Abstract
The recent finding of a novel Epstein-Barr virus-related lymphocryptovirus (CalHV-3) in a captive colony of common marmoset (Callithrix jacchus) in the United States modifies the view that the host range of lymphocryptovirus is restricted to humans and Old World primates. We investigated the presence of Epstein-Barr virus-related viruses in 79 samples of New World monkeys caught in the wild, including six species of the Cebidae family and one of the Callitrichidae, living in the rain forest of French Guiana. Using a degenerate consensus PCR method for the herpesvirus DNA polymerase gene, we identified three novel lymphocryptoviruses from golden-handed tamarin (Saguinus midas) of the Callitrichidae family and squirrel monkey (Saimiri sciureus) and white-faced saki (Pithecia pithecia) of the Cebidae family. With the CalHV-3 strain, these three novel viruses constitute a well-supported phylogenetic clade in the Lymphocryptovirus genus, which is clearly distinct from the lineage of Old World lymphocryptovirus, hosted by catarrhine monkeys and humans. In tamarins, the prevalence of the novel lymphocryptovirus was more than 50%, indicating that it circulates well in the wild population, perhaps due to specific ecoethological patterns such as confrontations and intergroup migration. The detection and partial molecular characterization of the polymerase gene of three novel Gamma-1-Herpesvirinae from New World monkeys caught in the wild clearly indicate that free-ranging populations of platyrrhine are natural hosts of lymphocryptoviruses. Further characterization of these novel viruses will provide new insight not only into the origin and evolution of Gammaherpesvirinae but also into their pathogenicity.
Herpesviruses are widespread in all vertebrate taxa (24). On the basis of molecular and biological patterns, the Herpesviridae family has been divided into Alpha-, Beta-, and Gammaherpesvirinae subfamilies (25), all of which are present in both humans and nonhuman primates. The diversity of New World mammals is very wide, but ecological constraints, low densities, and the small species numbers have limited knowledge of the diversity and the dynamics of several infectious agents in the Amazonian area. Nevertheless, some herpesviruses have been found in night monkeys and capuchins (Betaherpesvirinae), marmosets (Alpha- and Gammaherpesvirinae), squirrel monkeys (Gammaherpesvirinae), and spider monkeys (Alpha- and Gammaherpesvirinae) (8). Within the Gammaherpesvirinae subfamily, two genera are recognized, Lymphocryptovirus (or Gamma-1-Herpesvirinae), of which the Epstein-Barr virus (EBV) or human herpesvirus 4 (HHV4) is the human prototype (22), and the Rhadinovirus genus (Gamma-2-Herpesvirinae), including the Kaposi's sarcoma-associated herpesvirus or HHV8 (KSHV/HHV8) (27).
Viruses similar to EBV (Gamma-1-Herpesvirinae) have long been known to exist in several Old World primates, including apes (29), and have been partially characterized. In contrast, New World monkeys have until recently been found to harbor only Gamma-2-Herpesvirinae, namely, herpesvirus saimiri in the squirrel monkey (2, 4, 20) and herpesvirus ateles in the spider monkey (1, 19). The failure to detect EBV cross-reactive antibodies in New World primates suggested that lymphocryptoviruses were restricted to humans and nonhuman Old World primates (9). The general thinking was that the Old World-New World split resulted in dramatic changes in the evolution of Gammaherpesvirinae. The recent findings of KSHV/HHV8 in humans and, very recently, of EBV-like viruses naturally infecting New World primates have rewritten the old paradigm of Gamma-1 versus Gamma-2 host range restriction. Numerous molecular screenings have resulted in identification of Gamma-2-Herpesvirinae in several Old World monkey species, i.e., macaques (5, 26), African green monkeys (10), drills and mandrills (15), and apes (11, 16, 17), and Gamma-1-Herpesvirinae were recently reported in New World platyrrhine species in the Cebidae and Callitrichidae families (3). Callitrichine herpesvirus 3 (CalHV-3) was isolated from spontaneous lymphomas in captive common marmosets, Callithrix jacchus (21). The complete genome sequence has now been reported, providing further understanding of the phylogeny of herpesviruses (23). The three presently recognized Herpesvirinae subfamilies may have bifurcated before mammalian radiation, but diversification leading to the actual viruses may have occurred during recent evolution of the host species (18). Comparison of the complete genome sequences of three members of the lymphocryptovirus group, i.e., EBV, CalHV-3, and Cercopithecine herpesvirus 15 (hv15), showed different gene repertoires. While ancestral lymphocryptovirus genes are found in human and Old World and New World primate viruses, recently acquired lymphocryptovirus genes are not present in New World viruses (23). These accessory genes may explain some biological and epidemiological differences between Old and New World viruses, such as the prevalence in colonies.
