Abstract
Although iridoviruses vary widely within and among genera with respect to their host range and virulence, variation within iridovirus species has been less extensively characterized. This study explores the nature and extent of intraspecific variation within an emerging iridovirus of North American warm-water fishes, largemouth bass virus (LMBV). Three LMBV isolates recovered from three distinct sources differed genetically and phenotypically. Genetically, the isolates differed in the banding patterns generated from amplified fragment length polymorphism analysis but not in their DNA sequences at two loci of different degrees of evolutionary stability. In vitro, the isolates replicated at identical rates in cell culture, as determined by real-time quantitative PCR of viral particles released into suspension. In vivo, the isolates varied over fivefold in virulence, as measured by the rate at which they induced mortality in juvenile largemouth bass. This variation was reflected in the viral loads of exposed fish, measured using real-time quantitative PCR; the most virulent viral strain also replicated to the highest level in fish. Together, these results justify the designation of these isolates as different strains of LMBV. Strain variation in iridoviruses could help explain why animal populations naturally infected with iridovirus pathogens vary so extensively in their clinical responses to infection. The results of this study are especially relevant to emerging iridoviruses of aquaculture systems and wildlife.
Viruses of the family Iridoviridae are notable for their variability. Iridoviruses infect a diversity of vertebrate and invertebrate hosts, causing diseases that range in severity from subclinical to lethal (34). They are important economically because of their association with large-scale mortality in aquaculture systems (10, 34). They are important from the perspective of wildlife conservation because of their association with epidemic morbidity and mortality in wild animal populations, most notably as agents responsible for fish kills and amphibian declines (4, 6, 34).
The recent emergence of many new iridoviruses has drawn attention to the marked variation among the genera and species within this viral family (3). The nature and extent of variation within iridovirus species have, however, been less well characterized. Distinct intraspecific iridovirus variants may exist, but examples to date have been limited to incidental findings from studies of a few iridoviruses of amphibians (9, 37), fishes (5, 14, 17), and insects (18, 33, 35). Such studies have relied mainly on genotypic, rather than phenotypic, differences to characterize variants.
Phenotypic variation within iridovirus species could help explain the widely observed phenomenon that animal populations infected with particular iridoviruses often differ widely in the nature and magnitude of their clinical responses to infection (3, 34). Understanding the degree to which variation in the pathogen (rather than the host or the environment) underlies such differences would be critical for understanding the biology, evolution, and control of iridovirus diseases.
We have chosen to investigate an emerging iridovirus of wild North American warm-water fishes—largemouth bass virus (LMBV). Discovered in 1996 (26), LMBV is in the genus Ranavirus and is closely related genetically to iridoviruses of Southeast Asian ornamental fishes (22). A retrospective investigation has shown that LMBV has existed in the southeast United States since at least 1991 (12). The virus has subsequently been found throughout the eastern, southern, and midwestern United States (11, 36). Although LMBV can infect multiple species, it has been associated with epidemic mortality only in largemouth bass and Florida bass (Micropterus salmoides and M. floridanus, respectively) (11).
A notable feature of LMBV infection is its clinical variability. Certain populations of infected bass experience large-scale fish kills, while other infected populations remain apparently healthy (11). We hypothesize that differences among viruses might explain the variability in clinical response of host populations to LMBV infection. To address this hypothesis, we have examined in detail three LMBV isolates from three distinct sources, each associated with different clinical manifestations in the field. We have specifically endeavored to quantify differences among these isolates genetically and phenotypically in vitro and in vivo.
MATERIALS AND METHODS
Viruses and cells.
Three viral isolates from three separate origins were used in this study. The first isolate (SC) is the original type isolate recovered following a fish kill of approximately 1,000 adult bass in the Santee Cooper Reservoir, South Carolina, in 1996 (26). The second isolate (JW) was recovered from largemouth bass at the Jake Wolf State Fish Hatchery, Illinois, in 2001. Bass at the Jake Wolf hatchery have experienced intermittent health-related problems consistent with LMBV infection but compounded by infections with other pathogens. The third isolate (CL) was recovered from Cedar Lake, Illinois, in 2001. Cedar Lake has not experienced any known fish kills and has not been stocked with fish from the Jake Wolf hatchery.
Viruses used in the experiments described below were sixth-cell-culture passages of these three isolates. Fifth-passage viruses were inoculated at multiplicities of infection of 1.0 onto confluent monolayers of fathead minnow (FHM) cells in 150-cm2 tissue culture flasks containing 25 ml of Eagle's minimum essential medium with Hanks' balanced salt solution (HBSS), 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 0.25 μg of amphotericin B/ml, and 50 μg of gentamicin sulfate/ml and incubated at 30°C for 5 days or until maximum cytopathic effect was observed (25). Cell culture supernatants were harvested and filtered through 0.45-μm-pore-size filters to remove cellular debris. Titers of harvested viruses were determined by inoculating serial dilutions onto replicate FHM monolayers in 96-well plates. Viral concentrations (50% tissue culture infective doses per milliliter) were calculated according to the method of Karber (19). Concentrations of viral stocks were also determined using real-time quantitative PCR (qPCR) (see below).
AFLP analysis.
To screen the three LMBV isolates for genetic differences, we conducted amplified fragment length polymorphism (AFLP) analysis (21, 23). To our knowledge, AFLP has not previously been used to analyze heterogeneity within a viral system. Because standard restriction fragment profiles of LMBV do not differ substantially from those of other species within the genus Ranavirus (22), AFLP (which has greater discriminatory power) was chosen for measuring intraspecific variation in this system.
