished replication capacity associated with L74V and M184V. Several groups have linked the reduced replication capacities of viruses containing these substitutions to the reduced enzymatic processivity of the corresponding enzymes (4, 49, 50). In contrast, mutated enzymes containing the T215Y and K219Q TAMs showed increased processivity, in keeping with a possible compensatory effect of these two substitutions. However, the experimental conditions under which processivity is measured may not reflect physiologically relevant conditions, since reverse transcription takes place within the viral capsid that contains RT in large excess over the copackaged RNA genome. As a consequence, dissociated enzymes are likely to be rapidly replaced by others that continue the polymerization process. Thus, the effects of a moderate diminution in processivity may not contribute as much to reductions in DNA synthesis as do the rate limitations reported here in regard to initiation of both minus- and plus-strand synthesis of viral DNA.
The initiations of RNA-primed minus- and plus-strand DNA synthesis have recently been characterized as important rate-limiting steps that can reduce rates of DNA synthesis by 2 to 3 orders of magnitude relative to WT RT (19). The initiation of (−) ssDNA synthesis is accompanied by frequent pausing and is, in general, a distributive process that involves frequent dissociation of the enzyme from the template (23, 28). Here, we show that L74V- and M184V-containing enzymes cannot efficiently bypass these pausing events. Our data show a slow release from the +3 and +5 pausing sites by the doubly mutated enzyme compared to both WT RT and RTs containing only single mutations. These experiments were performed in the absence of a heparin trap and in the presence of an excess of enzyme over primer-template substrate to better mimic physiological conditions. We conclude that diminished rates of escape from pause sites during initiation of both minus- and plus-strand DNA synthesis are important in regard to low replication fitness, although enzymes that display a decrease in processivity are also unable to efficiently bypass early pause sites during RNA-primed DNA synthesis.
Prolonged therapy with ABC is associated with the additional emergence of the K65R or Y115F mutation or both mutations together. It is unlikely that K65R can compensate for the diminished viral fitness associated with L74V and M184V, since K65R is associated with diminished viral replication capacity and enzyme processivity. K65R also resulted in decreased incorporation of CBV-MP and other NRTIs, i.e., increased levels of drug resistance at a cost of decreased viral fitness (K. L. White, N. A. Margot, T. Wrin, C. J. Petropoulos, L. K. Naeger, and M. D. Miller, poster 3033, XIV International AIDS Conference, Barcelona, Spain, 7 to 12 July, 2002). We now wish to study whether other resistance-associated mutations in association with L74V and/or M184V lead to increased viral fitness, as well as increased rates of initiation of both minus- and plus-strand DNA synthesis.
Acknowledgments
We thank Maureen Oliveira and Mervi Deterio for technical assistance.
This research was supported by grants to M.G. and M.A.W. from the Canadian Institutes for Health Research and by Bristol-Myers Squibb Inc., Wallingford, Conn. We also thank Aldo and Diane Bensadown for generous support of our research program.
Footnotes
This paper is dedicated to the memory of James-Paul Marois, who was a graduate student in our laboratory.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arion, D., N. Kausshik, S. McConnick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant vira1 reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 3.Arts, E. J., X. Li, Z. Gu, M. A. Parniak, and M. A. Wainberg. 1994. Comparison of deoxyoligonucleotide and tRNALys3 as primers in an endogenous human immunodeficiency virus type 1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem. 269:14672-14680. [PubMed] [Google Scholar]
- 4.Back, N. K. T., M. Nijhuis, W. Keulen, C. A. B. Boucher, B. B. O. Essnik, A. B. P. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-l variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, C. A. B., N. Cammack, P. Schipper, R. Schuurman, P. Rouse, M. A. Wainberg, and J. M. Cameron. 1993. High-level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulerice, F., S. Bour, R. Geleziunas, A. Lvovich, and M. A. Wainberg. 1990. High frequency of isolation of defective human immunodeficiency virus type-l and heterogeneity of viral gene expression in clones of infected U-937 cells. J. Virol. 64:1745-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulbecco, R. 1988. Endpoint methods—measurement of the infectious titer of a viral sample, p. 22-25. In R. Dulbecco and H. S. Ginsberg (ed.), Virology, 2nd ed. J. P. Lippincott, Philadelphia, Pa.