Further work is therefore needed on free-ranging New World primates for further consideration of coidentification to species level of herpesviruses with their host lineages. The purpose of the present survey was to investigate the presence of herpesviruses in several New World (neotropical) monkeys species caught in the wild in French Guiana, by both serological and molecular screenings. Most of the previous findings were in clinical cases in captive units and therefore did not provide information on circulation of the virus in wild primate populations. The unusual opportunity to study a large number of free-ranging primates gave us a valuable opportunity to study the behavior of the virus in nature.
Investigations were conducted on 79 samples from seven monkey species, comprising six Cebidae species: 13 red howler monkeys (Alouatta seniculus), five tufted capuchins (Cebus apella), four white-faced sakis (Pithecia pithecia), two squirrel monkeys (S. sciureus), and one wedge-capped capuchin (Cebus olivaceus), one black spider monkey (Ateles paniscus), and 53 specimens of a Callitrichidae species, the golden-handed tamarin (Saguinus midas) (Table 1). The howlers, sakis, spider monkeys, and tamarins were captured in their flooded habitat (moist upland neotropical forest) during the filling of the Petit Saut hydroelectric dam reservoir (4°45′ to 5°04′N and 52°55′ to 53°15′W). The animals were anesthetized, examined clinically, and sampled and were then released into a nearby forest within 48 h (7). The squirrel monkeys were retrieved from animal dealers, but they had been captured in the wild a few days previously; the capuchins were kept as pets but had been captured in the wild and isolated from other primate species during their captivity.
TABLE 1.
Neotropical primates tested for herpesviruses by serological and molecular methods and survey results
| Family and species | No., distribution by sex, and age | No. with positive serological resultsa | No. with positive PCR results | Sequenced clones | Accession no. |
|---|---|---|---|---|---|
| Cebidae | |||||
| Red howler monkey (A. seniculus) | 13, 7 males, 6 females, adults | 0/13 | 5/13 | 5 clonesc | |
| Black spider monkey (A. paniscus) | 1 male, adult | 0/1 | 0/1 | ||
| Tufted capuchin (C. apella) | 5 males, adults | 0/5 | 0/5 | ||
| Wedge-capped capuchin (C. olivaceus) | 1 male, adult | 0/1 | 0/1 | ||
| White-faced saki (P. pithecia) | 4, 3 males, 1 female, adults | 0/3 | 2/4 | PpiLHV1-02 PpiLHV1-06 | AY166696AY166698 |
| Squirrel monkey (S. sciureus) | 2 males, adults | 0/2 | 1/2 | SscLHV1-01 | AY166697 |
| Callitrichidae | |||||
| Golden-handed tamarin (S. midas) | 53, 60% males, adults | 0/9b | 30/53 | SmiLHV1-011 | AY166691 |
| SmiLHV1-012 | AY166692 | ||||
| SmiLHV1-016 | AY166693 | ||||
| SmiLHV1-068 | AY166694 | ||||
| SmiLHV1-092 | AY166695 |
For both EBV and KSHV.
Only nine sera available.
PCR for five animals showed weakly positive amplification products of predicted size, but sequencing revealed genomic DNA amplification.
In a first step, we screened serologically for gammaherpesviruses. The KHSV immunoglobulin G immunofluorescence assay (Biotrin, Dublin, Ireland), specific for KSHV/HHV8, and the Platelia EBV EBNA immunoglobulin G enzyme-linked immunosorbent assay (Sanofi Pasteur Diagnostics, Marnes-la-Coquette, France), specific for EBV EBNA antigen, were used. As many other parasitological and virological investigations have been conducted with the serum bank, only nine sera were available from golden-handed tamarins, S. midas (Table 1). All the tested sera were negative for both KHSV/HHV8 and EBV.
