Abstract
Marek's disease (MD), a highly infectious disease caused by an oncogenic herpesvirus, is one of the few herpesvirus diseases against which live attenuated vaccines are used as the main strategy for control. We have constructed bacterial artificial chromosomes (BACs) of the CVI988 (Rispens) strain of the virus, the most widely used and effective vaccine against MD. Viruses derived from the BAC clones were stable after in vitro and in vivo passages and showed characteristics and growth kinetics similar to those of the parental virus. Molecular analysis of the individual BAC clones showed differences in the structure of the meq gene, indicating that the commercial vaccine contains virus populations with distinct genomic structures. We also demonstrate that, contrary to the published data, the sequence of the L-meq of the BAC clone did not show any frameshift. Virus stocks derived from one of the BAC clones (clone 10) induced 100 percent protection against infection by the virulent strain RB1B, indicating that BAC-derived viruses could be used with efficacies similar to those of the parental CVI988 vaccines. As a DNA vaccine, this BAC clone was also able to induce protection in 6 of 20 birds. Isolation of CVI988 virus from all of these six birds suggested that immunity against challenge was probably dependent on the reconstitution of the virus in vivo and that such viruses are also as immunogenic as the in vitro-grown BAC-derived or parental vaccine viruses. Although the reasons for the induction of protection only in a proportion of birds (33.3%) that received the DNA vaccine are not clear, this is most likely to be related to the suboptimal method of DNA delivery. The construction of the CVI988 BAC is a major step towards understanding the superior immunogenic features of CVI988 and provides the opportunity to exploit the power of BAC technology for generation of novel molecularly defined vaccines.
Herpesviruses are important pathogens associated with a wide range of diseases in humans and animals. Unlike a lot of human herpesvirus diseases for which safe attenuated vaccines are not available, control of many important herpesvirus diseases in veterinary medicine is dependent primarily on the use of vaccines. Most of the herpesvirus vaccines used in animals are derived from live viruses attenuated by classical methods. Although these vaccines are effective in inducing protection, there is a general desire to improve the effectiveness and safety of these vaccines, taking advantage of the advances in recombinant DNA technology. Recent applications of bacterial artificial chromosome (BAC) technology in cloning large DNA virus genomes have opened new avenues for reverse and forward genetics in basic herpesvirus research (1). While the use of BAC clones of herpesviruses is proving to be a powerful tool for the study of viral gene functions or for the identification of pathogenic determinants (22), this novel technology has not been examined for the improvement of herpesvirus vaccines. Recent success with the use of a BAC clone as an effective DNA vaccine in a mouse model of herpes simplex virus infection (16) validates the usefulness of this technology for the generation of novel herpesvirus vaccines. Such developments will have a huge impact and immediate application in veterinary medicine, where herpesvirus vaccines are already used extensively.
One of the most successful and widely used live herpesvirus vaccines is the one against Marek's disease (MD), a lymphoproliferative disease of poultry caused by a highly contagious oncogenic virus of the family Herpesviridae (20). In the last 30 years, these vaccines have dramatically reduced losses from MD by more than 99%. Today, the world poultry industry, with a yearly production of more than 30 billion birds, relies heavily on the sustained use of MD vaccines. Although largely successful, MD vaccines are under extreme pressure from severe virus challenges from the heavily contaminated poultry houses where tens of thousands of birds are housed together immediately after vaccination, even before the development of adaptive immune responses. Even though MD vaccines have been able to keep up with these challenges, the evolution of mutant viruses with greater virulence, as defined by their ability to break through the vaccine-induced protection, is threatening the sustainability of MD vaccines (26). The steady increase in disease severity is correlated with the increasing virulence of MD virus (MDV) strains, which are ranked into virulent (vMDV), very virulent (vvMDV), or very virulent plus (vv+MDV) pathotypes (23, 25). This increase in virulence has necessitated the introduction of newer generations of vaccines at regular intervals. These include the herpesvirus of turkey vaccine, introduced in 1970, the bivalent SB-1 and herpesvirus of turkey vaccine, introduced in the 1980s, and the CVI988 vaccine, introduced in the United States in the early 1990s (25).