Each viral isolate was inoculated onto confluent monolayers of FHM cells in three replicate 150-cm2 flasks as described above. High-quality genomic DNA was extracted from virions and cell-associated nucleocapsids by previously described methods (8, 28). Briefly, free viral particles and cell-associated nucleocapsids were isolated, concentrated using ultracentrifugation, digested with proteinase K, and subjected to phenol-chloroform DNA extraction. Concentrations and purities of extracted DNA were measured spectrophotometrically.
AFLP analysis was performed according to the method of Boumedine and Rodolakis (2), with slight modification. Briefly, 1.5 μg of viral DNA was digested with the restriction endonuclease MspI, and fragments were ligated to an AFLP adapter made by mixing equal molar amounts of oligonucleotides ADA1 (5′-CCAGGATCCT-3′) and ADA2 (5′-CGAGGATCCTGG-3′). Selective amplifications of ligated fragments were performed with a panel of 16 AFLP primers, consisting of base sequence P1 (5′-CCAGGATCCTCGG-3′) and variable bases at the 3′ end (P1-A, P1-C, P1-G, P1-T, P1-AA, P1-AC, P1-AG, P1-CA, P1-CC, P1-CT, P1-GA, P1-GG, P1-GT, P1-TC, P1-TG, and P1-TT).
PCRs were run in 25-μl reaction volumes containing 50 mM Tris-HCl, 1.5 mM MgCl2, 15 mM (NH4)2SO4, 0.1% Triton X-100, 0.2 mM deoxynucleoside triphosphates, 2 U of DyNAzyme EXT DNA polymerase (Finnzymes Oy), and 20 pmol of each AFLP primer. Amplifications were performed in an i-Cycler thermocycler (Bio-Rad Laboratories, Inc.). Reaction conditions consisted of an initial denaturation step at 94°C for 5 min, followed by 30 cycles of denaturation (94°C for 30 s), annealing (55°C for 30 s), and extension (72°C for 1.5 min) and a final extension step at 72°C for 4 min. Appropriate controls and replicate reactions were run according to the protocol of Boumedine and Rodolakis (2). To investigate the stability of AFLP banding patterns across viral passages in cell culture, the AFLP analysis was repeated using second-cell-culture-passage viruses available for the JW and CL isolates.
PCR products were heated to 72°C, mixed with 4 μl of agarose gel loading buffer containing 15% Ficoll, and stored at 4°C. Amplicons were visualized by electrophoresis of 3 μl of each reaction mixture in a gel consisting of 0.7% agarose and 0.7% Synergel (Diversified Biotech, Inc.), run at 40 V for approximately 5 h at 4°C, stained with ethidium bromide, and photographed under UV illumination.
PCR and DNA sequencing.
We targeted two viral gene regions for PCR and sequencing. The first was the highly conserved major capsid protein (MCP) gene, which differentiates reliably among iridovirus species and is useful for inferring broad patterns of iridovirus evolution (32, 33). Previously published oligonucleotide primers MCP-F (5′-GACTTGGCCACTTATGAC-3′) and MCP-R (5′-GTCTCTGGAGAAGAAGAA-3′) were used to amplify and sequence an approximately 0.5-kb fragment of the LMBV MCP gene (22).
Because the MCP is highly conserved, we hypothesized that a noncoding gene region might prove more variable and more informative for studies of intraspecific iridovirus variation. We aligned published sequences from the tiger frog virus (TFV), a newly described and completely sequenced Ranavirus (13) with homologous sequences from the distantly related Lymphocystis disease virus (31) to identify conserved coding regions flanking noncoding regions that would be suitable for primer design. Based on this analysis, we designed oligonucleotide primers 8RF (5′-CTATTCCTACTCTGACGACG-3′) and 9LR (5′-GGCAACGTCCGACGCCAGGC-3′) to amplify an approximately 0.5-kb region (NCR8/9) containing the 3′-terminal end of the putative DNA-dependent RNA polymerase largest subunit, the 3′-terminal end of the putative nucleoside triphosphatase I, and an approximately 0.35-kb intervening putative noncoding region of unknown function.
Viral DNA was extracted from cell culture supernatants with a commercial kit (QIAamp DNA Blood Mini kit; Qiagen, Inc.). PCRs were performed in 50-μl reaction volumes containing 50 mM Tris-HCl, 1.5 mM MgCl2, 15 mM (NH4)2SO4, 0.1% Triton X-100, 0.2 mM deoxynucleoside triphosphates, 2 U of DyNAzyme EXT DNA polymerase, and 5 pmol of upstream and downstream gene-specific oligonucleotide primers in an i-Cycler thermocycler. Reaction conditions consisted of an initial denaturation step at 94°C for 2 min, followed by 40 cycles of denaturation (94°C for 30 s), annealing (50°C for 30 s), and extension (72°C for 45 s) and a final extension step at 72°C for 7 min.
PCR products were visualized by gel electrophoresis in 1% agarose gels stained with ethidium bromide under UV illumination. Amplicons were excised from gels by using sterile razor blades and were purified from the agarose matrix by using a commercial kit (Zymoclean Gel DNA recovery kit; Zymo Research, Inc.). Purified amplicons were sequenced on ABI 377 automated fluorescent DNA sequencers (Applied Biosystems, Inc.), located at the University of Illinois W. M. Keck Center for Comparative and Functional Genomics, with the same gene-specific primers used for PCR amplification. Amplicons were sequenced in both directions until all ambiguous bases were resolved. Sequences were hand edited and aligned with respect to reference sequences by using the computer program Clustal X (30). All alignments were checked manually.
