Abstract
The left-hand or 3′-terminal hairpin of minute virus of mice (MVM) contains sequence elements essential for both viral DNA replication at the left-hand origin (oriL) and for the modulation of the P4 promoter, from which the viral nonstructural gene cassette is transcribed. This hairpin sequence has proven difficult to manipulate in the context of the viral genome. Here we describe a system for generating mutant viruses using synthetic hairpin oligonucleotides and a truncated form of the infectious clone. This allows manipulation of the sequence of the left-hand hairpin and examination of the effects in the context of the viral life cycle. We have confirmed the requirement for a functional parvovirus initiation factor (PIF) binding site and determined that an optimized PIF binding site, with 6 bases between the half-sites, was actually detrimental to viral growth. The distal PIF half-site overlaps a cyclic AMP-responsive element (CRE), which was shown to play an important role in initiating infection, particularly in 324K simian virus 40-transformed human fibroblasts. Interestingly, reducing the spacing of the PIF half-sites, and thus the affinity of the binding site for PIF, increased viral fitness relative to wild type in 324K cells, but not in murine A9 cells. These results indicate that the relative importance of factor binding to the CRE and PIF sites during the establishment of an infection differs markedly between these two host cells and suggest that the suboptimal spacing of PIF half-sites found in wild-type virus represents a necessary reduction in the affinity of the PIF interaction in favor of CRE function.
Parvoviruses differ from all known viruses in having single-stranded linear DNA genomes, which replicate by a unique rolling hairpin process, analogous to the rolling circle replication of single-strand coliphages and conjugative plasmids (29, 38). Minute virus of mice (MVM) is an autonomous parvovirus that encapsidates a single negative-sense strand containing two gene cassettes in a 4.8-kb coding region. The viral genes are bracketed by distinct palindromic telomeres that can fold back on themselves, forming imperfect duplex hairpins of 248 and 120 nucleotides at the right-hand (5′) and left-hand (3′) termini, respectively (1). In rolling hairpin replication, a unidirectional, leading-strand-specific replication fork travels down the linear genome until it reaches a terminal hairpin. The hairpin is sequentially unfolded, copied once, and then melted, freeing the newly synthesized complementary strand to fold back on itself, reforming the hairpin. This process, dubbed hairpin transfer, reverses the direction of the replication fork, redirecting it back along the genome so that it copies the strand that was just synthesized while displacing its previous complement (39). This process is continuous and leads to multimeric duplex molecules from which unit-length negative-sense genomes are subsequently excised (19). Excision is mediated by the site-specific single-strand nickase activity of NS1, a pleiotropic virally coded protein which nicks at unique origin sequences generated from each terminal hairpin sequence (15, 17).
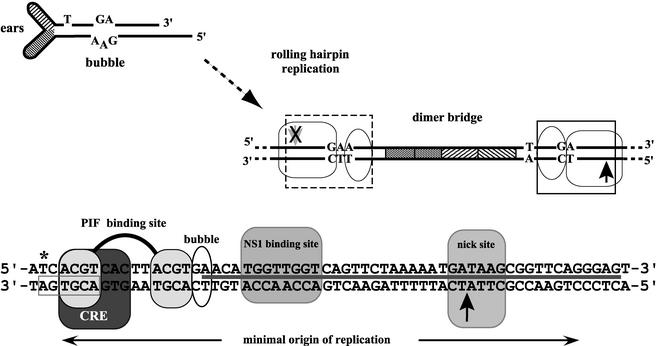
However, sequences from the left-hand terminus do not serve as an origin in the hairpin configuration. Instead, an active origin is generated only when rolling hairpin replication has extended and copied the hairpin to create the fully base-paired copy of the hairpin's palindromic sequence, termed the dimer junction, which spans adjacent genomes in the dimer replicative intermediate, as shown in Fig. 1. Small asymmetries in the hairpin, diagrammed in Fig. 1, such as an extra-helical T and a GA dinucleotide that opposes a GAA triplet in the “bubble” sequence, lead to slight differences in the sequence on either side of the axis of symmetry. One side of the axis, containing the dinucleotide bubble sequence, serves as an active origin of replication, oriLTC, while the analogous sequence on the side containing the bubble trinucleotide, oriLGAA, is completely inactive. The bubble sequence separates specific binding sites for NS1 and parvovirus initiation factor (PIF), a host cell cofactor required for activating the nickase activity of NS1 at the left-hand origin (10-12, 18). Individually, PIF and NS1 bind equally well to both forms of oriL; however, in combination, they show synergistic binding to oriLTC but not to oriLGAA, leading to asymmetric initiation of DNA replication across the dimer junction (10). In vitro replication assays have shown that the sequence composition of either the bubble dinucleotide or trinucleotide were relatively unimportant in determining the active or inactive origins, respectively (17). Asymmetric resolution of the dimer junction results in the conservation of a single sequence orientation, designated flip, in the left-hand telomere of all viral DNA molecules (1).
FIG. 1.
The dimer junction and left-hand minimal origin of replication. The left-hand hairpin of MVM is shown in diagrammatic form. Asymmetries such as the small ears, extra-helical T, and bubble sequence are indicated. The fully duplex, dimer junction, generated by rolling hairpin replication, is shown on the right. The short, palindromic sequences derived from the hairpin ears are represented by cross-hatched boxes. The minimal active origin is boxed, with an arrow indicating the nick site. The equivalent sequence generated on the GAA side of the bubble is also boxed; however, the arrow at the potential nick site is crossed out to indicate that it is not an active origin. Sequence details of the active left-hand origin are shown at the bottom of the figure. The minimal sequence required for origin activity is indicated by the double-headed arrow. Sequences of the bubble and the PIF, CREB, and NS1 binding sites are indicated, as is the NS1 nick site (12). An asterisk indicates the position of the extra-helical T, now base paired, and the gray box below it indicates the 7-of-8 match to the palindromic CRE consensus discussed in the text. The solid line between the complementary strands indicates the NS1 footprint.
PIF is a heteromeric complex containing two subunits, of 96 and 79 kDa (13, 27, 40, 41), which was simultaneously identified as glucocorticoid-modifying element binding protein, a factor responsible for modifying transcription of the glucocorticoid-responsive tyrosine transamidase promoter (13, 40). Human PIF96 and PIF79 are proteins of 563 and 530 amino acids, respectively, and are members of an emerging family of nuclear proteins containing a DNA binding SAND domain (Sp-100, AIRE-1, NucP41/75, and DEAF-1 [6, 20]). The intrinsic ability of both PIF subunits to transactivate reporter gene expression in mammalian one-hybrid assays (13) suggests that the normal function of the PIF complex in uninfected host cells is as a transcription factor.
The PIF binding site in the left-hand origin consists of two ACGT half-sites spaced five bases apart, with the proximal half-site juxtaposed to the bubble. Mutational analysis of the binding site revealed that while the first and fourth positions can vary in one of the half-sites, the core CpG dinucleotides in each are invariant. In contrast, the sequence of the spacer region between the half-sites is of minimal importance, and spacer sequences of 1 to 9 bases are all recognized (10, 12). PIF therefore has the unusual property of recognizing flexibly spaced half-sites, although dissociation rate analysis established that the optimum spacer length was 6 bp (7). Thus, the left-hand origin of MVM actually contains a suboptimal PIF binding site.