In a second step, we performed molecular screening of all 79 samples by nested PCR (nPCR). Peripheral blood mononuclear cell DNA from all monkeys was first extracted in a classical phenol-chloroform procedure and was then submitted to nPCR with degenerated consensus primers targeted to the highly conserved amino acid sequences of the herpesvirus polymerase gene (26) under PCR cycling conditions described elsewhere (15). nPCR with degenerated primers revealed five positive amplification products of the predicted size in the first eight tamarin samples tested, in one from a squirrel monkey, in two from white-faced sakis, and in five from howlers (Table 1). No positive amplification was observed in samples from the two capuchin species or the spider monkeys. The amplification products of the predicted size were gel purified, cloned, and sequenced by the Big Dye terminator technique. Analysis of the five clones obtained from the howlers showed that these sequences were genomic DNA and were not related to any herpesvirus.
BLAST searches showed that the three newly recognized sequences belonged to herpesviruses of the Lymphocryptovirus genus. The virus hosted by the golden-handed tamarin was named SmiLHV1, that hosted by the white-faced saki was named PpiLHV1, and that hosted by the squirrel monkey was named SscLHV1.
To obtain the nucleotide sequence extending upstream of the VYGA region (26), a nested set of gene-specific nondegenerate primers (Smi-1H, 5′GCA GTG TTC CCT CGG GAT TGA ATG ACA3′; Ssc-1H, 5′CAG CGA CCC CGC TGG GTT TAG GC3′; and Ppi-1H, 5′GCA GAG TGC CCT CCC CAT TAA GTG CCA3′) was derived from the complementary sequences of the small fragments and was used in a nPCR amplification with the DFASA primer pool (26), which allowed amplification of overlapping fragments of 426 bp. These upstream nPCR products were subsequently cloned and sequenced, as described previously (15).
The DNA sequences obtained from the five SmiLHV1 isolates were 98 to 99% identical at the nucleotide level, and the two PpiLHV1 sequences obtained from two saki monkeys were identical. As shown in Table 2, comparisons of the nucleotides in these newly characterized viruses with previously described New and Old World gamma-1 and gamma-2 herpesviruses showed that the sequences were more closely related to the gamma-1 herpesvirus (genus Lymphocryptovirus), with nucleotide homologies with EBV of 71, 73, and 77% for SmiLHV1, PpiLHV1, and SscLHV1, respectively. Homologies with KHSV/HHV8, the human prototype of the gamma-2 genus, were much weaker: 58, 58, and 62%, respectively. Within the Lymphocryptovirus genus, sequence identity analysis showed that SmiLHV1 is closely related to CalHV-3 (83% nucleotide homology), as are the two other New World lymphocryptoviruses, but at a lower level (nucleotide homologies of PpiLHV1 and SscLHV1 with CalHV-3, 79 and 74%). Amino acid homologies showed comparable relationships, with high homologies in the group of New World viruses (91, 86, and 82%, respectively, amino acid identity of SmiLHV1, PpiLHV1 and SscLHV1 with CalHV-3).
TABLE 2.
