Abstract
Lytic-cycle replication of Kaposi's sarcoma-associated herpesvirus (KSHV) in PEL cells causes G1 cell cycle arrest mediated by the virus-encoded replication-associated protein (RAP) (or K8 protein), which induces high-level expression of the cellular C/EBPα and p21 proteins. Here we have examined the mechanism of this induction at both the transcriptional and posttranslational levels. RAP proved to bind very efficiently to both C/EBPα and p21 and stabilized them by up to 10-fold from proteasome-mediated degradation in vitro. Cross-linking revealed that RAP itself forms stable dimers and tetramers in solution and forms higher-order complexes but not heterodimers with C/EBPα. Cotransfection of RAP with C/EBPα cooperatively stimulated both the C/EBPα and p21 promoters in luciferase reporter gene assays. Only the basic/leucine zipper region of RAP was needed for interaction with and stabilization of C/EBPα, but both the N-terminal and C-terminal domains were required for transcriptional augmentation. In vitro-translated RAP interfered with DNA binding by C/EBPα in electrophonetic mobility shift assay (EMSA) experiments but did not itself bind to the target C/EBPα sites or form supershifted bands. However, in endogenous chromatin immunoprecipitation (ChIP) assays with tetradecanoyl phorbol acetate-induced PEL cells, RAP proved to specifically associate with the C/EBPα promoter in vivo, but only in a C/EBPα-dependent manner, implying an in vivo piggyback interaction with DNA-bound C/EBPα. Expression of exogenous RAP (Ad-RAP) caused G1/S cell cycle arrest in human dermal microvascular endothelial cells and also induced both the C/EBPα and p21 proteins, which formed punctate nuclear patterns that colocalized with RAP in PML nuclear bodies. In the presence of RAP, C/EBPα was also efficiently recruited into viral DNA replication compartments in both infected and cotransfected cells. In support of a direct role for this interaction in viral DNA replication, three C/EBPα binding sites were identified by in vitro EMSA experiments within a 220-bp core segment of the duplicated KSHV Ori-Lyt region, and although RAP did not bind to Ori-Lyt DNA directly in vitro, both endogenous RAP and C/EBPα were found to be associated with the Ori-Lyt region by ChIP assays in lytically induced PEL cells. Finally, we found that the KSHV lytic cycle could not be triggered by either synchronizing KSHV latently infected PEL cells in G1 phase or inducing p21 in a C/EBPα-independent process.
Kaposi's sarcoma-associated herpesvirus (KSHV), a close relative of Epstein-Barr virus (EBV), is thought to be the causative agent of classical, endemic, and AIDS-associated forms of Kaposi's sarcoma (KS) (6), as well as of primary effusion lymphoma (PEL) in AIDS patients (45). KSHV infections convert cultured primary human dermal microvascular endothelial cells (DMVEC) into KS-like spindle cells in culture (11). Like all herpesviruses, KSHV undergoes two phases of infection, namely, a LANA1-positive quiescent latent state in both DMVEC spindle cells and in PEL cell lines and a tetradecanoyl phorbol acetate (TPA)-induced productive lytic cycle. KSHV and herpesviruses in general encode many of their own viral DNA replication and nucleotide synthesis machinery components, which partially obviate the need for cellular S-phase-associated replication machinery (48, 51).
Recent studies have shown that the KSHV lytic cycle induces host G1 cell cycle arrest in PEL cells (47). This process appears to be mediated by the KSHV lytic cycle replication-associated protein (RAP) (or K8 protein), which is related to bZIP family transcription factors and is known to associate efficiently with viral DNA replication compartments (RC) (48). Introduction of exogenous RAP leads to increased levels of expression of both the C/EBPα and p21 proteins in PEL and HF cells, which results in host G1 cell cycle arrest. Furthermore, the ability of RAP to induce p21 is dependent on the presence of C/EBPα, because RAP could not induce either p21 or cell cycle arrest in mouse C/EBPα knockout cells. In addition, C/EBPα transcriptionally activates the RAP promoter in cooperation with KSHV replication and transcription activator (RTA) and therefore establishes a positive self-reinforcing loop together with RAP during the early stages of the lytic cycle (43).
The ability to block the cell cycle in G0/G1 appears to be an important early step in the lytic cycle for most herpesviruses (14), and the EBV ZTA protein is also known to trigger host cell cycle arrest during the EBV lytic cycle (5, 14). ZTA also stabilizes p53 and p27 and represses the expression of c-MYC (31). We have recently shown that ZTA-mediated cell cycle arrest also involves the upregulation of both C/EBPα and p21 proteins (49). Furthermore, in addition to the transcriptional activation of the C/EBPα promoter, ZTA interacts with C/EBPα directly and stabilizes it from proteasome-mediated degradation. However, for KSHV, it has not yet been demonstrated whether RAP, an evolutionary homologue of EBV ZTA, also interacts directly with C/EBPα. Unlike ZTA, RAP is known not to bind directly to similar DNA target motifs such as ZRE or AP1 sites (C.-J. Chiou et al., unpublished data), nor does it induce the viral lytic cycle or transactivate viral promoters directly (29). Furthermore, despite parallel gene organization, splicing patterns, and overall size and structure, RAP (237 amino acids) and ZTA (245 amino acids) lack any significant amino acid homology (24, 52).
The C/EBPα, C/EBPβ, and CHOP-10 proteins belong to the family of bZIP DNA binding nuclear transcription factors that also includes c-Jun, c-Fos, cyclic AMP-responsive element binding protein, and both KSHV RAP and EBV ZTA (20). The C/EBPα gene encodes a 42-kDa protein that can positively autoregulate its own gene promoter (10, 36, 38), and all three C/EBP family proteins play important roles as determining factors for adipocyte, granulocyte, and neutrophil differentiation (12, 21, 44, 50). C/EBPα controls differentiation and G1 cell cycle arrest through multiple mechanisms including upregulating the expression of the Cdk inhibitor p21CIP, stabilizing the p21 protein through direct interaction (39, 40), inhibiting E2F transcription leading to c-MYC downregulation (33), and directly inhibiting CDK2 and CDK4 (17, 42). The endogenous full-length 42-kDa form of C/EBPα is detectable in KSHV-positive PEL cell lines as well as in Burkitt's lymphoma cell lines, but very little C/EBPβ protein is detectable in the same cells (47). On triggering of the KSHV lytic cycle, KSHV RAP levels are significantly induced by 24 h, and its expression persists throughout the lytic cycle. Increased expression of both C/EBPα and RAP also proved to correlate with strongly heightened expression of p21 between 24 and 72 h in TPA-induced PEL cultures, but only in the subset of cells expressing RAP (47). A novel and previously unrecognized region within the p21 promoter was identified that harbors multiple C/EBP binding sites and responds to C/EBPα-mediated transcriptional activation (49).
Because EBV ZTA interacts with and stabilizes C/EBPα, we wished to evaluate whether RAP could also physically interact with and affect the stability of either the C/EBPα or p21 proteins, as well as enhance C/EBPα-mediated upregulation of the C/EBPα or p21 gene promoters in transient reporter gene assays. Recent reports suggest that PML oncogenic domains (PODs) play an important role in cell cycle regulation and tumor suppression (16). Cell cycle regulators such as Rb and p53 are important for arresting cells at G1, and new data have suggested that only the active forms of Rb (nonphosphorylated version) and p53 accumulate in PODs (16). Furthermore, PML also increases p53 transcriptional activity (15). Earlier studies revealed that KSHV RAP targets to PML nuclear bodies (PODs) and forms nuclear punctate patterns without disrupting them (19, 48). Therefore, we asked whether C/EBPα and p21 also target to PODs or DNA replication compartments in KSHV-infected cells.
Finally, because RAP-positive TPA-induced PEL cells induce host G1 cell cycle arrest (47), questions arose about, whether the converse might also be true, i.e., that cell cycle arrest induced by drugs or occurring naturally in host cells might in turn trigger the KSHV lytic cycle, and whether viral interleukin-6 (vIL-6) expression might interfere with RAP functions in TPA-induced cells. To address some of these questions, we both synchronized KSHV-positive PEL cells in G1 phase and induced p21 in the absence of C/EBPα induction and monitored them for KSHV lytic cycle, as well as examining G1 arrest and p21 induction in vIL-6-positive TPA-induced cells.
MATERIALS AND METHODS
Cells and viruses.
The PEL cell line BCBL1 containing latent KSHV (4) and KSHV-negative DG75 B lymphoblasts were grown in RPMI 1064 plus 10% fetal bovine serum. HeLa, Vero, and Hep3B(−p53) cells (9) were grown in Dulbacco minimal essential medium plus 10% fetal bovine serum. Human primary DMVECwere grown and infected as described previously (11).
Recombinant adenovirus Ad-RAP(1-237) was described previously (47). Human DMVEC were seeded at 60% confluence in two-well slide chambers and infected with Ad-RAP at a multiplicity of infection (MOI) of 0.5 to 2 as described previously (47). Induction of the KSHV lytic cycle in BCBL1 cells with TPA (20 ng/ml) was performed as described previously (11). Synchronization of BCBL1 cells in the G1 cell cycle was carried out by incubating 0.5 × 106 cells with 0.5 mM mimosine for 30 h, and induction of p21 expression was carried out by incubating 0.5 × 106 cells with 200 ng of doxorubicin per ml for 30 h.
Reporter and effector plasmids.
Plasmids encoding Myc-tagged SV2-RAP (pFYW01) (48). CMV-C/EBPα (pSEW-C01) (43), and Flag-SV2-C/EBPα (pHC125c) are mammalian expression vectors for intact KSHV RAP, rat C/EBPα, and human C/EBPα, respectively. The GST-ZTA(1-245) fusion protein encoded by plasmid pDH237 contains the full-length EBV-encoded ZTA cDNA (22). The glutathione S-transferase (GST)-RAP(1-237) fusion protein in plasmid pFYW07 was generated by inserting full-length RAP cDNA flanked by 5′ and 3′ BamHI sites into the pGH416 background. The 714-bp insert was obtained by PCR amplification from pFYW01 as the template with primers LGH3773 (5′-CGCGGATCCAAAT GCCCAGAA TGAAGGAC-3′) and LGH3774 (5′-CGCGGATCCTCAGGGAGAATGTGACT GATC-3′).
Two sets of RAP deletion mutants were constructed by PCR amplification from pFYW01 as the template as follows: RAP(62-237), 525-bp fragment with primers LGH 3769 (5′-CGCGGATCC AACAGCCCCCGGCCATACCG-3′) and LGH3774; RAP(120-237), 351-bp fragment with LGH3770 (5′-CGCGGATCCAACAACAGCTTCCAACTCGC-3′) and LGH3774; RAP(135-237), 306-bp fragment with LGH3771 (5′-CGCGGATCCAATTTGAAGAGGAACGCTTA-3′) and LGH3774; RAP(1-219), 657-bp fragment with LGH3773 and LGH3777 (5′-CGCGGATCCTCACAGCATGTCGCGAAGGAA-3′); RAP(1-186), 558-bp fragment with LGH3773 and LGH3776 (5′-CGCGGATCCTCACTGTCTTGTGTATGCTTT-3′); RAP(1-168), 504-bp fragment with LGH3773 and LGH3775 (5′-CGCGGATCCTCAGGGAGAATGTGACTGATC-3′); and RAP(169-237), 204-bp fragment with LGH3772 (5′-CGCGGATCCAAA CGCGAAAGCAAGGCAGA-3′) and LGH3774. These fragments, all flanked by 5′ and 3′ BamHI sites, were inserted either into the BamHI site of the pGH416 vector to generate GST fusion plasmids or into the BamHI site of the pSG5-Myc vector to generate tagged mammalian expression plasmids. The GST fusion proteins used included wild-type GST-RAP(1-237) in pFYW07 and deletion mutants GST-RAP(62-237) in pFYW15, GST-RAP(120-237) in pFYW16, GST-RAP(135-237) in pFYW17, GST-RAP(1-219) in pFYW21, GST-RAP(1-186) in pFYW20, GST-RAP(1-168) in pFYW19, and GST-RAP(169-237) in pFYW18.
The mammalian effector plasmids used include SV2-Myc-RAP(61-237) in pFYW08, SV2-Myc-RAP(120-237) in pFYW09, SV2-Myc-RAP(1-168) in pFYW12, and SV2-Myc-RAP(169-237) in pFYW11. Fusion proteins generated by enhanced black beetle virus leader-driven in vitro translation included BBV-RAP(1-237) in pCJC514, BBV-p53 and BBV-PML encoding the intact cDNA sequences for human p53 and PML in plasmids pCJC313 and pSV2-PML(560), BBV-C/EBPα (1-358) in pYNC172 (7), and BBV-ZTA(1-245) in pYNC100 (7). Similarly, BBV-p21 in pFYW45 was constructed by inserting a 570-bp human p21 cDNA fragment between 5′ BamHI and 3′ EcoRI sites to replace the ZTA cDNA in the pYNC100 plasmid. The p21 cDNA fragment was PCR amplified from pCMV-p21 (a gift from B. Vogelstcin) with primers LGH4904 (5′-CAGTGGAT CCAATGTCAGAAC CGGCTGGGGA-3′) and LGH4905 (5′-GACTGAATTCTTAGGG CTTCCTCTTGGAGAA-3′). pGEM2-JAC1 is an in vitro translation plasmid for full-length c-JUN protein (26).
C/EBPα-LUC is a luciferase reporter gene driven by the 490-bp C/EBPα promoter in pSEW-CP1 (49). The p21-LUC reporter gene contains 2.4 kb of the WAF1/p21CIP-1 promoter in plasmid pWWP(a gift from B. Vogelstein, Johns Hopkins School of Medicine). Mammalian expression genes for the six KSHV core DNA replication machinery proteins, SV2-Flag-POL in pJX8, SV2-SSB in pJX3, SV2-PAF in pJX5, SV2-HEL in pJX7, SV2-PRI in pJX4, and SV2-PPF in pJX2 were described previously (48).