Overlapping the distal PIF half-site is a cyclic AMP-responsive element (CRE) which has been shown to contribute to basal levels of P4 promoter activity and to the upregulation of P4 in ras-transformed cells (32, 33). These coincident binding sites in the hairpin are thus required for both viral DNA replication and transcription from the P4 promoter. We surmised that these processes would compete with one another during infection, and we wished to investigate the relative importance of these overlapping sites by creating viruses in which individual elements, such as the PIF binding site and CRE, were mutated. However, sequences in the terminal hairpin have proven difficult to manipulate in the context of the viral genome because their palindromic nature eliminates unique restriction sites and makes the design of PCR primers problematic. The GC-rich terminal “ears,” which are essential for viability, are also highly prone to deletion during the PCR process. Thus, parvovirus terminal palindromes have resisted conventional methods of mutagenesis. Here we describe a system for the facile generation of viruses with mutant telomeres, by combining synthetic hairpin oligonucleotides with a truncated form of the viral genome, and use this system to assess the relative importance of the two overlapping transcription factor binding sites.
MATERIALS AND METHODS
Cell cultures.
A9ouabR11 cells are a mouse L-cell derivative, and 324K cells are a simian virus 40 (SV40)-transformed newborn human kidney fibroblast cell line. All cell types were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum.
Construction of pChopII.
The plasmid pChopII was created by excising the left-hand hairpin sequence of MVM in pdBMVp (28) with PmeI and AluNI and replacing it with an AluNI-to-PmeI fragment from pUC19 generated by PCR using mutagenic primers. The PCR product was engineered so that the vector sequence ended with a BsaI site followed by 35 bases of the MVM genome, ending at the PmeI site.
Generation and purification of mutant viruses.
The sequences of synthetic oligonucleotide forms of the left-end hairpin are shown in Fig. 2. Each oligonucleotide was phosphorylated using T4 polynucleotide kinase (New England Biolabs), purified through a Sephadex G-50 spin column (Sigma), and adjusted to 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol before heating to 95°C for 2 min. The oligonucleotides were then cooled to room temperature, allowing them to anneal intramolecularly, forming hairpin secondary structures. pChopII was digested with BsaI and the 6-kb fragment was gel purified. Ligation reactions were performed using 5 μg of purified pChopII fragment and 0.5 μg of hairpin oligonucleotide in a 75-μl reaction volume, with a final concentration of 4% polyethylene glycol 8000, 0.6 mM ATP, 1× ligase reaction buffer, and 2,000 U of ligase (reaction buffer and ligase from New England Biolabs). Following overnight incubation at 16°C, DNA was extracted with phenol and then chloroform before ethanol precipitation. Transfections of A9 monolayers were performed using SuperFect (Qiagen, Valencia, Calif.) as directed by the manufacturer. Individual plaques were picked and grown in A9 cells to generate clonal virus stocks as previously described (36). The left-hand hairpin sequences from these viral stocks were cloned into bacterial plasmids. For this, low-molecular-weight parvovirus DNA from infected A9 cells was isolated using a modified Hirt procedure (25, 37) 24 h after infection. Supernatant DNA was digested with EcoRI, and the 1-kb fragment containing the fully duplex-extended form of the left-end terminal hairpin was gel purified and ligated into SmaI/EcoRI-double-digested pUC19. Following transformation into Sure cells (Invitrogen), clones were screened for inserts in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and sequenced using an MVM-specific primer that binds to the genome between nucleotides 227 and 253 and primes toward the left terminus.
FIG. 2.
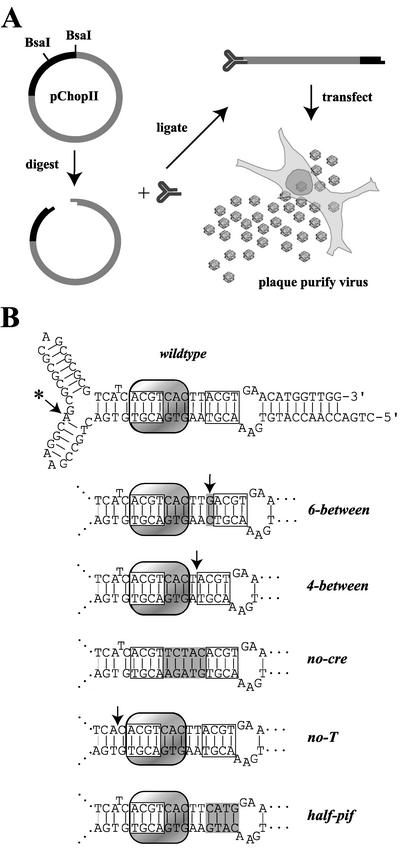
The MVM-Chop system for generating mutations in the left-hand terminal hairpin. (A) A map of pChopII, indicating the two BsaI restriction sites, is shown in the upper left, where gray indicates MVM sequence and black indicates bacterial vector. Digestion with BsaI and subsequent gel purification produced a 6-kb linear DNA with noncohesive 5′ overhangs at each end. Synthetic oligonucleotides that regenerate the terminal hairpin were ligated to the linearized plasmid. This construct was transfected into permissive cells from which mutant virus could be plaque purified for future analysis. (B) The sequence of the wild-type oligonucleotide is shown in hairpin form, with the two PIF half-sites and the CRE indicated by open and shaded boxes, respectively. The arrow with an asterisk indicates the identification of an A residue which differs from the previously published MVMp sequence, as discussed in the text. The sequences of the central regions of mutant hairpin oligonucleotides used in this study are shown below the wild-type hairpin. Altered and deleted sequences are indicated by gray boxes and arrows, respectively. Ellipses (…) indicate continuation of the wild-type sequence.
Virus was purified on 5-to-20% sucrose gradients centrifuged in a Beckman SW41 rotor at 35,000 rpm for 2.5 h at 4°C. Fractions were collected from the bottom of the tube and were analyzed for virus by hemagglutination assays as previously described (34). Packaged viral DNA was quantitated by incubating samples sequentially with micrococcal nuclease, proteinase K, and EDTA. Samples and duplex genomic standards purified from a plasmid form of the virus (1, 4, and 20 ng) were run on a 1.4% agarose-0.5 M NaOH denaturing gel for 16 h at 100 V. DNA was detected by Southern blotting and quantitated with a Molecular Dynamics PhosphorImager SI.
Immunofluorescence assays.
Immunofluorescence assays were performed essentially as described previously (16). Teflon-coated spot slides (Cell-Line Associates Inc.) were seeded with 2,500 A9 cells per spot in a total volume of 50 μl and then infected with 200, 1,000, 5,000, or 10,000 viral genomes per cell. Infection was limited to one round by the addition of neuraminidase (0.05 U/ml) added 6 h postinfection, and cells were fixed at 24 h postinfection. Infected cells were counted following indirect immunofluorescent staining with rabbit antiserum directed against the NS1 C-terminal 15 amino acids, detected with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories Inc.). MVM capsids were detected using a mouse monoclonal anti-capsid antibody, which only detects intact particles, followed by Texas Red-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories Inc.).
One-step growth-curve assays.
