Abstract
Certain major histocompatibility complex class I (MHC-I) alleles are associated with delayed disease progression in individuals infected with human immunodeficiency virus (HIV) and in macaques infected with simian immunodeficiency virus (SIV). However, little is known about the influence of these MHC alleles on acute-phase cellular immune responses. Here we follow 51 animals infected with SIVmac239 and demonstrate a dramatic association between Mamu-A*01 and -B*17 expression and slowed disease progression. We show that the dominant acute-phase cytotoxic T lymphocyte (CTL) responses in animals expressing these alleles are largely directed against two epitopes restricted by Mamu-A*01 and one epitope restricted by Mamu-B*17. One Mamu-A*01-restricted response (Tat28-35SL8) and the Mamu-B*17-restricted response (Nef165-173IW9) typically select for viral escape variants in early SIVmac239 infection. Interestingly, animals expressing Mamu-A*1 and -B*17 have less variation in the Tat28-35SL8 epitope during chronic infection than animals that express only Mamu-A*01. Our results show that MHC-I alleles that are associated with slow progression to AIDS bind epitopes recognized by dominant CTL responses during acute infection and underscore the importance of understanding CTL responses during primary HIV infection.
Several lines of evidence suggest that cellular immunity is critically important in the immune control of human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV). Strong antigen-specific CD8+ and CD4+ T-lymphocyte responses are correlated with improved clinical outcome in HIV-infected individuals (7, 33, 49), while depletion of CD8+ lymphocytes in SIV-infected macaques accelerates disease progression (20, 30, 55). Additionally, expression of certain major histocompatibility complex class I (MHC-I) and MHC-II alleles, such as HLA-B*27 and HLA-B*57, is associated with unusually favorable clinical outcomes (long-term nonprogression) (5, 8, 12-14, 17, 21-24, 35, 50, 53, 57, 58).
Chronic-phase cellular immune responses in HIV long-term nonprogressors expressing HLA-B*27 and -B*57 have been characterized in detail. Immunodominant responses directed against epitopes within Gag p24 are restricted by each of these molecules (11, 12, 15, 16, 42, 43). In addition, strong cytotoxic T lymphocyte (CTL) responses restricted by these molecules have been mapped in Rev, Env, and Pol (25, 56, 59). Most previous studies of long-term nonprogressors have emphasized the circumstantial link between these dominant chronic-phase CTL and delayed disease progression.
However, these chronic-phase CTL may be only one component of the vigorous chronic-phase immune response that characterizes long-term nonprogression. CD8+ T cells of nonprogressors proliferate, produce perforin, and elaborate soluble virus inhibitors (29, 34, 63, 68). Additionally, strong HIV-specific CD4+ proliferative responses are frequently detected in nonprogressors (49). It is not clear why these antigen-specific responses occur predominantly in individuals with certain MHC-I genotypes. One possibility is that the acute-phase CTL responses initially contain viral replication, allowing for the development of a coordinated and robust antiviral response. In support of this theory, plasma viral load following acute infection is predictive of disease outcome in HIV-infected individuals (viral setpoint) (10, 31, 32). Acute-phase cellular immune responses, which arise coincidently with the decline in primary viremia (65), likely play a role in determining this viral setpoint.
Unfortunately, it is difficult to study these immune responses during acute HIV infection (66). Synthetic peptides for ex vivo CTL assays need to be tailored to each infected individual, since consensus peptides do not accurately reflect the genotypic variability among infecting virus strains (62). Moreover, most HIV-infected individuals do not know precisely when they were infected, and many remain asymptomatic throughout acute infection. By the time that HIV infection is verified, MHC-I alleles are identified, and autologous viral peptides are synthesized, the narrow window of opportunity for studying the immune responses that are restricted by a patient's MHC-I alleles that might delay disease progression during acute HIV infection is usually closed. Complicating the situation, immune escape variants can be detected as early as 3 weeks postinfection (45) and can dominate the circulating virus population by 4 weeks postinfection (3). Studying the responses that select for these variants requires an exquisite knowledge of the precise timing of infection that is lacking in most cases of HIV.
The hypothesis that important acute-phase CTL responses influence long-term clinical outcome is more easily tested in macaques experimentally infected with SIV, since the precise timing of the acute phase, as well as the genetic composition of the infecting virus, are carefully controlled. We reasoned that the delayed disease progression associated with particular MHC-I alleles results from effective control of viral replication during acute infection. By evaluating acute-phase CTL responses prior to the emergence of viral escape variants, we eliminate the systematic bias against potentially important, but ephemeral, CTL that rapidly select for escape variants. In this study, we identify an association between three MHC-I alleles and positive outcome following challenge with highly pathogenic SIVmac239. We then show that the MHC-I alleles associated with improved control bind epitopes recognized by the dominant CTL responses during acute infection.
MATERIALS AND METHODS
Animals, viruses, and infections.
Fifty-one macaques were infected with a molecularly cloned virus, SIVmac239. Five stocks of virus were used, namely, stocks 1 to 3 (SIVmac239/nef open expanded on rhesus peripheral blood lymphocytes), stock 4 (SIVmac239/nef stop expanded on rhesus peripheral blood lymphocytes), and stock 5 (SIVmac239/nef open expanded on CEMx174). SIVmac239/nef stop differs from SIVmac239/nef open by a stop codon present in the nef gene (52). There is an intense selection for full-length Nef protein in vivo, and the nef open reading frame is restored within a few weeks of infection. Animals infected with SIVmac239/nef open received 10 50% monkey infectious doses, comprising approximately 3,000 50% tissue culture infective doses (TCID50), intrarectally. Animals infected with SIVmac239/nef stop received 1,000 TCID50 intrarectally. Animals infected with SIVmac239/nef open expanded on CEMx174 cells received 20 ng of p27 SIVmac239 (3,000 TCID50) intrarectally. Twenty-one of the 51 macaques, including SIV-controlling (controller) animals 95061 and 95096, were vaccinated as part of previous studies that failed to ameliorate disease progression (1, 2, 19, 61).
Stock 1 was used to challenge 11 animals (including Mamu-A*01-, -B*17-, and -B*29-positive animal 95061), stock 2 was used to challenge 8 animals, and stock 3 was used to challenge 22 animals (including Mamu-A*01-, -B*17-, and -B*29-positive animals 2065, 2095, and 2129). The SIVmac239/nef open expanded stock 4 was used to challenge 6 animals (including Mamu-A*01-, -B*17-, and -B*29-positive animals 1937 and 95096), while the SIVmac239/nef stop stock was used to challenge 7 animals. SIV-infected animals at the University of Wisconsin were cared for in accordance with an experimental protocol approved by the University of Wisconsin Research Animal Resource Committee.
Viral load analysis.
Viral loads were quantitated by using the SIV branched-chain DNA assay (Chiron Diagnostics, Emeryville, Calif.) and the Taqman kinetic reverse transcriptase PCR assay (18, 64). SIV viral RNA levels were measured every week for the first 4 weeks and biweekly thereafter.
Statistical analysis.