Immunofluorescence.
Procedures for bromodeoxyuridine (BrdU) labeling and fixation for double-label indirect fluorescent-antibody assay (IFA) and fluorescence microscopy were all performed as described in detail elsewhere (48). For PEL cell IFA, BrdU pulse-labeling (45 min) of BCBL-1 cells was carried out at 48 h after TPA induction. For IFA after Ad-RAP infection, BrdU pulse-labeling of DMVEC cells was performed for 45 min at 30 h postinfection. Sheep anti-BrdU primary antibody (Ab) (20-BS17; Fitzgerald Co., Concord, Mass.) was used to detect BrdU incorporation, and various secondary donkey or goat-derived fluorescein isothiocyanate (FITC)- or rhodamine-conjugated immunoglobulin G (Jackson Pharm., West Grove, Pa.) were used to detect primary Abs. Mounting solution with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Shield) was used to visualize cellular DNA. Primary Abs included rabbit anti-RAP antiserum (N-terminal epitope) (48), rabbit anti-vIL-6-antiserum (30), mouse anti-human p21 monoclonal antibody (MAb) (PharmaMingen), mouse anti-p16 MAb (NA29-100UG; Oncogene Laboratories), mouse anti-p27 MAb (Oncogene Laboratories), rabbit anti-C/EBPα antiserum (36), and goat anti-C/EBPα polyclonal antibody (PAb) (sc-9314; Santa Cruz Biotechnology).
DNA transfection and luciferase assays.
Lipofectamine (Invitrogen) DNA transfection was performed as described in the Invitrogen protocols. Vero, HeLa or Hep3B(−p53) cells were split at 5 × 105 cells/ml and transfected with a total of 2.5 μg of DNA (adjusted with pSG5 vector plasmid as carrier) per well in a six-well plate (0.2 μg of luciferase reporter gene, 1 μg of C/EBPα, and/or 1 μg of RAP or RAP mutants), and the cells were harvested 48 h posttransfection. DG75 cells were transfected using an electroporation method by procedures described elsewhere (49). The assembly of KSHV DNA replication compartments in Vero cells by cotransfection was done exactly as described previously (48). The luciferase assay system (Promega) was used for luciferase assays, and all procedures were carried out as described by the Promega protocol. Luciferase activity was measured for 10 s in a Lumat LB9501 luminometer (Berthold Systems, Inc.) after injection of 100 μl of 1 mM luciferin (Invitrogen).
Coimmunoprecipitation and Western immunoblot analysis.
Induction of the KSHV lytic phase in BCBL1 cells was performed with 500 ml of TPA-induced BCBL1 cells (5 × 105 cells/ml). For coimmunoprecipitation analysis, detailed procedures were described previously (49). Each nuclear extract aliquot (500 μl) was precleared with 5 μl of goat preimmune serum and 100 μl of 50% protein A/G-Sepharose beads for 1 h. After removal of the protein A/G-Sepharose beads from the extract, 3 μg of anti-C/EBPα goat PAb or 3 μg of goat preimmune serum was added to each nuclear extract aliquot and incubated at 4°C with gentle rocking for 2 h. Subsequently, 100 μl of 50% protein A/G-Sepharose beads was added, and the mixture was incubated for 3 h at 4°C. The beads were washed five times at 3-min intervals with cold immunoprecipitation buffer, and resuspended in 30 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. The supernatant was boiled for 3 min before being loaded onto SDS-10% polyacrylamide gels. A 50-μl aliquot from the original nuclear extract (500 μl) was also separated by SDS-PAGE. For Western immunoblot detection of coimmunoprecipitated endogenous RAP, rabbit RAP PAb was used at a 1:1,000 dilution.
GST protein affinity assay.
GST and GST fusion proteins were purified from DNA plasmid-transformed Escherichia coli (strain-BL21) as described elsewhere (22), and detailed procedures for the GST-affinity assay were described previously (43). The supernatant was loaded onto an SDS-10% polyacrylamide gel and separated by electrophoresis. After the gel was dried, the [35S]methionine-labeled proteins were visualized by autoradiography with Kodak film.
Cross-linking assay.
The in vitro-translated or cotranslated 35S-labeled proteins (4 μl each) were mixed with 9 μl of cross-linking buffer (10 mM potassium phosphate buffer [pH 8.0], 10 mM dithiothreitol [DTT]) and 1 μl of freshly diluted 0.1% glutaraldehyde (26). After incubation for 1 h at 20°C, the cross-linking mixture was boiled in 2× loading buffer and fractionated by electrophoresis on an SDS-8% polyacrylamide gel. The gel was fixed and dried, and the 35S isotope was detected by autoradiography after overnight exposure on Kodak X-ray film.
Nondenaturing protein size analysis.
The 4% separating gel for nondenaturing PAGE used 0.7 ml of 30/0.8% acrylamide/bisacrylamide, 1.25 ml of 4× buffer (pH 8.9) (18.2 g of Tris-HCl per 100 ml, pH adjusted with HCl), 3 ml of H2O, 50 μl of 10% ammonium persulfate and 5 μl of N,N,N′,N′-tetramethylethylenediamine (TEMED). The 3.5% stacking gel used 580 μl of 30/0.8% acrylamide/bisacrylamide, 710 μl of 7× buffer (pH 6.7) (5.7 g of Tris-HCl per 100 ml, pH adjusted with H3PO4), 3,710 μl of H2O, 50 μl of 10% ammonium persulfide, and 5 μl of TEMED. The 1× running buffer was diluted from 50× running buffer (pH 8.4) (7.5 g of Tris-HCl and 36 g of glycine per 250 ml). The in vitro-translated 35S-labeled proteins (4 μl each) were mixed with 3× sample buffer (3 ml of 100% glycerol, 0.6 ml of 50× running buffer, 2 mg of bromophenol blue, 6.4 ml of H2O), loaded onto a nondenaturing gel, and separated by electrophoresis for 2 h at 150 to 200 V. The 35S-labeled proteins were detected by autoradiography after the gel was dried
EMSA.
The in vitro-translated protein used in these studies was synthesized with the TNT Quick Coupled transcription-translation system (Promega) by adding either 2.0 μg of the BBV-C/EBPα plasmid DNA (pYNC172) or 2.0 μg of BBV-C/EBPα and 2.0 μg of BBV-RAP (pCJC514) to 40 μl of TNT Quick Master Mix-1 μl of RNase inhibitor-2 μl of unlabeled methionine. The mixture was incubated at 30°C for 90 min and stored subsequently at −80°C. For verification of protein expression, 2 μl of [35S]methionione (Amersham) was added in place of nonradioactive methionine and the reaction mixture was separated by SDS-PAGE and detected by autoradiography.
For electrophoretic mobility shift assay (EMSA), 2 μl of the in vitro-translated protein was used for each reaction and added to a 19-μl reaction mixture containing 1× binding buffer (10 mM HEPES [pH 7.5], 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100, 5% glycerol), and 2 μg of poly(dI)-poly(dC) (Sigma). After the mixture was incubated for 5 min at 20°C, 1 μl of [32P]dCTP-labeled oligonucleotides (50,000 cpm) was added to the mixture and incubated for 1 h. For competition assays, 0, 0.1, 0.2, 0.3, 0.5, 1, or 2 μl of in vitro-translated RAP or c-Jun was added to the binding mixture. For supershift experiments, the reaction mixture was incubated at 20°C for 30 min, and 0.5 μl of anti-C/EBPα PAb was added to the mixture and then incubated for 30 min.
Procedures involved in 32P-end labeling of oligonucleotides and the preparation and autoradiography of 4.5% polyacrylamide gels for EMSA were carried out as described elsewhere (9). The binding efficiency was measured by using an Instant Imager (Packard Instrument Co.) and its accompanying software.
Oligonucleotides were purchased from the Invitrogen Primer Synthesis Facility. LGH4276 (5′-GATCGAGGCGGTGGGCGTTGCGCCGCGGCCTGCTGG-3′) and LGH4277 (5′-GATCCCAGGCAGGCCGCGGCGCAACGCCCACCGCCTC-3′) were annealed to form the C/EBPα-Promoter probe. LGH3859 (5′-GATCCGCTGATTGGTTCCCGCTCTGGGCCAATCAGCA-3′) and LGH3860 (5′-GATCTGCTGATTGGCCCAGAGCGG GAACCAATCAGCG-3′) were annealed to form the Ori-L1 probe; LGH3861 (5′-GATCCTTTGATTGACGGCTGGGCGTCCAATGGAA-3′) and LGH3862 (5′-GATCTTCCATTGGACGCCCCAGCCGTCAATCAAAG-3′) were annealed to form the Ori-L2 probe; LGH3863 (5′-GATCCCGAGATTGGTCGGCCGGATGGGCCAATGGCGA-3′) and LGH3864 (5′-GATCTCGCCATTGGCCATCCGGCCGACCAATCTCGG-3′) were annealed to form the Ori-L3 probe; and LGH3865 (5′-GATCCTTTGATTGACGGGCCGGCGGACCAATGGGA-3′) and LGH3866 (5′ GATCTCCCATTGGTCCGCCGGCCCGTCAATCAAAG-3′) were an-nealed to form the Ori-L4 probe. LGH3878 (5′-GATCCGCTGATTCCTTCCCGCTCT GGGCCAATCAGCA-3′) and LGH3879 (5′-GATCTGCTGATTGGCCCAGAGCGGG AAGGAATCAGCG-3′) were annealed to form the Ori-L1-M1 probe; LGH3880 (5′-GATCCGCTGATTGGTTCCCGCTCTGGGGGAATCAGCA-3′) and LGH3881 (5′-GATCTGCTGATTCCCCCAGAGCGGGAACCAATCAGCA-3′) were annealed to form the Ori-L1-M2 probe; LGH3882 (5′-GATCCGCTGATTGGTTCCCGCTGGGCCAATCAGCA-3′) and LGH3883 (5′-GATCTGCTGATTGGCCCAGCGGAACCAATCAGCG-3′) were annealed to form the Ori-L1-M3 probe; and LGH3884 (5′-GATCCGCTGATTGGTTCCCCGTCT GGGCCAATCAGCA-3′) and LGH3885 (5′-GATCTGCTGATTGGCCCAGACGGGGGAACCA ATCAGCG-3′) were annealed to form the Ori-L1-M4 probe.
Using a KSHV(BCBL-R) genomic plasmid clone from the DSR region as template (pDJA63, 6-kb insert), a 310-bp Ori-L I probe was obtained by PCR amplification with primers LGH2504 (5′-GGGGTAGT CCGCTGGTATTCC-3′) and LGH 2083 (5′-CCCTAGAACTCCCAAGCTG-3′), a 255-bp Ori-L II probe was obtained with primers LGH5238 (5′-CAGCTTGGGAGTTCTAG GG-3′) and LGH2074 (5′-GGACCGTGAGCGACTCGA-3′), a 166-bp Ori-L III probe was obtained with primers LGH5250 (5′-TCGAGTCGCTCACGGTCC-3′) and LGH5239 (5′-TAAATGTGTGGATGGCCCTGC-3′), and an 82-bp Ori-L IV probe was obtained with primers LGH 2073 (5′-GGATTA TGGGGGATTAGC-3′) and LGH 2074. LGH5255 (5′-GATCGGATTATGGGGGATTAGCAAATTCAAGATGG-3′) and LGH5256 (5′-GATCCCATCTTGAATTTGCTAATCCCCCATAATCC-3′) were annealed to form the 35-bp Ori-L V probe; LGH5257 (5′-GATCCGGCGCCCATGAAATGGCCAAAAAT-3′) and LGH5258 (5′-GATCATTTTTGGCCATTTCATGGGCGCCG-3′) were annealed to form the 29-bp Ori-L VI probe; and LGH5252 (5′-GATCTAGGGATAGGGGCCAATGGGAGGCCTC-3′) and LGH5254 (5′-GATCGAGGCCTCCCATTGGCCCCTATCCCTA-3′) were annealed to form the 31-bp Ori-L VII probe.
Chromatin immunoprecipitation assay (ChIP).
KSHV-positive JSC1-PEL cells (5 × 106 cells in 20 ml) were induced by treatment with TPA for 40 h. Harvesting and chromatin extraction were performed by detailed procedures described elsewhere (43). For immunoprecipitation, 1 μg of CHOP-10 mouse MAb (Santa Cruz), C/EBPα rabbit PAb, or RAP rabbit PAb was used with 500 μl of extract. Depletion of endogenous proteins with specific Abs in the extracts was carried out by incubating the appropriate Abs attached to Sepharose beads with the extracts for 24 h at 4°C and subsequently collecting the Ab-cleared supernatant for a new round of PCR assays. For detection of the immunoprecipitated C/EBPα promoter region, two primers, LGH4273 (5′-ACTTCGG TACCGCTACCGACCACGTGGGCG-3′) and LGH4275 (5′-GTGAACTCGAGC ACCTCCGGGTCGCGAATGG-3′), specific for a 280-bp region in the cellular C/EBPα promoter that encompasses the C/EBP site were used for PCR amplification. For detection of nonrelevant cellular DNA sequences, two primers, LGH4268 (5′-ATTCACTCGAGG ATCCCCATGAGCGCGCTGAAGGGG CTG-3′) and LGH4270 (5′-GTGAACT CGAGGTACCTCACGCGCAGTTGCCCAT-3′), specific for the C/EBPα coding region (amino acids 251 to 358) were used as a negative control. For detection of the KSHV Ori-Lyt (R), two primers, LGH3859 and LGH3866, specific for the 200-bp region (positions 119468 to 119665) containing multiple tandemly repeated CTF binding sites were used for PCR amplification. The PCR products were analyzed on a 2.5% agarose gel, and the PCR products were quantitated in the MultiImage Light Cabinet (Alpha-Innotech Corp.) with the accompanying FluorChem (version 1.02) software.