The CVI988 strain (also called Rispens strain), developed from a natural isolate of low oncogenicity (11), is the most widely used MD vaccine, either alone or in combination with other vaccines. It is thought that approximately 5 billion doses of the CVI988 vaccine are used annually worldwide (Ian Tarpey, Intervet UK, personal communication). In spite of its widespread usage and exceptional protective efficacy, concerns do exist that the continued evolution of MDV towards greater virulence could lead to the failure of CVI988 vaccine in the future, in the same way that the evolution of MDV has reduced the useful life of the previous generations of MD vaccines. Since it is the only effective vaccine against many of the recent vvMDV and vv+MDV pathotypes (24), the emergence of MDV strains that can break through the CVI988 vaccine-induced immunity can have devastating effects on the poultry industry.
In order to meet the challenges raised by the continuing increase in virulence of MDV strains, an understanding of the determinants for the superior immunogenic properties of vaccines such as CVI988 is important. As a first step towards dissecting the functional properties of this most widely used vaccine, we have cloned the genome of the CVI988 strain as a stable infectious BAC clone. The virus rescued from the BAC provided complete protection against virulent MDV, demonstrating that the BAC-derived virus is biologically similar to the parental virus. Furthermore, we show that the use of BAC clone DNA as a vaccine capable of inducing protection, possibly through the reconstitution of live virus in vivo, represents a new generation of DNA-based vaccines against herpesviruses.
MATERIALS AND METHODS
Virus and cells.
Commercial MD vaccine strain CVI988 (11), obtained from Fort Dodge Animal Health, was used in this study. Both CVI988 and challenge virus stocks of the highly oncogenic MDV strain RB1B (13) were propagated in chicken embryo fibroblast (CEF) cultures prepared from 10-day-old specific-pathogen-free chicken embryos (12). In order to maintain the virulence of the challenge virus stocks, a low (fourth) passage stock of RB1B virus was used. We have used this virus stock consistently to induce a high incidence (more than 80%) of MD in susceptible lines of chickens.
Construction of CVI988 BAC.
The construction of CVI988 BAC was carried out essentially as described previously by the insertion of the F plasmid in the US2 region (14). Briefly, secondary CEF were cotransfected by the calcium phosphate precipitation method with CVI988 genomic DNA and transfer vector pDS-pHA1 (14). Genomic DNA extracted from infected CEF after four rounds of selection in medium containing mycophenolic acid, xanthine, and hypoxanthine was electroporated into Escherichia coli DH10B cells. Chloramphenicol-resistant colonies were analyzed for infectivity by transfecting 1 to 2 μg of DNA into CEF and examining for virus growth (12).
DNA analyses.
The analysis of the DNA was carried out by PCR as well as Southern blotting. Genomic regions of CVI988 viral and BAC DNA were amplified under standard PCR conditions using the following oligonucleotide primer pairs (5′-3′): Meq, GCACTCTAGAGGTGTAAAGAGATGTCTCAG and TAACTCGAGGAGAAGA AACATGGGGCATAG; pp38, TTGTCTTTCTGCCCGCACCG and TCGCACTGCT GGGCAAGCTC; 132-bp region, TACTTCCTATATAGATTGAGACGT and GAGAT CCTCG TAAGGTGTAATATA; gpt, ATGAGCGAAAAATACATCGTC and TTAGCG ACCGGAGATTGGCGG.
For Southern blot hybridization, BamHI and PstI restriction enzyme digests of the DNA from parental CVI988 virus or the pCVI988 clones were separated on 0.8% agarose gels and transferred to Duralon-UV membranes (Stratagene Europe, Amsterdam, The Netherlands). Digoxigenin-labeled (Roche Diagnostics, Lewes, East Sussex, United Kingdom) 0.5-kbp PstI-BamHI fragment of the PCR-amplified meq gene from pCVI988 clone 10 was used as the probe.
Vaccination and virus challenge.
The vaccination efficacy of pCVI988 clone 10 was tested in 1-day-old specific-pathogen-free HPRS Rhode Island Red chicks. In the first group, the protective efficacy of the BAC-derived virus stocks was examined. For this, CEF transfected with the BAC DNA was subcultured for three to four passages to allow the virus to grow to sufficient titers. One-day-old chicks (n = 19) were injected intra-abdominally with 1,000 PFU of pCVI988-derived virus. Control birds (n = 19) were injected with an equal number of uninfected CEF or tissue culture medium. Vaccine contact birds (n = 16) were kept in the same cages as the vaccinated birds. In the second group, the ability of the naked DNA of the pCVI988 clone 10 to protect against challenge was examined. BAC DNA diluted in phosphate-buffered saline (PBS) was inoculated intramuscularly into 1-day-old chicks at doses of 5 μg (n = 10) or 10 μg (n = 9). Control birds were injected with 5 μg (n = 9) or 10 μg (n = 8) of the transfer vector pDS-pHA1. BAC DNA contact birds (n = 9) were kept in the same cages as the pCVI988-injected birds. Seven days after vaccination, all birds were injected intra-abdominally with a fourth CEF passage stock of virulent RB1B strain (1,000 PFU) and monitored for 8 weeks. The numbers of birds that developed MD on the basis of gross and histological lesions in different groups were used to calculate the cumulative disease or survival rates, which were used as a measure of the protective efficacy of the vaccines.