Nucleotide and amino acid identities between the novel gammaherpesviruses (SmiLHV1, PpiLHV1, and SscLHV1) and other primate gammaherpesvirusesa
| Virus | % Identityb with:
|
|||||
|---|---|---|---|---|---|---|
| SmiLHV1
|
PpiLHV1
|
SscLHV1
|
||||
| Nucleotide | Amino acid | Nucleotide | Amino acid | Nucleotide | Amino acid | |
| Gamma-1-Herpesvirinae | ||||||
| SmiLHV1 | 98-99 | 97-100 | 79-80 | 86-87 | 77 | 80 |
| PpiLHV1 | 80 | 86-87 | 100 | 100 | 75 | 83 |
| SscLHV1 | 77 | 80 | 75 | 83 | ||
| CalHV-3 | 83 | 91 | 79 | 86 | 74 | 82 |
| Cercopithecine hv15 | 68 | 78 | 68 | 82 | 78 | 79 |
| EBV | 71 | 80 | 73 | 82 | 77 | 79 |
| Old World Gamma-2-Herpesvirinae (RV1) | ||||||
| KSHV | 58 | 60 | 58 | 62 | 62 | 61 |
| PanRHV1a/PtRV1 | 60 | 60 | 59 | 62 | 61 | 61 |
| PanRHV1b | 54 | 58 | 57 | 60 | 58 | 57 |
| GorRHV1 | 61 | 60 | 60 | 62 | 64 | 61 |
| RFHVMn | 63 | 60 | 62 | 63 | 65 | 62 |
| RFHVMm | 57 | 58 | 60 | 61 | 61 | 61 |
| MndRHV1 | 62 | 58 | 62 | 60 | 66 | 59 |
| ChRV1 | 60 | 60 | 60 | 63 | 60 | 61 |
| Old World Gamma-2-Herpesvirinae (RV2) | ||||||
| PanRHV2 | 59 | 57 | 58 | 60 | 59 | 59 |
| ChRV2 | 59 | 59 | 61 | 63 | 63 | 59 |
| MndRHV2 | 60 | 57 | 59 | 58 | 63 | 57 |
| MneRV2 | 58 | 57 | 60 | 60 | 63 | 59 |
| MGVMn | 58 | 57 | 60 | 60 | 63 | 59 |
| MGVMf | 57 | 58 | 60 | 60 | 63 | 58 |
| MGVMm | 57 | 57 | 60 | 60 | 62 | 58 |
| RRV | 57 | 57 | 60 | 60 | 62 | 58 |
| New World Gamma-2-Herpesvirinae | ||||||
| HVS | 54 | 56 | 55 | 59 | 54 | 58 |
| HVA3 | 55 | 57 | 58 | 60 | 55 | 59 |
Numbers refer to values obtained in comparison with the 426-bp fragment of the conserved DNA polymerase gene that is available for all viruses.
Bold values indicate sequences with the highest identities.
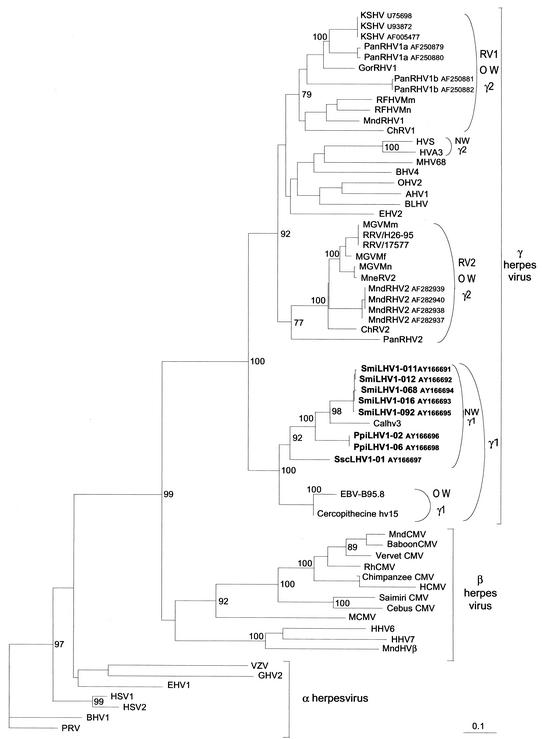
Phylogenetic analysis of nucleotide sequences with various methods (neighbor joining and DNA maximum parsimony) clearly placed these three novel neotropical primate viruses in the lymphocryptovirus genus (Fig. 1). A maximum bootstrap value of 100 supports the gamma-1-gamma-2 herpesvirus lineages. Further sublineages of Old and New World viruses within the two clusters are also supported by high bootstrap values. In the New World primate virus cluster, the tamarin sequences were clustered together and with the CalHV-3 sequence identified from Callithrix jacchus. Sequences from saki (PpiLHV1) and the squirrel monkeys (SscLHV1) also clustered (bootstrap values of 100 and 92, respectively) with this Callitrichidae lineage. All these neotropical primate virus sequences branch off from the Old World monkey group, which contains EBV and Cercopithecine hv15.
