Abstract
RNA replicons derived from flavivirus genomes show considerable potential as gene transfer and immunization vectors. A convenient and efficient encapsidation system is an important prerequisite for the practical application of such vectors. In this work, tick-borne encephalitis (TBE) virus replicons and an appropriate packaging cell line were constructed and characterized. A stable CHO cell line constitutively expressing the two surface proteins prM/M and E (named CHO-ME cells) was generated and shown to efficiently export mature recombinant subviral particles (RSPs). When replicon NdΔME lacking the prM/M and E genes was introduced into CHO-ME cells, virus-like particles (VLPs) capable of initiating a single round of infection were released, yielding titers of up to 5 × 107/ml in the supernatant of these cells. Another replicon (NdΔCME) lacking the region encoding most of the capsid protein C in addition to proteins prM/M and E was not packaged by CHO-ME cells. As observed with other flavivirus replicons, both TBE virus replicons appeared to exert no cytopathic effect on their host cells. Sedimentation analysis revealed that the NdΔME-containing VLPs were physically distinct from RSPs and similar to infectious virions. VLPs could be repeatedly passaged in CHO-ME cells but maintained the property of being able to initiate only a single round of infection in other cells during these passages. CHO-ME cells can thus be used both as a source for mature TBE virus RSPs and as a safe and convenient replicon packaging cell line, providing the TBE virus surface proteins prM/M and E in trans.
Subgenomic replicons of positive-stranded RNA viruses contain all of the genetic elements needed to amplify themselves in susceptible host cells but lack some or all of the genes coding for structural proteins. Consequently, these RNAs are replicated in cells but are not packaged into viral particles. Replicons have proven to be valuable tools for studying replication independently of virion assembly and maturation (4, 20, 30). Moreover, they have great potential as molecular tools for gene expression and as vectors for therapeutic and prophylactic purposes (15, 19, 40). The delivery of replicon RNAs to host cells can be achieved in three different ways: (i) transfection with in vitro-transcribed replicon RNA, (ii) transfection with plasmid DNA encoding replicon sequences under the control of a cellular RNA polymerase II promoter, and (iii) infection with virus-like particles (VLPs) generated by encapsidation of replicon RNA with viral structural proteins provided in trans. VLPs are capable of initiating a single round of infection, providing an efficient and easy way to deliver the replicon vector into specific host cells, and are therefore particularly useful tools in gene therapy or for immunization purposes (19).
Flaviviruses, members of the genus Flavivirus (family Flaviviridae), are positive-stranded RNA viruses and include important human pathogens such as yellow fever virus, the four serotypes of dengue virus, Japanese encephalitis virus, West Nile virus, and tick-borne encephalitis (TBE) virus (45). Mature flavivirus virions are spherical in shape and consist of a nucleocapsid surrounded by a lipid bilayer that contains two viral proteins, the glycosylated large surface protein E (envelope; approximately 54 kDa) and the small, nonglycosylated protein M (membrane; approximately 8 kDa) (29). In infected cells, the latter is found as a glycosylated precursor protein (prM; approximately 27 kDa) which is cleaved during the process of virion maturation in the trans-Golgi network by the cellular protease furin (43). The nucleocapsid is composed of a single capsid protein (C; approximately 11 kDa) and contains the viral genome, an unsegmented positive-stranded RNA of approximately 11 kb. The only open reading frame is flanked by 5′ and 3′ noncoding regions and gives rise to a polyprotein that includes the three structural proteins and the seven nonstructural proteins in the order C-prM/M-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 (29).
A common characteristic of the flavivirus surface proteins prM/M and E is their ability to assemble into subviral particles, which are formed in variable amounts as a by-product of flavivirus infection (23, 29). These subviral particles are smaller than infectious virions and do not contain a nucleocapsid. Recombinant subviral particles (RSPs) can also be generated by coexpression of proteins prM/M and E in the absence of protein C (2, 26) and have been shown to resemble whole virions with respect to their structural and functional organization of the surface proteins (8, 39).
Replicons of flaviviruses have been constructed by the introduction of in-frame deletions removing the coding regions for proteins prM and E and in some cases also the majority of protein C, and some of these constructs have been used for heterologous gene expression. Replicons derived from Kunjin virus, a close relative of West Nile virus, represent the most extensively characterized system, and this has been used successfully both to study flavivirus replication and to develop a promising new tool for heterologous gene expression (19, 20, 46, 47). Replicons have also been derived from West Nile virus, dengue virus, and yellow fever virus (35, 36, 41).
An interesting potential advantage of flavivirus replicons is that they seem to be naturally noncytopathic (reviewed in reference 19). Packaging of flavivirus replicons into VLPs has so far only been achieved in the Kunjin virus system by providing the structural proteins in trans by transient expression from an alphavirus replicon (21). However, this procedure yielded considerably lower VLP titers than can be achieved in packaging of alphavirus replicons. Additional limitations were imposed by the fact that this packaging protocol required two successive transformations of the producing cells and by the cytopathogenicity of the alphavirus replicons that were used to supply the structural proteins in trans. The availability of a packaging cell line could significantly facilitate the production and enhance the yields of flaviviral VLPs, but to date no such cell line has been described.
TBE virus is a well-characterized member of the genus Flavivirus. Specific mutagenesis of the TBE virus genome by means of an infectious cDNA clone has been performed in several studies during past years (7, 23, 31), but no replicons of this virus have so far been generated. RSPs of TBE virus derived by transient expression have been extensively characterized with respect to their structural, functional, and immunochemical properties (1, 8, 11, 39), but as yet there is no stable cell line for the production of such particles.