Nucleotide sequence accession numbers.
GenBank accession numbers for sequences determined in this study were AY164639 (S-meq) and AY164640 (L-meq).
RESULTS
pCVI988-derived viruses show characteristics similar to those of parental CVI988 virus.
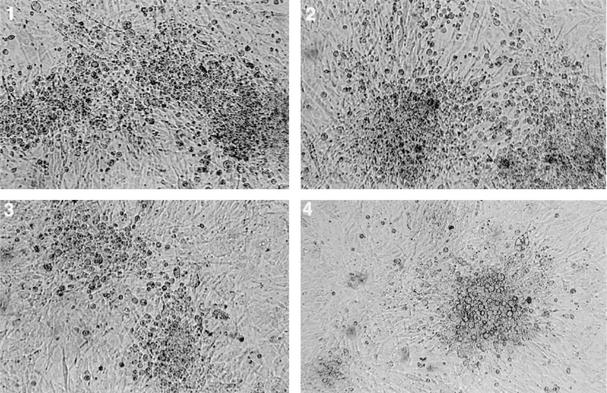
Transfection of CVI988 viral DNA together with pDS-pHAI DNA into CEF and subsequent growth in selection medium showed evidence of virus infection, indicating the presence of recombinant viruses. Electroporation of the DNA extracted from these CEF into E. coli DH10B cells produced several chloramphenicol-resistant colonies. Three of these colonies (pCVI988 clones 6, 10, and 19) that contained high-molecular-weight DNA were selected for further analysis. Transfection of BAC DNA from these clones into primary CEF produced MDV-specific plaques visible from about 3 days, demonstrating their infectivity. The morphology and size of the plaques from each of the pCVI988 BAC clones were indistinguishable from those produced by the parental CVI988 strain (Fig. 1). Analysis of the in vitro replication of the parental CVI988 or the viruses derived from the BAC clones by plaque titration or by measuring the amount of viral DNA using a real-time quantitative PCR (Taqman) assay did not show significant differences (data not shown). Virus stocks from BAC clone 10 were examined for their stability by continuous passage in vitro in CEF, and DNA extracted from infected cells at passage levels 5 and 10 were electroporated into E. coli. The appearance of chloramphenicol-resistant colonies indicated that the recombinant BAC MDV clones were stable at these passage levels. In addition, the effect of in vivo passage of the virus in birds vaccinated with pCVI988 (clone 10) was examined by inoculating lymphocytes collected from the spleen, either 7 days (one bird) or 48 days (three birds) after vaccination onto CEF. Virus plaques reminiscent of those induced by parental CVI988 virus could be seen at 4 to 5 days after inoculation.
FIG. 1.
Morphology of plaques induced by parental CVI988 (panel 1) and viruses derived from pCVI988 clones 6, 10, and 19 (panels 2, 3, and 4, respectively).
Molecular characteristics of pCVI988 clones.
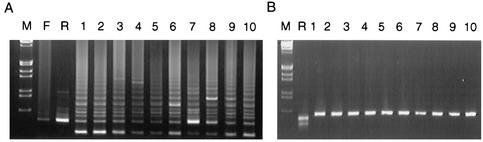
DNA isolated from pCVI988 clones 6, 10, and 19 exhibited identical BamHI digestion patterns that resembled the digestion pattern of the GA strain of MDV (6), indicating that the three BAC clones contained the entire CVI988 genome. Positive PCR tests for the gpt gene for the BAC vector further confirmed the specificity of the pCVI988-derived viruses. PCR analysis of the pp38 gene also gave products of identical sizes from both parental and pCVI988-derived viruses. Compared with the other serotype 1 MDV strains, the pp38 gene of CVI988 strain has a single G-to-A base substitution at coding position 320 that eliminates an epitope designated H19 in the protein sequence (3). We have noted that this mutation also eliminates a unique TspRI restriction site. PCR products of the pp38 gene from both parental and BAC-derived CVI988 viruses, unlike that of the oncogenic RB1B strain, shared this polymorphism (Fig. 2B). Another characteristic feature of CVI988 virus, used widely as a tool for the molecular identification of attenuated MDV strains such as CVI988, is the increase in the number of units of 132-bp tandem repeats in the BamHI-D/H region (4). Agarose gel separation of PCR products amplified from the 132-bp repeats showed multiple banding patterns, indicating expansion of the 132-bp repeat region in both the parental and pCVI988-derived viruses (Fig. 2A). Compared to this, the virulent RB1B strain and a field MDV isolate showed a major band representing two copies of these repeat sequences, although weak bands representing expansion of the region could also be seen (Fig. 2A). Sequence analysis of the BamHI-D/H region of the pCVI988 clone 10 showed eight tandem 132-bp repeats (unpublished data). Identical banding patterns of the 132-bp PCR on pCVI988 clone 10-derived virus stocks passaged in CEF as well as birds (Fig. 2A) indicated that the numbers of these repeat sequences remain stable after in vitro and in vivo passages.