Real-time qPCR.
We designed a real-time qPCR (15, 16) to measure viral concentrations in cell culture supernatants and viral loads in fish tissues. qPCR was performed in triplicate for each sample in 25-μl reaction volumes containing 12.5 μl of 2× TaqMan Universal PCR Master mix (Applied Biosystems, Inc.), 2.5 μl of template DNA, 900 nmol each of primers LMBVMCPQF (5′-TTCCGTAGGAGTGCCCAGGT-3′) and LMBVMCPQR (5′-CCGCCAGCAGTTTAATCTGAGG-3′), and 250 nmol of fluorescent probe LMBVMCPProbe (5′-6FAM-ATGTGCTCAACTCTTGGCTGGTCCTC-MGBNFQ-3′), in 96-well optical PCR plates on an ABI Prism 7000 Sequence Detection system (Applied Biosystems, Inc.). Primers and probe were designed to anneal to and amplify a 107-bp target sequence within the LMBV MCP gene (22).
The qPCR was standardized by using DNA samples of known concentrations. An approximately 500-bp fragment of the LMBV MCP gene, containing the full 107-bp target sequence, was PCR amplified according to published protocols (22). The amplicon was cloned into a plasmid vector for subsequent transformation of Escherichia coli, with use of a commercial kit (pGEM-T Vector system; Promega, Inc.). Plasmid DNA was purified from E. coli cells with a commercial kit (QIAprep Miniprep system; Qiagen, Inc.), and the DNA concentration was determined using a SmartSpec spectrophotometer (Bio-Rad Laboratories, Inc.). Serial dilution was used to prepare standards that ranged in concentration from 101 to 108 copies per μl. qPCR was performed on these standards as described above, and linear regression was used to create a standard curve for interpolating the concentrations of unknown samples. Variability in the qPCR was measured as the square of Pearson's product moment correlation coefficient (r2) describing the relationship between the known concentrations of standards and their concentrations inferred by qPCR.
Analysis of viral replication in vitro.
To analyze replication rates of the three LMBV isolates in vitro, we inoculated confluent monolayers of FHM cells in three replicate 25-cm2 tissue culture flasks per isolate, each containing 10 ml of medium as described above, at multiplicities of infection of 1.0, plus two uninoculated flasks to serve as controls. Every 24 h, 0.25 ml of cell culture supernatant was removed from each flask, and viral DNA was extracted using a commercial kit (QIAamp DNA Blood Mini kit). Viral concentrations (genomes per milliliter) were determined by real-time qPCR for each isolate at each time point.
Experimental challenge of juvenile bass.
To analyze the comparative virulence of the three LMBV isolates in vivo, we conducted a challenge experiment using juvenile largemouth bass. Bass used for the experiment were offspring of brood fish collected from the Kaskaskia River basin in Illinois. This population has been maintained in a closed experimental pond complex for over 7 years. A representative sample of these fish were tested for LMBV by virus isolation on FHM cells prior to the initiation of the experiment and were found to be negative.
Juvenile bass (<1 year old) were collected from holding ponds, and 12 fish were placed in each of 10 replicate 20-gallon (75.6-liter) aquaria, for a total of 120 fish. Tanks had separate aeration systems and were kept in a closed environmental chamber under constant ambient temperature (25°C) and photoperiod (12 h). Fish were allowed to equilibrate to indoor housing conditions for 1 week prior to initiation of the experiment. Fish were fed bloodworms (mosquito larvae) once daily (0.7 g per fish) but were subjected to fasting for 24 h prior to inoculation with LMBV.
Suspensions of each viral isolate were prepared for injection of fish by dilution of viral stocks into HBSS. Concentrations of viral stocks were quantified five times independently in cell culture and five times independently by qPCR to minimize measurement errors. The ratio of 50% tissue culture infective dose per milliliter to viral genomes per milliliter was similar for all three isolates (1:101.80, 1:101.82, and 1:101.77 for the SC, JW, and CL isolates, respectively), indicating that the isolates did not differ from each other significantly in the number of infectious particles per viral genome.
Three randomly selected tanks were chosen for inoculation with each of the LMBV isolates, and the remaining tank served as a control. On day 0 of the experiment, fish were netted from tanks and placed into temporary holding containers. Experimental fish were captured by hand and exposed to 107.8 viral genomes of virus via intraperitoneal injection of 0.1 ml of HBSS containing 108.8 viral genomes per ml. Initial experiments indicated that this dose is the approximate 50% lethal dose of the SC isolate at 14 days postexposure (unpublished data). Control fish were sham injected with 0.1 ml of virus-free cell culture supernatant. Injections were performed using 1-ml tuberculin syringes and 27-gauge 1/2-in. needles inserted into the peritoneal cavity along the ventral midline. Fish were returned to their respective tanks immediately following injection. Intraperitoneal injection was chosen over other routes of inoculation (e.g., bath immersion) because it allowed precise dosing of individual fish and because it has previously been shown to induce clinical disease in juvenile largemouth bass (27).
Tanks were monitored at 24-h intervals for 14 days postexposure. During this observation period, all dead and moribund fish were removed. At the end of the experiment, all surviving fish were euthanized. Moribund fish and fish that survived to the end of the experiment were euthanized by immersion in a solution containing clove oil (24, 29).
