Abstract
Crimean-Congo hemorrhagic fever (CCHF) virus is a tick-borne member of the genus Nairovirus, family Bunyaviridae. The mature virus glycoproteins, Gn and Gc (previously referred to as G2 and G1), are generated by proteolytic cleavage from precursor proteins. The amino termini of Gn and Gc are immediately preceded by tetrapeptides RRLL and RKPL, respectively, leading to the hypothesis that SKI-1 or related proteases may be involved (A. J. Sanchez, M. J. Vincent, and S. T. Nichol, J. Virol. 76:7263-7275, 2002). In vitro peptide cleavage data show that an RRLL peptide representing the Gn processing site is efficiently cleaved by SKI-1 protease, whereas an RKPL peptide representing the Gc processing site is cleaved at negligible levels. The efficient cleavage of RRLL peptide is consistent with the known recognition sequences of SKI-1, including the sequence determinants involved in the cleavage of the Lassa virus (family Arenaviridae) glycoprotein precursor. These in vitro findings were confirmed by expression of wild-type or mutant CCHF virus glycoproteins in CHO cells engineered to express functional or nonfunctional SKI-1. Gn processing was found to be dependent on functional SKI-1, whereas Gc processing was not. Gn processing occurred in the endoplasmic reticulum-cis Golgi compartments and was dependent on an R at the −4 position within the RRLL recognition motif, consistent with the known cleavage properties of SKI-1. Comparison of SKI-1 cleavage efficiency between peptides representing Lassa virus GP2 and CCHF virus Gn cleavage sites suggests that amino acids flanking the RRLL may modulate the efficiency. The apparent lack of SKI-1 cleavage at the CCHF virus Gc RKPL site indicates that related proteases, other than SKI-1, are likely to be involved in the processing at this site and identical or similar sites utilized in several New World arenaviruses.
Crimean-Congo hemorrhagic fever (CCHF) viruses are members of the genus Nairovirus of the family Bunyaviridae. The nairoviruses are predominantly tick-borne viruses. The two most important serogroups are the CCHF group, which includes CCHF and Hazara viruses, and the Nairobi sheep disease group, which includes Nairobi sheep disease and Dugbe viruses (14). The RNA genome of viruses of the family Bunyaviridae consists of three negative-sense segments, S, M, and L, which minimally encode the virus nucleocapsid, glycoproteins, and L polymerase proteins, respectively (17). The M segment generally encodes two structural glycoproteins, hereafter referred to as Gn and Gc, based on their location relative to the N and C termini of the polyprotein. The pathways involved in the biosynthesis and maturation of CCHF virus glycoproteins have remained largely unresolved. Recently it was demonstrated that during CCHF virus infection, the mature Gn (37-kDa) and Gc (75-kDa) proteins form the predominant structural glycoprotein components of the virus (16). However, virus protein expression studies with virus-infected cells and the virus M segment open reading frame containing plasmids showed that there are additional glycoprotein-specific proteins within the cell and in the extracellular media. Following extensive analyses, a 140-kDa protein was identified as the precursor of Gn (PreGn, previously referred to as PreG2) and an 85-kDa protein as the precursor of Gc (PreGc, previously referred to as PreG1) (16). A 160-kDa protein, which was secreted into the media along with mature Gn and Gc, was also observed (16). Sequence analyses showed that the CCHF virus M-segment-encoded polyprotein contained several novel features: (i) the N-terminal 243 amino acids of the polyprotein represents a mucin-like domain; (ii) the junction of the mucin-like domain and the remainder of the protein is marked by the presence of a typical furin cleavage site (amino acids 244 to 247 are RSKR); (iii) the amino termini of mature Gn and Gc are generated by proteolytic cleavage following amino acid positions 519 and 1040, respectively, after the RRLL and RKPL tetrapeptides; (iv) lack of typical hydrophobic sequences upstream to the amino termini of the mature Gn and Gc; and (v) the full-length glycoprotein is predicted to traverse the membrane multiple times (16).
Viral glycoproteins undergo a proteolytic processing during their biosynthesis and transport through the secretory pathway. In most cases, such activation is necessary for proper assembly and release of the infectious virus. Several previous studies have demonstrated the role of furin in the proteolytic activation of many human viral glycoproteins (5, 6, 9-11, 20, 25, 26). However, a recent study provided evidence for involvement of a novel protease, namely SKI-1 (18), belonging to a pyrolysin group of subtilases, in Lassa virus glycoprotein processing (12). One of the hallmarks of SKI-1-mediated processing is the unique ability of the protease to cleave peptides following a consensus motif, Arg (Arg/Lys) X (hydrophobic) Z, where Z can be preferably Leu, Phe, Lys, or Thr and excludes Val, Pro, Glu, Asp, or Cys (7). Accordingly, Lassa virus glycoprotein processing to yield GP1 and GP2 was shown to occur following the tetrapeptide RRLL (13). Interestingly, similar sequences (RKLL and RTLL) had also been reported to be cleavage sites for Pichinde and Tacaribe virus glycoproteins, respectively (4). A more recent study suggested the presence of possible SKI-1 target sequences at the predicted cleavage site in several Old and New World arenaviruses; these sequences include RRLL in the glycoproteins of Old World arenaviruses, such as Lassa and Mopeia, and RKPL in the glycoprotein of a New World arenavirus, namely Guanarito virus (19). However, it remains to be seen if SKI-1 is involved in the processing of other arenavirus glycoproteins in addition to Lassa virus. Given the presence of identical or similar tetrapeptide sequences at the known or proposed cleavage sites of glycoproteins of CCHF virus and New and Old World arenaviruses, our studies will have broad implications as to the nature of proteases involved in the glycoprotein processing of these viruses.