Viral load differences between groups infected with SIVmac239 were tested for statistical significance by using t tests after log transformation of the data to improve normality and homoscedasticity. In addition, Levene's test for homoscedasticity was conducted, and if differences were found to be significant, the Welch correction for unequal variances was employed. Finally, to further examine the robustness of the results, a nonparametric test, the Mann-Whitney U test, was performed. The P values for the nonparametric tests were calculated by exact methods. All the P values are two tailed.
In determination of the extent of variability in Tat28-35SL8, mean and median proportions of nonsynonymous nucleotide differences per nonsynonymous site (pN) in the Tat28-35SL8 epitope in comparisons between viruses from monkeys and the inoculum were calculated by using the direct virus sequence shown in Fig. 5B. Proviral sequence from 95061 and 95096 and plasma sequence from the other 16 animals were weighted equally in this analysis. Polymorphic nucleotides were weighted equally in computation of mean pN between the sequences of virus from each monkey and the inoculum.
FIG. 5.
Variation in dominant CTL epitopes in chronic SIVmac239 infection. The three epitopes were directly sequenced and conceptually translated approximately 1 year postinfection. Codons with identity to the wild-type sequence are shown as dots. Complete amino acid replacements are shown as uppercase single-letter amino acid codes; mixed populations are shown as lowercase single-letter amino acid codes. PBMC proviral sequences were analyzed in animals 95061 and 95096, since repeated attempts to isolate plasma virus RNA from these animals failed. Shown are results for Gag181-189CM9 (A), Tat28-35SL8 (B), and Nef165-173IW9 (C).
MHC-I typing by PCR-SSP.
Using the technique of sequence-specific DNA amplification (PCR-SSP), we performed molecular typing of MHC-I alleles which bind SIV-derived peptides, namely, Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B∗01, Mamu-B*03, Mamu-B∗04, Mamu-B*17, and Mamu-B*29. The 3′-terminal region of PCR-SSP primers targeted nucleotide polymorphism unique to these eight Indian rhesus MHC-I alleles. PCR was performed as previously described (26), and PCR products were electrophoresed on 2% agarose gels (0.5× Tris-borate-EDTA). The agarose gels were analyzed for the presence of an internal control product and each of the specific allele amplicons.
MHC-I and -II cloning and sequencing.
From 1 to 5 million herpesvirus papio-transformed B cells or peripheral blood mononuclear cells (PBMC) were used to isolate total cellular RNA with the Qiagen total RNEasy kit (Qiagen, Valencia, Calif.) following the manufacturer's instructions. cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, Calif.). The cDNA was amplified with Pwo DNA polymerase (Roche, Mannheim, Germany) and MHC-I and -II 5′ and 3′ sequence-specific primers (sequences available upon request). The amplified cDNA was purified from an agarose gel by using a gel extraction kit (Qiagen) and was ligated into pCR-Blunt by using the Invitrogen Zero Blunt cloning kit following the manufacturer's protocol. Ninety six clones from each animal were miniprepped and sequenced with T7 and M13 primers on an ABI 377 automated sequencing machine (Applied Biosystems, Foster City, Calif.). Sequences were analyzed using software from Applied Biosystems and Accelrys (Burlington, Mass.).
Neutralization of SIVmac239 by SIV control sera.
Serum neutralization experiments determined the antibody titer needed to protect 50% of cells from virus-induced killing, as previously described (19, 36). Additional enzyme-linked immunosorbent assays for p27 production were performed as previously described (47) to confirm these results.
Enumeration of SIV-specific CD8+ and CD4+ responses by intracellular IFN-γ cytokine staining.
Intracellular staining for gamma interferon (IFN-γ) production was performed as previously described (37).
SIV sequencing.
SIV sequencing from blood plasma was performed as previously described (44).
SIV sequences from animals 95061, 95096, 96072, and 96093 were derived from PBMC provirus. PBMC were lysed, and the genomic DNA (gDNA) was isolated by DNeasy spin column purification (Qiagen). PCR amplicons spanning the entire SIV epitopes of interest (3) were generated with the HotStarTaq master mix kit (Qiagen) and 500 ng of gDNA. At least four independent amplifications were performed for time points for animal 95061. The amplified products were purified using QIAquick PCR purification kits (Qiagen) and then directly sequenced as previously described (3). Mixed-base sites within single amplicons and single-site differences in the sequences of multiple amplicons from the same time point were both considered polymorphic. Nucleotide sequences were conceptually translated and aligned to SIVmac239 gene products by using MacVector 7.0 trial version (Accelerys, San Diego, Calif.).
RESULTS
Identification of SIV controllers.
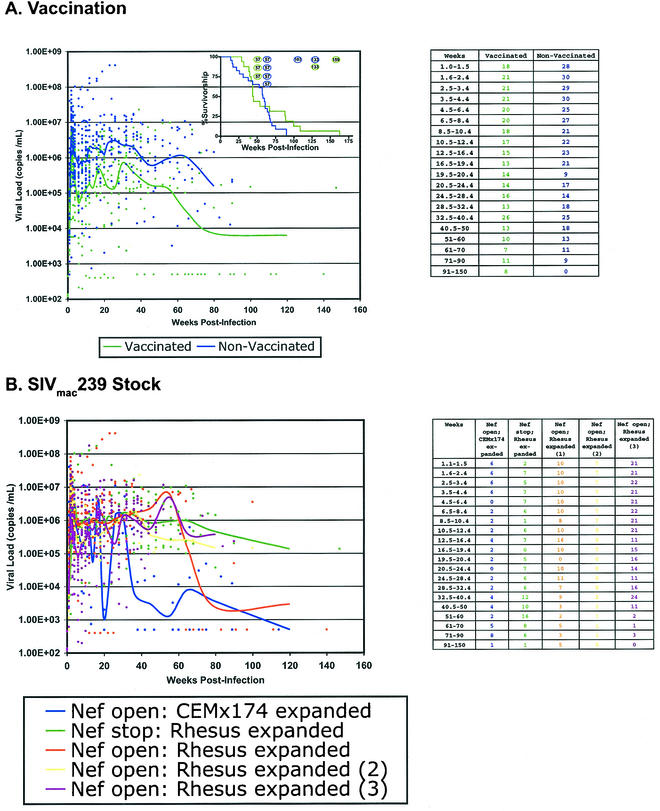
We first identified MHC-I alleles that are associated with delayed disease progression in rhesus macaques infected with SIVmac239. A cohort of 51 rhesus macaques was infected with the same highly pathogenic, molecularly cloned strain of SIV (SIVmac239) (41). The animals were infected between 1998 and 2002 in five SIV vaccine trials. The vaccine immunogens were: lipopeptide encoding the Gag181-189CM9 CTL epitope (animal 95096), DNA and modified vaccinia virus Ankara (MVA) encoding the Gag181-189CM9 CTL epitope (eight vaccinated animals, including animal 95061) (1), DNA and MVA encoding Tat (three vaccinated animals) (2), DNA and MVA encoding seven of the nine SIV proteins (eight vaccinated animals) (19), and DNA and MVA encoding Tat, Rev, and Nef (three vaccinated animals) (T. Vogel, unpublished data). The cohort also included 29 vaccine-naive animals (1, 2, 19, 37) that initially served as controls in these vaccine studies. The geometric mean viral load in the vaccinated animals was approximately 10-fold lower than the geometric mean viral loads in the vaccine-naive controls (Fig. 1A). These differences in viral load did not appear to affect survivorship, as the vaccinated animals had a lower median survivorship than the vaccine-naive controls (Fig. 1A, inset). However, the spectrum of survivorship was broader in the animals that were vaccinated, with several vaccinated animals surviving more than 18 months after SIV challenge. Since only 51 animals were used for these comparisons, it is possible that meaningful effects of the vaccination on suppression of viral replication or disease-free survivorship have been masked.
