Abstract
We have previously shown that the nonstructural (NS) proteins NS1 and NS2 of bovine respiratory syncytial virus (BRSV) mediate resistance to the alpha/beta interferon (IFN)-mediated antiviral response. Here, we show that they, in addition, are able to prevent the induction of beta IFN (IFN-β) after virus infection or double-stranded RNA stimulation. In BRSV-infected MDBK cells upregulation of IFN-stimulated genes (ISGs) such as MxA did not occur, although IFN signaling via JAK/STAT was found intact. In contrast, infection with recombinant BRSVs lacking either or both NS genes resulted in efficient upregulation of ISGs. Biological IFN activity and IFN-β were detected only in supernatants of cells infected with the NS deletion mutants but not with wild-type (wt) BRSV. Subsequent analyses of IFN-β promoter activity showed that infection of cells with the double deletion mutant BRSV ΔNS1/2, but not with BRSV wt, resulted in a significant increase in IFN-β gene promoter activity. Induction of the IFN-β promoter depends on the activation of three distinct transcription factors, NF-κB, ATF-2/c-Jun, and IFN regulatory factor 3 (IRF-3). Whereas NF-κB and ATF-2/c-Jun activities were readily detectable and comparable in both wt BRSV- and BRSV ΔNS1/2-infected cells, phosphorylation and transcriptional activity of IRF-3, however, were observed only after BRSV ΔNS1/2 infection. NS protein-mediated inhibition of IRF-3 activation and IFN induction should have considerable impact on the pathogenesis and immunogenicity of BRSV.
Virus infection of mammalian cells leads to the activation of signaling pathways that result in the expression of host genes involved in the establishment of an antiviral state. Essential in this early host defense mechanism against viral intruders is the family of alpha/beta interferons (IFN-α/β). Once produced, these secreted proteins induce gene expression in neighboring cells by binding to cell surface cytokine receptors and activating JAK-STAT signaling pathways. Activated STAT-1/STAT-2 heterodimers, together with IFN-stimulated gene (ISG) factor 3γ (IFN regulatory factor 9 [IRF-9] or p48), bind to IFN-stimulated response elements found in numerous IFN-responsive genes, such as the double-stranded RNA (dsRNA)-activated protein kinase, resulting in the induction of proteins that impair viral replication (for reviews see references 18, 20, and 35).
The regulation of IFN gene expression has been the subject of intense investigation, and several transcription factors that bind IFN gene promoters have been identified. Best characterized is the virus-inducible promoter-enhancer of the IFN-β gene (reviewed in reference 18). This promoter includes an overlapping set of regulatory elements designated positive regulatory domains (PRD) I to IV, which interact with several signal-responsive transcription factors including NF-κB, ATF-2/c-Jun heterodimers, and IRF-3. Together with the chromatin-associated high-mobility group I(Y) proteins, these transcription factors form a transcriptional enhancer complex that stimulates the high-level, transient activation of IFN-β transcription. IRF-3 is expressed constitutively and displays a unique response to viral infection. Phosphorylation of latent IRF-3 on serine and threonine residues in the C-terminal region by a virus-activated kinase (VAK) activity leads to dimerization, association with the nuclear p300/CBP coactivators, and binding to PRD I and PRD III elements in the IFN-β promoter (23, 28, 30, 37, 39, 48). Activation of IRF-3 by RNA viruses is generally assumed to be triggered by dsRNA, which may represent a by-product of virus replication (reviewed in reference 24). However, recent studies revealed that virus proteins also can directly lead to induction of IFN expression (43).
Bovine respiratory syncytial virus (BRSV), a member of the Paramyxoviridae family, subfamily Pneumovirinae, is a major etiological agent of respiratory tract disease in calves resulting in substantial economic loss (38, 47). The pathology and immune response in calves mimic symptoms caused by human RSV, which remains the leading cause of bronchiolitis in infancy and of serious lower respiratory tract infections throughout childhood (8). A characteristic of all members of the Pneumovirus genus is the presence of two nonstructural (NS) protein genes, NS1 and NS2. Both are located at the 3′ end of the negative-strand RNA genome (3′-NS1-NS2-N-P-M-SH-G-F-M2.1/M2.2-L-5′) and do not have counterparts in other Paramyxoviridae genera (8, 9). The abundantly expressed NS proteins (13) are nonessential for virus replication in vitro; however, deletion of either NS gene severely attenuates RSV in vivo, indicating important defects in virus-host interplay (6, 7, 25, 36, 41, 42, 49). We have previously reported that the NS proteins of BRSV and of human RSV are able to cooperatively antagonize the IFN-induced cellular antiviral response in a species-specific manner (6, 36).
Here, we show that the NS proteins of BRSV are in addition able to block induction of IFN-β and the subsequent establishment of a cellular antiviral state by specifically preventing the activation of IRF-3. This ability of the RSV NS proteins certainly has an influence on viral pathogenesis and immunogenicity and is therefore of importance to the development of efficacious live attenuated RSV vaccines.
MATERIALS AND METHODS
Cells and viruses.