With the identification of RRLL sequences located N terminal to mature Gn of CCHF virus (16), we initiated an investigation to determine the potential role of SKI-1 in the processing of CCHF virus glycoproteins. A synthetic peptide containing Gn cleavage site (RRLL) sequence was efficiently cleaved by human SKI-1. On the other hand, a peptide containing the Gc cleavage site (RKPL) was a very poor substrate, suggesting a role for SKI-1 only in Gn processing. To support this finding, we have used stable cell lines which express functional or defective SKI-1 and have analyzed the CCHF virus glycoprotein processing profiles. Results obtained from the mutational analysis of the potential SKI-1-dependant processing sites and cleavage of synthetic peptides confirm that SKI-1 is the protease involved in the processing of the CCHF virus Gn protein and that the RRLL tetrapeptide sequence represents the recognition site.
MATERIALS AND METHODS
Peptide synthesis.
Two intramolecularly quenched fluorogenic substrates containing the proposed conserved processing sites of Gn and Gc proteins of CCHF virus were designed for CCHF virus (Matin strain). The amino acids are indicated by single-letter codes as follows, with arrows indicating cleavage sites: Q-CCHFV-A, Abz-S-S-G-S-R-R-L-L ↓ S-E-E-S-Y (NO2)-Ala-NH2; and Q-CCHFV-B, Abz-A-L-V-L-R-K-P-L ↓ F-L-D-S-Y (NO2)-Ala-NH2, where Abz stands for 2-amino benzoic acid (fluorogenic group) and Y(NO2) stands for 3-nitro tyrosine (fluorescence quench group). The synthesis of these peptides was accomplished by using the Fmoc-based solid-phase chemistry protocol as reported earlier (1). The synthesis of the second peptide is accompanied by the formation of a large amount of des-Abz peptide [A-L-V-L-R-K-P-L ↓ F-L-D-S-Y(NO2)-Ala-NH2] and very little of the correct peptide (Q-CCHFV-B). Therefore, most of the in vitro SKI-1 digestions were performed on des-Abz Q-CCHFV-B and Q-CCHFV-A peptides.
Enzyme digestions.
The recombinant human SKI-1 (hSKI-1) enzymatic preparation was identical to that previously reported, and digestions were performed with recombinant hSKI-1 in a solution of 25 mM Tris, 25 mM morpholinepropanesulfonic acid (MES), 2.5 mM CaCl2 (pH 7.4) as described previously (2, 21).
Cells.
SW13 cells were maintained in Dulbecco's modified eagle medium supplemented with 10% fetal calf serum. CHO SRD-12B-derived SKI-1(+) and SKI-1(−) cells were developed in N. Seidah's laboratory. These cells were maintained in specialized media as described by Rawson et al. (15).
Antibodies and reagents.
Hyperimmune mouse ascites fluid (HMAF) for CCHF virus proteins was kindly provided by T. Ksiazek, Centers for Disease Control and Prevention. HMAF for CCHF virus was made by following a previously described protocol (3). Briefly, mice were inoculated (by subcutaneous and intraperitoneal routes) with CCHF virus-infected mouse brain homogenate emulsified with complete Freund's adjuvant. These mice were then injected with Sarcoma TG-180 cells to induce the production of large volumes of ascitic fluids (T. Ksiazek, personal communication). Antipeptide antibodies were generated by Research Genetics Inc., Huntsville, Ala. The antipeptide antibody used in this study was raised in rabbits against keyhole limpet hemocyanin-conjugated peptide sequences present in the mature Gn (amino acids 540 to 551 of CCHF virus 10200 strain: EIHGDNYGGPGD). Brefeldin A (BFA) was purchased from Roche Applied Science, Indianapolis, Ind.; NuPAGE ready-made gels (10% or 3 to 8%) and recommended buffers were purchased from Invitrogen, Carlsbad, Calif.
Mutagenesis of the tetrapeptide motifs.
Mutagenesis of the Gn and Gc processing sites was performed by using a QuikChange XL site-directed mutagenesis kit, following the suggested protocol (Stratagene, La Jolla, Calif.). A pair of mutagenic primers representing the P-4 Arg to Ile mutation was synthesized (Invitrogen). CCHF-516RRLL>IRLL(F), TCAACCGGCTCTATAAGATTGCTTTCA, and CCHF-516RRLL>IRLL(R), TGAAAGCAATCTTATAGAGCCGGTTGA; CCHF-804RKLL>IKLL(F), CTTTGGGTTGTAATAAAGCTGTTGCAG, and CCHF-804RKLL>IKLL(R), CTGCAACAGCTTTATTACAACCCAAGG; CCHF-1037RKPL>IKPL(F), GCTCTAGTACTCATAAAGCCTTTATTC, and CCHF-1037RKPL>IKPL(R), GAATAAAGGCTTTATGAGTACTAGAGC. For construction of IRLL, IKLL, and IKPL mutants, pcDNA3.1 TOPO vector (Invitrogen) containing the complete glycoprotein gene of CCHF virus strain 10200 was used as the template. The numbers mentioned in the primers indicate the amino acid position subjected to the mutation. For the Gc-IKPL plasmid, the RKPL>IKPL primer combination was used with TOPO-Gc (mentioned as TOPO-G1 in reference 16) as the template. Transformation was done in STBL2-competent cells (GIBCO-BRL), and the presence of introduced mutations was confirmed by DNA sequencing.
Transfection and protein analysis.
Plasmid DNA for the transfections was prepared by using Qiafilter kits purchased from Qiagen. The expression of CCHF virus wild-type (wt) and mutant glycoproteins were analyzed by using the vaccinia virus T7-RNA polymerase-based transient expression system (8) following transfection conditions described previously (22). Briefly, cells were seeded onto 6-well plates a day before transfection and were infected for 1 h with vaccinia virus expressing T7 RNA polymerase. Upon removal of the virus, cells were transfected with plasmid DNA encoding the wt or mutant CCHF virus glycoproteins. After 12 h, cells were labeled for 30 min with [35S]cysteine and were chased for the indicated periods. For experiments involving BFA (10 μg/ml), aliquots of the drug were added 15 min before labeling and during labeling and chase periods. In some cases, the media were collected and the CCHF virus glycoproteins secreted or shed were analyzed. Immunoprecipitated proteins were resolved in NuPAGE gels, and proteins were visualized by autoradiography.