FIG. 1.
Influence of prechallenge vaccination and SIVmac239 stock on viral loads. Viral loads were measured using kinetic real-time reverse transcriptase PCR or branched-chain DNA assays. Each data point used to calculate the viral load trend line is indicated as a colored dot. Trend lines were obtained by grouping viral loads into time point intervals and then determining the geometric mean of each interval. The number of samples from each interval is shown in the tables to the right. (A) Prechallenge vaccination does not influence postchallenge viral load. With the exception of the vaccinated macaques 95061 and 95096, all other vaccinated animals have viral loads in excess of 10,000 copies/ml of blood plasma.The inset shows survivorship of the vaccinated and naive macaques. Each open circle represents one animal that remains alive as of this writing. (B) Dose of SIVmac239 challenge does not influence viral load. While all 51 animals were infected with SIVmac239, different stocks of this virus have been used during the 3-year period when these animals were infected. All stocks of SIVmac239 establish infection, maintain a persistent viral burden in excess of 10,000 copies/ml, and lead to progressive immunodeficiency.
Nonetheless, viral loads in both groups were typically greater than 105 copies of virus per ml of blood plasma during the first year of infection. Animals that survived for more than 1 year postinfection tended to have lower viral loads, a trend that was noticed in both the vaccinated and control groups. Based on this data, we decided that including vaccinated animals in our analysis was reasonable.
Though all of the animals in this study were infected with SIVmac239, different SIVmac239 stocks were used in the different vaccine studies. To verify that the virus stock did not systemically affect chronic-phase viral load, we compared the viral loads of the animals challenged with each of the five SIVmac239 stocks (Fig. 1B). In each challenge, peak viral load exceeded 106 copies of virus per ml of plasma, with chronic-phase viral setpoints in excess of 105 copies of virus per ml of plasma. All four of the SIVmac239 stocks expanded on rhesus macaque PBMC induced similar viral loads. The six animals infected with SIVmac239/nef open expanded on CEMx174 had lower viral loads during chronic infection. However, three of the six animals infected with this challenge stock developed simian AIDS within 30 weeks of infection and were sacrificed. Two of the remaining three animals infected with this stock were Mamu-A*01, -B*17, and -B*29 positive, possibly accounting for the lower viral loads observed with this virus stock.
All 51 SIV-infected animals were previously tested for the presence of Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B*01, Mamu-B*03, Mamu-B*04, Mamu-B*17, and Mamu-B*29 by PCR-SSP (26, 39). Notably, Mamu-B*17 and Mamu-B*29 appear to be closely linked, with 39 of 43 Mamu-B*29-positive animals also being positive for Mamu-B*17.
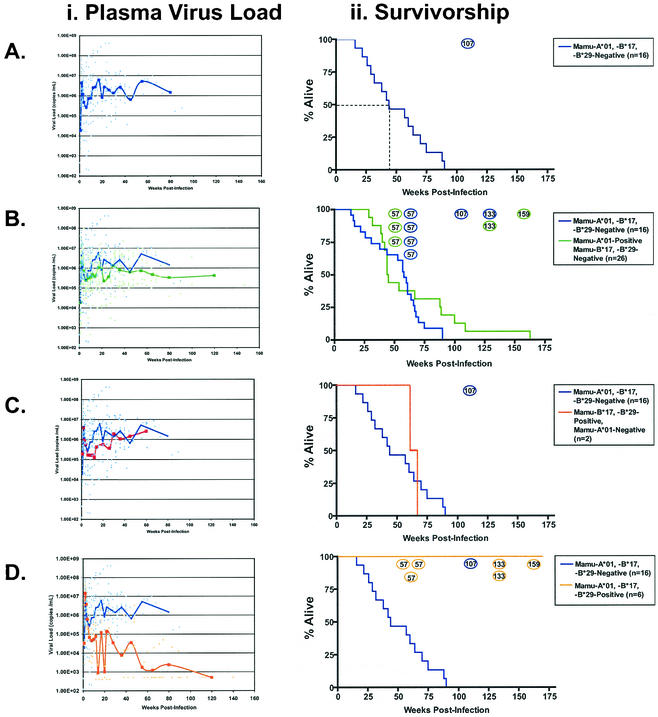
We grouped the animals according to their (PCR-SSP-deduced) MHC genotype and compared the viral loads and survivorship for each group (39). Of the animals that did not express Mamu-A*01, Mamu-B*17, or Mamu-B*29, median survivorship was less than 50 weeks, with only one animal surviving more than 100 weeks (Fig. 2A). In agreement with previous studies (39, 40, 46, 69), we observed that Mamu-A*01-positive animals controlled SIV replication more effectively than Mamu-A*01-negative animals following resolution of peak viremia (P = 0.001; t test) (Fig. 2B). However, these differences in viral load did not significantly prolong survivorship in Mamu-A*01-positive animals (P = 0.09) (Fig. 2B). Two vaccine-naive animals that expressed Mamu-B*17 and Mamu-B*29, but not Mamu-A*01, survived for 62 and 68 weeks postinfection, despite persistently high plasma viral loads (Fig. 2C).
FIG. 2.
Animals expressing Mamu-A*01, -B*17, and -B*29 have reduced viral loads and increased survivorship. Plasma viral loads and survivorship were followed in 51 SIVmac239-infected rhesus macaques. Viral load trend lines were calculated as described in the legend for Fig. 1. Survivorship curves include only animals that have been sacrificed. Animals that remain alive are shown as open circles. The number of weeks postinfection is shown inside each animal's open circle. There are seven Mamu-A*01-positive animals alive at 57 weeks postinfection, of which only three are Mamu-A*01, -B*17, and -B*29 positive. (A) In 16 Mamu-A*01-, -B*17-, and -B*29-negative macaques, median survivorship was 44 weeks, with a setpoint viral load in excess of 1,000,000 copies per ml of blood plasma. (B) Mamu-A*01-positive, Mamu-B*17-, and -B*29-negative animals have slightly improved outcomes relative to Mamu-A*01-, -B*17-, and -B*29-negative macaques in terms of plasma viral load and survivorship. (C) The two Mamu-B*17- and -B*29-positive, Mamu-A*01-negative animals have slightly improved survivorship relative to the Mamu-A*01-, -B*17-, and -B*29-negative macaques. (D) All six Mamu-A*01-, -B*17-, and -B*29-positive macaques remain alive with significantly lower viral loads than the Mamu-A*01-, -B*17-, and -B*29-negative controls.