All fish were processed for quantification of viral load as soon as possible following death or euthanasia. Fish were individually weighed and measured (total length) and dissected with flame-sterilized forceps and scissors. External and internal gross lesions were recorded. Body condition was calculated as Fulton's index [(weight/length3) × 105], a common measure for comparing body condition among fish within a population (1).
The viscera of each fish (including the gastrointestinal tract, reproductive organs, swim bladder, kidney, spleen, liver, heart, and mesentery) were removed, weighed, and placed in 50 volumes of HBSS containing 100 U of penicillin/ml, 100 μg of streptomycin/ml, 0.25 μg of amphotericin B/ml, and 50 μg of gentamicin sulfate/ml, in a sterile plastic bag. Viscera were homogenized by placing the bag in a Stomacher 80 Biomaster laboratory blender (Seward, Ltd.) for 60 s at normal speed. Homogenates were stored at −80°C for subsequent DNA extraction.
Viral DNA was extracted from tissue homogenates, and qPCR was performed on extracted DNA as described above. Viral loads were expressed as log10 viral genomes per gram of tissue, averaged over the three qPCR replicates.
Statistical analyses.
Viral growth rate data were analyzed using linear regression analysis. Survival data were analyzed using Cox proportional hazard regression analyses (20). Viral load data were analyzed using multiple linear regression analyses in which predictor variables included contrasts between fish exposed to the different LMBV isolates, as well as variables representing the body condition score of the fish at the time of death, the state of the fish at the time of death (found dead, found moribund, or surviving to the end of the experiment), and the presence of internal or external gross lesions. Analyses were performed using the computer program SAS, version 8.2 (SAS Institute, Inc.). Associations were considered statistically significant at the α = 0.05 level.
Nucleotide sequence accession number.
The sequence of the LMBV NCR8/9 region was deposited in GenBank (accession number AY208993).
RESULTS
qPCR.
The relationship between the known concentrations of DNA standards and their concentrations inferred by qPCR was described by the equation log10 copy number = 11.691 − 0.30182(CT), where CT refers to the PCR cycle number at which an increase in reporter fluorescence above baseline was first detected. The fit of the data to this line was described by a squared Pearson's product moment correlation coefficient (r2) of 0.99, indicating that the qPCR was extremely precise. The sensitivity of the assay was high, in that it detected viral DNA even at the concentration of the most dilute standard (101 copies per μl). Viral DNA was detected only in inoculated cell cultures and experimentally infected fish and was never detected in control flasks or sham-injected fish, indicating that the specificity of the assay was adequate for the purposes of this study. The specificity of the qPCR was not determined with reference to other related viral species.
Genetic differences among isolates.
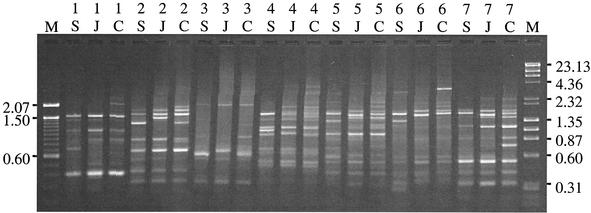
Different patterns of banding were observed among the three isolates for 7 of the 16 selective primers used for AFLP analysis (Fig. 1). These patterns were consistent across separate PCRs, confirming the reproducibility of the method (2). AFLP banding patterns were also consistent across viral passages; banding patterns for second-cell-culture-passage isolates were identical to those shown in Fig. 1 for sixth-cell-culture-passage isolates. This result indicates that differences in banding patterns do not represent random genetic changes accumulated during viral passage in cell culture but rather reflect stable genomic differences. From the gel shown in Fig. 1, a total of 99 band positions were identified (including both strong and weak bands). Genetic similarities between the isolate pairs, calculated as the proportion of total bands shared, were 26% (SC-JW), 22% (SC-CL), and 22% (JW-CL).
FIG. 1.
AFLP of three LMBV isolates. Lanes M are DNA length markers: left, 100-bp ladder (Invitrogen, Inc.); right, λ/HindIII plus φX174/HaeIII (Finnzymes OY). Lengths of specific marker bands (kilobases) are indicated by numbers to the right and left of the gel. Inner lanes show LMBV isolates Santee Cooper (S), Jake Wolf (J), and Cedar Lake (C). Numbers above isolate designations refer to specific primers (1, P1-AA; 2, P1-CA; 3, P1-CT; 4, P1-GA; 5, P1-TG; 6, P1-TT; 7, P1-T).
In all three LMBV isolates, PCR amplification of the specific gene regions MCP and NCR8/9 yielded amplicons of the predicted sizes. Nucleotide sequences of the MCP gene were identical among the SC, JW, and CL isolates. The MCP sequence of the isolates exactly matched that of the previously published LMBV MCP sequence (GenBank accession number AF080250) (22). Of nucleotide positions (376 of 495 positions) in the LMBV MCP, 76.0% were identical to the homologous region of the TFV Ranavirus (13).
Nucleotide sequences of the LMBV NCR8/9 region were identical among the SC, JW, and CL isolates. Of nucleotide positions (210 of 452, excluding gaps) in the LMBV NCR8/9 region, 46.5% were identical to the homologous region of the TFV Ranavirus (13).
In vitro differences among isolates.