Here we report on the generation of a stable CHO cell line (designated CHO-ME) that constitutively produces and releases mature TBE virus RSPs. Furthermore, the construction and characterization of subgenomic TBE virus replicons lacking the coding sequence for essentially all of the structural proteins (NdΔCME) or lacking the coding sequence for only the surface proteins prM/M and E (NdΔME) are described. We provide evidence that CHO-ME cells are able to specifically encapsidate replicon NdΔME into single-round infectious VLPs and thus can be used as a packaging cell line for TBE virus replicons.
MATERIALS AND METHODS
Virus.
The wild-type virus in all of the experiments was Western subtype TBE virus strain Neudoerfl, which has been characterized in detail, including the determination of its genomic sequence (GenBank accession no. U27495) (33, 34). Purified virions and RSPs used in control experiments were prepared according to established protocols (12, 39).
Cloning, sequencing, and RNA transcription.
The desired deletions were introduced into the genome of TBE virus strain Neudoerfl with the infectious cDNA clone system described in detail elsewhere (32). The mutations were first introduced into plasmid pTNd/5′, which carries cDNA corresponding approximately to the 5′-terminal one third of the TBE virus genome.
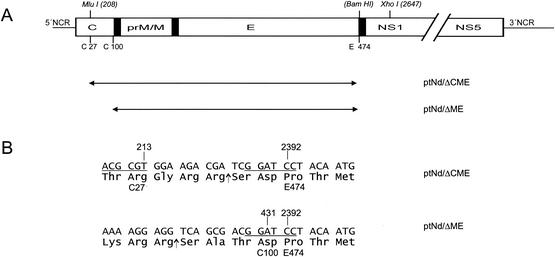
To construct plasmid pTNd/5′-ΔCME, a PCR fragment obtained with primers P-ΔCME1 and P-ΔCME2 (Table 1) was generated. Primer P-ΔCME1 was designed to introduce a deletion spanning from genome positions 213 to 2392 and to add the coding sequence for an artificial NS2B/3 protease cleavage site and a new BamHI site in place of the deleted sequence (Fig. 1). Taking advantage of the unique recognition sites for the restriction enzymes MluI and XhoI at genome positions 208 and 2647, respectively, the wild-type sequence was replaced with the PCR fragment. Plasmid pTNd/5′-ΔME is a derivative of pTNd/5′-ΔCME and was constructed by adding back all of the coding sequence for capsid protein C except for its 16 carboxy-terminal codons.
TABLE 1.
Primers used for the construction of cDNA clones
| Primer | Orientation | Sequence, 5′-3′a | Positionb |
|---|---|---|---|
| P-ΔCME1 | Sense | CCGACGCGTGGAAGACGATCGGATCCTACAATGTCCATGAGCTTTCT | 2392-2414 |
| P-ΔCME2 | Antisense | GTGACCGAGCTTCTCCACAT | 2659-2678 |
| P-ΔME1 | Sense | ACGTGTTGAGAAAAAGACAG | 91-110 |
| P-ΔME2 | Antisense | ATCGGATCCGTCGCTGACCTCCTTTTC | 411-431 |
Recognition sites for restriction enzymes are underlined (MluI, ACGCGT; BamHI, GGATCC). Additional nucleotides introduced for plasmid constructions are in italics.
Position numbers of matching nucleotides correspond to the wild-type virus genome sequence (GenBank accession no. U27495).
FIG. 1.
Structure of TBE virus replicons. (A) Schematic of the TBE virus genome (top) and the locations of the introduced deletions (below, double-headed arrows together with the corresponding plasmid designations). The open reading frame is represented by an open box, with only the structural protein coding region drawn to scale. Internal signal sequences are shown as black bars. Positions of cleavage sites of restriction enzymes used during cloning (nucleotide numbers refer to the wild-type genome of TBE virus) and amino acid residues flanking the deletions (numbering starts with the first amino acid residue of protein C or protein E) are indicated above and below the schematic, respectively. NCR, noncoding region. (B) Sequence details of the junction regions. Nucleotide and amino acid residues of the wild-type TBE virus sequence flanking the deletions are marked above and below the sequences, respectively. Recognition sites for restriction enzymes are underlined (MluI, ACG CGT; BamHI, GGA TCC). In the case of plasmid pTNdΔCME, 15 nucleotides coding for an artificial NS2B/3 protease cleavage site and creating the BamHI restriction site were introduced. The insertion of a T residue in plasmid pTNdΔME created a BamHI cleavage site (underlined) and maintained the authentic residue 100 (Asp) of protein C. Arrows depict the sites at which the amino acid sequences are presumed to be cleaved by the viral protease NS2B/3.
With primers P-ΔME1 and P-ΔME2 (Table 1), a PCR fragment harboring recognition sites for the restriction enzymes MluI (position 208) and BamHI (artificially introduced at the carboxy-terminal end of the deletion; see above) was generated and introduced into pTNd/5′-ΔCME DNA, resulting in a deletion spanning from genome positions 431 to 2392 and the introduction of a single nucleotide (T) which was necessary to maintain the correct BamHI site and reading frame (Fig. 1). These mutations were introduced into the full-length cDNA clone pTNd/c by swapping a fragment generated by cleavage at unique restriction sites for the enzymes SalI (site located upstream of the TBE virus 5′ end) and ClaI (position 3155), generating the final plasmids pTNd/ΔCME and pTNd/ΔME, respectively.
All plasmids were amplified in Escherichia coli HB101 and purified with commercially available systems (Qiagen). Sequence analysis of all PCR-derived fragments and surrounding recognition sites for restriction enzymes was performed with an automated DNA sequencing system (ABI) and confirmed the presence of only the desired mutations.
In vitro transcription of genomic RNA was performed with a commercially available system (Ambion) as reported previously (32).
Cell cultures.