Six of the 51 animals expressed Mamu-A*01, Mamu-B*17, and Mamu-B*29. Remarkably, all six animals remain alive as of this writing (Fig. 2D). The vaccination histories, SIV challenge strain, and current survivorship of these six animals are shown in Table 1. Three of these animals, namely, animals 95061, 95096, and 1937, controlled chronic phase viral replication below 5,000 vRNA copies/ml of plasma (Fig. 2D). Though animals 2065, 2095, and 2129 have chronic-phase viral loads in excess of 400,000 copies/ml (48 weeks), they have already exceeded the median survivorship of our cohort (Fig. 1D). The significantly increased survivorship (P < 0.001) and lower viral loads of Mamu-A*01-, -B*17-, and -B*29-positive animals infected with SIVmac239 correlate this MHC genotype with improved disease outcome.
TABLE 1.
Vaccination and challenge profiles of six Mamu-A*01-*B*17-, and -B*29-positive animals
| Animal no. | Vaccine | Challenge stocka | Peak viremia (wk) | Survivor- ship (wk) |
|---|---|---|---|---|
| 2065 | —b | SIVmac239 Nef open: rhesus expanded (stock 3) | 4.5 × 107 (2) | 57 |
| 2095 | — | SIVmac239 Nef open: rhesus expanded (stock 3) | 2.9 × 107 (3) | 57 |
| 2129 | — | SIVmac239 Nef open: rhesus expanded (stock 3) | 2.2 × 107 (2) | 57 |
| 1937 | — | SIVmac239 Nef open: CEMx174 expanded | 3.7 × 105 (2) | 133 |
| 95096 | Lipopeptide Gag181-189 CM9 CTL epitopec | SIVmac239 Nef open: CEMx174 expanded | 1.3 × 107 (2) | 133 |
| 95061 | DNA/MVA Gag181-189 CM9 CTL epitoped | SIVmac239 Nef open: rhesus expanded (stock 1) | 2.4 × 107 (2) | 159 |
Multiple stocks of SIVmac239 Nef open were used in this study; the arbitrary stock numbers correspond to the parenthetical stock numbers in Fig. 1.
—, vaccine naive.
Vaccine did not elicit detectable Gag181-189CM9-specific CTL in the periphery, and results were not published.
Vaccine results were published by Allen et al. (1).
We verified that Mamu-A*01, Mamu-B*17, and Mamu-B*29 were the only MHC alleles shared among these animals by sequencing individual cDNA clones from all six animals (Table 2). We also typed all of these macaques for their MHC-II DP, DQ, and DR alleles by denaturing gradient gel electrophoresis and direct sequencing. We did not identify any additional shared alleles (data not shown).
TABLE 2.
MHC profiles of Mamu-A*01-, -B*17-, and -B*29-positive animals
| MHC class | Profile for animal no.:
|
|||||
|---|---|---|---|---|---|---|
| 95061 | 95096 | 1937 | 2065 | 2095 | 2129 | |
| I | A*01 | A*01 | A*01 | A*01 | A*01 | A*01 |
| B*17 | B*17 | B*17 | B*17 | B*17 | B*17 | |
| B*29 | B*29 | B*29 | B*29 | B*29 | B*29 | |
| A*02 | A*11 | A*11 | A*1302 | A*1603 | A*26 | |
| B*64 | B*48 | B*12 | B*55 | B*66 | B*30 | |
| B*65 | ||||||
| II | DRB1*1003 | DRB*W101 | DRB*W2501 | DRB*W307 | DRB*W201 | Not tested |
| DRB*W2602 | DBR1*0303 | DRB*W2002 | DRB*W702 | DRB1*0309 | ||
| DRB*W602 | DRB1*1007 | DRB*W2602 | DRB1*0303 | DRB1*0701 | ||
| DRB1*0306 | DRB*W602 | DRB1*1007 | DRB3*0405 | |||
| DRB*W2501/04 | DRB5*0303 | |||||
| DRB6*0101 | ||||||
| DPB1*04 | DPB1*06 | DPB1*10 | DPB1*04 | DPB1*03 | Not tested | |
| DPB1*06 | DPB1*12 | DPB1*06 | ||||
| DQA1*01051 | DQA1*2601 | DQA1*01051 | DQA1*2601 | DQA1*0104 | DQA1*2302 | |
| DQA1*0102 | DQA1*2602 | DQA1*2603 | DQA1*2401 | |||
| DQB1*0602 | DQB1*1801 | DQB1*0602 | DQB1*1801 | DQB1*0601 | DQB1*1804 | |
| DQB1*0605 | DQB1*1804 | DQB1*1703 | DQB1*1811 | DQB1*1501 | DQB1*1810 | |
Humoral immune responses in SIV controllers.
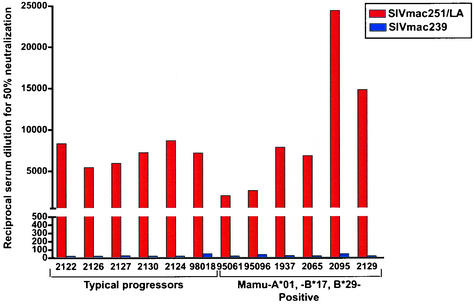
Effective immunity to viruses such as HIV and SIV likely requires the contribution of both cellular and humoral immunity. The rapid onset of disease in most SIVmac239-infected animals typically occurs in the absence of neutralizing antibodies (nAb) against SIVmac239 (70); moreover, SIVmac239 is largely insensitive to nAb. Sera from the Mamu-A*01-, -B*17-, and -B*29-positive animals, as well as sera from typical progressors, neutralized lab-adapted SIVmac251 samples (Fig. 3). In contrast, nAb against autologous SIVmac239 infection were rare in both groups of animals. Therefore, it is unlikely that viral control in the Mamu-A*01-, -B*17-, and -B*29-positive animals results from unusually potent nAb responses.
FIG. 3.
Sera from Mamu-A*01-, -B*17-, and -B*29-positive animals fail to neutralize SIVmac239. Dilutions of sera from the six Mamu-A*01-, -B*17-, and -B*29-positive animals were tested for the ability to neutralize SIVmac239 and lab-adapted SIVmac251. Data from six typical progressors is shown for comparison. No significant neutralization activity against SIVmac239 was detected in any of the animals.
Acute-phase cellular immune responses in Mamu-A*01-, -B*17-, and -B*29-positive animals.