FIG. 2.
(A) PCR of the 132-bp region from BAC-derived and wild-type CVI988 viruses. (B) TspRI restriction digestion profile of pp38 PCR product. Lane M, HindIII-EcoRI-digested lambda DNA molecular weight markers; lane F, field MDV isolate; lane R, RB1B virus; lane 1, BAC6; lane 2, BAC19; lane 3, BAC10; lane 4, BAC10, fifth passage; lane 5, BAC10, 10th passage; lanes 6 to 9, viruses isolated from birds 241 (BAC virus-infected), 932, 933, and 941 (BAC DNA-infected), respectively; lane 10, wild-type CVI988 virus.
CVI988 virus stocks contain subpopulations differing in the meq gene.
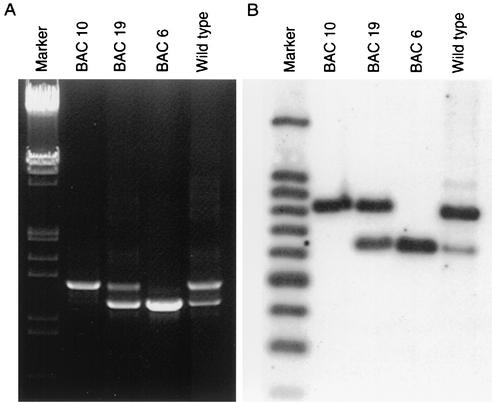
PCR amplification of the meq gene from the parental CVI988 virus gave two products, including a normal 1.0-kbp small meq (S-meq) product and an additional, larger, 1.2-kbp large meq (L-meq) product that contained a 178-bp insertion (8). However, it was unclear whether the two forms of the meq gene were located in the repeat regions of the same viral genome or whether they represented independent virus subpopulations. PCR tests showed differences in the meq gene between the parental CVI988 and the BAC clones (Fig. 3A). BAC clone 6 gave only a single 1.0-kbp product of S-meq, while clone 10 gave a single 1.2-kbp L-meq product. PCR on clone 19 gave products representing both S-meq and L-meq, indicating that the copies of meq in the internal and terminal repeat regions of virus clone 19 are different in relation to the insertion. The presence of the two different meq gene structures in the three BAC clones was also confirmed by Southern blot analysis. Hybridization of the PstI- and BamHI-digested DNA with the meq probe detected single bands representing L-meq and S-meq in pCVI988 clones 6 and 10, respectively, while pCVI988 clone 19 and the parental CVI988 showed both L-meq and S-meq hybridizing fragments (Fig. 3B). However, in comparison to BAC clone 19, where both S-meq and L-meq bands were present in equimolar amounts, the S-meq band in the parental CVI988 was much weaker than the L-meq band, suggesting that viruses harboring S-meq represented a minor population.
FIG. 3.
(A) Ethidium bromide-stained agarose gels showing 1.0- and 1.2-kbp PCR products of meq gene from the parental CVI988 or pCVI988-derived viruses. HindIII-EcoRI-digested lambda DNA is shown as marker. (B) Southern blot showing the L-meq and S-meq fragments. PstI-BamHI-digested DNA from CEF infected with parental and BAC-derived CVI988 viruses probed with digoxigenin-labeled meq-DNA probe is shown. One hundred-base-pair DNA ladder (Promega) was simultaneously hybridized with digoxigenin-labeled ladder to visualize size markers.
The insertion in meq does not disrupt the open reading frame.