Protein stability assay with proteasome extracts.
For the preparation of cellular proteasome extract, 200 ml of KSHV-latently infected BCBL-1 cells (5 × 105 cells/ml) were used. Detailed procedures for making proteasome extracts and conducting protein stability incubation assays were described previously (49). Samples were harvested at 0, 0.5, 1, 3, 6, or 24 h and boiled for 5 min after addition of 2× protein-loading dye. The samples were loaded onto an SDS-10% polyacrylamide gel and separated by electrophoresis. After the gel was dried, the [35S]-methionine-labeled proteins were visualized by autoradiography with Kodak film. The protein levels were quantitated in the MultiImage Light Cabinet with the accompanying FluorChem (version 1.02) software.
Flow cytometry analysis.
BCBL-1 cells were seeded at 0.5 × 106 cells/ml and treated with mimosine or doxorubicin for 30 h. After being washed with 1× phosphate-buffered saline (PBS) the cells were fixed in 2% paraformaldehyde for 20 min at room temperature. After being washed with 1× PBS, the cells were resuspended in Hoescht solution (10 ng of Hoescht per ml, 0.5% NP-40, 3.5% formaldehyde, 1× PBS) at 4°C. Flow cytometric analyses were performed with a BDLSR-Beckton Dickinson FACScan (10,000 FITC-positive events per sample), and the results were analyzed with Cell Quest software.
RESULTS
KSHV RAP interacts with and stabilizes C/EBPα.
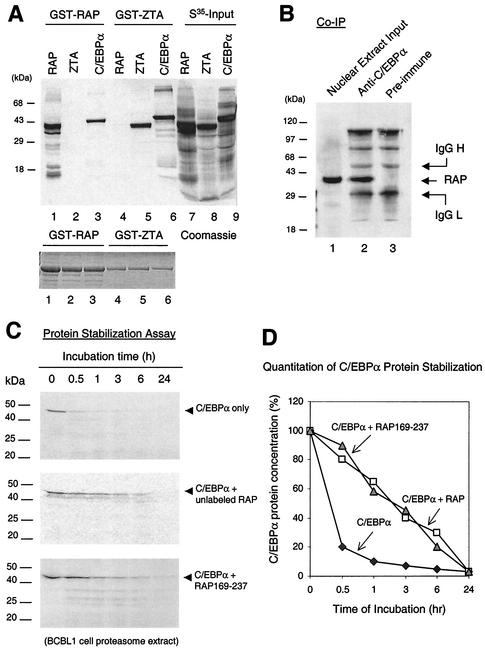
Because RAP expression correlates with elevated C/EBPα expression and because RAP and C/EBPα are both members of the bZIP family that form homodimers and heterodimers, we investigated whether RAP could physically interact with C/EBPα. For initial in vitro experiments, we performed GST affinity assays with GST-RAP and in vitro-translated 35S-labeled C/EBPα, RAP, and ZTA proteins (Fig. 1A). EBV ZTA, which is known to homodimerize efficiently (7) and also interacts strongly with C/EBPα (49), was used as a positive control. Both GST-RAP and GST-ZTA proved to self-interact very strongly with the [35S]RAP (35-kDa) or [35S]ZTA (37-kDa) protein, respectively. In addition, we found that [35S]C/EBPα (42 kDa) was able to interact almost as efficiently with both GST-RAP and GST-ZTA (Fig. 1A, lanes 3 and 5). Importantly, C/EBPα failed to interact with the negative control GST proteins, and we detected no interaction between GST-ZTA and [35S]RAP or between GST-RAP and [35S]ZTA (lanes 2 and 4), demonstrating that the C/EBPα-RAP and C/EBPα-ZTA interactions in the GST-affinity assay were highly specific.
FIG. 1.
KSHV RAP interacts with and stabilizes C/EBPα (A) In vitro GST affinity assay. (Upper panel) Lane 1, [35S]RAP binding to GST-RAP (self-interaction); 2, lack of [35S]ZTA binding to GST-RAP; 3, strong [35S]C/EBPα binding to GST-RAP; 3 and 4, binding of [35S]ZTA but not [35S]RAP to GST-ZTA; 5, strong binding of [35S]C/EBPα to GST-ZTA; 6 to 9, input [35S]-labeled in vitro-translated KSHV RAP, EBV ZTA, and C/EBPα. (Lower panel), Coomassie staining of the GST, GST-RAP, and GST-ZTA fusion proteins used. (B) Coimmunoprecipitation of RAP with C/EBPα from KSHV-positive BCBL-1 cells undergoing the KSHV lytic cycle after TPA induction. Detection was done by Western immunoblotting with anti-RAP PAb. Lanes: 1, positive control of RAP in the input cell lysate (10% of input sample); 2, recovery of RAP by immunoprecipitation using anti-C/EBPα PAb; 3, negative control, immunoprecipitation with preimmune goat serum failed to recover RAP. (C) In vitro protein stability assay using a BCBL-1 proteasome extract. (Top panel) Time course showing that the half-life of [35S]C/EBPα incubated alone was 30 min. (Middle) Parallel time course showing that preincubation of [35S]C/EBPα with unlabeled RAP increased the half-life of C/EBPα from 30 min to 3 h. (Bottom) Parallel time course showing that preincubation of [35S]C/EBPα with mutant RAP(169-237) also increased the half-life of C/EBPα to 3 h. (D) Graph showing quantitatively measured C/EBPα protein levels plotted against proteasome incubation time. IgG, immunoglobulin G.
To confirm that this interaction occurs in vivo, we attempted a coimmunoprecipitation experiment with KSHV-positive BCBL-1 cells. RAP expression in BCBL-1 cells was induced by TPA -treatment for 48 h. A nuclear extract was then prepared from induced BCBL-1 cells, and an immunoprecipitation experiment was performed with anti-C/EBPα goat polyclonal antibody. In both this and the GST affinity experiment above, we also included DNase I incubation and added ethidium bromide to the immunoprecipitation reaction mixture to destroy double-stranded DNA and prevent nonspecific protein-protein tethering via DNA binding. The immunoprecipitate was analyzed by SDS-PAGE and Western blotting with anti-RAP antibody, which detected a 37-kDa band corresponding to coprecipitated RAP protein (Fig. 1B, lane 2). A negative control immunoprecipitation performed with preimmune rabbit serum yielded no RAP signal (lane 3). Therefore, endogenous C/EBPα and RAP evidently interact strongly with each other in BCBL-1 cells.
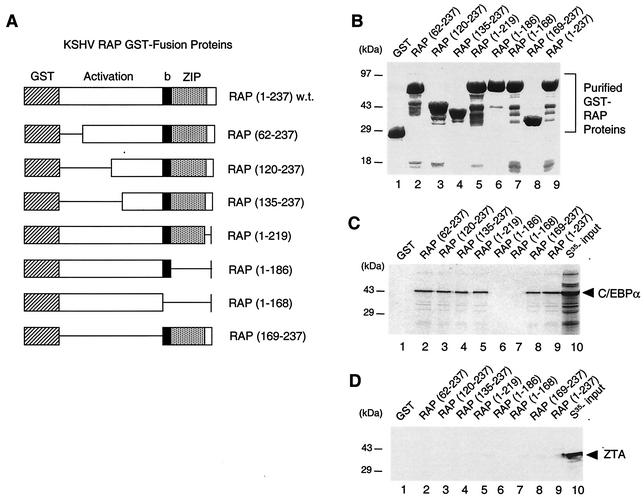
To map which domain of RAP interacts with C/EBPα in vitro, we performed GST affinity assays with [35S]C/EBPα and seven deleted GST-RAP fusion proteins (Fig. 2A and B). The results revealed that C/EBPα interacts specifically with the bZIP domain of RAP (Fig. 2C), because two truncated GST-RAP mutants that lacked the bZIP domain (amino acids 187 to 219) failed to bind to C/EBPα whereas deletion across the N-terminal region from positions 1 to 135 or positions 220 to 237 failed to interfere with binding (Fig. 2C). Negative controls performed with [35S]ZTA showed no interaction with any of the seven GST-RAP fusion proteins (Fig. 2D).
FIG. 2.
The bZIP domain of RAP mediates in vitro interactions with C/EBPα. (A) Diagram of the wild-type GST-RAP fusion and GST-RAP deletion mutants. (B) Relative sizes of the purified GST products from plasmid-transformed E. coli (strain BL21). (C) In vitro GST affinity binding assay showing that in vitro-translated [35S]C/EBPα binds only to GST-RAP fusion proteins that contain the bZIP domain (the leucine zipper maps to positions 187 to 219). (D) Negative control showing that EBV [35S]ZTA fails to bind to any of the GST-RAP fusions.
We then investigated whether RAP may play a role in maintaining the stability of the C/EBPα protein. The half-life of C/EBPα is relatively short (39), and a protein stability assay carried out by incubation with a BCBL-1 proteasome extract showed that the 42-kDa [35S]C/EBPα had an in vitro half-life of less than 30 min, with 90% degradation occuring by 1 h (Fig. 1C and D). However, when we preincubated the [35S]C/EBPα protein with unlabeled in vitro-translated RAP for 30 min before adding proteasome extract, the protein stability assay showed that the half-life of C/EBPα was increased nearly 10-fold, with 70% degradation at 6 h. Importantly, preincubation of C/EBPα with a deleted RAP(169-237) protein, which lacks the N-terminal putative transactivation domain but still interacts with C/EBPα, also stabilized C/EBPα by nearly 10-fold. Therefore, the high-level induction of C/EBPα expression by RAP might be partially attributable to RAP-mediated stabilization of the C/EBPα protein. Furthermore, this suggested that the N-terminal putative transactivation domain of RAP is not needed to stabilize C/EBPα and that it probably plays a transcriptional role in augmenting C/EBPα autoregulation of its own promoter (see below).
The oligomerization state of RAP is different from that of C/EBPα and EBV ZTA.
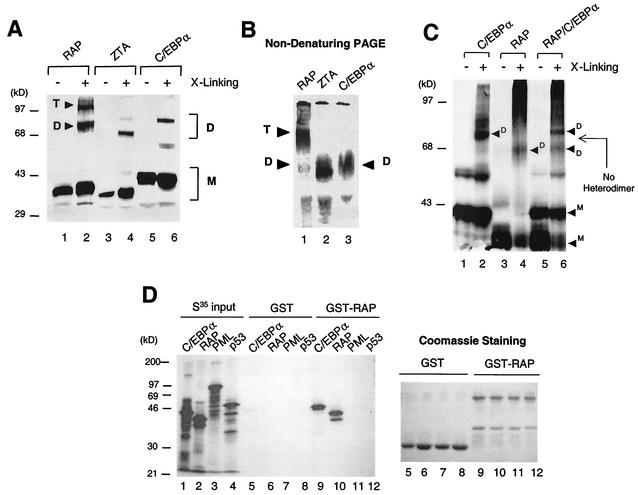
Although RAP is known to self-interact very strongly (24) (Fig. 1A and 3D), the oligomerization state of RAP was not known. Both ZTA and C/EBPα homodimerize through their bZIP domains, whereas c-Jun and c-Fos form the AP1 DNA binding factor only as stable heterodimers (7, 20, 23, 26). In cross-linking experiments involving 0.1% glutaraldehyde and subsequent SDS-PAGE separations, we detected three forms of RAP, namely, a monomer, a dimer, and a higher oligomer (probably tetramers), whereas the positive controls, C/EBPα and EBV ZTA, showed only the two expected forms, namely, monomers and dimers (Fig. 3A, lanes 2, 4, and 6). The same experiment was repeated at high DTT concentration (100 mM) to disrupt nonspecific aggregation of proteins, but RAP still proved to form a higher oligomer (data not shown). To further attempt to confirm this finding, nondenaturing PAGE was carried out to detect native forms of in vitro-translated RAP, C/EBPα, and ZTA. Although all three proteins have similar molecular weights and similar isoelectric pHs, the native gel electrophoresis showed that RAP migrated at a much slower (i.e. higher-molecular-weight) position compared to C/EBPα and ZTA (Fig. 3B), thus providing further evidence that the predominant form of RAP in solution may be a tetramer or higher oligomeric form, not a dimer.
FIG. 3.
The oligomerization state of RAP differs from that of C/EBPα and EBV ZTA. (A) Cross-linking assay showing that in vitro-translated [35S]C/EBPα and [35S]ZTA both form homodimers whereas [35S]RAP forms both homodimers and homotetramers after separation by SDS-PAGE. (B) Nondenaturing gel electrophoresis showing that the native form [35S]RAP migrates more slowly than the native forms of [35S]C/EBPα and [35S]ZTA. (C) Cross-linking of in vitro-cotranslated [35S]C/EBPα and [35S]RAP showing that they fail to form any novel heterodimer bands migrating at an intermediate position, compared to the homodimer bands in control samples of each translated alone. (D) Lack of in vitro interaction between RAP and PML or p53 in a GST affinity assay to compare the binding affinity of in vitro-translated [35S]C/EBPα, [35S]RAP, [35S]PML, and [35S]p53 to GST-RAP. (Left) Autoradiograph after SDS-PAGE showing input 35S-labeled proteins (30% of sample; lanes 1 to 4) and bound bands recovered from either GST beads (lanes 5 to 8) or GST-RAP beads (lanes 9 to 12). (Right) Samples of the stained GST and GST-RAP proteins that were loaded onto the beads in lanes 5 to 12.