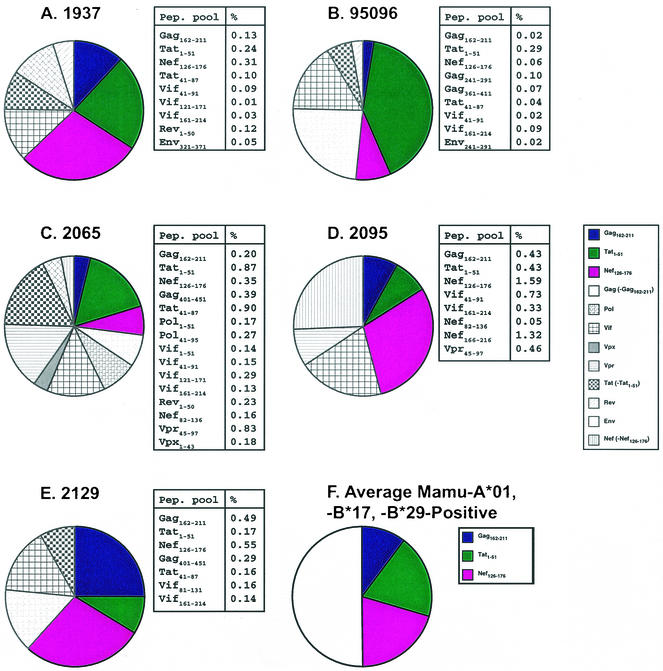
We evaluated the CD8+ T-lymphocyte response in five of the six Mamu-A*01-, -B*17-, and -B*29-positive animals at 4 weeks postinfection (37) by detecting intracellular cytokine production following PBMC stimulation with peptide pools spanning the SIVmac239 proteome. Since animal 95061 was infected with SIVmac239 before this method was introduced, we were unable to evaluate its acute-phase cellular immune response. All five animals tested responded to the peptide pools containing the Mamu-A*01-restricted epitopes Gag181-189CM9 (4) and Tat28-35SL8 (3) (Fig. 4). These responses are always seen in Mamu-A*01-positive animals, irrespective of their rate of disease progression (37). These peptide pools are not consistently recognized in Mamu-A*01-, -B*17-, and -B*29-negative macaques (see Fig. 1 in online supplemental data, http://yakui.primate.wisc.edu/people/watkins/pdf/SIVcontrollerSupp_Data.pdf).
FIG. 4.
Peptide pools containing Gag181-189CM9, Tat28-35SL8, and Nef165-173IW9 are well recognized by CD8+ T-lymphocytes from Mamu-A*01-, -B*17-, and -B*29-positive animals during acute infection. (A to E) Acute-phase CD8+ T-lymphocyte responses were monitored in the Mamu-A*01-, -B*17-, and -B*29-positive macaques (identified by animal number) by intracellular staining for IFN-γ production. Percentages of CD3+, CD8+, and IFN-γ-positive PBMC that were stimulated to produce IFN-γ by peptide pools are shown in the tables to the right of each chart. The Gag162-211 pool (containing the Gag181-189CM9 epitope), the Tat1-51 pool (containing the Tat28-35SL8 epitope), and the Nef126-176 pool (containing the Nef128-135IW9 epitope) were recognized in all animals. (F) On average, the three dominant responses constituted 50% of the total acute-phase CD8+ T-lymphocyte response. The acute-phase responses from 1937 and 95096 were previously reported (37).
All five Mamu-A*01-, -B*17-, and -B*29-positive animals also recognized the peptide pool containing the Mamu-B∗17-restricted Nef165-173IW9 (9, 38) epitope (Fig. 4). The same Nef peptide pool was weakly recognized by two Mamu-A*01-, -B*17-, and -B*29-negative macaques (macaques 92050 and 97086) at 3 weeks postinfection (see Fig. 1 at http://yakui.primate.wisc.edu/people/watkins/pdf/SIVcontrollerSupp_Data.pdf). However, this peptide pool contains epitopes restricted by molecules other than Mamu-B*17 (9, 44), including the Mamu-A*02-restricted Nef159-167YY9 epitope (44, 60). Both animals 92050 and 97086 are Mamu-A*02 positive, strongly suggesting that the reactivity is directed against Nef159-167YY9 and not Nef165-173IW9. Nonetheless, we verified the specificity of Nef165-173IW9 recognition in the Mamu-A*01-, -B*17-, and -B*29-positive animals by demonstrating reactivity against the synthetic Nef165-173IW9 9-mer (data not shown). Additionally, the peptide pool Vif41-91 was recognized in four of the five Mamu-A*01-, -B*17-, and -B*29-positive animals (Fig. 4). Recent work has shown that this peptide pool contains three Mamu-B*17-restricted CTL epitopes (38). Unfortunately, we cannot verify that the Vif reactivity is due to Mamu-B*17-restricted responses, since our acute-phase samples from these animals are exhausted.
On average, responses against the peptide pools containing Gag181-189CM9, Tat28-35SL8, and Nef165-173IW9 comprised over 50% of the acute-phase cellular immune responses in the Mamu-A*01-, -B*17-, and -B*29-positive animals. This is, to our knowledge, the first demonstration that MHC alleles associated with delayed disease progression bind epitopes recognized by the dominant CD8+ T-lymphocyte responses during acute infection.
CTL escape from Tat28-35SL8 in SIV controllers.
Escape from HIV- and SIV-specific CD8+ T-lymphocyte responses has been widely observed (3, 9, 12, 14, 44, 48, 51). Gag181-189CM9-specific CTL can select viral escape variants in animals challenged with SIVmac239 (Fig. 5) and SHIV89.6P (6). We reasoned that strong selective pressure exerted by Gag181-189CM9-specific CTL in the Mamu-A*01-, -B*17-, and -B*29-positive animals might accelerate viral escape from this response. While 3 of 11 Mamu-A*01-positive, Mamu-B*17-, and -B*29-negative animals exhibit epitope variation in Gag181-189CM9 by approximately 1 year postinfection, only one of the Mamu-A*01-, -B*17-, and -B*29-positive animals has variation in this epitope (Fig. 5A). It is unlikely that this difference is biologically significant, and it is more likely the result of vagaries in the rate of escape from the Gag181-189CM9 response.
In striking contrast to the sporadic escape that is observed from Gag181-189CM9 responses, viruses from most Mamu-A*01-positive animals escape recognition by Tat28-35SL8-specific CTL by 4 weeks postinfection. Since weak responses to this epitope were detectable during chronic-phase infection of Mamu-A*01-, -B*17-, and -B*29-positive animals (data not shown), we hypothesized that the maintenance of these responses might be correlated with reduced Tat28-35SL8 epitope diversity. We attempted to sequence Tat28-35SL8 from plasma virus obtained 1 year postinfection. Unfortunately, repeated attempts to amplify plasma virus from animals 95096 and 95061 failed, presumably because of the exceptionally low concentration of virus in these plasma samples. However, plasma SIV from the other four SIV controllers was successfully sequenced. Though evidence for Tat28-35SL8 variation was observed, the extent of variation was less dramatic than in typical Mamu-A*01-positive animals (P < 0.014) (Fig. 5B). By 1 year postinfection, viruses from most Mamu-A*01-positive macaques have accumulated at least two dominant nucleotide substitutions within Tat28-35SL8. In the Mamu-A*01-, -B*17-, and -B*29-positive animals, only a proline-for-serine substitution at the first position of the epitope was observed. This particular epitope variant binds Mamu-A*01 and is cross-reactive with Tat28-35SL8-specific CTL ex vivo (3). In contrast, the double replacements at P1/P5 and P1/P8 that occurred in 8 of the 12 typical progressors do not bind to Mamu-A*01 and are not recognized by Tat28-35SL8-specific CTL ex vivo (data not shown and reference 3).