In order to further characterize the structures of the meq gene in the different pCVI988 clones, we determined the sequences of S-meq and L-meq from pCVI988 clones 6 and 10, respectively. The S-meq sequence showed only four nucleotide changes, including an insertion of three nucleotides compared with the published S-meq sequences in the CVI988 virus (accession no. AF493555). In the L-meq gene of the parental CVI988 virus, it has been reported that the 178-bp insertion caused a frameshift in the coding region of the proline-rich repeat region of the transactivation domain (8). In addition, this also shifted the stop codon 13 nucleotides upstream, thereby truncating the protein. The sequence of L-meq of the pCVI988 BAC clone 10 showed seven nucleotide differences (Fig. 4) from the published CVI988 sequence (accession no. AB033119). One of these differences was a single G deletion from a stretch of GGGGG at positions 609 to 613 of the published L-meq sequence. The deletion of this single G, leaving a stretch of only GGGG at that position, restores the reading frame despite the insertion of 59 amino acids.
FIG. 4.
Alignment of the L-meq sequences from pCVI988 clone 10 with the published CVI988 (accession no. AB033119) sequences. The 178-bp insertion in L-meq is shown in bold. The position of the deletion of the single G residue in the pCVI988 sequence is shown by vertical arrow. Frameshift in the CVI988 sequence is underlined.
BAC-derived CVI988 virus induces protection against virulent MDV infection.
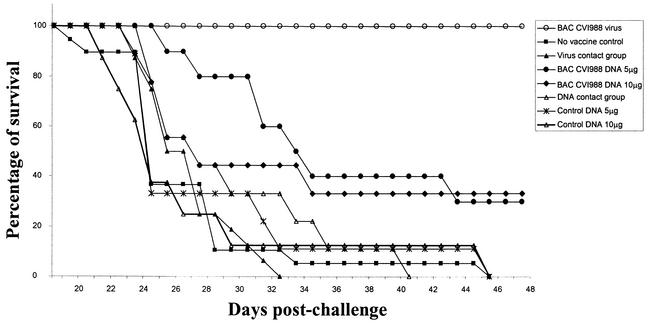
Since the CVI988 vaccine provides the most effective protection against MD, we examined the protective ability of the pCVI988-derived virus stocks against the virulent RB1B strain. Evidence of MD could be observed in the unvaccinated control birds from 3 weeks after infection, and postmortem examination revealed atrophic changes of the bursa and thymus, enlargement of peripheral nerves, and tumors of visceral organs. Histological examination demonstrated lymphoid infiltration in the nerves and neoplastic lymphoid proliferation in visceral organs, such as the liver, spleen, ovary, and kidney (not shown). All the birds in the unvaccinated control and contact exposure group developed the disease, demonstrating that the virulent RB1B strain induced MD in 100 percent of nonimmunized birds (Fig. 5). Compared with this, none of the birds that were vaccinated with the BAC-derived CVI988 virus developed MD during this period. These results demonstrated that the pCVI988-derived virus is similar to the parental vaccine virus in inducing protection against challenge infection with virulent MDV.
FIG. 5.
Protection induced by pCVI988 (clone 10) against virulent RB1B infection. Groups of 1-day-old birds immunized with pCVI988-derived viruses or DNA were challenged with the virulent RB1B strain of MDV. The cumulative number of birds that developed the disease was used to calculate the percent survival rates for different groups.
Protection by BAC DNA vaccine shows correlation with virus replication.
Having demonstrated the ability of pCVI988-derived virus to induce protection against MD, we examined whether pCVI988 BAC could be used as a DNA vaccine to induce protection against RB1B virus challenge. Birds injected with control DNA developed MD with characteristic gross and histological lesions. In addition, all birds in the pCVI988 DNA contact exposure group also developed the disease, with the same kinetics seen in nonimmunized birds (Fig. 5). In the two groups of birds that were injected with pCVI988 DNA in doses of 5 or 10 μg, 7 out of 10 and 6 out of 9 birds, respectively, developed MD during the course of the experimental period. The three remaining birds in each of these two groups did not develop MD during this period, and postmortem examination did not reveal any gross or histological lesions in these birds. These studies thus demonstrate that CVI988 BAC injected as naked DNA in doses of 5 to 10 μg induced protection in a proportion (30 to 33.3%) of birds against virulent MDV challenge.