FIG. 1.
Phylogenetic tree resulting from analysis of selected 426-bp fragments of herpesvirus DNA polymerase gene (primers QAHNA and GDTD1B) (26), which is available for all viruses. The phylogeny was derived by the neighbor-joining method applied to pairwise sequence distances calculated by the Kimura two-parameter method (transition-to-transversion ratio set at 2). Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacements per site. Numbers at each node indicate the percentage of bootstrap samples (out of 100) in which the cluster to the right is supported. Brackets on the right indicate previously defined subfamily and genus herpesviral classification. Previously published sequences included and their accession numbers are as follows: HSV1 (X04771); HSV2 (M16321); VZV (X04370); EBV (V01555); HCMV (M14709); HHV6A (X83413); HHV7 (U43400); KSHV (U75698, U93872 and AF005477); H. saimiri HVS (M31122); HVA3 (AF083424); ChRV1 (AJ251573); ChRV2 (AJ251574); RFHVMn (AF005478); RFHVMm (AF005479); RRV/17577 (AF083501); RRV/H26-95 (AF029302); MneRV2 (AF204167); Macaca gamma virus strains from Macaca mulatta (AF159033), Macaca fascicularis (AF159032), and Macaca nemestrina (AF159031), called here MGVMm, MGVMf and MGVMn, respectively; PanRHV1a (AF250879 and AF250880); PanRHV1b(AF250881 and AF250882); PanRHV2 (AF346490); GorRHV1 (AF250886); MndRHV1 (AF282943); MndRHV2 (AF282937 to AF282940); Cercopithecine hv15 (AY037858); CalHV-3 (AF319782); MndHVβ (AF282942); PRV (L24487); BHV1 (Z78205); EHV1 (M86664); GHV2 (L40431); MCMV (U68299); RhCMV (AF0033184); MndCMV (AF282941); baboon CMV (AF27664); vervet CMV (AY049066); Cercopithecine hv3 (AY049065); chimpanzee CMV (AF480884); cebus CMV (AF292067); S. sciureus CMV (AF292065); MHV68 (U97553); BHV4 (AF031811); EHV2 (U20824); BLHV (AF031808); AHV1 (AF005370); and OHV2 (AF031812).
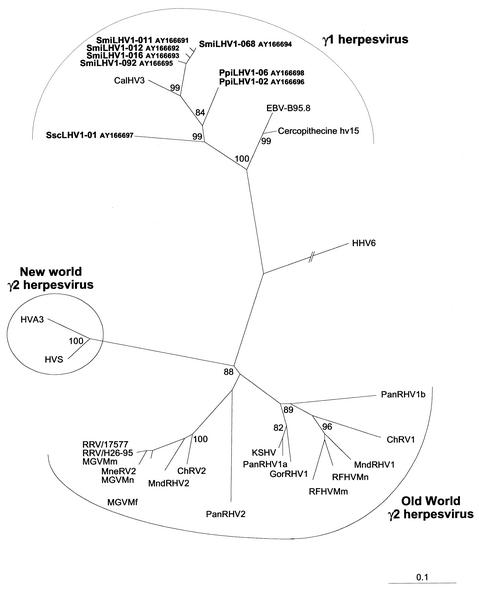
The neighbor-joining protein distance tree for the 142 amino acids encoded by the 426-bp fragment, comprising various gamma-1- and gamma-2 herpesviruses from New World and Old World monkeys and human herpesviruses (KHSV and EBV), clearly distinguished three groups with similar branchings to those obtained on nucleotide sequences: gamma-1 herpesvirus, gamma-2 herpesvirus from the New World, and gamma-2 herpesvirus from the Old World (Fig. 2). The Gamma-1-Herpesvirinae genus is organized into two clusters, the first comprising the three new viruses described in this study and CalHV-3 and the second comprising EBV and the Old World monkey lymphocryptovirus, Cercopithecine hv15.
FIG. 2.