RESULTS
Peptides encompassing the RRLL tetrapeptide are efficiently cleaved by SKI-1.
Previous studies indicated that mature Gn and Gc glycoproteins of CCHF virus were preceded by tetrapeptide sequences RRLL and RKPL, respectively (16). Since RRLL has been demonstrated as the processing site for Lassa virus (13) and is implicated in other arenaviruses (4, 19), and since SKI-1 was the protease involved in the processing of Lassa virus GP-C (12), we hypothesized that SKI-1 may be involved in the processing of CCHF virus glycoproteins. To address this question, quenched fluorogenic peptides containing the Gn processing site (RRLL↓) were synthesized and incubated with recombinant hSKI-1 as described in Materials and Methods. As shown in Fig. 1, Q-CCHFV-A peptide encompassing the Gn processing site was very efficiently cleaved in vitro by recombinant SKI-1 (2, 21) at the correct physiological site (RRLL↓). After incubation with the enzyme for 1 h at room temperature, the reverse-phase high-performance liquid chromatography (RP-HPLC) chromatogram of the crude digest exhibited two additional peaks at Rt values of 39.5 and 33.5 min in addition to the peak for undigested peptide [Rt = 41.45 min; m/z = 1704.1 (M+H)+]. The peaks were identified by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) (MALDI-TOF [MS]) as the highly fluorogenic N-terminal fragment Abz-S-S-G-S-R-R-L-L↓OH [m/z = 994.8 (M+H)+] and the nonfluorescent C-terminal peptide S-E-E-S-Y(NO2)-Ala-NH2 [m/z = 729.0 (M+H)+]. These data provided initial evidence that SKI-1 is directly involved in the processing of Gn following the RRLL site.
FIG. 1.
Peptides containing RRLL sequences are efficiently processed by hSKI-1 in vitro. (A and B) RP-HPLC chromatogram of the crude digest following 1 h of incubation at 37°C of 20 μg of Q-CCHFV-A [Abz-S-S-G-S-R-R-L-L↓S-E-E-S-Y(NO2)-Ala-NH2] with recombinant hSKI-1 (10 μl) in 25 mM Tris-25 mM MES-2.5 mM CaCl2, pH 7.4. The elution of the peaks was monitored in line by UV absorbance (absorbance unit full scale [AUFS] at a 214-nm wavelength) (A) as well as fluorescence (λex, 320 nm; λem, 420 nm) (B) detectors. The peaks eluting at retention times (Rt) 41.45, 39.5, and 33.5 min were characterized by MALDI-TOF (MS) as the undigested peptide [m/z = 1,704.1 (M+H)+], the highly fluorescent N-terminal (NT) Abz-S-S-G-S-R-R-L-L-OH [m/z = 994.8 (M+H)+], and the nonfluorescent C-terminal (CT) S-E-E-S-Y(NO2)-Ala-NH2 [m/z = 729.0 (M+H)+]. Note that both NT fragment and the undigested peptides are detectable by UV absorbance (A) as well as by fluorescence measurement (B) while the CT fragment is detectable by UV absorbance alone. Notice the loss of one or multiple 16 mass units (mu) (either NH2 or OH groups) or the addition of 22 mu (Na) in the observed mass spectra.
Peptides encompassing the RKPL tetrapeptide are poor substrates for SKI-1.
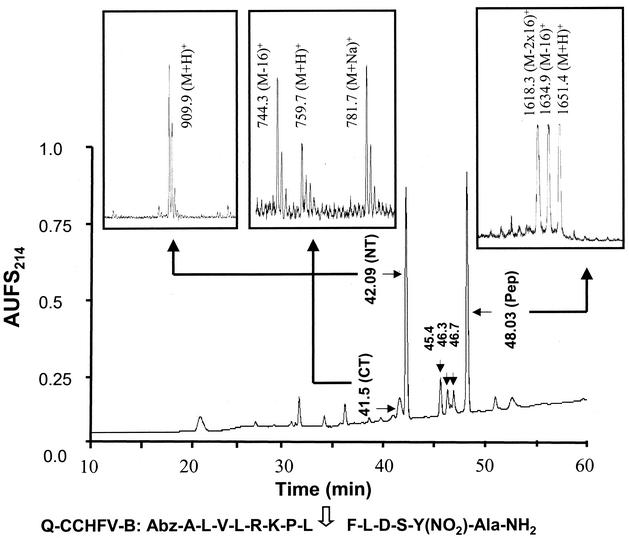
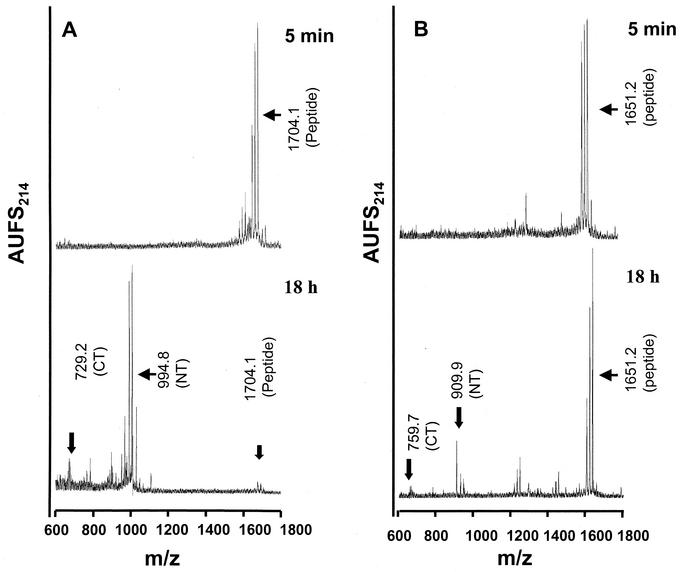
We also performed in vitro processing assays with synthetic quenched fluorogenic peptides encompassing the Gc processing site (RKPL↓). Our results indicated that desAbz Q-CCHFV-B containing the RKPL sequence was cleaved extremely inefficiently at the physiologically relevant site (RKPL↓) by recombinant hSKI-1 (Fig. 2). Indeed, incubation as long as 24 h with a fivefold excess of hSKI-1 was needed for the appearance of peaks at 42.09 and 41.5 min compared with that used for digestion of Q-CCHFV-A. Mass spectral analysis of all isolated peaks indicated that the peptide is also cleaved at two additional sites, R-K-P-L-F↓L↓D, suggesting that the cleavage of the Gc-specific site of CCHFV may be mediated by an enzyme other than SKI-1. The low abundance of the peak at Rt = 41.5 min for the C-terminal peptide is caused by its further degradation into smaller fragments eluted earlier on the RP-HPLC chromatogram (not identified). We also compared the cleavage efficiency and kinetics of the peptides containing RRLL and RKPL sequences. Samples were analyzed after 5 min and 18 h after incubation with hSKI, and the results of mass spectral analyses are depicted in Fig. 3. Peptides containing the RRLL sequences underwent >98% cleavage (Fig. 3A). In contrast, the RKPL containing peptides underwent only ∼0.5% cleavage even after 18 h of incubation (Fig. 3B). These results clearly provide evidence for the role of SKI-1 in Gn, but not Gc, processing.