Because pCVI988 DNA is infectious and contains the complete viral genome, the DNA-mediated protective responses could be induced either through the expression of the immunogenic viral genes or after replication of the reconstituted infectious virus. In order to examine the latter possibility for the induction of partial protection by the pCVI988 DNA, we attempted to isolate the virus from the spleens of all the six birds that survived RB1B challenge. Inoculation of spleen lymphocytes from these birds onto CEF produced CVI988 virus-like plaques in all cases. PCR tests that amplified the gpt gene, which identifies only pCVI988-derived virus and not the virulent RB1B challenge virus, confirmed that these isolates represented CVI988 virus reconstituted from the injected BAC DNA. This was also confirmed by the successful transformation of E. coli DH10B cells with the DNA extracted from the CEF infected with the recovered viruses.
DISCUSSION
Full-length infectious BAC clones of several herpesviruses are used as powerful tools for studying herpesvirus biology and pathogenesis. In the case of MDV, BAC clones of an attenuated MDV strain, 584Ap80C, have been used to investigate various gene functions (5, 14-15, 18). Among the different generations of MD vaccines, CVI988 (11) is the most widely used vaccine, mainly because of its exceptional protective efficacy, particularly against vvMDV and vv+MDV pathotypes (24). As a first step for studying the molecular basis for the unique features of this MDV strain as an effective vaccine, we have generated several infectious BAC clones of CVI988 from a commercially available vaccine. Transfection of the DNA prepared from three of these clones in CEF resulted in the rescue of infectious viruses with plaque morphology and growth kinetics indistinguishable from those of parental virus despite the presence of the BAC vector sequences. The original CVI988 isolate has undergone several passages, and the commercial vaccine is thought to be a pool of several viruses (7). Cloning of individual MDV genomes from these virus pools provides the opportunity to examine the properties of each of the viruses. Such studies could lead to the identification of virus strains with desirable characteristics, including higher-level growth or immunogenicity, that could be used as seed vaccine stocks. The pCVI988 clone showed remarkable stability during both in vitro and in vivo passages, despite insertion of the BAC vector into the genome. This was confirmed by the detection of the intact gpt gene by PCR on DNA samples from viruses passaged in cell culture and in birds. Furthermore, the DNA samples produced chloramphenicol-resistant colonies in E. coli, confirming the presence of mini-F-vector sequences in pCVI988-derived viruses. Although the BAC clones of many herpesviruses are stable (22), the influence of the vector sequences on the stability or growth rate varies with different viruses. For example, the presence of vector sequences had destabilizing effects on the BAC clones of pseudorabies virus (17) and mouse cytomegalovirus (21), resulting in spontaneous deletions of the BAC vector and neighboring viral sequences. However, since the vector sequences do not appear to affect the stability of the pCVI988 clones, they would be useful for maintaining as seed stocks.
Even though the individual BAC clones could not be differentiated based on BamHI restriction digestion pattern or by the PCR tests of pp38 or the 132-bp repeat sequences, they could be distinguished by the differences in the meq gene structure. In the parental CVI988 vaccine, two forms of the meq gene have been demonstrated (8). The major population was thought to contain L-meq, while a minor population contained either S-meq or both forms. Since L-meq was originally observed only in CVI988, it was thought to be associated with loss of oncogenicity (8). However, subsequent studies also detected the L-meq gene in oncogenic strains Md5 and RB1B, although its detection, unlike that in CVI988, was restricted to specific periods after infection (2). On this basis, it was speculated that a shift in the population of viruses with L-meq and S-meq sequences may play a role in establishing latency and/or tumorigenic transformation (2). Although such a hypothesis still needs to be proven, these studies further established that MDV strains do exist as pools of virus populations with distinct genomic structures and perhaps with distinct functions. Since each BAC clone represents individual viruses in these pools, they are valuable in delineating any functional differences between viruses. The three pCVI988 clones with distinct meq structures did not show significant differences in growth kinetics in vitro, demonstrating that meq variation probably is not important for replication in cultured cells. In birds infected with pCVI988 clone 10, only L-meq could be detected after in vivo passage, indicating that generation of virus pools containing both meq variants probably requires continuous passage over a long period of time. Even if such variations do arise during continuous passage, the stable BAC DNA offers the opportunity to generate new uniform virus stocks with the original characteristics.