Because RAP interacts strongly with C/EBPα, there appeared to be a strong possibility that the two might form RAP-C/EBPα heterodimers. Therefore, in vitro-cotranslated RAP and C/EBPα were cross-linked with 0.1% glutaraldehyde, separated by SDS-PAGE (Fig. 3C, lane 6), and compared in parallel with separately in vitro-translated and cross-linked RAP and C/EBPα samples (lanes 2 and 4). The results showed that after cross-linking, in addition to the higher states of RAP oligomerization typically formed in this type of assay, the RAP-C/EBPα cotranslated sample displayed just two rather than three dimeric forms. The higher-molecular-weight dimeric form exactly matched the migration of C/EBPα homodimers, and the lower-molecular-weight form exactly matched the migration of RAP homodimers, but no additional intermediate-sized forms were found to migrate in between the two homodimeric forms (Fig. 3C). Furthermore, a parallel cross-linking experiment done by mixing the individually translated C/EBPα protein with RAP protein also yielded the same result as the cotranslated and cross-linked sample (data not shown). These results clearly suggest that no heterodimerization occurred in RAP-C/EBPα cotranslated samples and imply that RAP interacts with C/EBPα via a dimer-dimer or higher-oligomer type of interaction.
Because RAP has been reported to interact with p53 (28), we wanted to compare the relative strengths of the interactions of RAP with C/EBPα and p53 in parallel assays. RAP has been observed to colocalize with PML nuclear bodies (48), raising the possibility that RAP might also interact directly with PML. Therefore, we performed GST affinity assays with GST-RAP and in vitro-translated 35S-labeled C/EBPα, RAP, PML, or p53 using ethidium bromide to disrupt any nonspecific DNA-mediated interactions (Fig. 3D). The results showed that although GST-RAP again interacted very strongly with both itself ([35S]RAP) (Fig. 3D, lane 10) and with [35S]C/EBPα (lane 9), it failed to interact with PML (lane 11) and interacted only very weakly with p53 (lane 12), with the level of the latter being at least 50-fold lower than the intensity of the interaction between GST-RAP and C/EBPα. The negative-control GST protein failed to interact with any of these 35S-labeled proteins (lanes 5 to 8), confirming that the binding detected between RAP and C/EBPα is highly specific.
RAP augments autoregulation of the C/EBPα promoter.
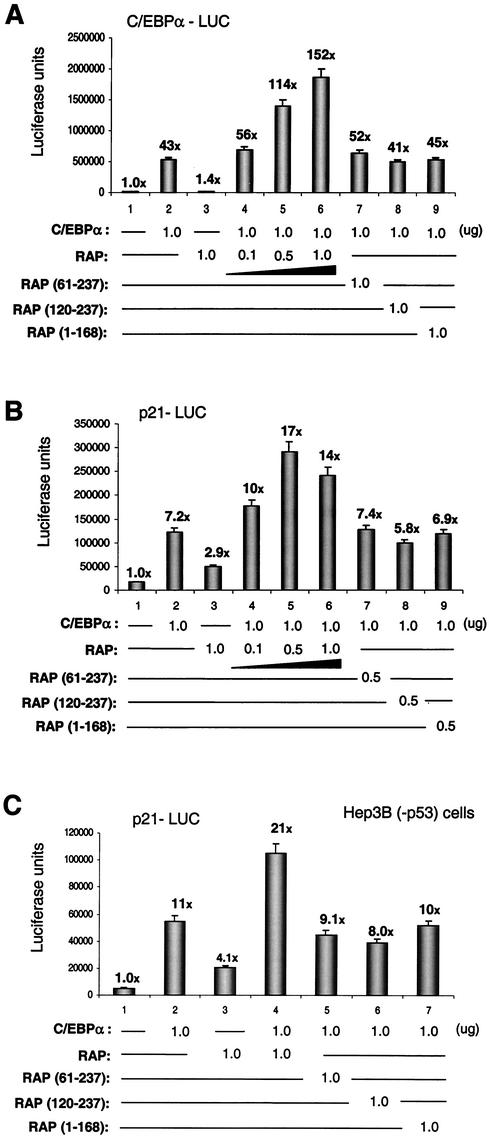
We have shown previously that transfection of SV2-RAP into HeLa cells or JSC1 cells leads to upregulation of both C/EBPα and p21 protein levels (47). Furthermore, Northern blot analysis of BCBL-1 cells revealed that C/EBPα mRNA is increased significantly during early lytic-cycle progression (43a), suggesting that one of the mechanisms leading to C/EBPα upregulation involves transcriptional effects. In addition, C/EBPα is known to positively autoregulate its own promoter (35-38), and we have demonstrated that cotransfected RAP augments C/EBPα upregulation of the KSHV RAP promoter in transient reporter gene assays (43). Therefore, we investigated whether the physical interaction with RAP may also play a role in autoregulation of the 490-bp C/EBPα promoter. The C/EBPα-LUC reporter gene was cotransfected with either the RAP or C/EBPα expression plasmids or both in Vero cells. Cotransfection of the C/EBPα-LUC target DNA (0.5 μg) with a mammalian expression plasmid encoding C/EBPα (1.0 μg) yielded 43-fold upregulation (Fig. 4A). In comparison, cotransfection of C/EBPα-LUC with an SV2-RAP expression plasmid (1.0 μg) did not result in any upregulation of the C/EBPα promoter. However, cotransfection of both the C/EBPα (1.0 μg) plus the wild-type RAP (1.0 μg) expression plasmids produced a 3.5-fold increase up to 152-fold activation of the C/EBPα promoter, and this effect occurred in a dose-responsive manner. Therefore, RAP can contribute to activation of the C/EBPα promoter, but only in the presence of the C/EBPα protein. Duplication of these experiments with both HeLa cells and electroporated DG75 B-lymphoblasts yielded similar results (data not shown). In parallel experiments, three large deletion mutants of the 237-amino-acid RAP, namely, RAP(61-237), RAP(120-237), and RAP(1-168), all failed to enhance C/EBPα autoregulation (Fig. 4A). Stable expression of each of these deleted proteins was confirmed by Western blot analyses (data not shown). These results indicate that transcriptional augmentation requires both N-terminal and C-terminal domains of RAP. Because the N-terminal segment of RAP was not required for stabilization of C/EBPα, we conclude that it probably contributes transcriptional activation functions, similar to those mapping to the N-terminus of ZTA.
FIG. 4.
RAP enhances C/EBPα autoregulation and C/EBPα-mediated p21 promoter upregulation. (A) Transient-expression assays showing the upregulation effects of cotransfected RAP and C/EBPα on a C/EBPα-LUC target reporter gene. Results of luciferase assays in which C/EBPα-LUC plasmid DNA (0.5 μg) was transfected into Vero cells either alone or in the presence of mammalian expression plasmids encoding C/EBPα, RAP, RAP(61-237), RAP(120-237), and RAP (1-168), as indicated, are shown. (B) Similar transient-cotransfection assay showing upregulation effects of RAP and C/EBPα on a p21-LUC target reporter gene. Results of luciferase assays in which p21-LUC plasmid DNA (0.2 μg) was transfected into Vero cells either alone orin the presence of mammalian expression plasmids encoding C/EBPα, RAP, RAP(61-237), RAP(120-237), and RAP(1-168), as indicated, are shown. (C) Similar transient-cotransfection assays with the p21-LUC target reporter gene showing that the upregulation involves a p53-independent pathway. Results of luciferase assays in which the p21-LUC target reporter gene was transfected into Hep3B (−p53) cells with the same effector gene combinations as in panel B are shown.
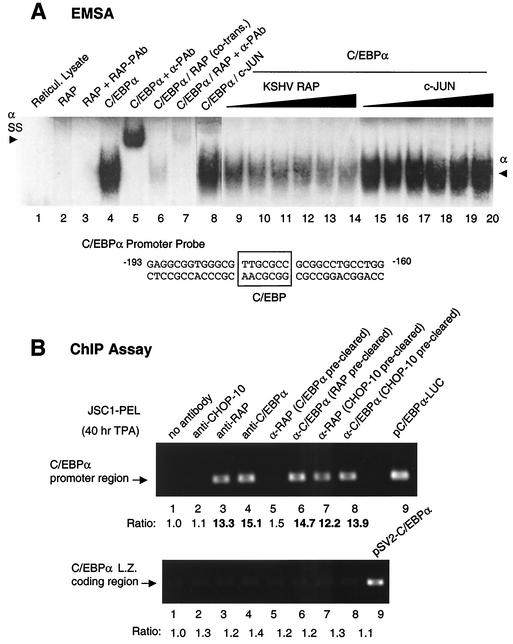
RAP affects the DNA binding activity of C/EBPα.
To examine whether interaction with RAP also affects C/EBPα binding to its known binding site at positions −173 to −179 in the C/EBPα promoter, we carried out EMSA analyses. First, in vitro-translated C/EBPα was confirmed to bind to a 37-bp oligonucleotide probe containing the C/EBPα autoregulation site, which was appropriately supershifted by addition of anti-C/EBPα PAb (Fig. 5A, lanes 4 and 5). In contrast, RAP did not bind to the C/EBPα probe by itself, and consequently no supershifted band was observed with the addition of RAP rabbit PAb (lanes 2 and 3). However, the binding of in vitro-translated C/EBPα to the probe was diminished in a dose-responsive fashion when increasing amounts of in vitro-translated RAP were added to the binding mixture (lanes 9 to 14). Similarly, cotranslation of C/EBPα and RAP greatly diminished C/EBPα binding to its own promoter site but without producing a supershifted band (lanes 6 and 7). As a negative control, cotranslation and addition of increasing amounts of in vitro-translated c-Jun protein with C/EBPα failed to affect the DNA binding activity of C/EBPα (lanes 8 and 15 to 20).
FIG. 5.
Evidence that C/EBPα mediates the binding of RAP to C/EBPα promoter DNA. (A) EMSA for binding to a 32P-labeled oligonucleotide probe containing the proximal upstream C/EBP binding site from the C/EBPα promoter: Lanes: 1, negative control with unprogrammed in vitro translation lysate; 2 and 3, in vitro-translated RAP does not bind to the C/EBP probe and cannot be supershifted by the addition of anti-RAP PAb; 4 and 5, positive control showing in vitro-translated C/EBPα protein binding to the C/EBP probe and being supershifted by the addition of anti-C/EBPα PAb; 6 and 7, cotranslation of RAP with C/EBPα interferes with C/EBPα binding but does not form a supershifted band; 8, cotranslation of c-Jun with C/EBPα does not interfere with C/EBPα binding; 9 to 14, addition of in vitro-translated RAP protein to the C/EBPα binding reaction mixture interferes with C/EBPα binding to the probe in a dose-responsive manner; 15 to 20, addition of increasing amounts of in vitro-translated c-Jun protein to the C/EBPα binding reaction mixture does not interfere with C/EBPα binding to the probe. The sequence of the C/EBP oligonucleotide probe from positions −193 to −160 in the C/EBPα promoter (the C/EBP site maps at position −180/−174) is shown below the EMSA results. (B) Endogenous ChIP assay showing a specific association of both the RAP and C/EBPα proteins with the C/EBPα promoter. A JSC1 cell extract prepared after a 40-h TPA treatment was used for cross-linking and immunoprecipitation. The results compare PCR DNA products obtained from the C/EBPα promoter to those obtained from the C/EBPα coding region. (Upper panel) Lanes: 1 to 4, ChIP assay results showing that C/EBPα promoter DNA was recovered from immunoprecipitates obtained with either the C/EBPα or RAP antibodies, but not from immunoprecipitates with the CHOP-10 antibody or in the absence of antibody; 5, removal of C/EBPα from the lysate (preclearing with C/EBPα antibody) abolished the recovery of C/EBPα promoter DNA from the RAP immunoprecipitate; 6, recovery of C/EBPα promoter DNA was not affected by preclearing with RAP antibody; 7 and 8, preclearing with CHOP-10 Ab failed to abolish the recovery of C/EBPα promoter DNA from the RAP or C/EBPα immunoprecipitates; 9, control PCR amplification product from the C/EBPα promoter plasmid (pC/EBPα-LUC) using the same primers (LGH4273 and LGH4273) to indicate the expected correct size of the PCR products. Measured values for band intensities are given below the lane numbers. (Lower panel) ChIP assay results showing that adjacent cellular DNA fragments did not coprecipitate nonspecifically with these antibodies. Lanes: 1 to 8, primers (LGH4268 and LGH4270) specific for the detection of C/EBPα coding region DNA failed to detect any positive signals above the basal level; 9, size control PCR DNA product from the C/EBPα expression vector.
Although this result provided further evidence for a strong physical interaction between C/EBPα and RAP, our implication that a piggyback interaction occurs with DNA-bound C/EBPα could not be proven directly because of the failure to detect a supershifted EMSA band in vitro. The observation of a dose-responsive inhibitory effect of RAP on C/EBPα binding without detection of a corresponding supershift band could have several possible explanations. First, RAP might inhibit C/EBPα binding to the recognition site, perhaps through formation of heterodimers. However, the cross-linking experiment using glutaraldehyde described above with in vitro-cotranslated C/EBPα and RAP revealed no evidence that heterodimers were formed in solution but, rather, suggested that the individual homodimeric interactions (and possible homotetrameric interaction for RAP) were very strong (Fig. 3C). Alternatively, binding may not be inhibited, but the RAP-C/EBPα protein-DNA complex is too large or not stably maintained in the in vitro EMSA system, or another unknown protein is required to stabilize the DNA-bound complex. Because RAP enhanced rather than inhibited C/EBPα autoregulation, it appears likely that DNA-bound RAP-C/EBPα protein-DNA complexes do form but that they are not stably maintained in our current in vitro EMSA system. In addition, we have shown elsewhere that both KSHV RTA and EBV ZTA also interact with C/EBPα and stimulate C/EBPα transcriptional activation of the RAP promoter; however, again, when either RTA or ZTA was added to the DNA-bound C/EBPα, only a dose-dependent loss of the C/EBPα-shifted band was observed by EMSA without detectable formation of any supershifted bands (43, 49). Therefore, for reasons that are not presently understood, stable binding of several different viral IE proteins piggybacked onto DNA-bound C/EBPα do not form supershifted bands.