We also examined viral escape from the Mamu-B*17-restricted Nef165-173IW9 response. Both Mamu-B*17- and -B*29-positive, Mamu-A*01-negative animals harbored escape variants in this epitope, in agreement with a previous study (9). Four of the Mamu-A*01-, -B*17-, and -B*29-positive animals also had variation in this epitope by 1 year postinfection (Fig. 5C).
Animals 95061 and 95096, the two Mamu-A*01-, -B*17-, and -B*29-positive animals with viral loads below 1,000 copies/ml of blood plasma, presented a unique opportunity to evaluate CTL escape in the context of undetectable plasma virus and durable immune control. Despite the lack of plasma virus, integrated proviruses from these animals could be consistently amplified from PBMC-derived genomic DNA. Though proviral sequence is not as representative of the replication-competent viral population as plasma virus (54), we determined that bulk provirus and plasma sequences are similar during chronic SIVmac239 infection (see Fig. 2 at http://yakui.primate.wisc.edu/people/watkins/pdf/SIVcontrollerSupp_Data.pdf). Wild-type Tat28-35SL8 and Nef165-173IW9 epitope sequences dominated in animals 95061 and 95096 throughout chronic infection (Fig. 5B and C).
To ensure that the wild-type virus in these animals remained replication competent, we cocultured CD8+-depleted PBMC from animal 95061 with uninfected CEMx174 cells for 3 weeks. Viral RNA was recovered from the supernatant and sequenced. Remarkably, the outgrown virus differed from wild-type SIVmac239 by only a single nucleotide (see Fig. 3 at http://yakui.primate.wisc.edu/people/watkins/pdf/SIVcontrollerSupp_Data.pdf). This data provides evidence that wild-type SIVmac239 is still present in animal 95061 and suggests that the ongoing immune responses are preventing SIVmac239 recrudescence.
DISCUSSION
For the first time, we have established a link between SIV survivorship, particular MHC-I alleles, and acute-phase CTL responses. Acute-phase CTL responses are extremely difficult to measure in HIV-infected individuals, since investigators rarely have synthetic peptides autologous to the infecting virus. Moreover, even closely related strains of HIV can differ in residues that are critical for CTL recognition. This problem may be exacerbated if viruses harboring variation in CTL responses are transmitted among individuals (12, 67). We overcame this issue by infecting all 51 animals in our cohort with the clonal virus SIVmac239 and by assaying immune responses during the acute phase with synthetic peptides identical to the infecting virus.
It is possible that other genes linked to Mamu-A*01 or Mamu-B*17 and -B*29 influence SIV viral replication during chronic infection. Unidentified gene products could exert their effect by reducing the susceptibility of host cells to HIV infection or by slowing the production of new HIV virions in infected cells. However, during the first 2 weeks of infection, prior to the development of the CTL response, SIVmac239 replicated to high titer in all six Mamu-A*01-, -B*17-, and -B*29-positive animals. Indeed, the levels of peak viremia in the Mamu-A*01-, -B*17-, and -B*29-positive animals were indistinguishable from those of animals that progress to simian AIDS with typical kinetics. If non-MHC host cell genes were contributing to the control of viral replication, we would have expected a reduction in peak viremia.
Alternately, it is possible that acute-phase immune responses that are first directed against epitopes bound by Mamu-A*01 and Mamu-B*17 have long-lasting effects on the chronic-phase immune response to SIVmac239. In five of the six SIV Mamu-A*01-, -B*17-, and -B*29-positive animals, SIV-specific CD4+ responses were detected during chronic infection (data not shown). HIV-infected individuals with low viral setpoints frequently mount strong HIV-specific CD4+ T-lymphocyte responses (49), while HIV-infected individuals with higher viral loads and macaques with normal SIV disease progression typically do not. These CD4+ responses may act in concert with the CTL responses by coordinating the immune response, suppressing viral replication, and increasing disease-free survival.
Vigorous acute-phase CTL responses in Mamu-A*01-, -B*17-, and -B*29-positive animals may also reduce the extent of viral escape during acute and chronic infection. If extremely effective CTL rapidly curb viral replication following peak viremia, the virus's capacity to spawn viable escape variants may be diminished. This is consistent with our observation that Mamu-A*01-, -B*17-, and -B*29-positive animals have less chronic-phase variation in the Tat28-35SL8 epitope than typical Mamu-A*01-positive animals. The reduction in diversification, in turn, may preserve the ability of CTL to combat the infection. In the longest-lived Mamu-A*01-, -B*17-, and -B*29-positive animal, animal 95061, low-level Tat28-35SL8 responses have been transiently detected throughout chronic infection by tetramer staining, IFN-γ ELISPOT, and IFN-γ intracellular cytokine staining (data not shown). Additionally, the rescue of virus from animal 95061 that differs from SIVmac239 by only a single nucleotide after 140 weeks of infection suggests that viral diversification has been reduced and that viral resurgence is actively prevented by ongoing immune responses. We hypothesize that containment of low-level SIV replication and reduction of viral diversification during chronic infection does not require the same CTL response magnitude that is required for initial viral containment during acute SIV infection. Results observed in animals challenged with SIVsmE660 and immediately treated with antiretroviral therapy indirectly support this hypothesis (27, 28). In these animals, undetectable plasma virus loads were associated with modest lymphoproliferative responses. Upon depletion of CD8+ cells by in vivo treatment with monoclonal antibody, viral replication could be detected in cultures of lymph node mononuclear cells.
Interestingly, the two SIV Mamu-A*01-, -B*17-, and -B*29-positive animals with the lowest viral loads were both vaccinated prior to SIV challenge. Animal 95061 was vaccinated with DNA and MVA encoding the nonameric Mamu-A*01-restricted CTPYDINQM peptide, whereas animal 95096 was vaccinated with the same epitope delivered as a lipopeptide. None of the other animals that received these minimal vaccines differed clinically from the vaccine-naive controls. However, our observed Mamu-A*01, -B*17, and -B*29 effect may have been amplified by vaccination. Vaccination alone, however, cannot account for the improved outcome of Mamu-A*01, -B*17, -B*29 animals, as the other four animals that expressed these two alleles were vaccine naive.
Understanding the precise role of Mamu-A*01, -B*17, and -B*29 in SIV control will require studies of additional Mamu-A*01-, -B*17, and -B*29-positive animals infected with SIVmac239 and other SIV strains. It is presently unclear whether the observed protective effect of Mamu-A*01, -B*17, and -B*29 positivity confers any benefit against closely related viruses such as SIVmac251 or SIV-HIV chimeras such as SHIV89.6P. Interestingly, we previously studied a Mamu-A*01-negative, Mamu-B*17-, and -B*29-positive macaque infected with a rhesus-passaged derivative of SIVmac239, SIVppm. Though we lacked the technology to fully evaluate the immune response in this animal at the time of the study, she lived for 127 weeks postinfection and was the longest lived animal in that five-animal cohort (9).