CHO-DG44 cells (44) were grown in Ham's F12 medium supplemented with 10% fetal calf serum. CHO-ME cells were grown in selective medium (minimal essential medium alpha supplemented with 10% dialyzed fetal calf serum). Vero cells were grown in Dulbecco's modified Eagle's medium with 4500 mg of glucose per liter and 5% fetal calf serum. BHK-21 cells were grown in minimal essential medium supplemented with 5% fetal calf serum.
Generation of cells continuously expressing TBE virus surface proteins.
The selection system employed in this experiment was the dihydrofolate reductase (DHFR) system described in detail elsewhere (18). Briefly, this system employs the cotransfection of DHFR-deficient CHO-DG44 cells with plasmid pSV2-dhfr (ATCC 37146), from which functional DHFR is expressed, and an expression plasmid harboring the desired gene(s). Plasmid pCMV-prM+E was constructed from the original expression plasmid SV-PEwt (2) as described in a previous study and contains the entire prM/E coding sequence of TBE virus strain Neudoerfl under the control of the cytomegalovirus immediate-early promoter-enhancer (1). Subconfluent CHO-DG44 cells were harvested from 75-cm2 tissue culture flasks, and approximately 107 cells were transfected with 10 μg of plasmid pCMV-prM+E (linearized with EcoRI) and 0.5 μg of pSV2-dhfr (linearized with PvuI) by electroporation with a Bio-Rad Gene Pulser (two subsequent pulses; setup values: 1.5 kV, 25 μF, and resistance [Ω] set to infinite). Transfected cells were grown for 24 h in Ham′s F12 medium, transferred to petri dishes (13.5-cm diameter; Nunclon), and selected for a DHFR+ phenotype for 14 days in selective medium (minimal essential medium alpha supplemented with 10% dialyzed fetal calf serum).
Single-cell clones were picked with sterile toothpicks and transferred to 2-cm2 wells for further propagation. Protein E-expressing cultures were identified by testing aliquots of the supernatants in a four-layer enzyme-linked immunosorbent assay (ELISA) (14). Two subcloning steps at limiting dilution conditions were performed to produce a clonal cell line, designated CHO-ME, with 100% of the cells expressing protein E, as determined by an immunofluorescence assay. For quantitation of protein E expression, 2 × 105 cells were seeded into 9.6-cm2 wells and grown to confluent monolayers corresponding to approximately 106 cells per well. The amount of protein E released into the supernatant was determined with a quantitative four-layer sodium dodecyl sulfate (SDS)-ELISA (13) 72 h after seeding of the cells.
Purification and analysis of particles released from CHO-ME cells.
To harvest subviral particles from CHO-ME cells, the culture medium of a subconfluent cell layer was replaced with serum-free Ultra-CHO medium (Bio-Whittaker), and cells were cultivated further. After 48 h the culture fluid was removed and cleared from insoluble material by low-speed centrifugation. Particles were precipitated with 10% polyethylene glycol (molecular weight 8,000) with protocols described elsewhere (8). To determine the buoyant density of recombinant subviral particles, the concentrate was subjected to rate zonal centrifugation (5 to 20% sucrose, Beckman SW40 rotor, 38,000 rpm, 4°C, 90 min) and a subsequent equilibrium density gradient centrifugation (20 to 50% sucrose, Beckman SW40 rotor, 38,000 rpm, 4°C, 15 h). All gradients were fractionated with an ISCO 640 gradient fractionator, and the concentration of protein E and hemagglutination (HA) activity in each fraction was determined. HA activity was determined with goose erythrocytes at pH 6.4 by the method of Clarke and Cassals (6). The sucrose density of the peak fraction of the equilibrium density gradient was measured in an Abbé refractometer (Atago) with corrections for temperature made with standard tables (ISCO).
The antigenic structure of RSPs was analyzed with a set of monoclonal antibodies as in previous studies (14, 39). For gel electrophoresis, RSPs were precipitated from the corresponding equilibrium density gradient fractions with deoxycholate and trichloroacetic acid and fractionated under denaturing conditions on 15% polyacrylamide (27). Protein bands were visualized with Coomassie PhastGel Blue R (Pharmacia) or by immunoblotting onto a polyvinylidene difluoride membrane with a Bio-Rad Trans-blot semidry transfer cell and immunoenzymatic detection as described previously (39).
RNA transfection, passaging, and infectivity titration experiments.
In vitro-transcribed RNA was introduced into cells by electroporation with a Bio-Rad Gene Pulser (BHK-21 cells: two subsequent pulses; setup values: 1.8 kV, 25 μF, and 200 Ω, CHO-DG44 and CHO-ME cells: one pulse at 0.45 kV, 960 μF, and resistance [Ω] set at infinite) as described in detail in a previous study (32). For infection studies, 200-μl aliquots of cleared supernatants were transferred to fresh cells in 24-well cluster plates. Expression of protein E and protein NS1 was visualized 72 h after transfection or infection by immunochemical staining as described below. For the quantitative determination of VLP titers, replicon RNA was introduced into CHO-ME cells by electroporation, and then 2 × 105 cells were seeded into a 2-cm2 surface area well of a 24-well cluster plate or 2 × 107 cells were seeded into a tissue culture flask with an enlarged surface area of 225 cm2 (Iwaki) and 1 ml or 35 ml of medium was added, respectively. Supernatants harvested 72 h after transfection were cleared, sequentially diluted in 0.5 log10 steps, and 200-μl aliquots of this dilution series were used to infect Vero cells grown in 24-well tissue culture plates. Cells were analyzed for NS1 expression by immunofluorescence 96 h later. Titers were determined by counting the number of positive cells in the last three positive wells and calculating an average.
Immunochemical staining.
To visualize intracellular expression of protein E or NS1, cells were fixed with acetone-methanol (1:1) and dried. After rehydration with phosphate-buffered saline, the monolayers were treated with a polyclonal rabbit anti-protein E serum or a monoclonal antibody directed against NS1 (17). Antibody-antigen reactions were visualized with fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin (Jackson Immune Research Laboratory) or anti-mouse immunoglobulin (Sigma) antibody.