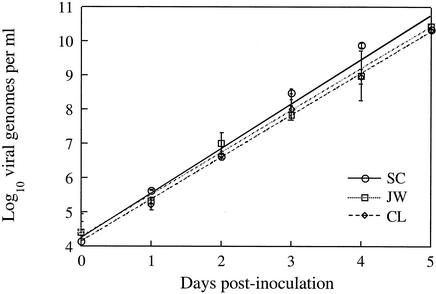
Rates of viral replication did not differ significantly among the three isolates (Fig. 2). All three isolates produced viral DNA in FHM cells at rates between 101.20 and 101.30 genomes per day. Rates of viral production were statistically indistinguishable among isolates but were extremely predictable, with r2 values greater than 0.98 describing the relationship between viral concentration and time in each case.
FIG. 2.
Rates of viral production for three LMBV isolates inoculated at multiplicities of infection of 1.0 onto monolayers of FHM cells, measured using real-time qPCR. Points represent average values across three replicate flasks inoculated with each isolate. Vertical bars represent standard errors of the means. Lines were fitted by the least-squares method and have statistically indistinguishable slopes and intercepts.
In vivo differences among isolates.
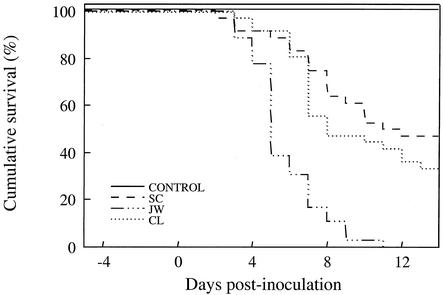
Significant differences were seen in the comparative virulence of the three LMBV isolates (Fig. 3). The median survival times of fish injected with the SC, CL, and JW isolates were 11.0, 7.5, and 4.5 days, respectively. Fish injected with the JW isolate experienced 100% mortality by day 11 postinoculation. These results were consistent across replicates (tanks). No morbidity or mortality was observed in sham-injected control fish.
FIG. 3.
Cumulative survival of juvenile largemouth bass injected intraperitoneally with equal doses of three LMBV isolates (SC, JW, and CL) or with virus-free cell culture supernatant (Control). Lines represent the combined survival data from three replicates for each isolate.
Rates of mortality of fish injected with the CL and SC isolates were not significantly different (hazard ratio = 1.41; chi-square test = 1.23; P = 0.27). The rate of mortality of fish injected with the JW isolate, however, was over five times higher than that of fish injected with the SC isolate (hazard ratio = 5.05; chi-square test = 26.07; P < 0.0001) and was over three times higher than that of fish injected with the CL isolate (hazard ratio = 3.63; chi-square test = 20.02; P < 0.0001).
The mean viral load of all fish found dead or moribund during the observation period (regardless of the isolate to which they were exposed) was 9.61 ± 0.05 viral genomes per g of tissue (Table 1). The mean viral load of all fish that survived to the end of the experiment was significantly lower (5.42 ± 0.32 viral genomes per g of tissue; t = 19.62; P < 0.0001; Table 1). Viral loads of fish found dead during the observation period did not differ significantly from those of fish found moribund (t = 0.18; P = 0.86). LMBV was not detected in sham-injected control fish.
TABLE 1.
Viral loads of juvenile largemouth bass exposed to equal doses of three LMBV isolates
| Isolate | State of fish
|
|||
|---|---|---|---|---|
| Found dead or moribund
|
Survived to end of experiment
|
|||
| n | Viral loada | n | Viral loada | |
| Santee Cooper | 19 | 9.43 ± 0.10 | 17 | 6.44 ± 0.32b |
| Jake Wolf | 36 | 9.80 ± 0.06b | 0 | |
| Cedar Lake | 24 | 9.45 ± 0.11 | 12 | 3.98 ± 0.31 |
Viral loads are expressed as log10 viral genomes per gram of tissue ± standard error.
Values indicated are statistically significantly higher than other values in that column.
The mean weight of all fish was 5.45 g (range of 2.38 to 10.31 g). External gross lesions included focal inflammation at the site of injection and abdominal distension. Internal gross lesions included pneumocystitis (inflammation of the swim bladder), exudative peritonitis, and discoloration (paleness) of the liver. Fifty-six percent of fish had neither internal nor external gross lesions. These results are consistent with previous descriptions of LMBV-associated gross pathology (38). Neither the presence of internal gross lesions (t = 0.86; P = 0.39) or external gross lesions (t = 0.38; P = 0.70) nor body condition score (t = 0.06; P = 0.95) was significantly associated with viral load.
Fish exposed to the JW isolate that were found dead or moribund during the observation period had significantly higher viral loads than did dead or moribund fish exposed to either the SC isolate (t = 3.08; P = 0.004; Table 1) or the CL isolate (t = 2.81; P = 0.008; Table 1). Fish exposed to the SC isolate that were found dead or moribund during the observation period did not differ significantly in their viral loads from dead or moribund fish exposed to the CL isolate (t = 0.14; P = 0.89; Table 1). No fish exposed to the JW isolate survived to the end of the experiment. Fish exposed to the SC isolate that survived to the end of the experiment had significantly higher viral loads than did fish exposed to the CL isolate that survived to the end of the experiment (t = 5.51; P < 0.001; Table 1).