If these results are generalizable to other virus strains besides SIVmac239, they may have important implications for preclinical vaccine testing. Because many CTL epitopes bound by Mamu-A∗01 have been previously identified, Mamu-A*01 animals are frequently selected for trials of vaccine efficacy. While the Mamu-A∗01 allele by itself offers only modest protection from disease progression, the selection of Mamu-A∗01-positive animals into vaccine cohorts will occasionally result in the inclusion of Mamu-A*01-, -B*17-, and -B*29-positive animals. If these animals are assigned to vaccine-naive control groups, a vaccine effect could be masked by uncharacteristically low viral loads in the controls. Perhaps more troubling, the inclusion of Mamu-A*01-, -B*17-, and -B*29-positive animals in groups of vaccinated animals could confuse vaccine-mediated viral control with genotype-mediated viral control.
One of the more interesting findings in this study is the dominance of cellular immune responses against targets bound by the alleles associated with slow progression, namely, Mamu-A*01, -B*17, and -B*29. Our association of dominance with Mamu-A*01 and Mamu-B*17 relied on our precise knowledge of strong responses directed against regions of Gag, Tat, and Nef. The three dominant responses restricted by Mamu-A*01 and Mamu-B*17 remained strong irrespective of the other MHC-I alleles present in these animals. Moreover, additional subdominant responses restricted by Mamu-A*01 and Mamu-B*17 may have further magnified the contribution of these alleles to the cellular immune response during acute infection.
Our results show, for the first time, that MHC-I alleles associated with slow disease progression bind epitopes recognized by dominant acute-phase CTL responses. In the two animals with the most effective containment of viremia, wild-type sequences persist in two CTL epitopes that normally escape CTL recognition during early infection. Thus, our study provides the first link between host immunogenetics, acute-phase CTL responses, reduction in CTL escape, and improved disease outcome following infection with a highly pathogenic immunodeficiency virus.
Acknowledgments
David H. O'Connor, Bianca R. Mothe, and Jason T. Weinfurter contributed equally to this work.
We thank Jody Helgeland and Jacque Mitchen for assistance with blood processing and viral coculture and Jacque Mitchen for coordinating all animal procedures.
This work was supported by NIH grants AI4920, RR1537, AI46366, AI45461, and RR00167. David I. Watkins is a recipient of an Elizabeth Glaser Award.
REFERENCES
- 1.Allen, T. M., P. Jing, B. Calore, H. Horton, D. H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B. R. Mothe, C. Emerson, N. Wilson, J. D. Lifson, I. M. Belyakov, J. A. Berzofsky, C. Wang, D. B. Allison, D. C. Montefiori, R. C. Desrosiers, S. Wolinsky, K. J. Kunstman, J. D. Altman, A. Sette, A. J. McMichael, and D. I Watkins. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., L. Mortara, B. R. Mothe, M. Liebl, P. Jing, B. Calore, M. Piekarczyk, R. Ruddersdorf, D. H. O'Connor, X. Wang, C. Wang, D. B. Allison, J. D. Altman, A. Sette, R. C. Desrosiers, G. Sutter, and D. I. Watkins. 2002. Tat-vaccinated macaques do not control simian immunodeficiency virus SIVmac239 replication. J. Virol. 76:4108-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 5.Balla-Jhagjhoorsingh, S. S., G. Koopman, P. Mooij, T. G. Haaksma, V. J. Teeuwsen, R. E. Bontrop, and J. L. Heeney. 1999. Conserved CTL epitopes shared between HIV-infected human long-term survivors and chimpanzees. J. Immunol. 162:2308-2314. [PubMed] [Google Scholar]
- 6.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 9.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 10.Gange, S. J., J. W. Mellors, B. Lau, R. Detels, J. P. Phair, A. Munoz, and J. B. Margolick. 2001. Longitudinal patterns of HIV type 1 RNA among individuals with late disease progression. AIDS Res. Hum. Retrovir. 17:1223-1229. [DOI] [PubMed] [Google Scholar]
- 11.Gillespie, G. M., R. Kaul, T. Dong, H. B. Yang, T. Rostron, J. J. Bwayo, P. Kiama, T. Peto, F. A. Plummer, A. J. McMichael, and S. L. Rowland-Jones. 2002. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS 16:961-972. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R. E. Phillips, and A. J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retrovir. 12:1691-1698. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., Y. Tang, S. I. Pelton, and B. D. Walker. 2000. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J. Virol. 74:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrer, T., E. Harrer, S. A. Kalams, P. Barbosa, A. Trocha, R. P. Johnson, T. Elbeik, M. B. Feinberg, S. P. Buchbinder, and B. D. Walker. 1996. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 156:2616-2623. [PubMed] [Google Scholar]
- 17.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, F. Schachter, D. Zagury, J. Rappaport, C. Winkler, G. W. Nelson, and J. F. Zagury. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942-6946. [PubMed] [Google Scholar]
- 18.Hofmann-Lehmann, R., R. K. Swenerton, V. Liska, C. M. Leutenegger, H. Lutz, H. M. McClure, and R. M. Ruprecht. 2000. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one- versus two-enzyme systems. AIDS Res. Hum. Retrovir. 16:1247-1257. [DOI] [PubMed] [Google Scholar]
- 19.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, X., X. Gao, M. Ramanathan, Jr., G. R. Deschenes, G. W. Nelson, S. J. O'Brien, J. J. Goedert, D. D. Ho, T. R. O'Brien, and M. Carrington. 2002. Human immunodeficiency virus type 1 (HIV-1)-specific CD8+-T-cell responses for groups of HIV-1-infected individuals with different HLA-B*35 genotypes. J. Virol. 76:12603-12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 23.Kaslow, R. A., C. Rivers, J. Tang, T. J. Bender, P. A. Goepfert, R. El Habib, K. Weinhold, and M. J. Mulligan. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 75:8681-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, M. R., S. H. van der Burg, E. Hovenkamp, A. M. Holwerda, J. W. Drijfhout, C. J. Melief, and F. Miedema. 1998. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J. Gen. Virol. 79(Pt 9):2191-2201. [DOI] [PubMed] [Google Scholar]
- 26.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A∗01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 27.Lifson, J. D., J. L. Rossio, R. Arnaout, L. Li, T. L. Parks, D. K. Schneider, R. F. Kiser, V. J. Coalter, G. Walsh, R. J. Imming, B. Fisher, B. M. Flynn, N. Bischofberger, M. Piatak, Jr., V. M. Hirsch, M. A. Nowak, and D. Wodarz. 2000. Containment of simian immunodeficiency virus infection: cellular immune responses and protection from rechallenge following transient postinoculation antiretroviral treatment. J. Virol. 74:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackewicz, C. E., H. W. Ortega, and J. A. Levy. 1991. CD8+ cell anti-HIV activity correlates with the clinical state of the infected individual. J. Clin. Investig. 87:1462-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72:164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellors, J. W., L. A. Kingsley, C. R. Rinaldo, Jr., J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 32.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 33.Migueles, S. A., and M. Connors. 2001. Frequency and function of HIV-specific CD8(+) T cells. Immunol. Lett. 79:141-150. [DOI] [PubMed] [Google Scholar]