A total of 7 × 105 A9 cells was plated in 60-mm dishes and infected with 5,000 genomes per cell. Six hours postinfection, the original medium was replaced with fresh medium containing neuraminidase (0.05 U/ml) in order to limit infection to a single round and to reduce the contribution of adsorbed input virions to the Southern blot signal. At 12, 24, and 36 h postinfection, cells were scraped into the medium, and both cells and medium were harvested separately following centrifugation. Cell pellets were resuspended in 400 μl of 20 mM Tris (pH 7.5), 150 mM NaCl, 10 mM EDTA. Medium was separated from the cell pellet and spun for 3 h at 4°C in a Beckman SW50 at 45,000 rpm. All but 400 μl of the medium was gently aspirated, and the pelleted virus was resuspended in this residual medium. Both viral and cellular resuspensions were digested with 80 μg of proteinase K and 1% sodium dodecyl sulfate for 30 min at 37°C in a final volume of 425 μl, followed by 4 h at 60°C. Samples were extracted twice with phenol-chloroform and twice with chloroform and then precipitated overnight with ethanol. DNA was resuspended and digested with BsaI (New England Biolabs). A 10-μl aliquot of each sample was run on a 1.4% agarose gel for 6 h at 120 V. Gels were stained with ethidium bromide and quantitated using the EDAS 290 Kodak gel imaging system and Kodak 1D 3.5.4 software. Viral DNA was detected by Southern blotting and quantitated using a Molecular Dynamics PhosphorImager SI.
Construction of pChopIII.
In order to allow competing viruses growing in the same cell cultures to be distinguished from one another, a marked version of the wild-type genome, pChopIII, was created from pChopII using mutagenic primers in a two-stage PCR to give the sequence shown below in Fig. 7A. The final PCR product was digested with BglII and used to replace the BglII fragment of pChopII, and the mutagenesis was confirmed by direct DNA sequencing. Marked virus was produced using the wild-type hairpin and BsaI-digested pChopIII as described above and was termed wild-type variant (wild-type-var).
FIG. 7.
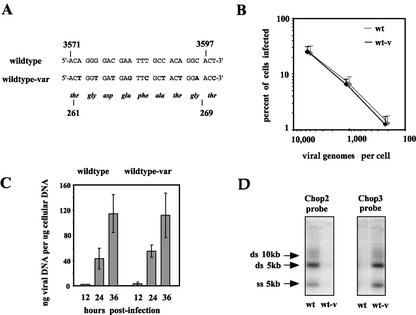
Design and validation of the virus competition system. (A) The nucleotide sequence of pChopII, numbered according to the method of Astell et al. (1), is indicated above the equivalent sequence of pChopIII, with altered nucleotides shown in bold. The predicted amino acid sequences of the VP proteins generated by both vectors are shown below the nucleotide sequences, numbered according to the amino acid sequence of VP2. Boxes indicate the sequences of the oligonucleotides Chop2 and Chop3 used as the discriminating probes in panel D. (B) Infection initiation assays performed with wild-type and wild-type-var viruses. Initiation efficiencies were determined as described in the legend for Fig. 4 by NS1 immunofluorescence, measured in rapidly dividing A9 cells. Infections were initiated at 200, 1,000, and 5,000 genomes per cell. For clarity, standard deviations from the mean are shown with the top error bars only, and the data points for wild-type-var (wt-v) are offset slightly to the left of those for wild-type (wt). (C) One-step growth-curve assays were performed with wild-type and wild-type-var viruses. Averaged results are from three experiments performed as described in the legend for Fig. 4. (D) Identical Southern blots of modified Hirt DNA extracts from wild-type- or wild-type-var-infected cells were probed with radiolabeled oligonucleotides, as for panel A above, specific for the relative viruses, as indicated above the autoradiograph. The major single- and double-stranded viral DNA species are indicated.
Competition experiments.
A total of 106 A9 or 324K cells was plated in 100-mm dishes and infected 24 h later with 1,000 genomes of ChopII virus per cell and 1,000 genomes of ChopIII virus per cell. At 72 h postinfection, cells were scraped into the medium, and cell pellets were prepared as before and resuspended in 200 μl of 10 mM Tris (pH 8.7), 0.1 mM EDTA. The amount of virus in a 5-μl sample was determined as described above for the purification of mutant viruses. Additional rounds of competition were performed exactly as for the first infection, using the virus generated by the previous round of competition at a multiplicity of 2,000 genomes per cell. The blots used to quantitate the amount of total virus were stripped by incubating the filter in 15 mM NaCl, 1.5 mM trisodium citrate for 20 min at 95°C and then assaying them for the relative amounts of pChopII- and pChopIII-derived genomes by Southern blotting. Discriminating oligonucleotide probes Ch2POS and Ch3POS were 5′-end labeled with [γ-32P]ATP using phage T4 polynucleotide kinase (New England Biolabs). The blot was probed with Ch2POS, stripped by incubating the filter in 15 mM NaCl, 1.5 mM trisodium citrate for 20 min at 95°C, and then probed with Ch3POS. Blots were quantitated using a Molecular Dynamics PhosphorImager SI. The sequences of Ch2POS and Ch3POS are shown below in Fig. 7.
RESULTS
The aim of this project was to create a reverse genetic system for introducing mutations into parvovirus hairpin telomeres by using an oligonucleotide containing the sequence of the left-hand hairpin to rescue a truncated MVM genome lacking this critical region. The power of this methodology lies in its ability to create mutant viruses in a system that has zero wild-type virus background. To this end, pChopII was derived from the MVMp infectious clone pdBMVp using mutagenic primers in a PCR to replace the sequence of the left-hand terminal hairpin with a BsaI restriction site. BsaI cleaves DNA downstream of its DNA recognition site (N1/N5), allowing for separation of viral sequence from that of the vector. Cleaving the pChopII plasmid with BsaI drops out 1 kb of vector sequence, allowing the truncated MVM genome to be recovered in a 6-kb fragment with 5′ overhangs, each four nucleotides long. These overhangs have different sequences at either end, as shown in Fig. 2A, which prevent its recircularization during ligation to the hairpin oligonucleotide. Likewise, the nonpalindromic nature of the overhang in the hairpin oligonucleotide prevents its self-ligation, allowing the ligation to be driven by high concentrations of phosphorylated oligonucleotide.
Development of the hairpin mutagenesis system.
Sequencing the left-hand hairpin region of the prototype MVM infectious clone, pdBMVp (28), in both directions with multiple primers revealed a single insertion in the previously published MVMp sequence (3). The corrected sequence contains an adenosine nucleotide opposite the thymidine found in the smaller terminal ear (Fig. 2B) and makes the left-end hairpin sequence of the prototype strain, MVMp (3), identical to that of the immunosuppressive strain, MVMi (2). The 92-base oligonucleotide used to reconstruct this corrected version of the wild-type left-hand terminal hairpin had a 5′ overhang complementary to that of the BsaI-truncated form of the left end of MVM in pChopII. The hairpin oligo was ligated onto the plasmid sequence, and the resulting DNA was transfected into permissive cells to generate wild-type virus. Using this system, mutations can be created in the left-hand terminal hairpin simply by altering the sequence of the synthetic oligonucleotide. pChopII lacks the sequence of the left-hand telomere, including the majority of the left-hand origin of replication, the terminal ears, and upstream elements of the P4 promoter, namely the CRE or CREB binding site, the E Box, and the Y Box (22, 32), and thus was predicted to be incapable of producing viable virus. Indeed, multiple transfections of the truncated plasmid alone have failed to produce virus, even after extensive blind passage.