DISCUSSION
The results of this study demonstrate that the three LMBV isolates examined differ genetically and phenotypically. The isolates displayed different banding patterns in 7 of 16 primers used in AFLP analysis and were roughly equidistant from one another genetically, as reflected in the proportion of AFLP bands shared between pairs. These genetic differences were not, however, reflected in the DNA sequences of the MCP and NCR8/9 gene regions. Although this result is not surprising for the highly conserved MCP region, the NCR8/9 region was clearly less conserved among viral species within the genus Ranavirus than was the MCP region. The LMBV MCP region differed from the homologous TFV MCP region at only 24.0% of its nucleotide positions. The LMBV NCR8/9 region, on the other hand, differed from the homologous TFV NCR8/9 region at more than twice that proportion of nucleotide positions (53.5%). Nevertheless, the three LMBV isolates were identical in sequence at NCR8/9, indicating that this locus may not be sufficiently variable for differentiating among closely related variants within iridovirus species. It may, however, prove useful for future studies of iridovirus evolution within genera and for finer resolutions of taxonomic relationships than those that can be obtained using MCP sequences.
The differences in AFLP banding patterns observed among the isolates must represent genetic differences at loci other than the MCP and NCR8/9 regions examined. Single-nucleotide changes, insertions, deletions, and/or chromosomal rearrangements at such loci could account for the observed differences in AFLP patterns. The possibility of insertions and/or deletions is especially likely, given the AFLP pattern shown in Fig. 1; minor “shifts” in the positions of adjacent bands appear to account for many of the differences observed. This pattern would be consistent with the previously documented observation that iridovirus genomes contain various amounts of repetitive DNA (34). The ability of AFLP analysis, but not DNA sequence analysis, to differentiate among these three isolates suggests that, for the Iridoviridae (as well as for other large, stable, double-stranded DNA viral families in which single-nucleotide genetic differences are probably widely dispersed throughout the genome), “genome screening” techniques such as AFLP analysis may be more informative than sequencing of specific gene regions for resolving close taxonomic relationships.
These genetic differences did not parallel the pattern of phenotypic differences observed among the three isolates in vitro. The three isolates, while between 22 and 26% different based on AFLP analysis, were indistinguishable statistically with respect to the rate at which they replicated in FHM cells. This measure was, however, based on the concentration of viral DNA recovered from cell culture supernatants. Rates of intracellular viral replication could conceivably still differ among these isolates, although this seems unlikely.
The phenotypes of the three isolates clearly differed in vivo. The JW isolate caused mortality in juvenile bass at a rate more than five times higher than that caused by the SC type isolate and more than three times higher than that caused by the CL isolate. This difference in virulence is far in excess of what would be expected at random or from small errors in the quantification of viral dose among the three isolates. Data from dose-response experiments (conducted prior to the initiation of this study) demonstrate that the median survival time of fish decreases at a rate of approximately 3 days per log10 increase in viral dose (unpublished data). Error in the measurement of viral dose would therefore have to be excessive (between approximately 1 and 3 orders of magnitude) to account for the observed greater virulence of the JW isolate. We conclude that the differences in virulence observed in this study reflect biological differences among the isolates.
The observation of increased virulence in the JW isolate is surprising for three reasons. First, the three isolates are roughly equidistant from one another genetically, as determined by AFLP analysis. Second, the CL isolate did not cause mortality at the same rate as did the JW isolate, despite the proximity of the geographic locations from which these two isolates were recovered. Third, the SC isolate, but not the JW isolate, has been associated with large-scale mortality in the field. Although fish at the Jake Wolf hatchery have experienced intermittent morbidity and mortality, this has not been severe. We conclude that factors other than the inherent virulence of the pathogen (such as environmental and host-related factors) must contribute significantly to the clinical manifestations of LMBV infection in the field.
Viral loads in challenged fish that died during the observation period were higher in fish exposed to the JW isolate than in fish exposed to the SC or CL isolate. The JW isolate may, therefore, achieve high virulence in part through an inherent ability to replicate to higher levels in fish and to consume host resources. The absolute difference in the viral loads of fish that died after exposure to the JW isolate and fish that died after exposure to the other isolates was, however, small (Table 1). We therefore hypothesize that the presence, absence, and/or differential expression of isolate-specific virulence factors may account for the differences in virulence observed among these isolates.
Among fish that survived to the end of the experiment, the SC isolate was recovered from fish tissues at concentrations 102.46 times higher than those of the CL isolate. This observation implies that the isolates may differ in the rates at which fish are able to clear infection. This difference could have important consequences for the long-term survival of fish or for the ability of LMBV to persist in natural populations.
Together, the results of this study imply that intraspecific variation in iridoviruses can occur and can manifest itself not only genetically but also phenotypically. In the specific case of LMBV, other factors in addition to pathogen virulence must contribute to the wide variation observed in the clinical responses of bass populations to LMBV infection. Factors such as temperature, water quality, and inherent genetic resistance/susceptibility of host populations may account for such variation (3, 11). The dramatic differences between viral loads of fish that died during the observation period and fish that survived to the end of the experiment (Table 1) demonstrate that individual fish also vary widely in their response to LMBV infection. We speculate that, given a longer observation period, some proportion of fish in our study may have cleared LMBV entirely and survived indefinitely.
Based on the genetic and phenotypic differences observed among the three LMBV isolates, we propose that these isolates represent different strains. Sequence identity among the isolates at the MCP and NCR8/9 loci demonstrates that SC, JW, and CL belong to the same species within the genus Ranavirus. Their variable genotypes and phenotypes nevertheless demonstrate that they possess biological differences significant enough to warrant some degree of taxonomic differentiation. We suggest that these isolates be differentiated using subscript notation indicating their geographic provenance, in this case the state and water body from which they were isolated. The isolates in this study would therefore be referred to as strains LMBVSCSC, LMBVILJW, and LMBVILCL. Future LMBV isolates with still different properties could be named accordingly.