RAP associates with the C/EBPα promoter in ChIP assays but only in the presence of C/EBPα.
As an alternative approach to confirm our model that piggyback C/EBPα-RAP complexes do bind to the C/EBPa promoter in vivo, we performed an endogenous ChIP assay with TPA-induced JSC1 PEL cells. First, the C/EBPα and RAP proteins were immunoprecipitated from the DNA-protein cross-linked cell lysates, and after removal of all proteins, the recovered DNA was ethanol precipitated and amplified by PCR using primers specific for the C/EBPα promoter (LGH4273 and LGH4275). C/EBPα promoter DNA was indeed found to be associated with each of the C/EBPα and RAP immunoprecipitates (Fig. 5B, lanes 3 and 4), whereas a negative control without antibody failed to precipitate any C/EBPα promoter DNA (lane 1).
CHOP-10 is related to C/EBPα but lacks a functional DNA-binding domain and acts as a negative repressor of C/EBPα through inhibition of C/EBPα DNA binding by direct heterodimer formation (34, 37, 46). As a result, CHOP-10 can repress C/EBPα transcriptional activation of downstream target promoters including the C/EBPα promoter. Therefore, as a control, we also attempted to immunoprecipitate CHOP-10-bound DNA from the DNA-protein cross-linked PEL cell lysate. However, we could not detect any immunoprecipitated C/EBPα promoter DNA in the CHOP-10 immunoprecipitate (Fig. 5B, lane 2). This control experiment suggested that unlike the interaction of RAP with C/EBPα, which apparently does form a DNA-bound complex in vivo, any CHOP-10 heterodimers formed with C/EBPα also lacked the ability to bind to DNA in vivo, resulting in the absence of any immunoprecipitated C/EBPα promoter DNA.
To address whether RAP and C/EBPα may bind to the C/EBPα promoter DNA cooperatively, we precleared the C/EBPα protein from the lysate with anti-C/EBPα PAb overnight and then reimmunoprecipitated RAP with RAP antiserum in a second round. The results showed that C/EBPα promoter DNA could no longer be recovered from the RAP immunoprecipitate in the second round (Fig. 5B, lane 5), suggesting that in the absence of C/EBPα protein, almost no RAP was bound directly to the C/EBPα promoter DNA. A parallel control second immunoprecipitate with C/EBPα confirmed that over 95% of the C/EBPα-bound DNA was removed (data not shown).
In contrast, removal of RAP by Ab preclearing did not significantly affect the level of association of the C/EBPα protein with C/EBPα promoter DNA (Fig. 5B, lane 6), suggesting that although almost all of the RAP present was complexed to C/EBPα, there must be at least an equal amount of excess DNA-bound C/EBPα present that was not in a complex with RAP. Again, preclearing with anti-CHOP-10 antibody did not significantly affect the association of either C/EBPα or RAP to the C/EBPα promoter DNA (lanes 7 and 8).
Importantly, a negative-control PCR assay using a pair of primers that specifically detected only coding-region DNA sequences from within the C/EBPα gene (LGH4268 and LGH4270) failed to yield any positive signals beyond the background level (Fig. 5B, lower panel), confirming that our ChIP assay was specific and that only recovered promoter region DNA sequences were bound by these proteins. Therefore, the ChIP assay data strongly support a model in which the association of RAP with the C/EBPα promoter requires a piggyback interaction with promoter-bound C/EBPα.
RAP enhances C/EBPα-mediated upregulation of the p21 promoter cooperatively by a p53-independent pathway.
Northern blot analysis of total RNA harvested from BCBL1 cells has shown that the mRNA level of p21 increases significantly after TPA induction of the KSHV lytic cycle (data not shown), suggesting that at least part of the mechanisms for p21 upregulation in lytically induced PEL cells is also transcriptional. C/EBPα itself is known to both stabilize the levels of the p21 protein and to upregulate the p21 promoter at the transcriptional level (39, 40). Furthermore, we showed previously that the latter effect requires the presence of multiple upstream C/EBP binding sites and that EBV ZTA is able to enhance the C/EBPα-mediated transactivation of the p21 promoter through these sites (49). Therefore, we tested whether KSHV RAP was also able to augment the transcriptional activation of the 2.4-kb human p21 promoter mediated by C/EBPα in luciferase reporter gene assays.
Transfection of the p21-LUC target reporter gene with the C/EBPα expression plasmid alone in Vero cells gave sevenfold upregulation, whereas addition of the RAP expression plasmid alone gave threefold direct activation (Fig. 4B). However, cotransfection with both the C/EBPα and RAP effector plasmids together resulted in up to 17-fold activation of p21-LUC. This observation implies that RAP(1-237) activates the p21 promoter cooperatively with C/EBPα. Again, none of the deletion mutants RAP(Δ1-61), RAP(Δ1-119), and RAP(Δ169-237) enhanced C/EBPα-mediated upregulation, suggesting that the effect is specific only for full-length RAP. Duplication of the same experiments in HeLa and DG75 cells yielded similar results (data not shown). Therefore, induction of p21 expression by RAP is likely to include a transcriptional component through enhancing C/EBPα-mediated p21 promoter upregulation, similar to its effects on the C/EBPα promoter.
Induction of p21CIP-1 has traditionally been linked to transcriptional activation by the p53 protein (13), and although there are no known interactions between the C/EBPα and p53 proteins, KSHV RAP has been reported to interact with the p53 protein (28). Therefore, despite our evidence that RAP interaction with C/EBPα is over 50-fold stronger than that with p53, we needed to verify whether RAP enhancement of C/EBPα-mediated p21 promoter upregulation could occur in a p53-independent pathway. When the p21-LUC reporter gene assay was repeated with Hep3B(−p53) cells, which contain double deletions of the p53 gene (9), C/EBPα still upregulated the p21 promoter 11-fold (Fig. 4C). Furthermore, RAP was still able to enhance the p21 promoter up to 21-fold in cooperation with C/EBPα, suggesting that p53 does not play a role in this pathway.
RAP and p21CIP-1 colocalize in PODs and interact in vitro.
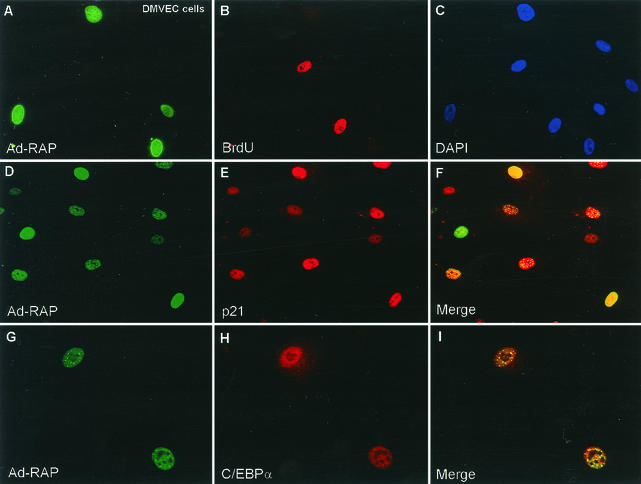
S-phase progression in KSHV-infected PEL cells is inhibited during the viral lytic cycle, and a defective adenovirus vector expressing RAP (Ad-RAP) can induce both the C/EBPα and p21 proteins as well as G1 cell cycle arrest in infected human diploid fibroblasts (HF cells) (47). Because KS involves endothelial cells, we considered that it was important to confirm that this process also occurs in primary human DMVEC, which produce KS-like spindle cells when infected by KSHV (11). Indeed, after Ad-RAP infection of DMVEC cells, all cells that were positive for RAP failed to progress into S-phase as revealed by double-label IFA using the BrdU-incorporation assay (Fig. 6A to C). Furthermore, Ad-RAP infection also induced both C/EBPα and p21 expression in RAP-positive DMVEC (Fig. 6D to I).
FIG. 6.
Ad-RAP also induces C/EBPα and p21 protein expression in human DMVEC. (A and B) G1 arrest as judged by double-label IFA showing the absence of BrdU incorporation (rhodamine, red) in RAP-positive cells (FITC, green) but not in RAP-negative cells of a DMVEC culture after infection with Ad-RAP(1-237) for 40 h at an MOI of 1.0. (C) DAPI nuclear staining of all DMVEC cells in the same field as panels A and B. (D and E) Double-label IFA showing the expression of RAP (green) and induced p21 (red) in the same subpopulation of cells infected with Ad-RAP. (F) Merge of panels D and E, showing that p21 is induced only in RAP-positive cells. (G and H) Double-label IFA illustrating the expression of RAP (green) and upregulation of C/EBPα (red) in the same subpopulation of cells after Ad-RAP infection. (I) Merge of panels G and H illustrating coexpression of C/EBPα only in RAP-positive cells.
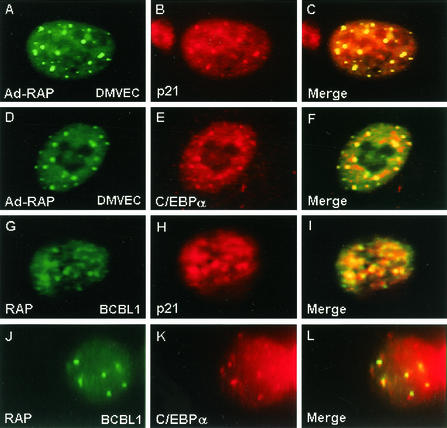
During these experiments, we observed that some p21 and C/EBPα perfectly colocalized with RAP within 10 to 20 small punctate nuclear domains in both DMVEC and lytically-induced PEL cells (Fig. 7). We already knew that RAP itself targets to PODs and forms punctate nuclear patterns in both DNA-transfected Vero cells and KSHV-infected PEL cells and DMVEC (19, 48). Because RAP interacts with C/EBPα, our IFA results suggested that RAP might recruit C/EBPα into the PODs. Similarly, because p21 is also known to interact with C/EBPα, it might have accumulated within the PODs through its association with C/EBPα. However, the colocalization of p21 protein with RAP and PML was highly efficient (over 90% of the nuclear punctate patterns formed by the two proteins colocalized perfectly), leading us to suspect that a direct physical interaction might also occur between p21 and RAP.
FIG. 7.
The C/EBPα and p21 proteins colocalize with RAP in punctate PML nuclear bodies (PODs). High-magnification immunofluorescence microscopy in Ad-RAP and KSHV-infected cells. (A and B) Ad-RAP-infected DMVEC culture. A RAP-positive cell (FITC, green) shows that the punctate POD-associated domains also colocalize with the induced p21 punctate pattern (rhodamine, red). (C) Merging of the two frames in panels A and B. (D and E) Ad-RAP-infected DMVEC culture. A RAP-positive cell (FITC, green) shows that the punctate domains also colocalize with the induced C/EBPα punctate pattern (rhodamine, red). (F) Merging of the two frames in panels D and E. (G and H) A KSHV-infected TPA-treated BCBL-1 cell shows that the induced p21 (red) forms a nuclear punctate pattern that colocalizes with the RAP punctate domains (green), which are known to colocalize with cellular PML in PODs. (I) Merging of the two frames in panels G and H. (J and K) A KSHV-infected TPA-treated BCBL1 cell shows that induced C/EBPα (red) forms punctate patterns that perfectly colocalize with RAP punctate structures (green). (L) Merging of the two frames in panels J and K.
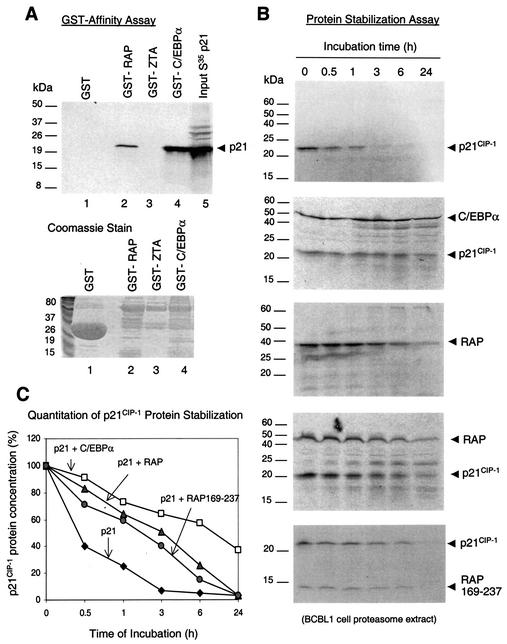
Therefore, we performed GST affinity assays with GST-RAP, GST-ZTA, and GST-C/EBPα to examine binding to the 35S-labeled p21 protein. The results revealed that p21 does interact with GST-RAP in vitro (Fig. 8A, lane 2), although the efficiency of binding was two to threefold lower than with C/EBPα (lane 4). In contrast, p21 failed to interact with the GST control protein (lane 1), and we detected no interaction between GST-ZTA and p21 (lane 3), demonstrating that the RAP-p21 interaction in the GST affinity assay is highly specific. Although EBV ZTA is evolutionarily related to RAP and induces C/EBPα and p21 protein expression (49), it disrupts PODs and does not form nuclear punctate structures (1, 3). Therefore, unlike ZTA, RAP proved to bind strongly to p21 as well as to C/EBPα in vitro, which correlates with the IFA evidence for colocalization between the three proteins in coexpressing cells.
FIG. 8.