FIG. 2.
Peptides containing RKPL sequences are poor substrates for hSKI-1. RP-HPLC chromatogram of the crude digest following 24 h of incubation at 37°C of 20 μg of desAbzQ-CCHFV-B [A-L-V-L-R-K-P-L↓F-L-D-S-Y(NO2)-Ala-NH2] with excess recombinant hSKI-1 (50 μl) in 25 mM Tris-25 mM MES-2.5 mM CaCl2, pH 7.4. The elution of the peaks was monitored in line by UV (214-nm wavelength) absorbance. The peaks eluting at retention times 48.03, 42.09, and 41.5 min were characterized by MALDI-TOF (MS) as the undigested peptide [m/z = 1,651.2 (M+H)+], the N-terminal (NT) A-L-V-L-R-K-P-L-OH [m/z = 909.9 (M+H)+], and the C-terminal (CT) F-L-D-S-Y(NO2)-Ala-NH2 [m/z = 759.7 (M+H)+]. The three minor peaks eluting at 45.4, 46.3, and 46.7 min were identified by MS as A-L-V-L-R-K-P-L-F-OH (m/z = 1,057.2), A-L-V-L-R-K-P-L-F-L-D-OH (m/z = 1,286.1), and A-L-V-L-R-K-P-L-F-L-OH (m/z = 1,169.5).
FIG. 3.
Comparison of the in vitro processing kinetics of peptides containing RRLL and RKPL sequences. desAbz Q-CCHFV-A (A) or desAbz Q-CCHFV-B (B) was incubated with SKI-1 (10 μl) for 5 min and 18 h in 25 mM Tris-25 mM MES-2.5 mM CaCl2, pH 7.4. Note the near disappearance of the peak at an m/z value of 1,704 for the 18-h digest of Q-CCHFV-A peptide, whereas the desAbz Q-CCHFV-B exhibited only a minor cleavage even after 18 h of digestion.
The measured apparent Vmax and Km values of SKI-1-mediated cleavage of Q-CCHFV-A were found to be 80.3 μmol/h and 5.01 μM, respectively, with Vmax/Km = 16.02 h−1 liter−1. These values suggested that Q-CCHFV-A is cleaved approximately threefold less efficiently than the corresponding fluorogenic peptide derived from Lassa virus glycoprotein Q-GP-C251-263 [Abz-D-I-Y-I-S-R-R-L-L↓G-T-F-T-Y(NO2)-Ala-NH2], which exhibited an apparent Vmax/Km value of 43.8 h−1 liter−1 (2). These results showed that the kinetics of proteolytic processing vary depending on the flanking amino acids, although the recognition motif (RRLL) was identical.
Expression of CCHF virus glycoproteins in SKI-1-deficient cell lines abolishes only Gn processing.
Having established the selective in vitro processing of Gn by SKI-1, we wanted to confirm the phenomenon during protein expression in mammalian cells. To that effect, we performed virus glycoprotein expression studies in SKI-1-deficient CHO cells, which have been engineered to express either a functional SKI-1 [referred to as SKI-1(+)] or a nonfunctional SKI-1 [SKI-1(−)]. The mutation in the nonfunctional SKI-1 is a histidine to alanine at amino acid position 249 at the active site of SKI-1. This mutation results in a SKI-1 protein, which remains in the endoplasmic reticulum (ER) as proSKI-1 and is defective in proteolytic functions (7). Cells were transfected with plasmid encoding the wild-type glycoproteins of CCHF virus, and glycoprotein-specific proteins were immunoprecipitated by using an HMAF generated against CCHF virus proteins, resolved by using NuPAGE (10% gels; Invitrogen) with the buffer conditions recommended by the manufacturer and visualized by autoradiography. In SKI-1(+) cells, PreGn was visualized as a 140-kDa protein during the 30-min pulse period (Fig. 4A, lanes 1 to 3). During the 2- and 3-h chase periods, the PreGn underwent processing resulting in the appearance of mature Gn (37 kDa). Similarly, PreGc, resolved as an 85-kDa protein, was processed into mature Gc (75 kDa). In parallel, we performed transfection in the SKI-1(−) cells to analyze the processing pattern and to specifically investigate the need for functional SKI-1 in the glycoprotein processing. Unlike in SKI-1(+) cells, the PreGn synthesized in the SKI-1(−) cells during the 30-min pulse did not undergo proteolytic processing during the chase periods to generate Gn (lanes 4 to 6). Prolonged exposure of autoradiographs failed to show even traces of Gn, confirming a defect in Gn processing (data not shown). The processing of Gc was not significantly affected in SKI-1(−) cells. In all our experiments, the level of protein expression in SKI-1(−) cells was lower than that observed in SKI-1(+) cells. Another feature we observed in the experiment in SKI-1(−) cells was the appearance of a higher-molecular-size protein, approximately 180 to 190 kDa. The identity of this protein is unknown, but it is not thought to be the precursor of the PreGn and PreGc proteins. The protein was not observed during experiments with a panel of mucin or Gn domain-specific antibodies to detect virus proteins expressed in SW13 cells infected with CCHF virus or transfected with expression plasmids encoding the CCHF virus full-length glycoproteins (16). This 180- to 190-kDa protein is only observed in the SKI-1-deficient CHO cells but is not specific to defects in Gn processing, as the protein is also observed in cells expressing Gn mutant proteins with no defect in processing. Another argument against this protein representing the precursor of PreGn and PreGc proteins is that the size of such a precursor would be predicted to be as large as 225 kDa.