We acknowledge, however, that multiple strains of LMBV may exist simultaneously in the same geographic locations and that strain variation in LMBV may not be related strictly to geography. Previous studies (17, 35) have demonstrated that restriction fragment profiles of iridoviruses can vary widely within iridovirus species and within geographic locations. The implications of this degree of genetic variation for the clinical variability of iridoviruses in the field are unknown. Iridovirus species in nature may exist as genotypically and phenotypically variable populations, akin to the “quasispecies” distributions more commonly used to describe populations of viruses with RNA genomes (7).
We believe that ours is the first prospective, systematic study of genetic and phenotypic strain variation in a viral species within the Iridoviridae. Intraspecific variation is a hallmark of viruses with RNA genomes (7) but is not generally considered a property common to large, complex DNA viruses. This paradigm may be incorrect. LMBV may have been introduced into North America only recently (11, 22). If so, then LMBV strain variation must have evolved quickly. Alternative explanations, such as multiple introductions of genetically and phenotypically different LMBV progenitors, would be less parsimonious. To the extent that the results of this study can be generalized to other iridoviruses, this conclusion implies a degree of evolutionary plasticity greater than that typically attributed to species within this viral family.
Regardless of the genesis of the variability observed, its existence could help explain why certain populations of infected fish experience large-scale mortality while others do not. More generally, strain variation in the Iridoviridae could help explain why populations of hosts infected with other iridoviruses often vary widely in their clinical responses to infection.
Understanding the basis of such variation is critical both for maintaining the health of animals in aquaculture systems and for conserving threatened wildlife populations. Future studies of strain variation in iridoviruses are clearly warranted. Paramount among the goals of such studies should be the identification and characterization of strain-specific virulence factors and the elucidation of host-related and ecological factors that may have led to the evolution of strains of different virulence.
Acknowledgments
We thank John Grizzle for kindly providing the original type isolate (SC) of LMBV and Andy Noyes for advice on propagating the virus. Mike Conlin, Larry Willis, and Steve Shultz (Illinois Department of Natural Resources) generously provided material from which the JW and CL viruses were isolated, as well as information about the bass populations from which they came. Robert Bakal, Bruce Shupp, and the Bass Anglers Sportsman Society (B.A.S.S.) offered valuable advice and discussions. We thank Gail Scherba and Anne Readel for help with laboratory genetic analyses and in vivo experiments, respectively.
This research was funded by grants from the Illinois Council on Food and Agricultural Research and the Conservation Medicine Center of Chicago.
REFERENCES
- 1.Bolger, T., and P. L. Connolly. 1989. The selection of suitable indices for the measurement and analysis of fish condition. J. Fish Biol. 34:171-182. [Google Scholar]
- 2.Boumedine, K. S., and A. Rodolakis. 1998. AFLP allows the identification of genomic markers of ruminant Chlamydia psittaci strains useful for typing and epidemiological studies. Res. Microbiol. 149:735-744. [DOI] [PubMed] [Google Scholar]
- 3.Chinchar, V. G. 2002. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch. Virol. 147:447-470. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham, A. A., T. E. Langton, P. M. Bennett, J. F. Lewin, S. E. Drury, R. E. Gough, and S. K. Macgregor. 1996. Pathological and microbiological findings from incidents of unusual mortality of the common frog (Rana temporaria). Philos. Trans. R. Soc. Lond. B Biol. Sci. 351:1539-1557. [DOI] [PubMed] [Google Scholar]
- 5.Darai, G., K. Anders, H. G. Koch, H. Delius, H. Gelderblom, C. Samalecos, and R. M. Flugel. 1983. Analysis of the genome of fish lymphocystis disease virus isolated directly from epidermal tumours of pleuronectes. Virology 126:466-479. [DOI] [PubMed] [Google Scholar]
- 6.Daszak, P., L. Berger, A. A. Cunningham, A. D. Hyatt, D. E. Green, and R. Speare. 1999. Emerging infectious diseases and amphibian population declines. Emerg. Infect. Dis. 5:735-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo, E., C. Escarmís, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 8.Esposito, J. J., R. Condit, and J. F. Obijeski. 1981. The preparation of orthopoxvirus DNA. J. Virol. Methods 2:175-179. [DOI] [PubMed] [Google Scholar]
- 9.Essani, K., and A. Granoff. 1989. Amphibian and piscine iridoviruses: proposal for nomenclature and taxonomy based on molecular and biological properties. Intervirology 30:187-193. [DOI] [PubMed] [Google Scholar]