RAP interacts with and stabilizes the p21 protein. (A) In vitro GST affinity binding assay. (Upper panel) Lanes: 1, [35S]p21 does not interact with purified GST protein; 2, [35S]p21 binds to GST-RAP; 3, [35S]p21 does not bind to GST-ZTA; 4, positive control, [35S]p21 interacts strongly with GST-C/EBPα; 5, input [35S]p21 protein. (Lower panel) Coomassie staining of the GST, GST-RAP, GST-ZTA, and GST-C/EBPα fusion proteins used. (B) In vitro protein stability assay with a BCBL-1 proteasome extract. (Top panel) PAGE analysis of the results of time course incubation shows that the half-life of [35S]p21 protein is less than 30 min. (Second panel from top) positive control shows that cotranslation with C/EBPα stabilized p21 in the same proteasome extract. (Third panel from top) The half-life of RAP is between 3 and 6 h. (Fourth panel from top) Cotranslation of p21 with RAP increased the half-life of p21 from 30 min to 3 h, whereas the half-life of RAP was unaffected by p21. (Bottom panel) Cotranslation of p21 with mutant RAP(169-237) increased the half-life of p21 from 30 min to almost 3 h. (C) Graph plotting quantitatively measured levels of p21 protein against proteasome incubation time.
RAP stabilizes the p21CIP-1 protein.
The Cdk inhibitor p21CIP-1 is an unstable protein with a short half-life, and the degradation of p21 is proteasome dependent, as shown by pharmacological inhibition of proteasome functions, which leads to accumulation of p21 (32). Because both RAP and ZTA proved to stabilize C/EBPα, we investigated whether RAP may also stabilize p21. Protein stability assays performed by incubation of 35S-labeled p21 protein with a BCBL-1 proteasome extract (Fig. 8B, top panel) showed that the half-life of p21 protein in the absence of RAP was less than 30 min, with 90% degradation occuring within 3 h of incubation in the proteasome extract (Fig. 8C). As a positive control, because C/EBPα is known to stabilize p21 and vice versa (39), we examined the stability of in vitro-translated p21 in the presence and absence of cotranslated C/EBPα. Indeed, cotranslation of p21 and C/EBPα increased the half-lives of both proteins from less than 30 min to well over 6 h (Fig. 8B, second panel from top). A similar experiment with the cotranslated RAP and p21 proteins revealed that the p21 half-life was also increased by RAP from less than 30 min to 3 h in the proteasome extract incubation assay (Fig. 8B, fourth panel from top). Cotranslation of p21 with the deleted RAP(169-237), which lacks the N-terminal putative transactivation domain, also prolonged and stabilized the p21 half-life to almost 3 h (Fig. 8B, bottom panel), suggesting that the N terminus of RAP is not needed for p21 stabilization. A parallel negative control experiment showed that RAP, which does not interact with CHOP-10 in vitro, also did not stabilize CHOP-10 protein (half-life of 30 min) in this assay (data not shown). Furthermore, RAP itself had a half-life of 3 to 6 h (Fig. 8B, third panel from top), which was unaffected by the presence of p21 (Fig. 8B, fourth panel from top) or by either C/EBPα or CHOP-10 (data not shown).
Cytoplasmic vIL-6 expression in TPA-treated PEL cells does not block RAP functions.
We have previously reported that only 1% of BCBL-1 cells express the p21 protein as detected by IHC prior to lytic induction, whereas after TPA induction, up to 18% of the cells expressed nuclear p21 and a majority of the p21-positive cells colocalized with RAP-positive cells (47). These cells could not progress into S-phase, as shown by the BrdU incorporation assay (47). Significantly, p16 and p27, two other Cdk inhibitor proteins associated with G1 cell cycle arrest, were absent as assayed both by Western blotting and IHC or IFA in TPA-induced BCBL-1 cells, including cells that were positive for RAP and C/EBPα. Therefore, possibly unlike EBV ZTA, which reportedly induces both p21 and p27 protein expression (5), the KSHV RAP protein may induce G1 cell cycle arrest only through the C/EBPα-plus-p21 pathway (47). However, KSHV lytically infected cells also express vIL-6, and a recent study by Chatterjee et al. (8) has suggested that alpha interferon induction and secretion of vIL-6 in PEL cells (which does not induce the lytic cycle) might lead to C/EBPβ-dependent downregulation of p21 and a subsequent increase in S-phase and cell proliferation in the culture.
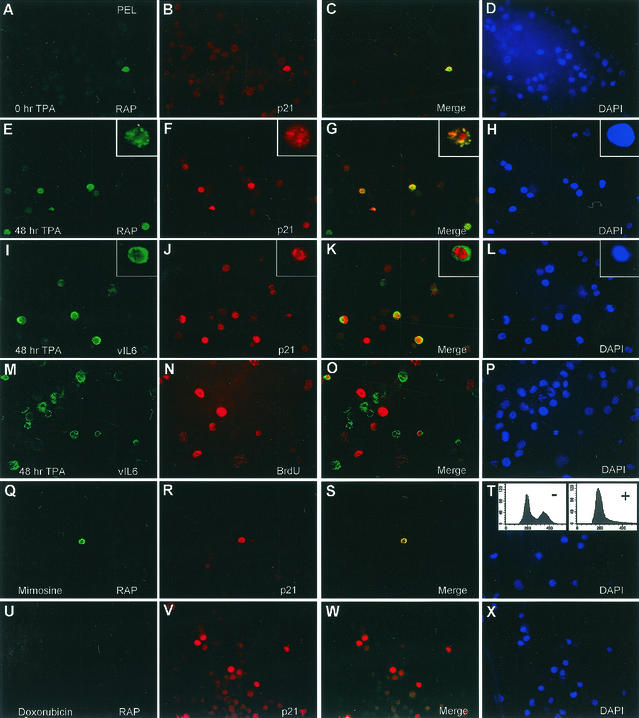
Because of these potentially contradictory effects, we wanted to address the p21 status of TPA-induced cells that express vIL-6. Therefore, we repeated the previous PEL double-label IFA experiment but also examined vIL-6-positive cells in comparison to RAP-positive cells (Fig. 9A to P). As before, both 1% of spontaneously RAP-positive uninduced cells (Fig. 9A to D) and the majority of the 15 to 20% of TPA-induced RAP-positive cells (Fig. 9E to H) coexpressed p21. Furthermore, vIL-6 expression is induced by TPA from 1% (spontaneously positive cells during latency) before treatment (data not shown) to 20% after TPA induction, and the majority of the TPA-induced cytoplasmic vIL-6-positive cells also coexpressed nuclear p21 (Fig. 9I to L). Virtually all vIL-6-positive cells were arrested in G1 as judged by the lack of BrdU incorporation (Fig. 9M to P). Therefore, if this proposed p21 shutoff pathway is induced by vIL-6 in TPA-treated, lytically infected PEL cells, it is evidently not sufficient to counteract RAP-induced p21 upregulation and cell cycle arrest in the same or adjacent cells that are either RAP or vIL-6 positive.
FIG. 9.
vIL-6 expression does not block RAP functions, and neither G1 cell cycle arrest nor p21 induction alone triggers the KSHV lytic cycle. (A to P) Double-label IFA experiments showing that TPA induction of vIL-6 during the lytic cycle in PEL cells does not interfere with RAP-mediated p21 induction and cell cycle arrest. (A to D) Uninduced BCBL-1 cells. Colocalized expression of both nuclear RAP (green) and p21 (red) proteins occurs in only a few spontaneously lytic cells. (E to L) TPA-induced BCBL-1 cells. Expression of p21 (red) occurs in a majority of the RAP-positive and vIL-6-positive cells (cytoplasmic, green). (M to P) TPA-induced BCBL-1 cells. S-phase BCBL-1 cells incorporating BrdU (red) do not colocalize with vIL-6-positive cells undergoing the lytic cycle. (Q to T) Uninduced BCBL-1 cells. Synchronization in G1 with mimosine does not trigger KSHV RAP expression. (Q) Detection of very few RAP-positive cells (green) in 30-h mimosine-treated BCBL1 cells. The inset FACS analysis shows the cell cycle profile of the same BCBL-1 cells before (−) and after (+) mimosine treatment. (R) Detection of p21-positive cells in the same field as in panel Q. (S) Merge of the two previous frames. (T) DAPI staining of the same field showing all cell nuclei. (U to X) Uninduced BCBL-1 cells. Doxorubicin-induced p21 expression does not trigger KSHV RAP expression in the same cells. (U) Detection of RAP-positive cells (green) after a 30-h doxorubicin treatment. (V) Detection of p21-positive cells in the same field. (W) Merge of the two previous frames. (X) DAPI stain showing all cell nuclei in the same field.
Synchronization of PEL cells in G1 phase and induction of p21 do not trigger KSHV lytic cycle.
Although the KSHV lytic cycle is associated with host cell cycle arrest and the isolated KSHV RAP can induce cell cycle arrest on its own even outside of the PEL cell environment (47), the converse might also be true, namely, that host cell cycle arrest might trigger the KSHV lytic cycle. To begin examining this question, we synchronized BCBL-1 cells in G1 phase with mimosine to create a host cell cycle arrest environment and then monitored for levels of KSHV RAP expression as a lytic-cycle marker. In addition, we used doxorubicin because it is known to cause cellular DNA damage and to trigger stress-related p53-dependent induction of the p21 protein (25). In contrast to doxorubicin, mimosine induces G1 arrest by a p21-independent mechanism (41) (Fig. 9R). For the mimosine treatment, the majority of the BCBL-1 cells in the culture were confirmed to be arrested in G1 phase through fluorescence-activated cell sorter analysis (Fig. 9Q, inset), and for the doxorubicin treatment, the BCBL1 cells were examined for p21 induction by IFA (Fig. 9V).
Interestingly, after treating the BCBL-1 cells with both drugs independently for 30 h, we found that neither cell cycle arrest nor p21 induction had any effect on triggering KSHV RAP expression. In untreated BCBL-1 cells, 2.1% of the cells were positive for RAP and p21 by IFA (Fig. 9A to D), which represents the typical basal level of spontaneous lytic cycle induction. In cells treated with mimosine, 2.3% of the cells were positive for RAP and 1.7% of the cells were positive for p21 (Fig. 9Q to T); however, in cells treated with doxorubicin, only 0.8% of the cells were found to be positive for RAP (Fig. 9U), even though 27% of the same cell population was now expressing the p21 protein (Fig. 9V). Neither treatment led to induction of C/EBPα expression (data not shown). Therefore, in contrast to TPA-triggered BCBL-1 cells, which showed lytic induction of RAP (as well as C/EBPα and p21 expression) in up to 20% of the cells at either 24 or 48 h (Fig. 9E and data not shown), neither G1 arrest nor induction of the p21 protein alone for 30 h was able to trigger RAP expression. These data strongly suggest that KSHV lytic-cycle induction precedes the onset of host cell cycle arrest and that the subsequent G1 arrest observed results directly from the expression of KSHV lytic cycle-associated proteins. However, questions about whether events that induce C/EBPα-mediated cell cycle arrest or whether the expression of exogenous p21 transfected into latent PEL cells can lead to KSHV lytic cycle induction still remain to be resolved.
Identification of a 200-bp region of KSHV Ori-Lyt that contains C/EBP binding sites and indirectly associated with KSHV RAP.
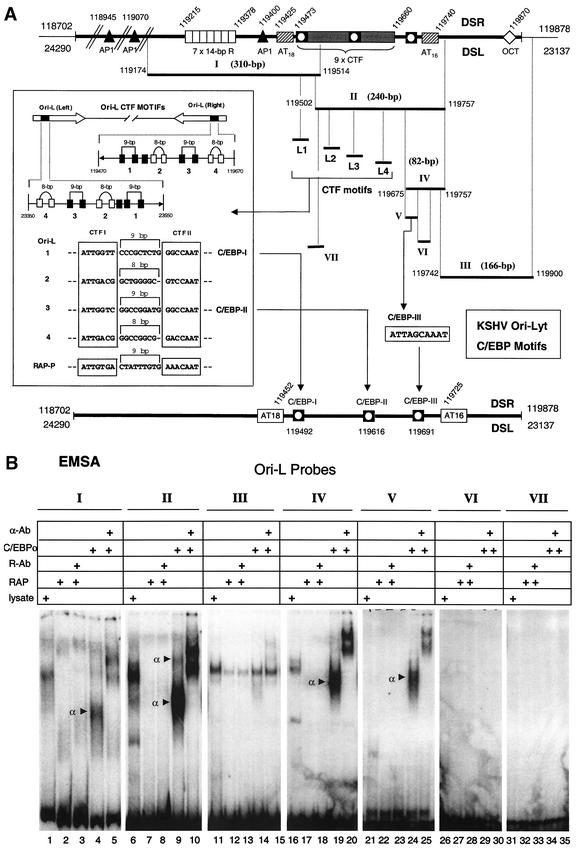
Two duplicate copies of a 1,000-bp region of KSHV DNA predicted to be the KSHV lytic origin of DNA replication [and referred to as DSL or Ori-Lyt (L) and DSR or Ori-Lyt (R)] were first described by Nicholas et al. (27) and subsequently confirmed functionally by both AuCoin et al. (2) and Lin et al. (24a). Lin et al. also found that the KSHV RAP protein was associated with a 500-bp essential core segment of the KSHV Ori-Lyt region (DSL), between nucleotide 23128 and 23635, using both antibody selection and ChIP assays. Because RAP is efficiently incorporated into the KSHV viral DNA replication compartments (48), this finding suggested that RAP may be the origin binding protein, similar to EBV ZTA.
To examine whether the RAP or C/EBPα protein binds to KSHV Ori-Lyt, we obtained three large DNA fragments encompassing the entire Ori-Lyt (R) region, namely, Ori-L I (310 bp), Ori-L II (255 bp), and Ori-L III (166 bp) (Fig. 10A) and conducted EMSA with them by using in vitro-translated RAP or C/EBPα proteins. The Ori-L I probe proved to contain at least one C/EBP site (one C/EBPα shift), and the Ori-L II probe contained at least two C/EBP sites (two C/EBPα shifts), whereas the Ori-L III probe contained no C/EBP sites (Fig. 10B, lanes 4, 5, 9, 10, 14, and 15). In contrast, RAP did not bind directly to any of the three DNA fragments (lanes 2, 3, 7, 8, 12, and 13). Therefore, this preliminary screening revealed the presence of at least three strong C/EBP binding sites within the KSHV Ori-Lyt.