Characterization of single-round infectious particles.
Supernatants of transfected CHO-ME cells were cleared from insoluble material by low-speed centrifugation, concentrated with polyethylene glycol as described above, and resolved on a 10 to 50% sucrose gradient (Beckman SW40 rotor, 38,000 rpm, 4°C, 180 min). Fractions of 0.6 ml were collected and analyzed for protein E by SDS-ELISA (13), HA activity (6), and amount of virus or replicon RNA by reverse transcription (RT)-PCR (see below).
Semiquantitative RT-PCR.
For RT-PCR analysis, RNA was extracted with a QIAamp viral RNA kit (Qiagen, Hilden, Germany) and reverse transcribed with 200 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Lofer, Austria) after addition of 50 ng of random primers (PE Applied Biosystems, Vienna, Austria) and 20 U of RNase inhibitor (Roche, Vienna, Austria). The reaction mixture was incubated at 42°C for 25 min and than heated to 99°C for 5 min. Then a real-time TaqMan PCR (10) was performed with the TaqMan universal PCR master mix (PE Applied Biosystems), and 25 pmol of both forward and reverse primers and 10 pmol of TaqMan probe. The sequences of the primers and probes were specific for a region of the E gene or the NS5 gene (Eforward, 5′-TCA GGG GAC TAC GAG GGT CA-3′; Ereverse, 5′-CGT CAA GCC ACA CAT CCA TT-3′; ETaqMan probe, 5′-FAM-CAT CCA CCC AGT TCC AGC ACC AAG-TAMRA-3′; NS5forward, 5′-GAA GCG GAG GCT GAA CAA CT-3′; NS5reverse, 5′-TTG TCA CGT TCC GTC TCC AG-3′; NS5TaqMan probe, 5′-FAM-CAT CCA CCC AGT TCC AGC ACC AAG-TAMRA-3′). Amplification and detection were done on the i-cycler iQ (Bio-Rad Laboratories, Hercules, Calif.) under the following conditions: 10 min at 95°C and 45 cycles of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Results were evaluated with a standard curve prepared from a log10 dilution series of a purified preparation of TBE virus.
RESULTS
Construction of subgenomic TBE virus replicons.
Replicons of TBE virus were constructed by introducing two different deletions into plasmid pTNd/c, which harbors full-length cDNA of prototypic strain Neudoerfl. In one case, essentially all of the coding sequence for proteins prM and E was deleted, yielding plasmid pTNd/ΔME, and in another construct, the coding sequences for proteins prM, E, and most of protein C were removed (plasmid pTNd/ΔCME). Specific details of the resulting constructs are illustrated in Fig. 1. To ensure correct membrane integration of the primary translation product of the corresponding RNAs, the last 23 amino acid residues of protein E, which function as an internal signal sequence for the subsequent protein NS1 (29), were retained in both constructs. In plasmid pTNd/ΔME, this sequence was amino-terminally fused to the protein C coding region lacking only the carboxy-terminal 16 codons specifying the hydrophobic internal signal sequence for protein prM. Notably, the authentic cleavage site recognized by the viral protease NS2B/3, which in a natural infection cleaves mature protein C from this carboxy-terminal hydrophobic domain (29), was preserved in this construct. In plasmid pTNd/ΔCME, only the first 27 codons of the protein C coding sequence were retained. In addition, an artificial NS2B/3 cleavage site was engineered between this short fragment of protein C and the signal sequence for protein NS1.
To obtain evidence for the ability of these constructs to replicate and translate viral proteins in cells, the corresponding in vitro-synthesized RNAs (NdΔME and NdΔCME) were introduced into BHK-21 cells by electroporation, and protein expression was monitored by immunofluorescence with a specific monoclonal antibody against protein NS1. Expression of protein NS1 was readily detected 24, 48, and 72 h posttransfection in both deletion mutants and the full-length wild-type RNA control (data from the 72-h time point are shown in Fig. 2). No significant cytopathic effect was observed in these cells even 72 h posttransfection. A replication-deficient negative control RNA, NdΔNS5, carrying a major deletion in the coding region of the viral polymerase NS5 did not express detectable amounts of protein NS1.
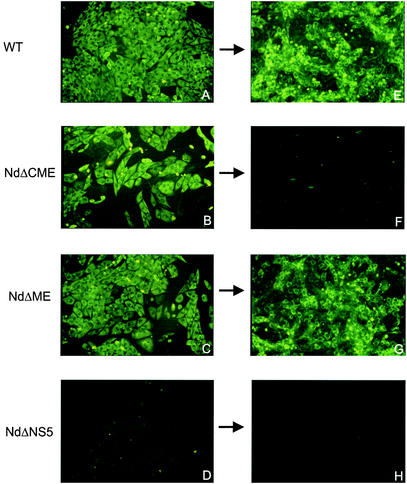
FIG. 2.
Protein expression (left column) and attempted cell culture passage (right column) of TBE virus replicons. Wild-type (WT, A), NdΔCME (B), NdΔME (C), and replication-deficient mutant NdΔNS5 (D) RNAs were synthesized in vitro and transfected into BHK-21 cells. Expression of protein NS1 was tested 72 h posttransfection by immunofluorescence. (E to H) Supernatants were passaged (as indicated by arrows) onto fresh BHK-21 cell cultures, and the expression of protein NS1 was determined 72 h postinoculation by immunofluorescence.