- 8.Eron, J. J., Y. K. Chow, A. M. Caliendo, J. Videler, K. M. Devore, T. P. Cooley, H. A. Liebman, J. C. Kaplan, M. S. Hirsch, and R. T. D'Aquila. 1993. Pol mutation conferring zidovudine and didanosine resistance with different effects in vitro yield multiply resistant human immunodeficiency virus type 1 isolates in vivo. Antimicrob. Agents Chemother. 37:1480-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1997. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 10.Gao, Q., Z. Gu, M. A. Parniak, X. Li, and M. A. Wainberg. 1992. In vitro selection of variants of human immunodeficiency virus type 1 resistant to 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine. J. Virol. 66:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, Q., Z. Gu, M. A. Parniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Götte, M., G. Maier, A. Mochi Onori, L. Cellai, M. A. Wainberg, and H. Heumann. 1999. Temporal coordination between initiation of HIV (+)-strand DNA synthesis and primer remova1. J. Biol. Chem. 274:11159-11169. [DOI] [PubMed] [Google Scholar]
- 13.Götte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of the human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis. J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Götte, M., and M. A. Wainberg. 2000. Biochemical mechanisms involved in overcoming HIV resistance to nucleoside inhibitors of reverse transcriptase. Drug Resist. Updates 3:30-38. [DOI] [PubMed] [Google Scholar]
- 15.Götte, M., M. Kameoka, N. McLellan, L. Cellai, and M. A. Wainberg. 2001. Analysis of efficiency and fidelity of HIV-1 (+)-strand DNA synthesis reveals a novel rate-limiting step during retroviral reverse transcription. J. Biol. Chem. 276:6711-6719. [DOI] [PubMed] [Google Scholar]
- 16.Gu, Z., Q. Gao, X. Li, M. A. Parniak, and M. A. Wainberg. 1992. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J. Virol. 66:7128-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, Z., Q. Gao, H. Fang, H. Salomon, M. A. Parniak, E. Goldberg, J. Cameron, and M. A. Wainberg. 1994. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistant to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 38:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrigan, P. R., C. Stone, P. Griffin, I. Nájera, S. Bloor, S. Kemp, M. Tisdale, B. Larder, and the CNA 2001 Investigative Group. 2000. Resistance profile of the human immunodeficiency virus type 1 reverse transcriptase inhibitor abacavir (1592U89) after monotherapy and combination therapy. J. Infect. Dis. 181:912-920. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J.-C., S. Zinnen, and P. Modrich. 1993. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J. Biol. Chem. 268:24607-24613. [PubMed] [Google Scholar]
- 20.Hsu, M., P. Inouye, L. Rezende, N. Richard, Z. Li, V. R. Prasad, and M. A. Wainberg. 1997. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 25:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 22.Isel, C., R. Marquet, G. Keith, C. Ehresmann, and B. Ehresmann. 1993. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 268:25269-25272. [PubMed] [Google Scholar]
- 23.Isel, C., J. M. Lanchy, S. F. Le Grice, C. Ehresmann, B. Ehresmann, and R. Marquet. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 15:917-924. [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kati, W. M., K. A. Johnson, L. F. Jerva, and K. S. Anderson. 1992. Mechanism and fidelity of HIV-1 reverse transcriptase. J. Biol. Chem. 267:25988-25997. [PubMed] [Google Scholar]
- 27.Krebs, R., U. Immendörfer, S. H. Thrall, B. M. Wöhrl, and R. S. Goody. 1997. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucleoside inhibitors zidovudine and 3TC. Biochemistry 36:10292-10300. [DOI] [PubMed] [Google Scholar]
- 28.Lanchy, J.-M., C. Ehresmann, S. F. J. Le Grice, B. Ehresmann, and R. Marquet. 1996. Binding and kinetic properties of HIV-1 reverse transcriptase markedly differ during initiation and elongation of reverse transcription. EMBO J. 15:7178-7187. [PMC free article] [PubMed] [Google Scholar]
- 29.Lanchy, J.-M., G. Keith, S. F. J. Le Grice, B. Ehresmann, C. Ehresmann, and R. Marquet. 1998. Contact between reverse transcriptase and the primer strand governs the transition from initiation to elongation of HIV-1 reverse transcriptase. J. Biol. Chem. 273:24425-24432. [DOI] [PubMed] [Google Scholar]
- 30.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 31.Le Grice, S. F., and F. Leith-Grüninger. 1990. Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem. 187:307-314. [DOI] [PubMed] [Google Scholar]
- 32.Li, X., J. Mak, E. J. Arts, Z. Gu, L. Kleiman, M. A. Wainberg, and M. A. Parniak. 1994. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J. Virol. 68:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, C., L. Rong, M. Götte, X. Li, Y. Quan, L. Kleiman, and M. A. Wainberg. 1998. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcriptase. J. Biol. Chem. 273:21309-21315. [DOI] [PubMed] [Google Scholar]
- 34.Mansour, T. S., H. Jin, W. Wang, E. Hooker, C. Ashman, N. Cammack, H. Salomon, A. R. Belmonte, and M. A. Wainberg. 1995. Anti-human immunodeficiency virus and anti-hepatitis-B virus activities and toxicities of the enantiomers of 2′-deoxy-3′-oxa-4′-thiocytidine and their 5-fluoro analogues in vitro. J. Med. Chem. 38:1-4. [DOI] [PubMed] [Google Scholar]