FIG. 4.
Processing of Gn is abolished in cells which are deficient in SKI-1 expression. Plasmid DNA encoding CCHF virus glycoproteins was transfected into SKI-1(+) and SKI-1(−) cells and was labeled with [35S]cysteine. Precleared cell lysates were immunoprecipitated by using HMAF specific to CCHF virus proteins. Proteins were resolved in 10% NuPAGE gels, and proteins were visualized by autoradiography. (A) Expression of CCHF virus glycoproteins in SKI-1(+) and SKI-1(−) cells. Cells were labeled for 30 min (lanes 1 and 4) and were chased for 2 h (lanes 2 and 5) and 3 h (lanes 3 and 6). The numbers at the right-hand side denote molecular size markers. PreGn refers to the 140-kDa Gn precursor, and Gn refers to the mature Gn protein. PreGc and Gc correspond to the precursors of Gc and mature Gc, respectively. (B) Processing of the glycoproteins in the presence of BFA. SW13 cells were transfected with plasmid expressing the wt protein, and the treatments are indicated on the top. Proteins were resolved by using 3 to 8% gradient NuPAGE. P160 is likely to be a higher-molecular-weight protein secreted into the medium. The intracellular accumulation is due to BFA treatment.
Overall, the data above confirm that SKI-1 protease is needed to facilitate the processing of mature Gn, and the mutant inactive SKI-1 was unable to functionally participate in Gn processing. The Gc processing appears to occur in both SKI-1(+) and SKI-1(−), suggesting that this process occurs independent of SKI-1. Attempts were made to infect these SKI-1(−) CHO cells with CCHF virus in order to determine if SKI-1 cleavage of Gn is required to get release of virions and whether such particles would be infectious. Unfortunately, these CHO-derived cells were refractory to infection.
Gn processing occurs in ER-cis Golgi compartments.
We further investigated the cellular compartment in which the processing of CCHF virus glycoproteins occurs. For this purpose, we made use of BFA, a fungal metabolite known to prevent the anterograde transport of proteins from the ER to the Golgi and cause the redistribution of cis, medial, and trans-Golgi membranes but not the trans-Golgi network. The expression was performed in SW13 cells, which we normally use to analyze CCHF virus infection. Briefly, SW13 cells were transfected with plasmid DNA expressing the full-length CCHF virus polyprotein. Cells were labeled with [35S]cysteine for 30 min, were chased for 4 h either in the absence or presence of BFA, and were immunoprecipitated with HMAF alone or in combination with an anti-peptide antibody specific to the Gn region of CCHF virus glycoprotein (Fig. 4B). In untreated cells, PreGn, PreGc, Gn, and Gc were visualized as 140-, 85-, 37-, and 75-kDa proteins, respectively. Interestingly, Gn processing in BFA-treated cells was not at all affected, as no appreciable difference was observed in the untreated and BFA-treated cells (lanes 2 and 3). The level of PreGc/Gc was slightly reduced, coupled with a decrease in PreGn. In addition, a prominent band (corresponding to the location of P160) above PreGn was observed in BFA-treated cells (lane 3). This protein most likely represents the accumulated form of P160, which otherwise would be secreted into the medium in the absence of BFA treatment. Lane 4 served as the control. The presence of Gn in BFA-treated cells suggests that Gn processing occurs in ER-cis Golgi compartments, consistent with the subcellular location in which SKI-1 is functionally active (18, 12). These results indicate that the major processing events resulting in the generation of Gn and Gc occur in ER-cis Golgi compartments of mammalian cells.
Mutation of a critical R in the context of RRLL tetrapeptide prevents the processing of CCHF virus Gn glycoprotein.
N-terminal sequencing of mature Gn had demonstrated previously that proteolytic processing occurs following the tetrapeptide sequence RRLL (amino acids 516 to 519) (16). In the present study, expression of CCHF virus glycoprotein in SKI-1(−) cells resulted in abrogation of processing of Gn (Fig. 4A). To investigate the sequence requirements for recognition of PreGn by SKI-1, we targeted the RRLL sequence located at the N termini to mature Gn. We made a DNA construct in which R at amino acid position 516 was altered to I. The introduced mutation was confirmed by DNA sequencing. Arginine at the P-4 in the context of the RRLL tetrapeptide was chosen, since substitution of this amino acid for other amino acids had been shown to be sufficient to abrogate the SKI-1-mediated processing activity in the ER and Golgi (12, 13). Infection and transfection were performed as described for Fig. 4. The 35S-labeled cells were immunoprecipitated, and proteins were resolved in 10% NuPAGE (Fig. 5). When the IRLL mutant was expressed in SKI-1(+) cells, we observed the PreGn, PreGc, and Gc proteins but not the mature Gn (37 kDa) during the 2- and 3-h chase periods (lanes 6 to 8). This observation is in contrast to distinct Gn processing during wt glycoprotein expression (lanes 3 to 5). Although the cells express functional SKI-1, generation of mature Gn was completely abolished because of the mutation in the SKI-1 target sequences on the CCHF virus glycoprotein. Thus, we confirmed that SKI-1-mediated processing of Gn is dependent on the integrity of the RRLL tetrapeptide sequences located N terminal to the mature Gn amino terminus.