One of the critical limiting steps in creating virus with the pChop system is the efficiency of the ligation reaction. To maximize virus yield, the ligation reaction was optimized using polyethylene glycol 8000 and ATP. The extent of ligation was monitored both by restriction digestion and by measuring the incorporation of 32P-labeled oligonucleotide into genome-length molecules, and it was reproducibly found to yield attachment of oligonucleotide to >95% of the truncated vector (data not shown).
As shown in Fig. 2B, mutant oligonucleotides were designed to investigate the roles of various sequence elements found in the left-hand hairpin. The hairpin 6-between contains a GC base pair inserted between the PIF half-sites and generates optimal spacing for PIF binding (7), while in 4-between an AT base pair was deleted from the sequence separating the PIF half-sites. Both 6-between and 4-between alter the spacing of the PIF half-sites without disrupting the CRE and, based on a previous dissociation rate analysis (7), are predicted to increase and decrease, respectively, the affinity of PIF binding compared to that with the wild-type sequence. In no-T, the conserved extra-helical T was deleted. In no-cre and half-pif, the sequences between the PIF half-sites and that of the proximal PIF half-site have been scrambled, without altering the base composition of the hairpin, to disrupt the CREB and PIF binding sites, respectively.
Inactivation of the PIF binding site.
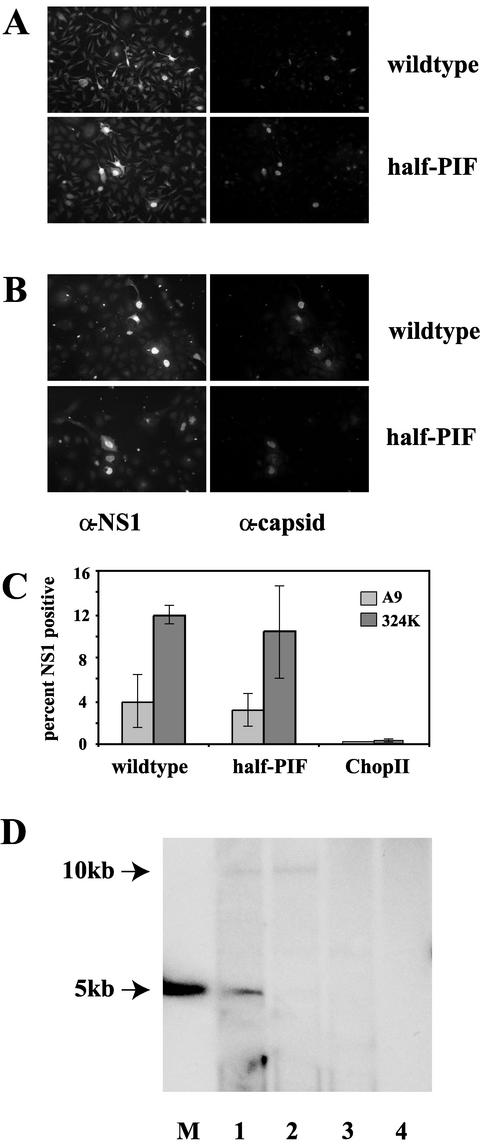
All of the mutant constructs, except half-pif, yielded virus after transfection, with efficiencies similar to the wild type. In six separate experiments, half-pif failed to generate viable mutant virus. However, immunofluorescent staining in A9 and 324K cells confirmed that transfection of the half-pif construct was successful and that transfected cells were capable of producing NS1 and the viral capsid proteins, indicating that the half-pif genome was capable of supporting both early and late viral gene expression (Fig. 3A). The efficiency of transfection as measured by the percentage of NS1-positive nuclei for both the wild-type and half-pif constructs was indistinguishable in both cell types tested (Fig. 3B). Cells transfected with BsaI-cut pChopII ligated in the absence of a hairpin oligo showed a 30- to 60-fold reduction in transient-expression efficiency compared to wild type, indicating that the P4 promoter is greatly reduced in its ability to initiate viral gene expression when dissociated from its upstream hairpin elements. In all instances, cells transfected with wild-type succumbed to infection and developed cytopathic effect within the first or second passage. In contrast, cells transfected with half-pif were split up to 10 times, without generating any detectable virus, and remained phenotypically normal. These cells remained susceptible to MVM infection and were indistinguishable from unmanipulated A9 cells when infected with wild-type virus (data not shown). Plasmids containing the right-hand terminal hairpin sequence of MVM have been shown to replicate in vivo when NS1 is supplied in trans (19). Because half-pif contains the right-hand terminal hairpin sequence and is capable of expressing NS1, we predicted that half-pif would be able to replicate its DNA utilizing the right-hand origin but would be unable to resolve unit-length genomes from the resulting 10-kb dimer molecules. In order to test this hypothesis, we analyzed modified Hirt DNA extracts from transfected 324K cells by Southern blotting, as shown in Fig. 3C. The dominant replicative intermediate in extracts from cells infected with wild-type virus was a duplex 5-kb species (lane 1), although a 10-kb double-stranded dimer band was also apparent. However, in extracts of cells transfected with half-pif, only the 10-kb dimer band was present (lane 2), indicating that a functional PIF binding site is required for resolution of the dimer intermediate in vivo as well as in vitro. Transfections of pChopII without a hairpin oligonucleotide attached failed to generate detectable levels of replicating viral DNA and gave results equivalent to those for untransfected cells (compare lanes 3 and 4).
FIG. 3.
Transfected half-pif expresses NS1 and capsid proteins. (A and B) Fluorescence microscopy was used to detect NS1 and VP expression during a single-round infection in A9 (A) or 324K (B) cells transfected with either wild-type or half-pif ligation products, as indicated. Cells were fixed 48 h posttransfection and then stained for NS1 (fluorescein isothiocyanate) and capsids (Texas Red), as indicated above the panels. (C) Bar graph showing the average percentage of NS1-positive cells, determined by counting 10 fields equivalent to those shown in panel A, following transfection with either wild-type or half-pif or with pChopII vector in the absence of hairpin oligonucleotide. Error bars represent 1 standard deviation from the mean. (D) DNA extracted by the modified Hirt procedure from 324K cells at48 h posttransfection was digested to completion with DpnI, run on native agarose gels, blotted, and then probed for MVM DNA. A double-stranded version of the MVM genome used as a marker is shown in the lane labeled M. Lanes 1 and 2 contain DNA extracted from wild-type and half-pif transfections, respectively, while DNA extracted from pChopII vector alone in transfected and untransfected cells is shown in lanes 3 and 4, respectively.
The viable mutants differ in intrinsic infectivity.
Using the MVM-Chop system, wild-type, 6-between, 4-between, no-T, and no-cre viruses were generated, and their left-hand hairpins were verified by sequencing. Viral stocks of each were purified on sucrose gradients and analyzed for packaged viral DNA by probing Southern blots of alkaline agarose gels as described in Materials and Methods. Stocks were adjusted to 1 μg of packaged viral DNA/ml, and these concentrations were verified by a further round of gel electrophoresis and Southern blotting.