Vero, MDBK, and HEp-2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and antibiotics. The human embryonic kidney 293 (HEK 293) cell line was propagated in DMEM containing 10% FCS and antibiotics.
Recombinant BRSV (rBRSV) was derived from BRSV strain A51908 (American Type Culture Collection) variant Atue51908 (GenBank accession no. AF092942) (7) and was grown in Vero cells as described previously (6). Generation of the NS gene deletion mutants rBRSV ΔNS1, rBRSV ΔNS2, and rBRSV ΔNS1/2 has been described previously (7, 36). For preparation of virus stocks, 80% confluent Vero cell monolayers were infected at a multiplicity of infection (MOI) of 0.1 in serum-free DMEM (Gibco). After 1 h of adsorption, the inoculum was removed and the cells were incubated at 37°C in DMEM supplemented with 2.5% FCS (Gibco) in a 5% CO2 atmosphere until an extensive cytopathic effect was observed. Virus was released by freezing and thawing. Virus titers were determined on Vero cells by limiting dilution in microwell plates and counting of infected cell foci after indirect staining with an antibody recognizing the RSV fusion protein.
Infections and Western blot analysis.
For Western blot analyses, MDBK or HEp-2 cells were mock infected or infected in suspension with rBRSV, rBRSV ΔNS1, rBRSV ΔNS2, or rBRSV ΔNS1/2 at an MOI of 0.2 for 1 h in serum-free DMEM and subsequently seeded into 12-well dishes with DMEM plus 2.5% FCS. When indicated, mock-infected or wild-type (wt) BRSV-infected MDBK or HEp-2 cells were treated with 2,000 U of IFN-α A/D per ml (PBL Biomedical Laboratories) directly after seeding. Cells were lysed with cell lysis buffer (10 mM Tris HCl [pH 7.5], 100 mM NaCl, 10 mM EDTA, 0.5% Triton X-100, 0.5% deoxycholate) at indicated time points, and equivalent amounts of cell extracts (determined with Coomassie blue protein assay reagent [Pierce]) were loaded onto a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel. Proteins were transferred to a nitrocellulose membrane (Schleicher & Schuell) with a semidry transfer apparatus (OWL Scientific). After incubation with blocking solution (5% dry milk and 0.05% Tween 20 in phosphate-buffered saline [PBS-T]) at room temperature for 1 h, membranes were incubated overnight with the respective antibodies. The antibody recognizing PCNA was obtained from Santa Cruz Biotechnology, Inc.; anti-RSV serum (polyclonal) was purchased from Biogenesis; and anti-MxA antibody M134 was kindly provided by Otto Haller, Freiburg, Germany. The blot was then incubated for 2 h with peroxidase-conjugated anti-rabbit, anti-goat, or anti-mouse immunoglobulin G (1:10,000 in PBS-T; Dianova), and proteins were visualized by chemifluorescence (Renaissance; NEN).
For Western blot analysis of phosphorylated IRF-3, 2 × 106 HEK 293 cells were mock infected or infected in suspension with BRSV wt or BRSV ΔNS1/2 at an MOI of 0.25 for 1 h and then seeded in six-well dishes. Ten hours postinfection cells were harvested and nuclei were prepared as follows: cells were resuspended in NP-40 buffer (10 mM HEPES [pH 7.9], 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) and incubated on ice for 40 min. After centrifugation, supernatants were kept as cytoplasmic extracts and pellets were washed once with PBS. Nuclei were then lysed with cell lysis buffer containing 1% Triton X-100, and equivalent amounts of extract were subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred, and membrane was probed with IRF-3 antibody (Santa Cruz Biotechnology, Inc.) as described above.
For detection of IFN-β in cell culture supernatants, cell culture supernatants from infected or noninfected HEp-2 cells were harvested 37 h postinfection and subjected to trichloroacetic acid (TCA) precipitation. One volume of 100% (wt/vol) TCA was added to 4 volumes of cell supernatant and incubated for 1 h at 4°C. After centrifugation, protein pellets were washed twice with cold (−20°C) 100% acetone and dried for 5 to 10 min at 95°C. SDS-PAGE and blotting were performed as described above. IFN-β was detected with rabbit polyclonal antibody (1:1,000) to human IFN-β (Abcam; Cambridge, United Kingdom).
RNA analysis.
RNA from cells infected as described above for Western blot analyses was isolated with the RNeasy Mini kit (Qiagen). Northern blotting and hybridizations with [α-32P]dCTP-labeled cDNAs were performed as described previously (10). Hybridization signals were quantitated by phosphorimaging (Molecular Dynamics Storm).
Supernatant IFN assays.
Supernatants of infected MDBK and HEp-2 cells were harvested at the indicated time points. Supernatants were incubated at 56°C for 30 min to eliminate replication-competent RSV and mixed with MDBK cells previously infected with rabies virus (RV) at an MOI of 10, and then cells were seeded into 12-well plates in DMEM containing 2.5% FCS. RV titers were determined 2 days postinfection by limiting dilution and counting of infected cell foci after immunostaining with a fluorescein isothiocyanate conjugate (Centocor) recognizing RV N protein. Rabies SAD L16 virus stocks were prepared on BSR cells as described previously (14).