FIG. 5.
Mutation of critical R at the P-4 position in the context of RRLL at the SKI-1 recognition site abolishes Gn processing. SKI-1(+) cells were transfected with plasmid encoding CCHF virus glycoprotein with a mutation at the potential processing site (516R of the RRLL tetrapeptide to I or 804R of RKLL tetrapeptide to I). Transfected cells were labeled with [35S]cysteine for 30 min and were chased for 2 and 3 h. Proteins were immunoprecipitated by using CCHF virus-specific antibodies and were resolved in 10% NuPAGE. The numbers at the top indicate the periods of sampling, and those on the right-hand side indicate the position of molecular size markers. PreGn and PreGc denote precursors of Gn and Gc, respectively. A darker exposure of the proteins ranging from 50 kDa to the bottom of the gel is presented as the bottom panel to better visualize the Gn protein.
Since mature Gn is a glycoprotein with a molecular size of 37 kDa, we hypothesized that there should be another cleavage event to generate the Gn C terminus prior to the cleavage generating the mature Gc (RKPL) amino terminus. An RKLL tetrapeptide (amino acids 804 to 807) is present in the predicted cytoplasmic tail region of Gn. Interestingly, RKLL is the known glycoprotein processing site for Pichinde virus GPC (4) and the proposed processing site for Pirital and Parana viruses, all being New World arenaviruses. In an attempt to determine if RKLL is the C-terminal processing site of Gn and if it is mediated by SKI-1, we mutated the P-4 R (amino acid 804) in the context of RKLL to I. The expression and processing of Gn were analyzed in SKI-1(+) cells (Fig. 5, lanes 9 to 11). During the 30-min pulse period PreGn was observed, which was processed to mature Gn during the 3-h chase period, albeit at a slightly reduced level. The darker exposure of the gel (between 50 kDa and the bottom of the gel) is presented in the bottom panel to better visualize mature Gn. The fact that the mutation did not affect the Gn processing led us to conclude that the RKLL tetrapeptide is not the C-terminal processing site of Gn or that SKI-1 is not involved in the C-terminal processing. In addition, we examined the role of a stretch of basic amino acids (KKRKK) at positions 940 to 944 as the C terminus processing site. Individual or multiple mutations of these amino acids did not disrupt the processing of Gn (data not shown), indicating that these amino acids do not constitute the C terminus processing site. We are performing further studies to identify the C terminus processing steps.
Status of Gc processing.
Mature Gc is processed after the tetrapeptide RKPL, and this motif is conserved in all the CCHF virus isolates characterized to date (16). Interestingly, RKPL is presumed to be the cleavage site for the GPC of Guanarito virus, a New World arenavirus (19). As the P-1 and P-4 amino acids were L and R, respectively, we initially expected that SKI-1 might also be involved in the processing of mature Gc. Contrary to our expectations, the in vitro data as well as data obtained from SKI-1(−) cells led us to conclude that SKI-1 is not involved in Gc processing. Further evidence for this conclusion was obtained by mutating the RKPL tetrapeptide to IKPL. The Gc processing of the IKPL mutant was compared with the processing of Gc from the wt full-length protein. SW13 cells expressing the wt or mutant Gc-IKPL were analyzed for Gc processing from PreGc (Fig. 6). In cells expressing the wt glycoprotein, PreGc and mature Gc were visualized during the 30-min pulse, and levels of mature Gc continued to increase during the 3-h chase periods (lanes 1 and 2). As shown previously, mature Gn is seen only during the chase periods. Analysis of the IKPL mutant showed a similar processing profile with only a marginal variation in PreGc/Gc ratio (lanes 3 and 4), suggesting the lack of a distinct effect in Gc processing attributable to the IKPL mutation. Furthermore, we also examined the clone expressing Gc in the absence of Gn as previously described (16) and a variant carrying the IKPL mutation. Autoradiographs confirmed the presence of P85 (PreGc) and mature Gc from the Gc clone (lanes 5 and 6). If Gc is processed by SKI-1 and the process is mediated by virtue of the RKPL tetrapeptide, we would expect only the PreGc in cells expressing the Gc-IKPL mutant. However, Gc was generated from the Gc-IKPL plasmid as a 75-kDa protein at similar levels (lanes 7 and 8). Thus, Gc processing from PreGc appears to be relatively independent of SKI-1. Processing of mature Gc from the IKPL mutants further confirmed that a protease other than SKI-1 is involved in the maturation of Gc and that it was not dependent on an R at the −4 position. Extensive analyses will be required to clearly define the exact protease responsible for cleavage following the RKPL site and generation of the mature Gc amino terminus.
FIG. 6.
Processing of Gc and effects of RKPL>IKPL mutation. SW13 cells were transfected with plasmids encoding the wt glycoprotein or a mutant (1037 IKPL) at the Gc processing site (1037R in the RKPL tetrapeptide to I) and Gc or a mutant form of Gc (Gc-IKPL). Proteins were immunoprecipitated with HMAF and were resolved in a 10% NuPAGE gel. In each batch, cells were labeled for 30 min (designated 0) and were chased for 3 h (designated 3). Lanes 1 and 2, wt protein; lanes 3 and 4, 1037 IKPL mutant in the context of full-length protein; lanes 5 and 6, Gc; lanes 7 and 8, IKPL mutant in the context of Gc. PreGn and PreGc denote the precursors of Gn and Gc, respectively. The numbers at the right-hand side indicate the position of molecular size markers.