To confirm that the deletions introduced in the structural region of the TBE virus genome abolished the ability to produce infectious particles, supernatants of BHK-21 cells transfected with either NdΔCME, NdΔME, NdΔNS5, or wild type virus RNA were cleared from insoluble material by low-speed centrifugation and transferred to fresh BHK-21 cells. Immunofluorescence analysis revealed no detectable NS1 expression in these new cells in the cases of NdΔCME, NdΔME, and NdΔNS5 (Fig. 2). As expected, the wild-type virus control efficiently infected the new cell cultures. These results indicated that NdΔME and NdΔCME represented functional TBE virus replicons capable of replication and protein expression but lacking the ability to form infectious particles.
Establishment of a cell line constitutively expressing mature TBE viral RSPs.
For the generation of a cell line constitutively expressing the surface proteins of TBE virus, we took advantage of the established CHO-DG44/DHFR selection system (18). This approach depended on the cointegration of two expression plasmids, plasmid pSV2-dhfr, encoding DHFR, and plasmid pCMV-prM+E, encoding the TBE virus envelope proteins prM/M and E, into the genome of CHO-DG44 cells, which are deficient for the enzyme DHFR. Two weeks after transfection of the two plasmids into CHO-DG44 cells, 36 viable cell colonies were isolated and cultivated further. As revealed by immunochemical methods, 11 of these 36 cell cultures expressed protein E, but none of the cultures contained more than 70% protein E-expressing cells. The amount of protein E released into the cell culture supernatants was quantified by SDS-ELISA, and those cells showing the highest expression level were subjected to two rounds of limiting dilution and isolation of single-cell-derived cell colonies. As summarized in Fig. 3, this procedure allowed the expression level to be increased from 0.6 μg initially to 1.5 μg of protein E released from approximately 106 cells and generated a cell line that consisted exclusively of protein E-expressing cells (Fig. 4A). This cell line, designated CHO-ME, was shown to maintain high-level expression of protein E for at least 15 additional passages.
FIG. 3.
Monitoring of protein E release (A) and proportion of protein E-producing cells (B) during the derivation of CHO-ME cells. Protein E was quantified 72 h postseeding of cells by SDS-ELISA. Protein E-producing cells were identified by immunofluorescence staining. 1, cells derived from a single-cell colony after transfection of CHO-DG44 cells with the expression plasmids; 2, cells after the first subcloning step; 3, CHO-ME cells as derived after a second subcloning step.
FIG. 4.
Characterization of CHO-ME cells and released particles. (A) Cell morphology and protein E expression of low-passage cells (left) and after 15 passages (right) as examined by immunofluorescence. (B) Determination of buoyant density of secreted particles. Particles were precipitated from supernatants of CHO-ME cells purified by rate zonal centrifugation and then subjected to sucrose density gradient centrifugation. Individual fractions were tested for their protein E content by SDS-ELISA (open circles) and the presence of hemagglutination activity (open boxes). The buoyant density, as determined for the peak fraction, is indicated above the graph. (C) Protein analysis of purified particles by fractionation on a 15% denaturing polyacrylamide gel. Proteins were visualized by Coomassie blue staining (left) or immunoblot analysis (right). 1, CHO-ME derived particles; 2, wild-type virus. The positions of viral structural proteins are indicated on the right.
The physical form in which viral proteins were released from CHO-ME cells was analyzed and compared with the established properties of TBE viral RSPs produced in COS-1 cells by transient expression. CHO-ME cells were grown for 48 h in serum-free medium, and particles were harvested from the supernatants by polyethylene glycol precipitation and purified by rate zonal centrifugation. These particles were then subjected to equilibrium density gradient centrifugation, and individual fractions were assayed for the presence of protein E by quantitative SDS-ELISA and for hemagglutination (HA) activity (Fig. 4B), which is a characteristic of mature flavivirus particles and subviral particles (39, 43). The buoyant density derived from this analysis (1.14 g/cm3) was exactly the same as that determined previously for RSPs produced in COS-1 cells (39). Furthermore, purified particles derived from CHO-ME cells were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were visualized by staining with Coomassie blue or Western blotting (Fig. 4C). This analysis revealed that these particles contained proteins M and E but provided no evidence for the presence of uncleaved precursor protein prM.
Analysis of the antigenic structure by means of a panel of monoclonal antibodies is an established method to check the structural integrity of TBE viral and subviral particles and to distinguish between mature and immature (prM-containing) particles (13, 39). The reaction profile obtained for particles purified from CHO-ME cells was found to be identical to the one observed with mature RSPs prepared from COS-1 cells (data not shown). In summary, all of these results indicated that CHO-ME cells released mature RSPs that were indistinguishable from previously characterized RSPs prepared by transient expression.
Production of single-round infectious particles by trans-complementation.
To address the question of whether CHO-ME cells were able to package TBE virus replicons, the in vitro-synthesized replicon RNAs NdΔCME and NdΔME were introduced into these cells by electroporation. Full-length wild-type RNA and NdΔNS5 were transfected in parallel as positive and negative controls, respectively. Expression of protein NS1 was detected in approximately 60% of the cells transfected with either NdΔCME, NdΔME, or the wild-type control 24 h posttransfection (data not shown). In the cases of NdΔME and the wild-type control, the percentage of NS1-expressing cells determined at 72 h posttransfection (shown in Fig. 5) had increased to approximately 90% and 100%, respectively, whereas no increase in the number of expressing cells was observed for replicon NdΔCME.
FIG. 5.
Formation of infectious particles in CHO-ME cells transfected with in vitro-synthesized wild-type (A), NdΔCME (B), NdΔME (C), and NdΔNS5 (D) RNAs. Expression of protein NS1 was visualized by immunofluorescence 72 h posttransfection. (E to H) Supernatants were passaged onto fresh Vero cell cultures as indicated by the arrows, and the expression of protein NS1 was determined 96 h postinoculation.