- 35.Mansour, T. S., H. Jin, W. Wang, D. M. Dixit, C. A. Evans, H. L. A. Tse, B. Belleau, J. W. Gillard, E. Hooker, C. Ashman, N. Cammack, H. Salomon, A. R. Belmonte, and M. A. Wainberg. 1995. Structure-activity relationships among a new class of antiviral heterosubstituted 2′,3′-dideoxynucleoside analogues. Nucleosides Nucleotides 14:627-635. [Google Scholar]
- 36.Martin, J. L., J. E. Wilson, R. L. Haynes, and P. A. Furman. 1993. Mechanism of resistance of human immunodeficiency virus type 1 to 2′,3′ dideoxyinosine. Proc. Natl. Acad. Sci. USA 90:6135-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer, P. R., S. E. Matsuura, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant RN-1 reverse transcriptase. Mol. Cel1 4:35-43. [DOI] [PubMed] [Google Scholar]
- 38.Miller, V., M. Stürmer, S. Staszewski, B. Gröschel, K. Hertogs, M.-P. de Béthume, R. Pauwels, P. R. Harrigan, S. Bloor, S. D. Kemp, and B. A. Larder. 1998. The M184V mutation in HIV-1 reverse transcriptase (RT) conferring lamivudine resistance does not result in broad cross-resistance to nucleoside analogue RT inhibitors. AIDS 12:705-712. [DOI] [PubMed] [Google Scholar]
- 39.Miller, V., M. Ait-Khaled, C. Stone, P. Griffin, D. Mesogiti, A. Cutrell, R. Harrigan, S. Staszewski, C. Katlama, G. Pearce, and M. Tisdale. 2000. HIV-1 reverse transcriptase (RT) genotype and susceptibility to RT inhibitors during abacavir monotherapy and combination therapy. AIDS 14:912-920. [DOI] [PubMed] [Google Scholar]
- 40.Oude Essink, B. B., N. K. Back, and B. Berkhout. 1997. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 25:3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan, Y., Z. Gu, X. Li, C. D. Morrow, and M. A. Wainberg. 1996. Mutated HIV-1 M184V reverse transcriptase displays resistance to the triphosphate of (−)2′,3′-dideoxy-3′-thiacytidine (3TC) in both endogenous and cell-free enzyme assays. J. Virol. 70:5642-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray, A. S., Z. Yang, J. Shi, A. Hobbs, R. F. Schinazi, C. K. Chu, and K. S. Anderson. 2001. Mechanistic studies to understand the inhibition of wild type and mutant HIV-1 reverse transcriptase by carbovir-triphosphate. Nucleosides Nucleotides Nucleic Acids 20:1247-1250. [DOI] [PubMed] [Google Scholar]
- 43.Ray, A. S., A. Basavapathruni, and K. S. Anderson. 2002. Mechanistic studies to understand the progressive development of resistance in HIV-1 reverse transcriptase to abacavir. J. Biol. Chem. 43:40479-40490. [DOI] [PubMed] [Google Scholar]
- 44.Reardon, J. E. 1993. Human immunodeficiency virus reverse transcriptase. A kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J. Biol. Chem. 268:8743-8751. [PubMed] [Google Scholar]
- 45.Rittinger, K., G. Divita, and R. S. Goody. 1995. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA 92:8046-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarafianos, S. G., K. Das, J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem. Biol. 6:137-146. [DOI] [PubMed] [Google Scholar]
- 47.Schinazi, R. F., R. M. Lloyd, Jr., M. H. Nguyen, D. L. Cannon, A. McMillan, N. Ilksoy, C. K. Chu, D. C. Liotta, H. Z. Bazmi, and J. W. Mellors. 1993. Characterization of human immunodeficiency virus resistant to oxathiolane-cytosine nucleosides. Antimicrob. Agents Chemother. 37:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Shipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, C. Christopherson, and C. A. B. Boucher. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 17l:1411-1419. [DOI] [PubMed]
- 49.Shah, F. S., K. A. Curr, M. E. Hamburgh, M. Parniak, H. Mitsuya, J. G. Arnez, and V. R. Prasad. 2000. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 275:27037-27044. [DOI] [PubMed] [Google Scholar]
- 50.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 71:8846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma, P. L., and C. S. Crumpacker. 1999. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: a comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J. Virol. 73:8448-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.St. Clair, M. B., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 53.Thrall, S. H., R. Krebs, B. M. Wöhrl, L. Cellai, R. S. Goody, and T. Restle. 1998. Pre-steady-state kinetic characterization of RNA-primed initiation of transcription by HIV-1 reverse transcriptase and analysis of the transition to a processive DNA-primed polymerization mode. Biochemistry 37:13349-13358. [DOI] [PubMed] [Google Scholar]
- 54.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tisdale, M., T. Alnadaf, and D. Cousens. 1997. Combination of mutations in human immunodeficiency virus type 1 reverse transcriptase required for resistance to the carbocyclic nucleoside 1592U89. Antimicrob. Agents Chemother. 41:1094-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wainberg, M. A., W. C. Drosopoulos, H. Sa1omon, M. Hsu, G. Borkow, M. A. Parniak, Z. Gu, Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 217:1282-1285. [DOI] [PubMed] [Google Scholar]
- 57.Wei, X., C. Liang, M. Gotte, and M. A. Wainberg. 2002. The M184V mutation in HIV-1 reverse transcriptase reduces the restoration of wild-type replication by attenuated viruses. AIDS 16:2391-2398. [DOI] [PubMed] [Google Scholar]
- 58.Winters, M. A., R. W. Robert, A. Jellinger, G. Mamtora, T. Gingeras, and T. C. Merigan. 1997. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob. Agents Chemother. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]