The published sequence of L-meq from parental CVI988 virus indicated that the 178-bp insertion resulted in a frameshift of the C-terminal region of the Meq protein (8). Since this region contains an important transactivation domain (10), the frameshift could interfere with the function of Meq as a transcription factor. However, the deletion of G in the poly(G) tract (at positions 609 to 613) of the pCVI988 BAC clone 10 restores the reading frame even though it introduces a 59-amino-acid repeat in the proline-rich region. It is not clear whether this expansion of the proline-rich region would modulate the functions of Meq. However, based on the observation that only one proline-rich repeat is necessary for the transactivation function of Meq (9), one could speculate that the insertion of the 59-amino-acid repeat sequence is unlikely to influence Meq function. The inserted sequence, which is almost identical to a tandem repeat observed in the meq gene of the GA and Md5 strains, increases the number of tandem repeats to three in the CVI988 genome. Similar expansions of tandem repeat sequences, such as those seen with the 132-bp repeat, are thought to occur during viral replication in cultured cells.
Virus derived from pCVI988 (clone 10) is as highly immunogenic as the parental vaccine, since it induced 100 percent protection against virulent RB1B challenge, demonstrating that MDV BAC clones could be developed as commercial vaccines. In addition, BAC DNA was able to induce protection in 6 of 20 birds even at relatively low doses of 5 to 10 μg, proving that it retains immunogenicity even as a DNA vaccine. However, unlike the virus stock derived from the BAC clone, which induced 100 percent protection, the BAC clone used as a DNA vaccine was able to induce protection only in a proportion of birds. Although the reasons for this difference are not clear, this is most likely to be related to the suboptimal DNA delivery methods, since the immunogenicity of DNA vaccines is dependent on the efficiency of uptake into the cells. Administration of higher doses or adopting more efficient DNA immunization strategies could further improve the efficiency of these novel DNA vaccines. Advantages of stability and the ease of handling would make such vaccines very attractive compared to the currently used MD vaccines, which require the mass production of primary CEF cultures and the maintenance of the liquid nitrogen cold chain for their transportation. We were able to isolate BAC-derived viruses from the spleens of all birds that were protected by the DNA immunization, suggesting that replication of the reconstituted virus is probably a requirement for protection. However, since we did not examine the tissues of all the DNA-vaccinated birds for evidence of virus replication, it is difficult to make such a conclusion. Furthermore, confirmatory evidence for the need for virus replication can be obtained only by examining whether the DNA from a replication-defective BAC clone of CVI988 could induce any protection. A recent study demonstrating the inability of a gE-deletion mutant BAC clone of MDV strain 584Ap80C to induce protection against virulent EU1 MDV challenge (19) supports the suggestion that protective immune responses against MD are dependent on virus replication.
The current vaccination strategy for control of MD is at a critical juncture. A major shift in virulence of MDV isolates in the last 30 years has been clearly documented (23), and the CVI988 strain is the only effective vaccine against many of the most recently emerged MDV pathotypes. Construction of infectious BAC clones of CVI988 is an important step in understanding the determinants for its superior immunogenic features. Additionally, it also provides the opportunity to develop this highly immunogenic virus as an expression vector for the delivery of immunomodulatory molecules or protective immunogens.
Acknowledgments
We thank F. de Palma and S. Uppal for assisting in the earlier part of the project.
The work was partly funded by BBSRC.
REFERENCES
- 1.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 2.Chang, K. S., S. I. Lee., K. Ohashi, A. Ibrahim, and M. Onuma. 2002. The detection of the meq gene in chicken infected with Marek's disease virus serotype 1. J. Vet. Med. Sci. 64:413-417. [DOI] [PubMed] [Google Scholar]
- 3.Cui, Z. Z., A. Qin., L. F. Lee, P. Wu, and H. J. Kung. 1999. Construction and characterisation of a H19 epitope point mutant of MDV CVI988/Rispens strain. Acta Virol. 43:169-173. [PubMed] [Google Scholar]
- 4.Davidson, I., R. Borenshtain, and Y. Weisman. 2002. Molecular identification of the Marek's disease virus vaccine strain CVI988 in vaccinated chickens. J. Vet. Med. 49:83-87. [DOI] [PubMed] [Google Scholar]