Infection initiation efficiencies of wild-type, 6-between, 4-between, no-T, and no-cre viruses, defined as the percentage of infected cells expressing NS1, were determined by immunofluorescence. A9 and 324K cells were infected with each virus at a wide range of multiplicities, as shown in Fig. 4. While 6-between, 4-between, and no-T were only slightly impaired in their abilities to initiate infection in A9 cells relative to that with wild-type, the no-T virus showed a relatively greater reduction in initiation efficiency in 324K cells. Significantly, while no-cre showed a 10-fold reduction in initiation efficiency in A9 cells, this mutant exhibited a more than 100-fold reduction in 324K cells, suggesting that a functional CRE plays a significant role in initiating infection in general but is especially important in 324K cells. Over the range of multiplicities tested, all of the viable mutant viruses gave dose-response curves with slopes of approximately one, implying single-hit kinetics, as reported previously for the plaque assay kinetics of the parental MVMp (36). Thus, even in the case of the extremely debilitated no-cre mutant in 324K cells, the mutant phenotype cannot be overcome by increasing the number of input genomes.
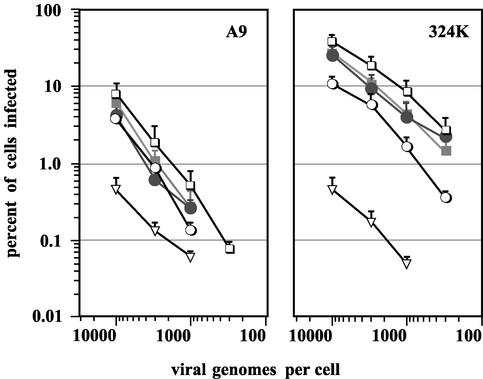
FIG. 4.
Relative efficiency of viral infection initiation. Infection initiation dose responses were determined by measuring the percentage of NS1-positive nuclei 24 h postinfection in rapidly dividing A9 or 324K cells, using immunofluorescence, over a 30-fold range of input multiplicities. Infections were initiated at 10,000, 3,300, 1,100, and 360 genomes per cell. Each data point is the average percentage of NS1-positive cells from at least 10 randomly chosen fields at a magnification of ×200 (approximately 200 A9 cells or 100 324K cells per field). □, wild-type; •, 6-between; ▪, 4-between; ▿, no-cre; ○, no-T. For clarity, standard deviations from the mean are shown with the top error bar only.
The abilities of wild-type and mutant viruses to form plaques in A9 and 324K cell monolayers were compared. Figure 5 shows the number of plaques formed in a 60-cm dish with an input of 5 × 106 viral genomes per dish (equivalent to 10 genomes per cell). These results are in agreement with previous findings in that wild-type virus formed plaques more efficiently in SV40-transformed human cells than in murine A9 cells (36). While 6-between and 4-between showed equivalent levels of plaque formation in both cell types relative to wild-type, no-T was reduced in its relative ability to form plaques in 324K cells, and while no-cre showed approximately 5-fold fewer plaques in A9 cells relative to wild-type, its ability to form plaques in 324K monolayers was reduced by greater than 99%, consistent with its much lower efficiency in initiating infection in these cells.
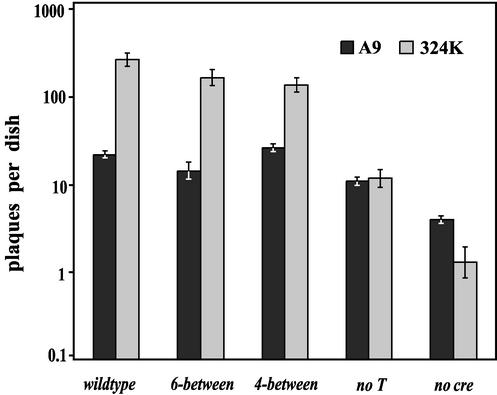
FIG. 5.
Plaque-forming abilities of mutant viruses in A9 and 324K cells. Plaque assays were performed in A9 and 324K cells by using serial dilutions of virus stocks adjusted to an equal genome concentration. Results were normalized to 5 × 106 viral genomes per dish and are plotted on a logarithmic scale. Error bars represent one standard deviation from the mean of three experiments.
Mutant DNA replication in single-cycle infections.
One-step growth conditions were used to compare mutant viral DNA production with that of the wild type in both A9 and 324K cells. These experiments were performed using 5,000 genomes per cell which, based on previous infection initiation studies, is a subsaturating level of input virus. Infection was limited to a single round by adding neuraminidase to the medium at 6 h postinfection in order to remove the cell surface receptor for the virus (4), and viral DNA production was measured at 12, 24, and 36 h postinfection. In Fig. 6, autoradiographs of Southern blots probed for MVM DNA are shown above the ethidium bromide-stained gels from which they were produced to give an indication of the amount of cellular DNA in each sample. For all five viruses used in this experiment, intracellular viral DNA contained single-stranded, genomic-length progeny DNA molecules (5 kb), as well as the 5- and 10-kb double-stranded replicative intermediates generated by rolling hairpin replication (Fig. 6A and C, lanes 2 and 3). Packaged single-stranded viral DNA, found in the medium, was not detectable until 36 h postinfection (lane 6).
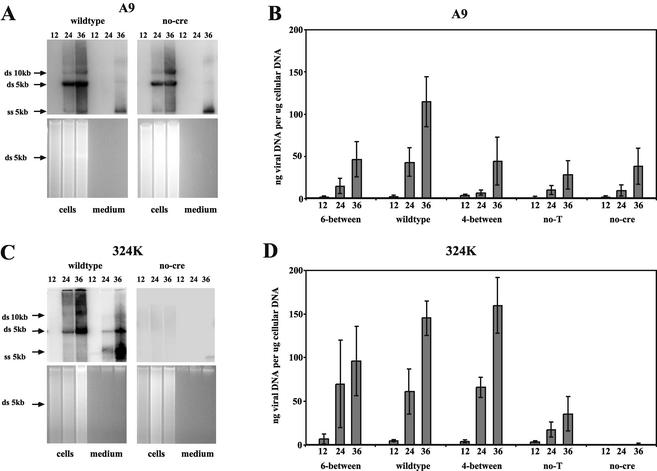
FIG. 6.
Relative DNA replication under one-step growth conditions. (A and C) DNA extracted from single-cycle infections using wild-type and no-cre viruses in A9 (A) and 324K (C) cells, with autoradiographs positioned above the gels from which they were created by Southern blotting. Individual lanes are aligned and contain total DNA extracted at 12, 24, and 36 h postinfection and then digested with BsaI. DNA extracted from equivalent amounts of both cells and medium are shown for each time point. The gels were stained with ethidium bromide, and the position of the double-stranded (ds) 5-kb band is shown. (B and D) On the Southern blots, the positions of 10-kb and 5-kb viral double-stranded bands, as well as those of 5-kb single-stranded (ss) progeny genomes, are indicated. Results for all five viruses, tested in A9 (B) and 324K (D) cells, are expressed in graphic form as the quantity of viral DNA (in nanograms), normalized to the amount of total DNA (in micrograms) in each sample. Each bar represents three independent experiments, with the standard deviations indicated. The numbers under each bar of the graph indicate hours postinfection.
Viral DNA detected on Southern blotting and total ethidium bromide-stained DNA detected in the gels were both quantitated as described in Materials and Methods and used to normalize the amount of viral DNA per infected culture. Averaged results from three separate experiments are shown in Fig. 6B and D. By 24 h postinfection in A9 cells, wild-type virus produced two- to threefold more DNA than the mutant viruses, a trend that continued out to 36 h postinfection. In 324K cells, 6-between, wild-type, and 4-between infection produced similar amounts of viral DNA at 24 and 36 h. Relative to wild-type, no-T showed a threefold decrease in viral DNA production at the later time points, while no-cre showed a 250-fold reduction in its ability to produce viral DNA by 36 h postinfection. When the amounts of packaged DNA found in the medium were expressed as a percentage of total viral DNA produced at a given time point, there was no significant statistical difference between the samples (P > 0.2), indicating that the amount of released, extracellular virus was proportional to the amount of intracellular replicating DNA for all of the viruses tested in both cell types (data not shown). This observation suggests that none of the viruses have a defect in packaging their DNA.