- 10.Essbauer, S., and W. Ahne. 2001. Viruses of lower vertebrates. J. Vet. Med. B Infect. Dis. Vet. Public Health 48:403-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg, T. L. 2002. Largemouth bass virus: an emerging problem for warmwater fisheries? Am. Fish. Soc. Symp. 31:411-416. [Google Scholar]
- 12.Grizzle, J. M., I. Altinok, W. A. Fraser, and R. Francis-Floyd. 2002. First isolation of largemouth bass virus. Dis. Aquat. Org. 50:233-235. [DOI] [PubMed] [Google Scholar]
- 13.He, J. G., L. L., M. Deng, H. H. He, S. P. Weng, X. H. Wang, S. Y. Zhou, Q. X. Long, X. Z. Wang, and S. M. Chan. 2002. Sequence analysis of the complete genome of an iridovirus isolated from the tiger frog. Virology 292:185-197. [DOI] [PubMed] [Google Scholar]
- 14.Hedrick, R. P., T. S. McDowell, W. Ahne, C. Torhy, and P. de Kinkelin. 1992. Properties of three iridovirus-like agents associated with systemic infections in fish. Dis. Aquat. Org. 13:203-209. [Google Scholar]
- 15.Higuchi, R., G. Dollinger, P. S. Walsh, and R. Griffith. 1992. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 10:413-417. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 17.Hyatt, A. D., A. R. Gould, Z. Zupanovic, A. A. Cunningham, S. Hengstberger, R. J. Whittington, J. Kattenbelt, and B. E. Coupar. 2000. Comparative studies of piscine and amphibian iridoviruses. Arch. Virol. 145:301-331. [DOI] [PubMed] [Google Scholar]
- 18.Jakob, N. J., R. G. Kleespies, C. A. Tidona, K. Muller, H. R. Gelderblom, and G. Darai. 2002. Comparative analysis of the genome and host range characteristics of two insect iridoviruses: Chilo iridescent virus and a cricket iridovirus isolate. J. Gen. Virol. 83:463-470. [DOI] [PubMed] [Google Scholar]
- 19.Karber, G. 1931. Beitrag zur kollekivan Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 20.Kleinbaum, D. G. 1996. Survival analysis: a self-learning text. Springer-Verlag, New York, N.Y.
- 21.Lin, J.-J., and J. Kuo. 1995. AFLP: a novel PCR-based assay for plant and bacterial DNA fingerprinting. Focus 17:66-70. [Google Scholar]
- 22.Mao, J., J. Wang, G. D. Chinchar, and V. G. Chinchar. 1999. Molecular characterization of a ranavirus isolated from largemouth bass Micropterus salmoides. Dis Aquat. Org. 37:107-114. [DOI] [PubMed] [Google Scholar]
- 23.Mueller, U. G., and L. L. Wolfenbarger. 1999. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 14:389-394. [DOI] [PubMed] [Google Scholar]
- 24.Peake, S. 1998. Sodium bicarbonate and clove oil as potential anesthetics for nonsalmonid fishes. N. Am. J. Fish. Manag. 18:919-924. [Google Scholar]
- 25.Piaskoski, T. O., J. A. Plumb, and S. R. Roberts. 1999. Characterization of the largemouth bass virus in cell culture. J. Aquat. Anim. Health 11:45-51.
- 26.Plumb, J. A., J. M. Grizzle, H. E. Young, A. D. Noyes, and S. Lamprecht. 1996. An iridovirus isolated from wild largemouth bass. J. Aquat. Anim. Health 8:265-270. [Google Scholar]
- 27.Plumb, J. A., and D. Zilberg. 1999. The lethal dose of largemouth bass virus in juvenile largemouth bass and the comparative susceptibility of striped bass. J. Aquat. Anim. Health 11:246-252. [Google Scholar]
- 28.Schnitzlein, W. M., N. Ghildyal, and D. N. Tripathy. 1988. Genomic and antigenic characterization of avipoxviruses. Virus Res. 10:65-76. [DOI] [PubMed] [Google Scholar]
- 29.Taylor, P. W., and S. D. Roberts. 1999. Clove oil: an alternative anaesthetic for aquaculture. N. Am. J. Aquac. 61:150-155. [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tidona, C. A., and G. Darai. 1997. The complete DNA sequence of lymphocystis disease virus. Virology 230:207-216. [DOI] [PubMed] [Google Scholar]
- 32.Tidona, C. A., P. Schnitzler, R. Kehm, and G. Darai. 1998. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16:59-66. [DOI] [PubMed] [Google Scholar]
- 33.Webby, R., and J. Kalmakoff. 1998. Sequence comparison of the major capsid protein gene from 18 diverse iridoviruses. Arch. Virol. 143:1949-1966. [DOI] [PubMed] [Google Scholar]
- 34.Williams, T. 1996. The iridoviruses. Adv. Virus Res. 46:345-412. [DOI] [PubMed] [Google Scholar]
- 35.Williams, T., and J. Cory. 1993. DNA restriction fragment polymorphism in iridovirus isolates from individual blackflies (Diptera: Simuliidae). Med. Vet. Entomol. 7:199-201. [DOI] [PubMed] [Google Scholar]
- 36.Woodland, J. E., A. D. Noyes, and J. M. Grizzle. 2002. A survey to detect largemouth bass virus among fish from hatcheries in the southeastern USA. Trans. Am. Fish. Soc. 131:308-311. [Google Scholar]
- 37.Zhan, Q. Y., F. Xiao, Z. Q. Li, J. F. Gui, J. Mao, and V. G. Chinchar. 2001. Characterization of an iridovirus from the cultured pig frog Rana grylio with lethal syndrome. Dis. Aquat. Org. 48:27-36. [DOI] [PubMed] [Google Scholar]
- 38.Zilberg, D., J. M. Grizzle, and J. A. Plumb. 2000. Preliminary description of lesions in juvenile largemouth bass injected with largemouth bass virus. Dis. Aquat. Org. 39:143-146. [DOI] [PubMed] [Google Scholar]