Neighbor-joining protein distance tree for the 142 amino acid residues encoded by the 426-bp fragment (primers QAHNA and GDTD1B) (26) of DNA polymerase. Sequences were aligned by ClustalW and were analyzed with the PROTDIST and NEIGHBOR programs in PHYLIP. One hundred replica samplings were subjected to bootstrap analysis (SEQBOOT). The branch lengths are proportional to the evolutionary distance (scale bar) between the taxa. Previously published sequences included and their accession numbers are as follows: HHV6A (X83413); HVS (M31122); HVA3 (AF083424); EBV-B95.8 (V01555); KSHV (U75698); ChRV1 (AJ251573); ChRV2 (AJ251574); RFHVMn (AF005478); RFHVMm (AF005479); RRV/17577 (AF083501); RRV/H26-95 (AF029302); MneRV2 (AF204167); Macaca gamma virus strains from M. mulatta (AF159033), M. fascicularis (AF159032), and M. nemestrina (AF159031), called here MGVMm, MGVMf, and MGVMn, respectively; PanRHV1a (AF250879); PanRHV1b (AF250881); PanRHV2 (AF346490); GorRHV1 (AF250886); MndRHV1 (AF282943); MndRHV2 (AF282937); Cercopithecine hv15 (AY037858); and CalHV-3 (AF319782).
In order to determine the viral prevalence of the novel SmiLHV1, the 53 DNA samples from tamarins were screened with the degenerated primers. A prevalence of 24 was found among the 53 animals. To confirm this prevalence, semi-nPCRs were carried out with the Smi-1H-specific antisense primer and newly designed specific sense primers, Smi-2S (5′AAA TCC TTC CTG GCC AGT CT3′) for the first round of PCR and Smi-3S (5′GAC CAT CCT GGA CAA GCA AC3′) for the second round of PCR. This analysis showed that six other animals were positive for SmiLHV1, demonstrating an overall prevalence of more than 50%, with 30 of 53 positive animals. Tamarins live in familial units (n = 4.7 ± 2.4 in our sample), and the prevalence of positive animals was not statistically significantly different between groups. Furthermore, the two sexes were equally infected.
We report here the detection and partial molecular characterization of the polymerase gene of three novel Gamma-1-Herpesvirinae subfamily members present in New World monkeys caught in the wild. This finding indicates clearly that free-ranging colonies of these animals are natural hosts of lymphocryptoviruses. In nonhuman primates, most herpesvirus infections are latent and asymptomatic (29) and some of the reported diseases (21) may be due to cross-species infection in captivity (8). In our collection of samples, basic clinical and hematological investigations showed no detectable abnormalities known to be associated with herpesvirus infection (7).
The first lymphocryptovirus reported here, SmiLHV1, was found in the red-handed tamarin (S. midas), a primate of the Callitrichidae family that is very common in the northern Amazon basin. We have shown that this novel virus is widespread in the wild population, as 30 of 53 tested animals were infected. Other EBV-related viruses, such as CalHV-3 and herpesvirus papio, are frequent in marmosets (13) and baboons (12) in captivity, respectively. Ecological and behavioral patterns may indeed favor horizontal transmission. Tamarins have large home ranges, and their social organization is characterized by frequent subadult and adult movements between groups, temporary aggregations (28), and territorial confrontations by both sexes (14).
Recently, a gamma-1 herpesvirus named SaHV-3 was identified in common squirrel monkeys kept in captivity in the United States (3). Nonetheless, as sequence comparisons indicated a 35% nucleotide divergence over a 135-bp common fragment, we conclude that the novel lymphocryptovirus SscLHV1 reported here, from wild species in French Guiana rain forests, is distinct from SaHV-3. The variation could be related to a difference in hosts at the subspecies level, since the phylogeny of squirrel monkeys is still unclear. Apart from the previously described Gamma-2-Herpesvirinae subfamily member herpesvirus saimiri, identification of a third virus in squirrel monkeys confirms that a single host species can be infected by at least two distinct viruses of the same Herpesvirinae subfamily (3, 15). Furthermore, molecular screening of a captive colony of squirrel monkeys in our breeding center (6) also showed that these two distinct viruses, SscLHV1 and herpesvirus saimiri, could be hosted by a single animal (B. de Thoisy et al., unpublished data).