In order to investigate the possibility that the mutant viruses may be defective in their ability to resolve the various intermediate forms of viral DNA produced during rolling hairpin replication, the relative amounts of the three major forms of viral DNA (double-stranded 10-kb, double-stranded 5-kb, and single-stranded 5-kb forms) seen in the A9 cell extracts were individually quantitated. No statistically significant differences (P > 0.2) were found between wild-type and mutant viruses in the ratios of the single-stranded 5-kb, double-stranded 5-kb, and double-stranded 10-kb intermediates produced (data not shown).
Measurement of mutant virus fitness by one-on-one competition.
In order to test if the defects in infection initiation and viral DNA replication seen with these mutant viruses would lead to a reduction in fitness, we designed an experimental system in which each mutant virus could be placed in direct competition with the wild type. Competition assays are capable of measuring the relative fitness of two viruses regardless of whether the exact effects of the mutation are known, provided one can distinguish between the two competing viruses. To this end, pChopII was marked by altering every third base in a 27-nucleotide-long region within the VP reading frame common to both VP1 and VP2, giving pChopIII. These mutations do not change the sequence of the nine amino acids encoded here (Fig. 7A) but allow derivation of viruses equivalent to, yet distinguishable from, those produced by pChopII. The common region of the VP gene was chosen because only a single reading frame encodes protein in this region of the virus. Additionally, the immunosuppressive strain of MVM (MVMi) contains a DNA sequence similar to that engineered into pChopIII, without apparently impairing its viability. Thus, we predicted that the DNA sequence changes in pChopIII would have negligible effects on the life cycle of a virus produced using this vector when compared to equivalent virus derived from pChopII. Using the wild-type sequence hairpin oligonucleotide and pChopIII vector, wild-type-var virus was produced and was compared to pChopII-derived wild-type virus in both infection initiation assays and one-step growth curves (Fig. 7B and C). There was no appreciable difference between wild-type and wild-type-var viruses in their ability to initiate viral infection, and the two viruses also showed equivalent levels of DNA replication in single-round infections. In order to confirm our ability to distinguish these two viruses, Southern blots of infected cell DNA extracts were probed with 5′-end-labeled oligonucleotides specific for the engineered sequence differences between wild-type and wild-type-var. As shown in Fig. 7D, the oligonucleotide probes bound specifically to the genome of the virus they were directed against, without significant nonspecific cross-reaction.
Competition assays were performed in A9 and 324K cells by infecting cells at a multiplicity of 2,000 genomes per cell with a mixture containing equal amounts of wild-type-var and one of the pChopII-derived viruses. Infected cells were incubated for 72 h to allow at least two complete rounds of the viral life cycle. Virus was then harvested, quantitated, and used to infect new cell cultures at the same initial multiplicity. Following four rounds of competition, samples from each round were assayed for the relative amounts of wild-type-var and pChopII-derived viral DNA by Southern blotting and differential oligonucleotide probing. The averaged results from three separate experiments are shown in Fig. 8.
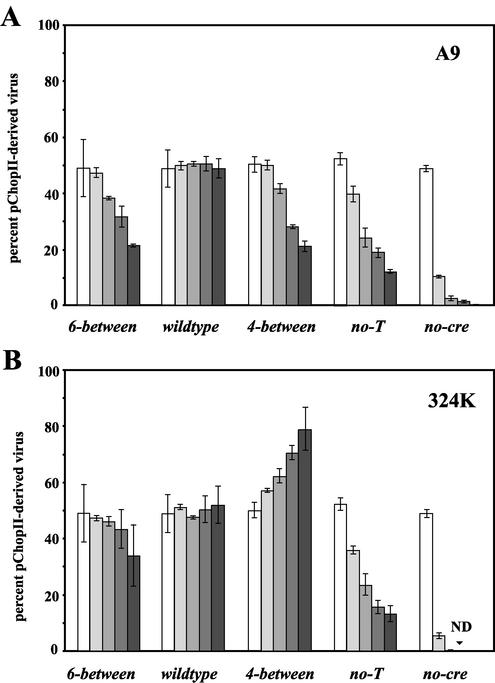
FIG. 8.
Relative fitness of each mutant virus in A9 (A) and 324K cells (B). Cells were simultaneously infected with wild-type-var virus and either 6-between, wild-type, 4-between, no-T, or no-cre pChopII-derived viruses, each at a multiplicity of 1,000 genomes per cell. Virus produced by each infection was used to infect new cells for an additional round of competition, as described in Materials and Methods. Average results from three experiments are given and are expressed as the percentage of mutant virus present in the total amount of virus detected in the sample. Each round of infection is represented by a bar of increasing shading, and the input prior to the first round of competition is represented by an unfilled bar. Error bars indicate one standard deviation from the mean.
As expected, the average amount of wild-type virus remained relatively constant in both cell types when placed in competition with wild-type-var. In A9 cells each of the mutant viruses showed reduced fitness relative to wild-type-var, in that the wild-type virus outperformed the mutant and comprised a greater fraction of the total virus produced in each successive round. 6-between and 4-between appeared to be competed out at similar rates, indicating that in A9 cells the wild-type spacing of 5 bases between the PIF half-sites is optimal. Of the mutant viruses, no-cre showed the weakest competition, being reduced by roughly 80% per round, relative to input. In 324K cells, while 6-between showed reduced fitness relative to wild-type-var, surprisingly 4-between out-competed the wild type, suggesting that, in this cell type, lower-affinity PIF binding sites lead to increased fitness. In 324K cells, no-cre virus was reduced by ≈90% per round and was undetectable by the third round of competition.
DISCUSSION
The PIF binding site is required for viral DNA replication.
Previous in vitro studies characterized PIF as a cofactor required for initiation of DNA replication at the viral left-hand end (10-12). We initially used the hairpin oligo-based mutagenesis system to ask whether PIF was also essential in vivo. Since the ACGT of the PIF half-site distal to the NS1 binding site is an invariant component of the CRE, we chose to eliminate the NS1-proximal half-site. Our inability to isolate mutant virus using the half-pif hairpin strongly supports the conclusion that a functional PIF site, and therefore the ability to recruit PIF to initiate replication at the left-hand end, is absolutely required for virus viability. There remained the possibility that we inadvertently disrupted the binding site of another unknown, but essential, transcription factor and that this lesion, rather than the inhibition of PIF binding, underlies our inability to recover viable virus from this mutant. However, half-pif transfections gave rise to infected cells with the same efficiency as wild-type transfections, and these expressed high levels of both NS1 and the viral capsid genes. In addition, these cells went on to display the typical morphological alterations, such as cytoplasmic extensions and rounding up, associated with NS1 cytotoxicity (9, 14, 31). This indicates that the P4 promoter in the half-pif genome was unaffected by the mutation and, interestingly, that PIF itself plays at most a minor role in driving nonstructural gene expression, at least as detectable by transfection assay. Since half-pif is predicted to lack a functional left-hand origin of replication, the mutant should be incapable of dimer junction resolution, and this is consistent with the observed buildup of 10-kb duplex genome dimers. This inability to resolve unit-length genomes from dimer intermediates would explain why half-pif fails to produce progeny virus capable of propagating infection and confirms that PIF plays an essential role in the replication of MVM in vivo.