- 34.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 35.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montefiori, D. C., T. W. Baba, A. Li, M. Bilska, and R. M. Ruprecht. 1996. Neutralizing and infection-enhancing antibody responses do not correlate with the differential pathogenicity of SIVmac239delta3 in adult and infant rhesus monkeys. J. Immunol. 157:5528-5535. [PubMed] [Google Scholar]
- 37.Mothe, B. R., H. Horton, D. K. Carter, T. M. Allen, M. E. Liebl, P. Skinner, T. U. Vogel, S. Fuenger, K. Vielhuber, W. Rehrauer, N. Wilson, G. Franchini, J. D. Altman, A. Haase, L. J. Picker, D. B. Allison, and D. I. Watkins. 2002. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J. Virol. 76:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mothe, B. R., J. Sidney, J. L. Dzuris, M. E. Liebl, S. Fuenger, D. I. Watkins, and A. Sette. 2002. Characterization of the peptide-binding specificity of mamu-b*17 and identification of mamu-b*17-restricted epitopes derived from simian immunodeficiency virus proteins. J. Immunol. 169:210-219. [DOI] [PubMed] [Google Scholar]
- 39.Mothe, B. R., J. Weinfurter, C. Wang, W. Rehrauer, N. Wilson, T. M. Allen, D. B. Allison, and D. I. Watkins. 2003. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 77:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhl, T., M. Krawczak, P. Ten Haaft, G. Hunsmann, and U. Sauermann. 2002. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 169:3438-3446. [DOI] [PubMed] [Google Scholar]
- 41.Naidu, Y. M., H. W. Kestler III, Y. Li, C. V. Butler, D. P. Silva, D. K. Schmidt, C. D. Troup, P. K. Sehgal, P. Sonigo, M. D. Daniel, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nietfield, W., M. Bauer, M. Fevrier, R. Maier, B. Holzwarth, R. Frank, B. Maier, Y. Riviere, and A. Meyerhans. 1995. Sequence constraints and recognition by CTL of an HLA-B27-restricted HIV-1 gag epitope. J. Immunol. 154:2189-2197. [PubMed] [Google Scholar]
- 43.Nixon, D. F., A. R. Townsend, J. G. Elvin, C. R. Rizza, J. Gallwey, and A. J. McMichael. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484-487. [DOI] [PubMed] [Google Scholar]
- 44.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 45.O'Connor, D. H., T. M. Allen, and D. I. Watkins. 2002. Cytotoxic T-lymphocyte escape monitoring in simian immunodeficiency virus vaccine challenge studies. DNA Cell Biol. 21:659-664. [DOI] [PubMed] [Google Scholar]
- 46.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 49.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 50.Polycarpou, A., C. Ntais, B. T. Korber, H. A. Elrich, R. Winchester, P. Krogstad, S. Wolinsky, T. Rostron, S. L. Rowland-Jones, A. J. Ammann, and J. P. Ioannidis. 2002. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res. Hum. Retrovir. 18:741-746. [DOI] [PubMed] [Google Scholar]
- 51.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 53.Saah, A. J., D. R. Hoover, S. Weng, M. Carrington, J. Mellors, C. R. Rinaldo, Jr., D. Mann, R. Apple, J. P. Phair, R. Detels, S. O'Brien, C. Enger, P. Johnson, R. A. Kaslow, et al. 1998. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. AIDS 12:2107-2113. [DOI] [PubMed] [Google Scholar]
- 54.Sabino, E., L. Z. Pan, C. Cheng-Mayer, and A. Mayer. 1994. Comparison of in vivo plasma and peripheral blood mononuclear cell HIV-1 quasi-species to short-term tissue culture isolates: an analysis of tat and C2-V3 env regions. AIDS 8:901-909. [PubMed] [Google Scholar]
- 55.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 56.Shankar, P., J. A. Fabry, D. M. Fong, and J. Lieberman. 1996. Three regions of HIV-1 gp160 contain clusters of immunodominant CTL epitopes. Immunol. Lett. 52:23-30. [DOI] [PubMed] [Google Scholar]
- 57.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang, J., C. M. Wilson, S. Meleth, A. Myracle, E. Lobashevsky, M. J. Mulligan, S. D. Douglas, B. Korber, S. H. Vermund, and R. A. Kaslow. 2002. Host genetic profiles predict virological and immunological control of HIV-1 infection in adolescents. AIDS 16:2275-2284. [DOI] [PubMed] [Google Scholar]
- 59.Van Baalen, C. A., M. Schutten, R. C. Huisman, P. H. Boers, R. A. Gruters, and A. D. Osterhaus. 1998. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J. Virol. 72:6851-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogel, T. U., T. C. Friedrich, D. H. O'Connor, W. Rehrauer, E. J. Dodds, H. Hickman, W. Hildebrand, J. Sidney, A. Sette, A. Hughes, H. Horton, K. Vielhuber, R. Rudersdorf, I. P. De Souza, M. R. Reynolds, T. M. Allen, N. Wilson, and D. I. Watkins. 2002. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J. Virol. 76:11623-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogel, T. U., H. Horton, D. H. Fuller, D. K. Carter, K. Vielhuber, D. H. O'Connor, T. Shipley, J. Fuller, G. Sutter, V. Erfle, N. Wilson, L. J. Picker, and D. I. Watkins. 2002. Differences between T cell epitopes recognized after immunization and after infection. J. Immunol. 169:4511-4521. [DOI] [PubMed] [Google Scholar]
- 62.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 63.Walker, C. M., D. J. Moody, D. P. Stites, and J. A. Levy. 1986. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science 234:1563-1566. [DOI] [PubMed] [Google Scholar]
- 64.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 65.Yasutomi, Y., K. A. Reimann, C. I. Lord, M. D. Miller, and N. L. Letvin. 1993. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J. Virol. 67:1707-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu, X. G., M. M. Addo, E. S. Rosenberg, W. R. Rodriguez, P. K. Lee, C. A. Fitzpatrick, M. N. Johnston, D. Strick, P. J. Goulder, B. D. Walker, and M. Altfeld. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J. Virol. 76:8690-8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, Z. Q., T. M. Fu, D. R. Casimiro, M. E. Davies, X. Liang, W. A. Schleif, L. Handt, L. Tussey, M. Chen, A. Tang, K. A. Wilson, W. L. Trigona, D. C. Freed, C. Y. Tan, M. Horton, E. A. Emini, and J. W. Shiver. 2002. Mamu-A*01 allele-mediated attenuation of disease progression in simian-human immunodeficiency virus infection. J. Virol. 76:12845-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhuge, W., F. Jia, E. B. Stephens, Z. Li, C. Wang, S. V. Joag, and O. Narayan. 1998. Failure of SIVmac to be neutralized in macrophage cultures is unique to SIVmac and not observed with neutralization of SHIV or HIV-1. AIDS Res. Hum. Retrovir. 14:1045-1051. [DOI] [PubMed] [Google Scholar]