DISCUSSION
Virus glycoproteins are synthesized as precursor proteins, undergo proteolytic processing to result in mature proteins, and are incorporated in the virions. To enable this process, most viruses utilize the host cellular machinery and proteases. Notable examples are the processing of human immunodeficiency virus type 1 glycoprotein precursor gp160 (5, 9, 10); influenza virus hemagglutinin (20, 25); and measles virus fusion protein (26). The processing of the glycoproteins of these viruses is mediated by furin and furin-like convertases. Among viruses associated with hemorrhagic manifestations, the proteases involved in the processing of Ebola, Marburg, and Lassa virus glycoproteins have been identified. In the case of Ebola and Marburg viruses, the processing of the glycoprotein precursor to GP1 and GP2 is mediated by cellular furin (23, 24). The involvement of SKI-1 was described for the Lassa virus glycoproteins, and the processing occurs after a RRLL tetrapeptide sequence (12). Almost all the viral glycoproteins described above involve a precursor protein undergoing proteolytic processing resulting in two subunits.
The processing of CCHF virus glycoprotein appears to be more complex than that of viruses in other genera of the family Bunyaviridae. Our working hypothesis is that PreGn (P140) and PreGc (P85) are products of a cotranslational processing from a full-length polyprotein precursor (Fig. 7). Furthermore, evidence obtained by employing Western blotting and immunoprecipitation by using domain-specific antibodies indicated that mature Gn (37 kDa) is generated from a 140-kDa PreGn protein, and mature Gc (75 kDa) is generated from the 85-kDa PreGc (16). Data presented so far confirmed that SKI-1 is the protease responsible for the processing of CCHF virus Gn and is mediated by the RRLL sequence, as evidenced by the lack of mature Gn processing in SKI-1(−) cells and in the case of mutant 516RRLL>IRLL. For the generation of a 37-kDa Gn protein, cleavage has to occur at least twice: once at Gn's N terminus following the tetrapeptide RRLL and a second cleavage at a yet undetermined location in the C terminus. The C terminus cleavage site is probably located somewhere before mature Gc. One possibility we examined was the RKLL tetrapeptide (amino acid 808). Analysis of a mutant (804RKLL>IKLL) indicated that the processing and size of Gn was not substantially affected due to the mutation. This result could mean that RKLL is not the C terminus processing site of Gn or that SKI-1 is not involved in the processing at the C terminus of Gn. In addition, hydrophobicity and membrane topology predictions suggested that the domain containing RKLL might occupy the cytoplasmic side of the protein (data not shown). It is unclear if SKI-1 is physiologically active in the cytoplasm of mammalian cells. Another potential location for the C terminus processing of Gn was amino acids 940 to 944, constituting a stretch of basic amino acids (KKRKK). Individual and multiple mutations of these amino acids failed to disrupt Gn processing, suggesting the lack of any role for this region in Gn processing. We also do not know if the cleavage events at the N and C termini of Gn occur simultaneously or in cascades or if they are independent.
FIG. 7.
Proposed model of CCHF virus glycoprotein processing in mammalian cells. CCHF virus glycoprotein is likely synthesized as a precursor (its molecular size is not yet known). Upon translation, PreGn and PreGc are generated possibly after the hydrophobic domain and before RKPL, and this cleavage may be mediated by signal peptidase. From PreGc, mature Gc is processed following the RKPL (amino acid 1040) site, and this process is mediated by a protease other than SKI-1. This process is rapid as mature Gc can be visualized even during 20- or 30-min pulse periods. Mature Gc (but not PreGc) is recovered from the extracellular medium and becomes the structural element of the virus. Gn is generated from PreGn following the RRLL (amino acid 519) site, and this process is mediated by SKI-1. The processing of Gn appears to be a rather slow process, as Gn is visualized only in 2- to 3-h chase samples. Processing of Gn and Gc are not affected by BFA treatment, suggesting their occurrence in the ER-cis Golgi compartments. A 160-kDa protein (PreGn with terminal O-glycosidic modifications) and Gn are recovered from the extracellular media. CCHF virus contains predominantly mature forms of Gn and Gc as the glycoprotein elements. It remains to be seen if furin-mediated cleavage occurs at the RSKR (amino acid 247) site. If that happens, it will result in the separation of a mucin-like region and a protein with an expected molecular size of approximately 35 kDa (P35). The predicted O-glycosylation sites are indicated by long Y bars, and the predicted N-glycosylation sites are indicated by short Y bars.
Mutational analysis (full-length IKPL and Gc-IKPL) suggests that the protease responsible for Gc processing does not require a target containing an arginine at the −4 position, dramatically contrasting with SKI-1 target tetrapeptides. The presence of proline at P-2 appears to alter the specificity for SKI-1 and support the possibility that yet another protease is responsible for Gc processing. The nature of the protease involved in Gc processing is still under investigation. Apart from this, we are extending our analyses to investigate if there is a furin-mediated processing following the RSKR site and its probable role in virus biology (Fig. 7).
Comparison of the in vitro kinetics of processing reveals that Lassa virus glycoprotein-derived Q-GP-C peptide (2) is the most potent in vitro fluorogenic substrate for SKI-1 found so far. It is possible that the presence of hydrophobic Ile252-Tyr-Ile at P7-P5 positions and the absence of negative Glu residues at the P2′ and P3′ residues as found in Q-GPC of Lassa virus but not in Q-CCHFV-A may explain the observed difference in kinetics of SKI-1-mediated cleavages of these peptides. Future mutagenesis studies should define the importance of these residues in the processing of CCHF virus at this site. As the in vitro analyses did reflect the in vivo processing events, it will be a reliable tool to rapidly screen several peptides encompassing potential SKI-1 target recognition sites.