The smallness of the sample of white-faced sakis, P. pithecia, another primate of the Cebidae family, precludes further epidemiological analysis. The groups of capuchins and spider monkeys are also not large enough for further conclusions. In contrast, the apparent lack of detection of Gammaherpesvirinae in howler monkeys could be explained by a natural low viral load in this species. Another possibility is that the virus eventually present in this species is highly divergent and is thus not detectable with the protocol used. This last assumption would imply full reconsideration of the parallel evolution of herpesviruses and hosts, which would not be supported by previous data and the present report. Further research on herpesviruses in many samples from various howler monkey species would be useful in clarifying this hypothesis.
The presence of these three novel herpesviruses, together with that of CalHV-3, demonstrates that New World monkey species are reservoirs for a large set of herpesviruses. As South America and Africa separated 100 million years ago (18) and since both Gamma-1 and Gamma-2 subgroups are reported in platyrrhine and catarrhine, the divergence between Gamma-1- and Gamma-2-Herpesvirinae may thus have occurred earlier. The finely detailed branching analysis of the lymphocrytovirus genus is consistent with cospecies evolution between hosts and viruses: Old World viruses, comprising a virus from Cercopithecidae (Cercopithecine hv15) and one from Hominidae (EBV), cluster together and branch independently of the New World viral strains.
Further characterization of these novel New World herpesviruses and comparative analyses of their genomic structures with those of the other known gammaherpesviruses will permit new insights into the origin and molecular evolution of such viruses and also understanding of their pathogenicity in both natural and experimental models.
Nucleotide sequence accession number.
The sequences described herein have been deposited in the National Center for Biotechnology Information database. The GenBank accession numbers are AY166691 to AY166695 for the SmiLHV1 sequences, AY166696 and AY166698 for the PpiLHV1 sequences, and AY166697 for the SscLHV1 sequence.
Acknowledgments
We thank the Ministry of Research (Programme de Recherche fondamentale en Microbiologie des Maladies Infectieuses et Parasitaires), the Association pour la Recherche contre le Cancer (ARC), and the Virus Cancer Prevention Association for financial support. Vincent Lacoste was supported financially by La Fondation pour la Recherche Médicale and the Virus Cancer Prevention Association. The wildlife rescue operation at the Petit Saut dam site, Sinnamary River, French Guiana, was funded by Electricité de France and by the Centre National d'Equipement Hydraulique, Le Bourget du Lac, France.
REFERENCES
- 1.Albrecht, J.-C. 2000. Primary structure of the Herpesvirus ateles genome. J. Virol. 74:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. C., and B. Fleckenstein. 1990. Structural organization of the conserved gene block of Herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology 174:533-542. [DOI] [PubMed] [Google Scholar]
- 3.Cho, Y. G., J. Ramer, P. Rivailler, C. Quink, R. L. Garber, D. R. Beier, and F. Wang. 2001. An Epstein-Barr-related herpesvirus from marmoset lymphomas. Proc. Natl. Acad. Sci. USA 98:1124-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniel, M. D., A. Karpas, L. V. Melendez, N. W. King, and R. D. Hunt. 1969. Isolation of herpes-T virus from a spontaneous disease in squirrel monkeys (Saimiri sciureus). Arch. Gesamte Virusforsch. 22:324-331. [DOI] [PubMed] [Google Scholar]
- 5.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Thoisy, B., and H. Contamin. 1998. The squirrel monkey breeding colony of the Pasteur Institute, Cayenne, French Guiana. Neotrop. Primates 6:14-18. [Google Scholar]
- 7.de Thoisy, B., I. Vogel, J. M. Reynes, J. F. Pouliquen, B. Carme, M. Kazanji, and J. C. Vié. 2001. Health evaluation of translocated free-ranging primates in French Guiana. Am. J. Primatol. 54:1-16. [DOI] [PubMed] [Google Scholar]