Supernatants from the transfected CHO-ME cells were cleared by low-speed centrifugation from insoluble material and transferred to fresh Vero cells to test for the presence of infectious particles. As shown in Fig. 5, only supernatants from CHO-ME cells transfected with either wild-type virus RNA or NdΔME RNA were able to initiate a further round of infection in these cells. No infectivity was detected in the case of NdΔCME. A second passage in which supernatants from the infected Vero cells were transferred to fresh Vero cells resulted in infection only in the case of the wild-type control but not NdΔME (data not shown), demonstrating that the particles produced by the CHO-ME cells transfected with the replicon had been capable only of initiating a single round of infection. Identical results were obtained when BHK-21 cells or CHO-DG44 cells were used instead of Vero cells in equivalent passaging experiments (not shown).
On CHO-ME cells, however, the NdΔME-derived infectious particles could be passaged repeatedly. Examination of the nucleotide sequence of NdΔME after five passages on CHO-ME cells by RT-PCR sequence analysis revealed no changes from the original replicon sequence. If aliquots from any of these passages were used to infect cells other than CHO-ME cells (Vero, CHO-DG44, or BHK-21), they consistently initiated only a single round of infection and could not be propagated any further. Taken together, these experiments demonstrated that CHO-ME cells were capable of complementing NdΔME replicons, resulting in the formation of single-round infectious particles. There was no evidence for spontaneous mutations or the regeneration of infectious virus in any of these cell culture assay systems even after several passages in the CHO-ME packaging cell line.
Infectivity titers of supernatants of CHO-ME cells transfected with NdΔME were determined by limiting dilution (in 0.5 log10 steps) infection experiments on Vero cells. This typically yielded a titer of 5 × 106 IU/ml if supernatants from cells grown in 24-well tissue culture plates were analyzed. If cells were seeded into tissue culture flasks with an increased surface-to-volume ratio (225 cm2 surface area, 35 ml of growth medium), supernatants containing up to 5 × 107 IU/ml were obtained. Infectivity titers could not be increased further by subsequent passages in CHO-ME cells.
The physical structure of the particles released from CHO-ME cells after transfection with NdΔME replicon RNA was analyzed by centrifugation through 10 to 50% sucrose gradients (Fig. 6). Different from the result obtained with untransfected CHO-ME cells, which released only particles corresponding to mature RSPs (see above), this sedimentation analysis revealed, in addition to RSPs, the presence of a second particle species that banded almost at the same position (maximum in fraction 13) as a purified wild-type virus control that was analyzed in a parallel gradient (maximum in fraction 14). To test for the presence of replicon RNA, all of the fractions were analyzed by semiquantitative PCR with primers that were specific for a region within the NS5 gene. This analysis indicated that RNA was significantly associated with the second (virus-like) particle species with approximately 50% of the total RNA detected in the peak fraction 13 (Fig. 6). Control PCRs performed with primers recognizing a region of the E gene were negative in all fractions, indicating that no detectable prM-E mRNA was packaged into any of the particles.
FIG. 6.
Sedimentation analysis of particles harvested from supernatants of CHO-ME cells transfected with in vitro-synthesized NdΔME replicon RNA. Particles were precipitated with polyethylene glycol and applied to a 10 to 50% sucrose gradient. The protein E concentration of individual fractions (open circles) was determined by SDS-ELISA. The presence of replicon RNA in individual fractions was assessed by a semiquantitative RT-PCR and is shown as a percentage of the total from all of the fractions (open diamonds). Control gradients loaded with purified preparations of RSPs and wild-type TBE virus were analyzed in parallel (peak fractions indicated by arrows).
In summary, the results suggest that CHO-ME cells are able to specifically package NdΔME RNA into VLPs that closely resemble wild-type TBE virus particles. The small difference (one fraction) in sedimentation behavior may simply reflect the somewhat lower density of VLPs caused by the fact that NdΔME RNA is approximately 20% smaller than the wild-type RNA and does not necessarily suggest a relevant structural difference between VLPs and wild-type virions.
DISCUSSION
This report presents a novel flavivirus encapsidation system consisting of a replicon of TBE virus that lacks the coding region for the two surface proteins prM and E (NdΔME) and stably transformed CHO cells providing these two proteins in trans (CHO-ME) that can be used as a packaging cell line. Several studies have demonstrated the suitability of flavivirus replicons as vectors for heterologous gene expression (22, 36, 41). The first flavivirus replicons were derived from Kunjin virus and have subsequently been developed into a well-characterized expression system for which a packaging protocol based on transient expression of the structural proteins has also been developed (21, 47). Although the currently available flavivirus vectors do not match the efficiency and utility of the widely used alphavirus systems, they nevertheless seem to exhibit certain features, such as their natural noncytopathogenicity, which enables a longer-lasting expression of the heterologous genes, and the apparent lack of spontaneous recombination events during packaging that might make them especially useful for some applications (19, 40). Drawbacks of the Kunjin flavivirus system have been that the transient packaging system yielded a relatively low titer (1.3 × 106 VLPs/ml) (21) compared to other systems, e.g., alphaviruses (19) and the unavailability of a convenient stable packaging cell line.
This work now for the first time describes a packaging cell line that represents a convenient encapsidation tool for a TBE virus-derived replicon. The VLP titers achieved with this system amounted to 5 × 106 VLPs/ml when the cells were seeded into 24-well tissue-culture plates but a titer of 5 × 107 VLPs/ml could be achieved when cells were seeded into tissue-culture flasks with an increased surface-to-volume ratio. This value still remains well below the maximum titers that can be achieved with alphavirus vectors (108 to 109 VLPs/ml) (5, 28). However, it approaches the level obtained with three-component alphavirus systems or alphavirus packaging cell lines designed to overcome the inherent problem of regeneration of infectious particles, which typically yield titers in the range of 107 to 108 VLPs/ml (9, 37, 38, 42). A comparison between the Kunjin and the TBE virus packaging systems based on the number of originally transfected cells indicates that the former yielded 1 × 107 VLPs per 2 × 106 cells whereas the TBE virus system, depending on the type of growth vessel used, gave between 5 × 107 and 2 × 108 VLPs per 2 × 106 cells. Additional work will certainly be necessary to further increase the yields of flavivirus packaging systems, but the CHO-ME cells investigated in this study represent an important step towards a more practical and efficient system.