- 5.Dorange, F., B. K. Tischer, J.-F. Vautherot, and N. Osterrieder. 2002. Characterisation of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuchi, K., A. Tanaka, L. W. Schierman, R. L. Witter, and M. Nonoyama. 1985. The structure of Marek's disease virus DNA: the presence of unique expansion of nonpathogenic viral DNA. Proc. Natl. Acad. Sci. USA 82:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooft van Iddekinge, B. J. L., L. Stenzler, K. A. Schat, H. Boerrigter, and G. Koch. 1999. Genome analysis of Marek's disease virus strain CVI-988: effect of cell culture passage on the inverted repeat regions. Avian Dis. 43:182-188. [PubMed] [Google Scholar]
- 8.Lee, S.-I., M. Takagi, K. Ohashi, C. Sugimoto, and M. Onuma. 2000. Difference in the meq gene between oncogenic and attenuated strains of Marek's disease virus serotype 1. J. Vet. Med. Sci. 62:287-292. [DOI] [PubMed] [Google Scholar]
- 9.Qian, Z., P. Brunovskis, F. Rauscher III, L. F. Lee, and H.-J. Kung. 1995. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 69:4037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian, Z., P. Brunovskis, L. F. Lee, P. K. Vogt, and H.-J. Kung. 1996. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J. Virol. 70:7161-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rispens, B. H., H. van Vloten, N. Mastenbroek, H. J. Mass, and K. A. Schat. 1972. Control of Marek's disease in the Netherlands. I. Isolation of an avirulent Marek's disease virus (strain CVI988) and its use in laboratory vaccination trials. Avian Dis. 16:108-125. [PubMed] [Google Scholar]
- 12.Ross, L. J. N. 1998. Recombinant vaccines against Marek's disease. Avian Pathol. 27:S65-S73. [Google Scholar]
- 13.Schat, K. A., B. W. Calnek, and J. Fabricant. 1982. Characterisation of two highly oncogenic strains of Marek's disease virus. Avian Pathol. 11:593-605. [DOI] [PubMed] [Google Scholar]
- 14.Schumacher, D., B. K. Tischer, W. Fuchs, and N. Osterrieder. 2000. Reconstitution of Marek's disease virus serotype 1 (MDV-1) from DNA cloned as bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 74:11088-11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumacher, D., B. K. Tischer, J.-P. Teifke, K. Wink, and N. Osterrieder. 2002. Generation of a permanent cell line that supports efficient growth of Marek's disease virus (MDV) by constitutive expression of MDV glycoprotein E. J. Gen. Virol. 83:1987-1992. [DOI] [PubMed] [Google Scholar]
- 16.Suter, M., A. M. Lew, P. Grob, G. J. Adema, M. Ackerman, K. Shortman, and C. Fraefel. 1999. BAC-VAC, a novel generation of DNA vaccines: a bacterial artificial chromosome (BAC) containing a replication-competent, packaging-defective virus genome induces protective immunity against herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 96:12697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tischer, B. K., D. Schumacher, M. Beer, J. Beyer, J. P. Teifke, K. Osterrieder, K. Wink, V. Zelnik, F. Fehler, and N. Osterrieder. 2002. The products of UL10 (gM) and the UL49.5 genes of Marek's disease virus serotype 1 are essential for virus growth in cultured cells. J. Gen. Virol. 83:997-1003. [DOI] [PubMed] [Google Scholar]
- 19.Tischer, B. K., D. Schumacher, M. Beer, J. Beyer, J. P. Teifke, K. Osterrieder, K. Wink, V. Zelnik, F. Fehler, and N. Osterrieder. 2002. A DNA vaccine containing an infectious Marek's disease virus genome can confer protection against tumourigenic Marek's disease in chickens. J. Gen. Virol. 83:2367-2376. [DOI] [PubMed] [Google Scholar]
- 20.van Regenmortel, M. H. V., C. M. Fauqet, D. H. L. Bishop, E. Carstens, M. K. Estes, S. Lemon, J. Maniloff, M. A. Mayo, D. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 21.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 23.Witter, R. L. 1997. Increased virulence of Marek's disease virus field isolates. Avian Dis. 41:149-163. [PubMed] [Google Scholar]
- 24.Witter, R. L. 1992. Safety and comparative efficacy of the CVI988/Rispens vaccine strain, p. 315-319. In 4th International Symposium on Marek's disease, 19th World's Poultry Congress, vol. 1. World's Poultry Science Association, Amsterdam, The Netherlands.
- 25.Witter, R. L. 2001. Protective efficacy of Marek's disease vaccines. Curr. Top. Microbiol. Immunol. 255:57-90. [DOI] [PubMed] [Google Scholar]
- 26.Witter, R. L. 2001. Marek’s disease virus vaccines—past, present and future (chicken vs. virus—a battle of the centuries), p. 1-9. In K. A. Schat, R. M. Morgan, M. S. Parcells, and J. L. Spencer (ed.), Current progress on Marek’s disease research. Proceedings of the 6th International Symposium on Marek’s disease. American Association of Avian Pathologists, Kennett Square, Pa.