In addition to confirming the role of SKI-1 in Gn processing of CCHF virus, our data provide insights about the commonalities involved in the processing of Gn of CCHF and glycoproteins of Lassa fever virus; that is, both CCHF virus and Lassa virus contain identical tetrapeptide sequences (RRLL) at their processing sites, and SKI-1 plays a major role in the processing in both viruses. The lack of SKI-1-mediated processing at the RKPL preceding CCHF virus mature Gc and the presence of RKPL and other related tetrapeptides of the known or proposed cleavage site of several arenaviruses leaves us to postulate that although CCHF virus Gc and some of the New and Old World arenaviruses contain similar tetrapeptide sequences, they may be processed by proteases other than SKI-1. Valuable insights should be obtained by future studies to understand the convergence and the divergence in the processing pathways of CCHF virus and New World arenaviruses, especially those associated with hemorrhagic manifestations.
Acknowledgments
M.J.V. is thankful to Thomas Ksiazek and Pierre Rollin for providing the training to work in the Biosafety Level 4 facility at the Centers for Disease Control and Prevention. We also acknowledge T. Ksiazek for providing antibodies to CCHF virus and for constant support, John O'Connor for critical reading of the manuscript, and Kent Wagoner for help in the preparation of the figures. We also thank J. L. Goldstein and R. B. Rawson for kindly providing the SRD-12B and SRD-13A cell lines.
This work was supported in part by a Canadian Institutes of Health Research (CIHR) group grant MGC-11474 and by the Protein Engineering Network of Excellence (PENCE).
REFERENCES
- 1.Basak, A., M. Zhong, J. S. Munzer, M. Chrétien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses - a comparative analysis using fluorogenic peptides. Biochem. J. 353:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak, A., M. Chrétien, and N. G. Seidah. 2002. A rapid fluorometric assay for the proteloytic activity of SKI-1/S1P based on the surface glycoprotein of the hemorrhagic fever Lassa virus. FEBS Lett. 514:333-339. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, W. E., E. L. Buescher, and F. M. Hetrick. 1967. Production and characterization of arbovirus antibody in mouse ascitic fluid. Am. J. Trop. Med. Hyg. 16:339-347. [DOI] [PubMed] [Google Scholar]
- 4.Burns, J. W., and M. J. Buchmeier. 1993. Glycoproteins of the arenaviruses, p. 17-35. In M. S. Salvato (ed.), The Arenaviridae. Plenum Press, New York, N.Y.
- 5.Decroly, E., M. Vandenbranden, J. M. Ruysschaert, J. Cogniaux, G. S. Jacob, S. C. Howard, G. Marshall, A. Kompelli, A. Basak, F. Jean, C. Lazure, S. Benjannet, M. Chrétien, R. Day, and N. G. Seidah. 1994. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-I SU) and gp41 (HIV-I TM). J. Biol. Chem. 269:12240-12247. [PubMed] [Google Scholar]
- 6.Decroly, E., S. Wouters, C. Di Bello, C. Lazure, J. M Ruysschaert, and N. G. Seidah. 1996. Identification of the paired basic convertases implicated in HIV gp160 processing based on in vitro assays and expression in CD4(+) cell lines. J. Biol. Chem. 271:30442-30450. [DOI] [PubMed] [Google Scholar]
- 7.Elagoz, A., S. Benjannet, A. Mammarbassi, L. Wickham, and N. G. Seidah. 2002. Biosynthesis and cellular trafficking of the convertase SKI-1/SIP. Ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277:11265-11275. [DOI] [PubMed] [Google Scholar]
- 8.Fuerst, T. R., P. L. Earl, and B. Moss. 1987. Use of a hybrid virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hallenberger, S., M. Moulard, M. Sordel, H.-D. Klenk, and W. Garten. 1997. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J. Virol. 71:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H.-D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 11.Klenk, H.-D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 12.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz, O., J. ter Meulen, H. Feldmann, H.-D. Klenk, and W. Garten. 2000. Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J. Virol. 74:11418-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 15.Rawson, R. B., D. Cheng, M. S. Brown, and J. L. Goldstein. 1998. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 273:28261-28269. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, A. J., M. J. Vincent, and S. T. Nichol. 2002. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 14:7263-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 18.Seidah, N. G., S. J. Mowla, J. Hamelin, A. M. Mamarbachi, S. Benjannet, B. B. Touré, A. Basak, J. S. Munzer, J. Marcinkiewicz, M. Zhong, J. C. Barale, C. Lazure, R. A. Murphy, M. Chretien, and M. Marcinkiewicz. 1999. Mammalian subtilase/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA 96:1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B viruses, utilize alpha-dystroglycan as their major receptor. J. Virol. 76:5140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stieneke-Grober, A., M. Vey, H. Angliker, H., E. Shaw, G. Thomas, C. Roberts, H.-D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endo-protease. EMBO. J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Touré, B. B., J. S. Munzer, A. Basak, S. Benjannet, J. Rochemont, C. Lazure, M. Chrétien, and N. G. Seidah. 2000. Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J. Biol. Chem. 275:2349-2358. [DOI] [PubMed] [Google Scholar]
- 22.Vincent, M. J., N. U. Raja, and M. A. Jabbar. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J. Virol. 67:5538-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volchkov, V. E., H. Feldmann, V. A. Volchkova, and H.-D. Klenk. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 95:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volchkov, V. E., V. A. Volchkova, U. Stroher, S. Becker, O. Dolnik, M. Cieplik, W. Garten, H.-D. Klenk, and H. Feldman. 2000. Proteolytic processing of Marburg virus glycoprotein. Virology 268:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Walker, J. A., S. S. Molloy, G. Thomas, T. Sakaguchi, T. Yoshida, T. M. Chamber, and Y. Kawaoka. 1994. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J. Virol. 68:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe, M., A. Hirano, S. Stenglein, J. Nelson, G. Thomas, and T. C. Wong. 1995. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 69:3206-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]