While 6-between and 4-between generated slightly lower numbers of NS1-positive cells than wild-type virus in the infection initiation assay (Fig. 4), these mutants were capable of producing equivalent numbers of plaques in A9 monolayers (Fig. 5). This indicates that increasing or decreasing the spacing of the PIF half-sites by 1 nucleotide does not significantly affect the ability of MVM to initiate infection or complete the viral life cycle in these host cells. However, wild-type virus was able to out-compete either 6-between or 4-between in coinfections of A9 cells, a result that is most likely explained by the reduction in relative viral DNA replication seen in the one-step growth-curve experiments (Fig. 6). Likewise, the ability of 4-between to out-compete wild-type virus in 324K cells correlates well with the relative increase in viral DNA replication observed for the mutant. Because 4-between did not show increased levels of infection initiation in 324K cells relative to wild type, the overall relative increase in viral DNA produced in a single round of infection by the mutant represents an increase in replication per infected cell. Our inability to detect statistically significant differences (P > 0.2) between wild-type, 6-between, and 4-between in their relative production of the various rolling hairpin intermediates (data not shown) suggests that these small changes in the spacing of the PIF half-sites alter the level to which viral replication intermediates accumulate without affecting their processing.
The CRE is necessary for efficient initial viral gene expression.
In infection initiation assays, no-cre virus generated 10-fold fewer NS1-positive cells than wild type in A9 cells and 100-fold fewer in 324K cells, at all concentrations of virus tested. This reduction in infection initiation accounts for the lower levels of no-cre viral DNA per unit of cellular DNA seen in the one-step growth curves, suggesting that once viral infection is initiated, viral DNA replication itself is not significantly impaired. Likewise, its reduced initiation rate correlated directly with its reduced plaquing efficiency, suggesting that this was due solely to its inability to induce expression of NS1 from the P4 promoter rather than to a deficiency in any subsequent step of the viral life cycle.
The CRE upstream of the MVM P4 promoter has been shown to contribute to basal levels of P4 promoter activity and to the upregulation of P4 in ras-transformed cells (32). These studies measured reporter gene activity following transfection of plasmid forms of the promoter that did not mimic the terminal structure in which the CRE is located in authentic viral genomic DNA. However, the debilitated phenotype shown here by the no-cre virus indicates that this element does indeed play a critical role in the establishment of infection and indicates that this role is especially important in 324K human fibroblasts. Since these cells are transformed by SV40, and not by ras, this system does not afford a strict parallel to the observations in ras-transformed cells. However, recent studies on the step-wise transformation of human fibroblasts with defined oncogenes (23, 24) show that the activation of ras is certainly one solution to a crucial step in the pathway to transformation by SV40 large and small T antigens. Further studies comparing wild-type and no-cre virus growth in human fibroblasts at distinct steps on this transformation pathway may establish if one particular oncogenic event makes the CRE necessary for efficient viral infection.
The observation that the CRE site is needed in cis for P4 promoter activation suggests the possibility that supplying NS-1 in trans complements the mutant phenotype. The results of infection initiation assays predict that no-cre should initiate infection at rates 10- and 100-fold lower than that for wild-type virus in A9 and 324K cells, respectively. Given that in each round of competition the virus can complete two rounds of its life cycle, the expected maximum production of no-cre virus relative to wild-type virus per round of competition would be 1% in A9 cells and 0.01% in 324K cells. However, no-cre was only reduced 80 and 90% per round of competition in A9 and 324K cells, respectively. This discrepancy between predicted and observed results indicates that, despite the stringency of its phenotype over a 100-fold range of input multiplicities, the reduced ability of no-cre to initiate infection may be partially rescued by coinfection with wild-type virus.
The extra-helical T may augment CRE function.
The CRE present in the MVM hairpin is conserved in canine, porcine, hamster (H3), and rat (H1) parvoviruses. These viruses also conserve the extra-helical T positioned on the outer arm of the palindrome, 1 bp outboard of the distal PIF half-site and 3 to 4 bp from the terminal ears. Conservation of this asymmetry suggests that it is likely to play an important role in the viral life cycle, and our results indicate that its activity may be related to that of the CRE. Like no-cre, no-T demonstrated a reduction in intrinsic infectivity, as measured in the infection initiation assay, responsible for the reduction in viral DNA replication seen in the one-step growth-curve experiments. Statistical analysis of these data revealed that, like no-cre, the reduction in viral DNA production shown by no-T can be explained by a reduced efficiency of infection initiation. Additionally, no-T showed a more dramatic reduction in infection initiation and plaque formation, relative to wild type, in 324K cells than it did in A9 cells, which would be expected if the extra-helical T contributes in some way to the function of the CRE. Also, in competition with wild-type, no-T generated 10-fold higher levels of virus than would have been predicted by the infection initiation assay, indicating that, like no-cre, no-T could be partially rescued by coinfection. As with the other mutants, no-T infection produced a ratio of the various forms of viral replicative intermediates and progeny genomes equivalent to that of wild type, suggesting that the extra-helical T is not involved in the processing of concatemeric viral DNA or the packaging of progeny single strands.
Sequencing degenerate oligonucleotides of the form N3CGN13 selected by binding to recombinant CREB protein, we previously found the preferred binding site for CREB to be ACGTCAC (8), an observation that supports the identification of this sequence as a functional CRE within the retinoblastoma gene promoter (21, 35). However, the consensus CRE sequence has been primarily reported in the literature as comprising the palindrome TGACGTCA (5, 26, 30). While the palindromic nature of a given CRE may not be absolutely required for ATF-CREB binding, the observation that many functional CREs are palindromic raises the possibility that this sequence arrangement may potentiate ATF-CREB interaction in vivo. Thus, it is possible that the extra-helical T is involved in optimizing the MVM CRE, as its presence creates a 7-of-8 match to the palindromic consensus, as shown in Fig. 1. We are currently using the hairpin oligo mutagenesis system to test the sequence requirements at, or in the vicinity of, this enigmatic nucleotide in order to determine whether the extra-helical T may contribute to NS1 expression levels in a CREB/ATF-dependent manner.
Infection initiation efficiency is influenced by competition between factors binding at the CRE and PIF binding sites.
The overlapping natures of the CRE and the distal PIF half-site suggest that the relative affinities of these two elements for their cognate factors reflect a balance between their respective importance in NS gene transcription and viral DNA replication. We have shown previously that the optimal spacing between the PIF half-sites is 6 bases and that the affinity of the site for PIF falls off approximately twofold for each nucleotide deleted between the half-sites (7). Interestingly, in 324K cells decreasing the spacing between the half-sites, and thus reducing affinity of the binding site for PIF, increases the fitness of 4-between relative to wild-type, an effect contrary to that of inhibition of ATF/CREB binding in no-cre, suggesting that the two mutant phenotypes result from tilting the balance between PIF and ATF/CREB binding in opposite directions, although this would be expected to enhance 4-between infection initiation in 324K cells compared to that with wild-type, a phenomenon which we did not detect.