FIG. 10.
Identification of a 200-bp region within KSHV Ori-Lyt that associates with the C/EBPα protein. (A) Schematic diagram showing the map location of all DNA probes used from the duplicated KSHV Ori-Lyt domain as well as the subregion containing multiple CTF motifs used for EMSA experiments and the identified C/EBP binding sites relative to the two AT-rich palindromes (bottom line). (B) Results of EMSA experiments testing the abilities of seven DNA probe fragments from across the Ori-Lyt domain to bind RAP or C/EBPα proteins. (C) Results of EMSA analysis to detect binding of C/EBPα or RAP to wild-type or mutated 32P-labeled oligonucleotide probes encompassing the four putative paired CTF motifs.
One of several striking structural features about the Ori-Lyt region pointed out by Nicholas et al. (27) is a 200-bp segment between nucleotide 119470 and 119670 in Ori-Lyt (R) and exactly duplicated in the inverted direction between nucleotide 23350 and 23550 in Ori-Lyt (L), which harbors nine consensus CTF motifs, some of which resemble unusually spaced palindromic NFI sites as well as potential C/EBP motifs (Fig. 10A). This region lies between two 16 to 18-bp AT-rich palindromes and also falls within the 500-bp RAP-associated region proposed by Lin et al. (24a) to be part of the functional Ori-Lyt domain. However, as shown above, we were unable to detect any direct binding by RAP to subsegments of the CTF repeat region in in vitro EMSA experiments.
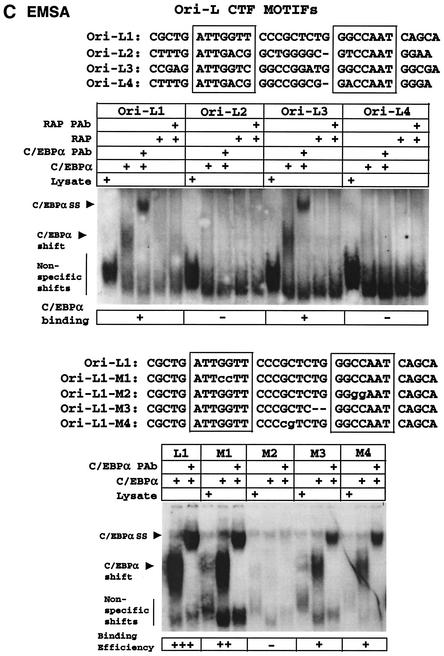
Interestingly, the four conserved paired CTF-like motifs present within both KSHV 200-bp Ori-Lyt subregions each mimic the organization of the functional paired C/EBP motifs defined previously within the KSHV RAP promoter (43). All five loci contain two adjacent head-to-head RRCCAAT-like core motifs, named the CTF I and CTF II boxes in this study (Fig. 10A), which in this case are separated by either 8 or 9 bp, in contrast to the somewhat related head-to-head XGCCAAX palindromic motifs with 3-bp spacings for functional high-affinity palindromic NF1 sites (18). Remarkably, one copy of this entire structure of nine consensus CTF motifs, including four head-to-head palindromic CTF pairs, as well as the adjacent AP1 and (AT)18 motifs are retained, despite considerable sequence divergence, between positions 117494 and 117792 in the related gamma-2 herpesvirus subclass genome of rhesus rhadinovirus 1 (31a). To evaluate the C/EBPα binding properties of each of these double CTF sites, four separate 48-bp oligonucleotide probes (referred to as Ori-L1, Ori-L2, Ori-L3, and Ori-L4) were used, and an EMSA experiment was conducted with in vitro-translated C/EBPα. Surprisingly, despite considerable similarities between the four putative paired sites, only two of them, Ori-L1 and Ori-L3, bound to C/EBPα (Fig. 10C).
To evaluate the specific nucleotide sequence requirements for functional C/EBP binding, the Ori-L1 probe was selected for mutagenesis. First, the CTF I box was mutated to generate the Ori-L1-M1 probe. Similar to the situation in the RAP promoter, the CTF I box proved to be largely dispensible for C/EBPα binding (10% loss of binding efficiency). However, when the CTF II box was mutated to generate the Ori-L1-M2 probe, C/EBPα binding was completely abolished, suggesting that this sequence is crucial (again similar to the situation in the RAP promoter). When the spacing between the CTF I and CTF II boxes was changed by 2-bp (Ori-L1-M3 probe), C/EBPα binding was reduced by 50%, and similarly a 2-bp transposition mutation introduced between the two motifs without changing the spacing (Ori-L1-M4 probe) also resulted in a 50% reduction in C/EBPα binding. Therefore, the multicopy CTF segment of Ori-Lyt does indeed bind to the C/EBPα protein, although only two of the potential eight CTF-like motifs tested appear to be directly involved.
In addition, a ninth CTF site is located across the junction between Ori-L1 and Ori-L2, but this is not palindromic and has only the CTF II box, not the CTF I box. Although the CTF II box here contains the identical core sequence to the CTF II boxes of Ori-L 1 and Ori-L 3 (GGCCAAT), this ninth CTF site (Ori-L VII probe) did not bind at all to C/EBPα (Fig. 10B, lanes 34 and 35), clearly suggesting that more than just the single core consensus CTF II box is needed for C/EBPα binding in EMSA experiments.
Finally, considering that two C/EBP shifts were observed with the 240-bp Ori-L fragment during EMSA, we deduced that a third, non-CTF-like C/EBP site may also reside within the adjacent 82-bp region from the right-hand side of the 240-bp Ori-L II fragment. Therefore, we generated this additional 82-bp fragment by PCR (Ori-L IV probe) for use in EMSA and found that at least one very strong C/EBP site did indeed reside within that region (Fig. 10B, lanes 19 and 20). Two additional smaller oligonucleotide probes were synthesized that harbored each of two potential C/EBP sites from within the 82-bp non-CTF region of the Ori-L II and Ori-L IV fragments. One of these probes, Ori-L V (ATTAGCAAAT [lanes 24 and 25]), but not Ori-L VI (AATGGCCAAA [lanes 29 and 30]), proved to bind strongly to C/EBPa, thus confirming the identity of the third C/EBP site within the KSHV Ori-Lyt.
Confirmation of the binding of C/EBPα and RAP to Ori-Lyt in PEL cell ChIP assays.
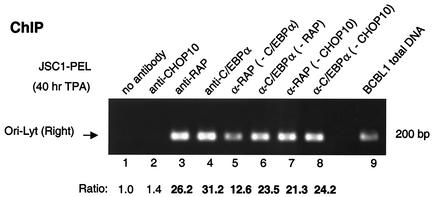
We next asked whether RAP could indeed associate with KSHV ORI-Lyt indirectly in vivo through C/EBPα, similar to our observations with the RAP promoter (43). A ChIP assay was conducted with the same recovered DNA samples used earlier (Fig. 4B) by using PCR analysis with primers (LGH3859 and LGH3866) specific for the 200-bp CTF repeat region from Ori-Lyt (R) (positions 119468 to 119665). Ori-Lyt DNA was indeed found to be strongly associated with both the C/EBPα and RAP immunoprecipitates (Fig. 11, lanes 3 and 4), whereas neither of the negative controls with PBS or CHOP-10 antibody precipitated any significant amounts of Ori-Lyt DNA (lanes 1 and 2). After C/EBPα had been precleared from the reaction mixture, the amount of DNA bound to RAP was reduced by 50%, but some RAP still efficiently associated with the Ori-Lyt region (lane 5). Preclearing of RAP had even less effect (25% reduced) on the binding of C/EBPα protein to Ori-Lyt (lane 6), whereas preclearing of CHOP-10 from the reaction mixture failed to significantly affect the association of either RAP or C/EBPα with the Ori-Lyt region (lanes 7 and 8). Therefore, these results suggest that RAP might also indirectly target this segment of Ori-Lyt DNA via another cellular and/or viral factor in addition to C/EBPα.
FIG. 11.
In vivo association of RAP and C/EBPα proteins with KSHV Ori-Lyt DNA. The ChIP experiment with JSC1 cell extracts after a 40-h TPA treatment showed that C/EBPα and RAP both associate with a 200-bp Ori-Lyt (R) region in infected cells and that the association of RAP with Ori-Lyt is only partially dependent on C/EBPα. Lanes: 1 to 4, ChIP assay results showing that the 200-bp Ori-Lyt region can be recovered by PCR from immunoprecipitates obtained with either C/EBPα or RAP Abs, but not from immunoprecipitates obtained with the CHOP-10 Ab or in the absence of Ab; 5, removal of C/EBPα from the lysate (preclearing with C/EBPα Ab) reduces the efficiency of RAP association with the Ori-Lyt by half; 6, association of C/EBPα with Ori-Lyt DNA is only slightly reduced by removal of the RAP protein; 7 and 8, association of RAP or 1 C/EBPα with Ori-Lyt DNA is not significantly affected by preclearing with CHOP-10 Ab; 9, control PCR amplification product from BCBL-1 total-cell DNA using the same primers (LGH3859 and LGH3866) to indicate the expected correct size of the Ori-Lyt PCR product. Measured values for band intensities are given below the lane numbers.
C/EBPα efficiently incorporates into cotransfection-assembled KSHV replication compartments only in the presence of RAP.
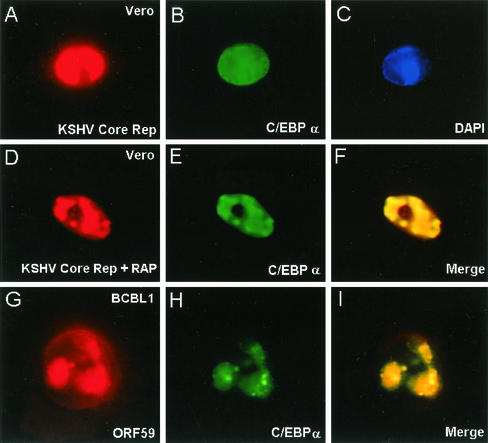
Because C/EBPα interacts strongly with RAP and was found to associate with Ori-Lyt, we asked whether C/EBPα can be recruited into assembled KSHV DNA RC formed after contransfection of the six KSHV core DNA replication genes with or without RAP (48). The results showed that in the absence of RAP, the RC excluded cotransfected C/EBPα (Fig. 12A to C), whereas when RAP was included in the cotransfection mixture, cotransfected C/EBPα became very efficiently incorporated into the RC even in the absence of Ori-Lyt plasmid DNA (Fig. 12D to F). Furthermore, in KSHV lytically infected cells, C/EBPα proved to be recruited into the viral RC in both BCBL-1 and DMVEC cells (Fig. 12G to I and data not shown).
FIG. 12.
C/EBPα is incorporated into KSHV DNA replication compartments in the presence of RAP. (Top and middle) Double-label IFA showing the association of C/EBPα with KSHV RC formed in transient-assembly assays in Vero cells. KSHV RC were assembled in Vero cells by cotransfecting KSHV Flag-POL, SSB, PPF, PRI, PAF, and HEL with C/EBPα in the presence or absence of RAP as described by Wu et al. (48). C/EBPα proved to be excluded from the RC when RAP was omitted but was efficiently incorporated into the RC when RAP was included in the cotransfection mixture. (Bottom), C/EBPα was also detected by double-label IFA in ORF59-positive KSHV RC in the nucleus of a single TPA-treated BCBL-1 cell.
DISCUSSION
In this study, we have demonstrated that KSHV RAP interacts strongly with both the C/EBPα and p21 proteins in vitro and that admixed RAP also stabilizes both the C/EBPα and p21 proteins from proteasome-mediated degradation. In KSHV-infected cells, induced p21 and C/EBPα were retargeted to nuclear PODs and colocalized there with both RAP and PML. In transient reporter gene expression assays, cotransfected RAP augmented C/EBPα-mediated activation of both the C/EBPα and p21 promoters. Moreover, endogenous ChIP assays with TPA-induced PEL cells revealed that RAP associates with the cellular C/EBPα promoter in vivo and that this association is almost totally dependent on the presence of C/EBPα Furthermore, both RAP and C/EBPα associated with KSHV Ori-Lyt DNA in in vivo ChIP assays and, similar to RAP, C/EBPα was also incorporated into viral DNA RC. Therefore, we conclude that the powerful induction of both C/EBPα and p21 protein levels in RAP expressing cells represents a combination of both cooperative transcriptional and posttranslational stabilization effects mediated by protein-protein interactions, including piggyback binding of RAP to C/EBPα when bound to specific C/EBP sites in promoter DNA. While many of the biochemical and functional studies described here used the isolated rat C/EBPα gene, we have since repeated both the GST binding and transactivation studies with human C/EBPα and obtained exactly the same results (data not shown).
The fact that the cooperative transactivation function requires the N-terminal segment of RAP as well as the C-terminal bZIP domain, whereas interaction with C/EBPα itself requires only the C-terminal bZIP region, suggests that, like ZTA, RAP possesses an N-terminal activation domain, although direct evidence for this from GAL4 domain transactivation assay is lacking. We also cannot exclude the additional possibility that, like ZTA, RAP may have negative postranscriptional effects on c-Myc levels, for example, that may also contribute to cell cycle arrest, although whether this effect of ZTA (or RAP) is also mediated by C/EBPα interaction or involves N-terminal domains remains to be resolved.