- 8.Eberle, R., and J. Hilliard. 1995. The simian herpesvirus. Infect. Agents Dis. 4:55-70. [PubMed] [Google Scholar]
- 9.Franck, A., W. A. Audiman, and G. Miller. 1976. Epstein-Barr virus and non-human primates: natural and experimental infection. Adv. Cancer Res. 23:171-201. [DOI] [PubMed] [Google Scholar]
- 10.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greensill, J., J. A. Sheldon, K. K. Murthy, J. S. Bessonette, B. E. Beer, and T. F. Schulz. 2000. A chimpanzee rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. AIDS 14:F129-F135. [DOI] [PubMed] [Google Scholar]
- 12.Jenson, H. B., Y. Ench, S. J. Gao, K. Rice, D. Carey, R. C. Kennedy, J. R. Arrand, and M. Mackett. 2000. Epidemiology of herpesvirus papio infection in a large captive baboon colony: similarities to Epstein-Barr virus infection in humans. J. Infect. Dis. 181:1462-1466. [DOI] [PubMed] [Google Scholar]
- 13.Jenson, H. B., Y. Ench, Y. Zhang, S. J. Gao, J. R. Arrand, and M. Mackett. 2002. Characterization of an Epstein-Barr virus-related gammaherpesvirus from common marmoset (Callithrix jacchus). J. Gen. Virol. 83:1621-1633. [DOI] [PubMed] [Google Scholar]
- 14.Kessler, P. 1995. Preliminary field study of the red-handed tamarin, Saguinus midas, in French Guiana. Neotrop. Primates. 3:184. [Google Scholar]
- 15.Lacoste, V., P. Mauclère, G. Dubreuil, J. Lewis, M.-C. Georges-Courbot, J. Rigoulet, T. Petit, and A. Gessain. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J. Virol. 74:11993-11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacoste, V., P. Mauclère, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151-152. [DOI] [PubMed] [Google Scholar]
- 17.Lacoste, V., P. Mauclère, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2001. A novel gamma 2-herpesvirus of the Rhadinovirus 2 lineage in chimpanzees. Genome Res. 11:1511-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. R. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 19.Melendez, L. V., R. D. Hunt, N. W. King, H. H. Barahona, M. D. Daniel, C. E. Fraser, and F. G. Garcia. 1972. Herpesvirus ateles, a new lymphoma virus in monkeys. Nat. New Biol. 235:182-184. [DOI] [PubMed] [Google Scholar]
- 20.Melnik, J. L., M. Midulla, I. Wimberly, J. B. Barrera-Oro, and B. M. Levy. 1964. A new member of the herpesvirus group isolated from South American marmosets. J. Immunol. 92:596-601. [PubMed] [Google Scholar]
- 21.Ramer, J. C., R. L. Garber, K. E. Steele, J. F. Boyson, C. O'Rourke, and J. A. Thomson. 2000. Fatal lymphoproliferative disease associated with a novel gammaherpesvirus in a captive colony of common marmosets. Lab. Anim. Sci. 60:59-68. [PubMed] [Google Scholar]
- 22.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Raven Press, Philadelphia, Pa.
- 23.Rivailler, P., Y.-g. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a New World primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055-12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roizmann, B., and P. E. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Raven Press, Philadelphia, Pa.
- 25.Roizmann, B., R. C. Desrosiers, B. Fleckenstein, C. Lopez, A. C. Minson, M. J. Studdert, et al. 1992. The family Herpesviridae: an update. Arch. Virol. 123:425-429. [DOI] [PubMed] [Google Scholar]
- 26.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C.-C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz, T. F. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 79:1573-1591. [DOI] [PubMed] [Google Scholar]
- 28.Snowdon, C. T., and P. Soini. 1988. The tamarins, genus Saguinus, p. 223-298. In R. A. Mittermeier, A. B. Rylands, A. Coimbra-Filho, and G. A. B. Fonseca (ed.), Ecology and behavior of neotropical primates, vol. 2. World Wildlife Fund, Washington, D.C.
- 29.Wang, F., P. Rivailler, P. Rao, and Y. Cho. 2001. Simian homologues of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B 356:489-497. [DOI] [PMC free article] [PubMed] [Google Scholar]