Since NS1 is required to initiate viral DNA replication, it is possible that the reduced DNA replication levels observed for 6-between in 324K cells result from the creation of a binding site in which PIF has a competitive advantage over ATF/CREB, thereby reducing the latter's ability to upregulate NS gene transcription through the CRE. Perros and colleagues (32, 33) showed that at least three different species of CREB/ATF dimers can bind the MVM CRE and that one of these is more common in extracts associated with increased P4 activation. Our results suggest that the activating CREB/ATF complexes found in transformed 324K cells may compete directly with PIF for the MVM CRE, a condition which would reduce the fitness of the virus in the context of an optimally spaced PIF binding site. If borne out by further studies, these observations will have important implications for optimizing the oncotropic properties of this group of autonomously replicating parvoviruses.
Acknowledgments
We thank Susan Cotmore and Jesper Christensen for valuable discussion and colleagues at the HHMI Biopolymer Laboratory and W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for DNA sequencing and synthesis of oligonucleotides used in this study.
This work was supported by U.S. Public Health Service grants AI26109 and CA29303 from the National Institutes of Health. E.B. was supported, in part, by T32 GM07499.
REFERENCES
- 1.Astell, C. R., M. B. Chow, and D. C. Ward. 1985. Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J. Virol. 54:171-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell, C. R., E. M. Gardiner, and P. Tattersall. 1986. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J. Virol. 57:656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell, C. R., M. Thomson, M. B. Chow, and D. C. Ward. 1983. Structure and replication of minute virus of mice DNA. Cold Spring Harbor Symp. Quant. Biol. 47:751-762. [DOI] [PubMed] [Google Scholar]
- 4.Ball-Goodrich, L. J., R. D. Moir, and P. Tattersall. 1991. Parvoviral target cell specificity: acquisition of fibrotropism by a mutant of the lymphotropic strain of minute virus of mice involves multiple amino acid substitutions within the capsid. Virology 184:175-186. [DOI] [PubMed] [Google Scholar]
- 5.Benbrook, D. M., and N. C. Jones. 1994. Different binding specificities and transactivation of variant CRE's by CREB complexes. Nucleic Acids Res. 22:1463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottomley, M. J., M. W. Collard, J. I. Huggenvik, Z. Liu, T. J. Gibson, and M. Sattler. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8:626-633. [DOI] [PubMed] [Google Scholar]
- 7.Burnett, E., J. Christensen, and P. Tattersall. 2001. A consensus DNA recognition motif for two KDWK transcription factors identifies flexible-length, CpG-methylation sensitive cognate binding sites in the majority of human promoters. J. Mol. Biol. 314:1029-1039. [DOI] [PubMed] [Google Scholar]
- 8.Burnett, E. D. 2002. Genetic and functional analysis of a parvoviral hairpin telomere. Ph.D. dissertation. Yale University, New Haven, Conn.
- 9.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, J., S. F. Cotmore, and P. Tattersall. 2001. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 75:7009-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J. Virol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J. Virol. 71:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, J., S. F. Cotmore, and P. Tattersall. 1999. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol. Cell. Biol. 19:7741-7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corbau, R., V. Duverger, J. Rommelaere, and J. P. Nuesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore, S. F., J. P. Nuesch, and P. Tattersall. 1992. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology 190:365-377. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177:477-487. [DOI] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., and P. Tattersall. 1994. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 13:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotmore, S. F., and P. Tattersall. 1992. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J. Virol. 66:420-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, T. J., C. Ramu, C. Gemund, and R. Aasland. 1998. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem. Sci. 23:242-244. [DOI] [PubMed] [Google Scholar]
- 21.Gill, R. M., P. A. Hamel, J. Zhe, E. Zacksenhaus, B. L. Gallie, and R. A. Phillips. 1994. Characterization of the human RB1 promoter and of elements involved in transcriptional regulation. Cell Growth Differ. 5:467-474. [PubMed] [Google Scholar]
- 22.Gu, Z., S. Plaza, M. Perros, C. Cziepluch, J. Rommelaere, and J. J. Cornelis. 1995. NF-Y controls transcription of the minute virus of mice P4 promoter through interaction with an unusual binding site. J. Virol. 69:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. A. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 26.Iguchi-Ariga, S. M., and W. Schaffner. 1989. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 3:612-619. [DOI] [PubMed] [Google Scholar]
- 27.Jimenez-Lara, A. M., M. J. Heine, and H. Gronemeyer. 2000. Cloning of a mouse glucocorticoid modulatory element binding protein, a new member of the KDWK family. FEBS Lett. 468:203-210. [DOI] [PubMed] [Google Scholar]
- 28.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg, A., and T. A. Baker. 1991. DNA replication, 2nd ed. W. H. Freeman, New York, N.Y.
- 30.Montminy, M. R., K. A. Sevarino, J. A. Wagner, G. Mandel, and R. H. Goodman. 1986. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl. Acad. Sci. USA 83:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mousset, S., Y. Ouadrhiri, P. Caillet-Fauquet, and J. Rommelaere. 1994. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J. Virol. 68:6446-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perros, M., L. Deleu, J. M. Vanacker, Z. Kherrouche, N. Spruyt, S. Faisst, and J. Rommelaere. 1995. Upstream CREs participate in the basal activity of minute virus of mice promoter P4 and in its stimulation in ras-transformed cells. J. Virol. 69:5506-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perros, M., F. Fuks, Z. Kherrouche, and J. Rommelaere. 1999. Atypical nucleoprotein complexes mediate CRE-dependent regulation of the early promoter of minute virus of mice. J. Gen. Virol. 80:3267-3272. [DOI] [PubMed] [Google Scholar]
- 34.Ron, D., P. Tattersall, and J. Tal. 1984. Formation of a host range mutant of the lymphotropic strain of minute virus of mice during persistent infection in mouse L cells. J. Virol. 52:63-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai, T., N. Ohtani, T. L. McGee, P. D. Robbins, and T. P. Dryja. 1991. Oncogenic germ-line mutations in Sp1 and ATF sites in the human retinoblastoma gene. Nature 353:83-86. [DOI] [PubMed] [Google Scholar]
- 36.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tattersall, P., L. V. Crawford, and A. J. Shatkin. 1973. Replication of the parvovirus MVM. II. Isolation and characterization of intermediates in the replication of the viral deoxyribonucleic acid. J. Virol. 12:1446-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tattersall, P., and D. C. Ward. 1976. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263:106-109. [DOI] [PubMed] [Google Scholar]
- 39.Willwand, K., E. Mumtsidu, G. Kuntz-Simon, and J. Rommelaere. 1998. Initiation of DNA replication at palindromic telomeres is mediated by a duplex-to-hairpin transition induced by the minute virus of mice nonstructural protein NS1. J. Biol. Chem. 273:1165-1174. [DOI] [PubMed] [Google Scholar]
- 40.Zeng, H., D. A. Jackson, H. Oshima, and S. S. Simons, Jr. 1998. Cloning and characterization of a novel binding factor (GMEB-2) of the glucocorticoid modulatory element. J. Biol. Chem. 273:17756-17762. [DOI] [PubMed] [Google Scholar]
- 41.Zeng, H., S. Kaul, and S. S. Simons, Jr. 2000. Genomic organization of human GMEB-1 and rat GMEB-2: structural conservation of two multifunctional proteins. Nucleic Acids Res. 28:1819-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]