Our experiments also confirmed the above-mentioned advantages of flavivirus systems, i.e., the replicons appeared to exert no significant cytopathic effect on their host cells, and we observed no spontaneous regeneration of infectious virus. In the Kunjin virus system, the possibility of unwanted recombination events was reduced by the use of an expression system (an alphavirus replicon) for the three structural proteins (C, prM/M and E) that was unrelated to the complemented virus, and in addition, the genes were arranged in the expression cassette in an order different from that of the native Kunjin virus sequence (21). In the TBE virus system, the proteins prM/M and E were translated from a mRNA produced by the packaging cells, whereas the capsid protein C was encoded by the replicon itself. Although the prM/E mRNA and replicon NdΔME contained overlapping elements of the viral genome that had the potential to promote homologous recombination, no generation of viral infectivity was observed in any of our experiments, even in the course of repeated passages of VLPs in CHO-ME cells. Genetic stability represents an important safety feature for practical applications, and future work will have to address this issue in further detail, with more sensitive host systems such as suckling mice and prolonged passaging conditions. In this context it is also relevant to note that PCR analysis did not provide any evidence for the packaging of the prM/E mRNA into VLPs or RSPs.
The VLPs produced in the Kunjin virus system were found to be smaller than native virus particles and to have a different sedimentation behavior (21). In contrast, sedimentation analysis of the TBE virus VLPs suggested that they closely resembled infectious virions and were structurally distinct from the smaller RSPs. Importantly, PCR testing of the fractions obtained from the gradient analysis revealed that replicon RNA was predominantly associated with VLPs, whereas no significant packaging of replicon RNA into smaller, capsidless RSPs was observed. The requirement of capsid protein C for packaging of replicon RNA was further demonstrated by the fact that no infectivity was generated after transfection of CHO-ME cells with replicon NdΔCME, which is identical to replicon NdΔME except for the lack of the majority of the capsid coding region. In summary, all of our results are consistent with the view that replicon NdΔME was specifically packaged into capsid-containing single-round infectious particles that closely resemble natural infectious virions.
Another relevant aspect of this work, in addition to replicon encapsidation, concerns the potential use of CHO-ME cells as a source of mature TBE virus RSPs. RSPs are valuable tools for structural and functional studies (8, 39) and may also be used for diagnostic and prophylactic applications as they are known to be excellent immunogens (11). RSP-producing cell lines have so far been reported for two other flaviviruses, Japanese encephalitis virus and dengue virus 2 (16, 24, 25). However, in the cases of these two viruses, it was difficult to obtain stable cell lines, apparently due to a severe cytotoxicity of the gene products, probably caused by the fusogenicity of mature flavivirus RSPs. In two studies, this problem was overcome by suppressing the fusogenic activity by specific mutagenesis of the furin cleavage site of protein prM, resulting in the production of immature, prM-containing RSPs (24, 25). With this modification, it was possible to harvest approximately 1 μg of protein E per 107 cells for at least five passages in the case of the Japanese encephalitis virus cell line and 40 ng of protein E per 107 cells for at least 25 passages in the case of the dengue virus 2 cell line. Interestingly, in the latter case it was also shown that coexpression of the antiapoptotic bcl-2 gene increased the yield of protein E to 75 ng per 107 cells.
In another case, the generation of a cell line stably producing mature RSPs of Japanese encephalitis virus was reported, but the level of expression and the genetic stability and viability of this cell line were not characterized in detail (16). We did not encounter a relevant cytotoxicity problem with the mature TBE virus surface proteins upon construction of the CHO-ME cells, and our data demonstrate that the expressed proteins retained their structural and functional integrity. The protein E expression level from CHO-ME cells (15 μg of protein E per 107 cells) was significantly higher than it had been observed for the Japanese encephalitis virus or dengue virus cell lines but still remained below the levels obtained from wild-type TBE virus infected cells or from transient expression of TBE virus RSPs, which typically range from 20 to 40 μg of protein E per 107 cells (3, 23).
CHO-ME cells will be useful both for the production of mature TBE virus RSPs and for the encapsidation of TBE virus replicons into single-round infectious VLPs in the course of the ongoing development of TBE virus-based expression vectors.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Melby Wilfinger, Angela Dohnal, and Claudia Kellner, and we thank Agnes Leitner for help with plasmid construction. We are very thankful to L. Chasin for providing CHO-DG44 cells and to C. S. Schmaljohn for the monoclonal anti-NS1 antibody.
This work was supported in part by the Austrian “Fonds zur Förderung der wissenschaftlichen Forschung,” FWF project number P16376-B04.