We also demonstrated that a recently proposed vIL-6 pathway to inhibit p21 and stimulate cell proliferation in alpha interferon-induced PEL cells does not overcome RAP-mediated p21 upregulation and G1 cell cycle arrest in TPA-induced PEL cells expressing both viral proteins. Our studies do not address whether paracrine effects of TPA-induced vIL-6 expression may promote the proliferation of adjacent latent or uninfected cells in the same cultures, but in the majority of lytically induced TPA-treated PEL cells that express either vIL-6 or RAP (or both), the increased levels of p21 and G1 cell cycle arrest were clearly evident. Finally, we showed that drug-mediated synchrony of BCBL1 cells either to induce G1 arrest or to induce p21 expression for up to 30 h both failed to trigger the KSHV lytic cycle (as assayed by IFA for RAP).
At first sight, the interaction of two members of the bZIP family might be expected to occur through classical heterodimerization. However, we were unable to detect simple heterodimers between RAP and C/EBPα in cross-linking cotranslation experiments; instead, we conclude that much larger complexes appear to be formed. Logically, because both C/EBPα and RAP form very strong homodimers (and perhaps homotetramers for RAP), it should be energetically and entropically unfavorable for the two strong homodimers, once folded, to disassemble and reform heterodimers. Both c-Jun and c-Fos form only very weak homodimers, which contributes to their heterodimeric interaction being energetically and entropically favorable. Our results suggest instead that C/EBPα-RAP dimer-dimer (or dimer-tetramer) types of hetero-oligomeric interactions occur that for unknown reasons do not form stable bands in our EMSA experiments. Additional evidence against the concept of simple heterodimers came when we found that KSHV RTA also interacts with C/EBPα and functions as a positive transactivator very much in the same way as RAP, again including interfering with C/EBPα gel shifts but without forming supershifts (43). However, although itself a dimer, RTA is not a bZIP family protein and would be extremely unlikely to form heterodimers rather than higher-order complexes with C/EBPα.
Functionally, heterodimer formation would be expected to result in RAP being a negative regulator of C/EBPα like CHOP-10, because RAP itself cannot bind to C/EBP sites or to any other known DNA targets. CHOP-10 homodimers cannot bind to DNA, and therefore CHOP-10-C/EBPα heterodimers lack C/EBP DNA binding activity and act as powerful inhibitors of C/EBPα transactivation of target promoters. However, the opposite result was observed for RAP, and despite our inability to detect DNA-bound C/EBPα-RAP supershift complexes by in vitro EMSA experiments, RAP nevertheless enhanced C/EBPα transactivation on several target promoters, including those for RAP, C/EBPα, and p21, and was found to associate with each of them only in the presence of DNA-bound C/EBPα protein in in vivo ChIP assays (43, 47, 49). Therefore, RAP exhibits the opposite behavior to that of CHOP-10, and presumbly the nature of the interaction with C/EBPα should differ. Perhaps there needs to be an additional component similar to HCF that acts as a scaffold for DNA-bound HSV VP16-OCT1 complexes that is still missing in our in vitro assays.
The possibilities that host cell cycle arrest might preferentially place KSHV in an environment favorable for lytic cycle induction or that TPA may function to induce the lytic cycle by first leading to cell cycle arrest do not appear to be valid for the case of KSHV latently infected BCBL-1 cells. From the data obtained in both this and our previous study (47), we conclude that during the KSHV lytic cycle, RAP stabilizes the C/EBPα protein and the accumulation of C/EBPα then leads to an upregulation of both the C/EBPα and p21 promoters, which is also enhanced cooperatively by RAP. Once a sufficient level of p21 protein is expressed, both RAP and C/EBPα can further stabilize the p21 protein from proteasome-mediated degradation. The summation of these events probably defines a pathway that leads to G1 cell cycle arrest in KSHV-infected host cells. Accumulating evidence that KSHV RTA shares many of these properties with RAP further enhances these effects (43, 43a).
Our studies do not exclude the possibility that KSHV may employ other RAP-independent pathways for cell cycle arrest, although clearly C/EBPα is essential for the RAP-dependent pathway in the cell types that we have examined. EBV ZTA mimics much of the same pathway as RAP in upregulating C/EBPα and p21 during the EBV lytic cycle, but there is at least one difference in that ZTA does not interact directly with p21 in in vitro assays. Moreover, for EBV ZTA, in addition to the C/EBPα and p21-mediated cell cycle arrest, other groups have reported that ZTA mediates the downregulation of c-Myc as well as the stabilization of the p27 and p53 proteins (14, 31). However, increased levels of the C/EBPα protein inhibit E2F transactivation of many of its downstream S-phase-related factors, and one of those target factors is c-Myc (33). Therefore, even this effect, if it occurs in KSHV RAP-expressing cells, might conceivably involve C/EBPα. Because we have provided strong evidence that the C/EBPα-plus-p21 pathway can proceed in cells lacking p53, a functional role, if any, for p53 in the KSHV lytic cycle remains unresolved. Earlier studies reported that RAP can interact with p53 in vitro and repress its transcriptional activity (28), but we were unable to confirm any significant interaction between the two proteins in GST binding assays, and more studies are needed to provide a more comprehensive picture on the subject. In addition, because RAP can be phosphorylated by cyclin-dependent kinases (29), an examination of its potential ability to interact with cellular cyclin-dependent kinases, cyclins, pRb, or E2F is worthy of investigation.
With regard to the question whether RAP, like ZTA in EBV, may function as the KSHV lytic origin binding protein (2, 13a, 24a, 48), we found that both C/EBPα and RAP associate with the 200-bp CTF subregion of KSHV Ori-Lyt in ChIP assays. In addition, C/EBPα was efficiently recruited into KSHV RC both in infected cells and in cotransfection assays, but only in the presence of RAP. Furthermore, three specific C/EBPα binding sites were mapped in this region centered at positions 119492, 119616, and 119691 between the two AT-rich palindromes in DSR and duplicated at positions 23523, 233999, and 23323 in DSL. However, only C/EBPα, and not RAP itself, bound directly to Ori-Lyt DNA by EMSA, suggesting that RAP partially associates with this segment of Ori-Lyt DNA via piggyback binding to C/EBPα but that the association probably involves other cellular or viral factors as well. Therefore, additional studies examining the abilities of KSHV RTA, cellular CTF, AP1, and other factors to bind to the complex KSHV Ori-Lyt domain, as well as the possible interaction of RAP with these proteins, should prove to be informative in the future.
Acknowledgments
These studies were funded by National Cancer Institute Research Grants (R01 CA73585 and RO1 CA81400) to G.S.H. from the National Institutes of Health. F.Y.W. was a graduate student at the Department of Pharmacology and Molecular Sciences and is currently a postdoctoral fellow in the Viral Oncology Program at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins School of Medicine. F.Y.W. was also partially supported by the Anti-Cancer Drug Development Training Program (T32 CA09243).
We thank Jae-Myun Lee, Gangling Liao, and Jian Huang for providing valuable advice and technical assistance.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, P., P. M. Lieberman, and G. G. Maul. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800-11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 74:10187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., J. Lee, Y. Wang, D. Huang, R. Ambinder, and S. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and ZTA interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christy, R. J., K. H. Kaestner, D. E. Geiman, and M. D. Lane. 1991. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 88:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciufo, D. M., J. S. Cannon, L. J. Poole, F. Y. Wu, P. Murray, R. F. Ambinder, and G. S. Hayward. 2001. Spindle cell conversion by Kaposi's sarcoma-associated herpesvirus: formation of colonies and plaques with mixed lytic and latent gene expression in infected primary dermal microvascular endothelial cell cultures. J. Virol. 75:5614-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darlington, G. J., S. E. Ross, and O. A. MacDougald. 1998. The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273:30057-30060. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 13a.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogal, V., M. Gostissa, P. Sandy, P. Zacchi, T. Sternsdorf, K. Jensen, P. P. Pandolfi, H. Will, C. Schneider, and G. Del Sal. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottifredi, V., and C. Prives. 2001. P53 and PML: new partners in tumor suppression. Trends Cell Biol. 11:184-187. [DOI] [PubMed] [Google Scholar]
- 17.Harris, T. E., J. H. Albrecht, M. Nakanishi, and G. J. Darlington. 2001. CCAAT/enhancer-binding protein-alpha cooperates with p21 to inhibit cyclin-dependent kinase-2 activity and induces growth arrest independent of DNA binding. J. Biol. Chem. 276:29200-29209. [DOI] [PubMed] [Google Scholar]
- 18.Jeang, K. T., D. R. Rawlins, P. J. Rosenfeld, J. H. Shero, T. J. Kelly, and G. S. Hayward. 1987. Multiple tandemly repeated binding sites for cellular nuclear factor 1 that surround the major immediate-early promoters of simian and human cytomegalovirus. J. Virol. 61:1559-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2001. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 20.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 21.Lane, M. D., Q. Q. Tang, and M. S. Jiang. 1999. Role of the CCAAT enhancer binding proteins (C/EBPs) in adipocyte differentiation. Biochem. Biophys. Res. Commun. 266:677-683. [DOI] [PubMed] [Google Scholar]
- 22.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation. dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus Ori-Lyt-dependent DNA replication: identification of the Ori-Lyt and association of K8 bZIP protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, L. A., L. Yang, E. S. Vazquez, M. C. Rodriguez-Vargas, M. Olive, J. T. Hsieh, C. J. Logothetis, and N. M. Navone. 2002. p21 modulates threshold of apoptosis induced by DNA-damage and growth factor withdrawal in prostate cancer cells. Carcinogenesis 23:1289-1296. [DOI] [PubMed] [Google Scholar]
- 26.Nakabeppu, Y., K. Ryder, and D. Nathans. 1988. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell 55:907-915. [DOI] [PubMed] [Google Scholar]
- 27.Nicholas, J., J. C. Zong, D. J. Alcendor, D. M. Ciufo, L. J. Poole, R. T. Sarisky, C. J. Chiou, X. Zhang, X. Wan, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J. Natl. Cancer Inst. Monogr. 23:79-88. [DOI] [PubMed] [Google Scholar]
- 28.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole, L., Y. Yu, Q. Zheng, J. Pevsner, and G. Hayward. 2002. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3395-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez, A., E. J. Jung, Q. Yin, C. Cayrol, and E. K. Flemington. 2001. Role of c-myc regulation in Zta-mediated induction of the cyclin-dependent kinase inhibitors p21 and p27 and cell growth arrest. Virology 284:159-169. [DOI] [PubMed] [Google Scholar]
- 31a.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheaff, R., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21CIP-1 does not require p21CIP-1 ubiquitination. Mol. Cell 5:403-410. [DOI] [PubMed] [Google Scholar]
- 33.Slomiany, B. A., K. L. D'Arigo, M. M. Kelly, and D. T. Kurtz. 2000. C/EBPalpha inhibits cell growth via direct repression of E2F-DP-mediated transcription. Mol. Cell. Biol. 20:5986-5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sylvester, S. L., C. M. ap Rhys, J. D. Luethy-Martindale, and N. J. Holbrook. 1994. Induction of GADD153, a CCAAT/enhancer-binding protein (C/EBP)-related gene, during the acute phase response in rats. Evidence for the involvement of C/EBPs in regulating its expression. J. Biol. Chem. 269:20119-20125. [PubMed] [Google Scholar]
- 35.Tang, Q. Q., M. S. Jiang, and M. D. Lane. 1999. Repressive effect of Sp1 on the C/EBPalpha gene promoter: role in adipocyte differentiation. Mol. Cell. Biol. 19:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, Q. Q., and M. D. Lane. 1999. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13:2231-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Q. Q., and M. D. Lane. 2000. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc. Natl. Acad. Sci. USA 97:12446-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timchenko, N., D. R. Wilson, L. R. Taylor, S. Abdelsayed, M. Wilde, M. Sawadogo, and G. J. Darlington. 1995. Autoregulation of the human C/EBP alpha gene by stimulation of upstream stimulatory factor binding. Mol. Cell. Biol. 15:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timchenko, N. A., T. E. Harris, M. Wilde, T. A. Bilyeu, B. L. Burgess-Beusse, M. J. Finegold, and G. J. Darlington. 1997. CCAAT/enhancer binding protein alpha regulates p21 protein and hepatocyte proliferation in newborn mice. Mol. Cell. Biol. 17:7353-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timchenko, N. A., M. Wilde, M. Nakanishi, J. R. Smith, and G. J. Darlington. 1996. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 10:804-815. [DOI] [PubMed] [Google Scholar]
- 41.Wang, G., R. Miskimins, and W. K. Miskimins. 2000. Mimosine arrests cells in G1 by enhancing the levels of p27 (Kip1). Exp. Cell Res. 254:64-71. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W. J. Roesler, and N. A. Timchenko. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell 8:817-828. [DOI] [PubMed] [Google Scholar]
- 43.Wang, S. E., F. Y. Wu, M. Fujimuro, J. C. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of the CCAAT/enhancer-binding protein-alpha (C/EBP a) in activation of the KSHV lytic replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. CCAAT/enhancer-binding protein-α is induced during the early stages of Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 44.Wang, X., E. Scott, C. L. Sawyers, and A. D. Friedman. 1999. C/EBPalpha bypasses granulocyte colony-stimulating factor signals to rapidly induce PU.1 gene expression, stimulate granulocytic differentiation, and limit proliferation in 32D c13 myeloblasts. Blood 94:560-571. [PubMed] [Google Scholar]
- 45.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, and A. S. Denton. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 46.Williams, S. C., C. A. Cantwell, and P. F. Johnson. 1991. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 5:1553-1567. [DOI] [PubMed] [Google Scholar]
- 47.Wu, F., Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. Lane, and G. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi's sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, F. Y., H. Chen, S. E. Wang, C. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein-α interacts with ZTA and mediates ZTA-induced p21CIP-1 accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, D. E., P. Zhang, N. D. Wang, C. J. Hetherington, G. J. Darlington, and D. G. Tenen. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA 94:569-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]