REFERENCES
- 1.Aberle, J. H., S. W. Aberle, S. L. Allison, K. Stiasny, M. Ecker, C. W. Mandl, R. Berger, and F. X. Heinz. 1999. A DNA immunization model study with constructs expressing the tick-borne encephalitis virus envelope protein E in different physical forms. J. Immunol. 163:6756-6761. [PubMed] [Google Scholar]
- 2.Allison, S. L., C. W. Mandl, C. Kunz, and F. X. Heinz. 1994. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes 8:187-198. [DOI] [PubMed] [Google Scholar]
- 3.Allison, S. L., K. Stadler, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, D. H. A. J. C. 1958. Techniques for hemagglutination and hemagglutination inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561-573. [DOI] [PubMed] [Google Scholar]
- 7.Elshuber, S., S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J. Gen. Virol. 84:183-191. [DOI] [PubMed] [Google Scholar]
- 8.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 9.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 11.Heinz, F. X., S. L. Allison, K. Stiasny, J. Schalich, H. Holzmann, C. W. Mandl, and C. Kunz. 1995. Recombinant and virion-derived soluble and particulate immunogens for vaccination against tick-borne encephalitis. Vaccine 13:1636-1642. [DOI] [PubMed] [Google Scholar]
- 12.Heinz, F. X., and C. Kunz. 1981. Homogeneity of the structural glycoprotein from European isolates of tick-borne encephalitis virus: comparison with other flaviviruses. J. Gen. Virol. 57:263-274. [DOI] [PubMed] [Google Scholar]
- 13.Heinz, F. X., K. Stiasny, G. Püschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 14.Heinz, F. X., W. Tuma, F. Guirakhoo, and C. Kunz. 1986. A model study of the use of monoclonal antibodies in capture enzyme immunoassays for antigen quantification exploiting the epitope map of tick-borne encephalitis virus. J Biol. Stand. 14:133-141. [DOI] [PubMed] [Google Scholar]
- 15.Hewson, R. 2000. RNA viruses: emerging vectors for vaccination and gene therapy. Mol. Med. Today 6:28-35. [DOI] [PubMed] [Google Scholar]
- 16.Hunt, A. R., C. B. Cropp, and G. J. Chang. 2001. A recombinant particulate antigen of Japanese encephalitis virus produced in stably transformed cells is an effective noninfectious antigen and subunit immunogen. J. Virol. Methods 97:133-149. [DOI] [PubMed] [Google Scholar]
- 17.Iacono-Connors, L. C., J. F. Smith, T. G. Ksiazek, C. L. Kelley, and C. S. Schmaljohn. 1996. Characterization of Langat virus antigenic determinants defined by monoclonal antibodies to E, NS1 and preM and identification of a protective, non-neutralizing preM-specific monoclonal antibody. Virus Res. 43:125-136. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman, R. J. 1990. Selection and coamplification of heterologous genes in mammalian cells. Methods Enzymol. 185:537-566. [DOI] [PubMed] [Google Scholar]
- 19.Khromykh, A. A. 2000. Replicon-based vectors of positive strand RNA viruses. Curr. Opin. Mol. Ther. 2:555-569. [PubMed] [Google Scholar]
- 20.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konishi, E., and A. Fujii. 2002. Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 20:1058-1067. [DOI] [PubMed] [Google Scholar]
- 25.Konishi, E., A. Fujii, and P. W. Mason. 2001. Generation and characterization of a mammalian cell line continuously expressing Japanese encephalitis virus subviral particles. J. Virol. 75:2204-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konishi, E., S. Pincus, E. Paoletti, R. E. Shope, T. Burrage, and P. W. Mason. 1992. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology 188:714-720. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 28.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology (N.Y.) 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 29.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, et al (ed.), Field's virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 30.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 31.Mandl, C. W., S. L. Allison, H. Holzmann, T. Meixner, and F. X. Heinz. 2000. Attenuation of tick-borne encephalitis virus by structure-based site-specific mutagenesis of a putative flavivirus receptor binding site. J. Virol. 74:9601-9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandl, C. W., M. Ecker, H. Holzmann, C. Kunz, and F. X. Heinz. 1997. Infectious cDNA clones of tick-borne encephalitis virus European subtype prototypic strain Neudoerfl and high virulence strain Hypr. J. Gen. Virol. 78:1049-1057. [DOI] [PubMed] [Google Scholar]
- 33.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166:197-205. [DOI] [PubMed] [Google Scholar]
- 34.Mandl, C. W., F. X. Heinz, E. Stockl, and C. Kunz. 1989. Genome sequence of tick-borne encephalitis virus (Western subtype) and comparative analysis of nonstructural proteins with other flaviviruses. Virology 173:291-301. [DOI] [PubMed] [Google Scholar]
- 35.Molenkamp, R., E. A. Kooi, M. A. Lucassen, S. Greve, J. C. Thijssen, W. J. Spaan, and P. J. Bredenbeek. 2003. Yellow fever virus replicons as an expression system for hepatitis C virus structural proteins. J. Virol. 77:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang, X., M. Zhang, and A. I. Dayton. 2001. Development of dengue virus replicons expressing HIV-1 gp120 and other heterologous genes: a potential future tool for dual vaccination against dengue virus and HIV. BMC Microbiol. 1:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polo, J. M., B. A. Belli, D. A. Driver, I. Frolov, S. Sherrill, M. J. Hariharan, K. Townsend, S. Perri, S. J. Mento, D. J. Jolly, S. M. Chang, S. Schlesinger, and T. W. Dubensky, Jr. 1999. Stable alphavirus packaging cell lines for Sindbis virus and Semliki Forest virus-derived vectors. Proc. Natl. Acad. Sci. USA 96:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389-401. [DOI] [PubMed] [Google Scholar]
- 39.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlesinger, S. 2000. Alphavirus expression vectors. Adv. Virus Res. 55:565-577. [DOI] [PubMed] [Google Scholar]
- 41.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 42.Smerdou, C., and P. Liljestrom. 1999. Two-helper RNA system for production of recombinant Semliki forest virus particles. J. Virol. 73:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urlaub, G., and L. A. Chasin. 1980. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc. Natl. Acad. Sci. USA 77:4216-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Regenmortel, M. H. V., C. M. Faquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses, p. 859-878. Academic Press, London, England.
- 46.Varnavski, A. N., and A. A. Khromykh. 1999. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology 255:366-375. [DOI] [PubMed] [Google Scholar]
- 47.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]