Transfections and luciferase assays.
All transfections were carried out with subconfluent HEK 293 cells grown in 35-mm-diameter dishes. Except for pAP-1Luc (Stratagene), all reporter construct plasmids used here were a generous gift from Takashi Fujita, Kyoto, Japan, and are described in reference 52. To assess IFN-β promoter activity, 1 μg of reporter construct p125luc per 35-mm-diameter dish was introduced into target cells with the FuGENE 6 transfection reagent (Roche Molecular Biochemicals). This plasmid contains the luciferase gene under the control of the entire IFN-β promoter (52) and was kindly provided by Takashi Fujita. When indicated, 1.5 μg of a plasmid encoding IRF-3 or a dominant-negative mutant ΔIRF-3 (pEF HA-IRF-3 and pEF HA-IRF-3 58-427, respectively) was cotransfected.
To assay activity of NF-κB, ATF-2/c-Jun (AP-1), or IRF-3, HEK 293 cells were transfected with 0.5 μg of p55A2Luc (luciferase gene under the control of repeated PRD II elements of the IFN-β promoter) (52), 0.5 μg of pAP-1Luc containing seven AP-1 binding motif repeats (Stratagene; Path-Detect Cis-Reporting Systems), or 1 μg of p55CIBLuc, in which the luciferase gene is under the control of repeated PRD I elements of the IFN-β promoter (52), respectively, by using the FuGENE transfection reagent. Ten hours posttransfection cells were infected in suspension with the indicated viruses at an MOI of 0.2 for 1 h in serum-free DMEM and seeded out into 24-well plates in DMEM supplemented with 2.5% FCS. Fourteen hours postinfection cells were lysed with cell lysis buffer (20 mM Tris [pH 7.8], 2 mM dithiothreitol, 2 mM trans-1,2-diamino-cyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100) and assayed for reporter gene activity. Cell extracts were mixed with luciferase buffer [20 mM Tricine, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM dithiothreitol, 1.07 mM MgCO3 · Mg(OH)2 · 5H2O] containing 470 μM luciferin, 270 μM acetyl coenzyme A, and 530 μM ATP, and light emission was measured as relative light units in a luminometer (Perkin-Elmer Wallac GmbH).
RESULTS
BRSV but not BRSV NS gene deletion mutants prevent upregulation of ISG expression.
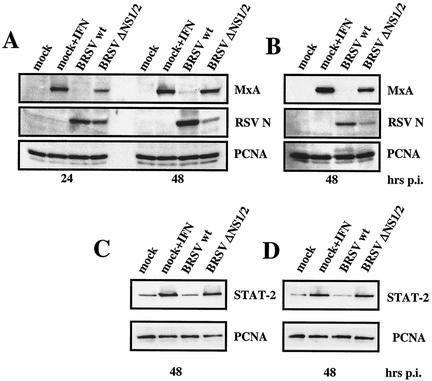
We have previously shown that BRSV NS proteins are required to protect virus from antiviral activity induced by exogenous IFN (36). A strikingly reduced growth of BRSV NS deletion mutants in MDBK cells even in the absence of exogenous IFN indicated that NS deletion mutants may efficiently trigger the activation of the cellular antiviral response. To investigate the response of cellular ISGs to infection with BRSV and BRSV lacking NS genes, MDBK cells were infected at an MOI of 0.2 with wt BRSV or the double gene deletion mutant BRSV ΔNS1/2. Cells were harvested 24 and 48 h postinfection, and equal amounts of cell extract were subjected to SDS-PAGE and Western blotting. The blots were probed with antibodies to MxA, PCNA as a loading control, and anti-RSV serum. In cells treated with exogenous IFN or infected with BRSV ΔNS1/2 a significant level of MxA was detectable already at 24 h postinfection, indicating that induction of IFN and the subsequent upregulation of ISGs were successfully triggered. However, in wt BRSV-infected cells, MxA was hardly detectable even at 48 h postinfection (Fig. 1A), demonstrating that upregulation of ISGs does not occur. Corresponding results were also obtained in human HEp-2 cells (Fig. 1B). As expected from earlier studies, due to the lack of IFN resistance mediated by the two NS proteins, viral protein expression of BRSV ΔNS1/2 was greatly reduced in MDBK cells compared to that of wt BRSV (Fig. 1A and B). In addition to MxA, other ISGs including dsRNA-activated protein kinase, p48, STAT-1 (data not shown), and STAT-2 (Fig. 1C and D) were found upregulated in BRSV ΔNS1/2-infected MDBK (Fig. 1C) and HEp-2 (Fig. 1D) cells compared to wt BRSV-infected cells.
FIG. 1.
BRSV NS deletion mutants but not wt BRSV induce ISGs. Shown are Western blot analyses of MxA expression in MDBK (A) and HEp-2 (B) cells and of STAT-2 in MDBK (C) and HEp-2 (D) cells. Cells were mock infected or infected at an MOI of 0.2 with BRSV wt and BRSV ΔNS1/2. As a control for upregulation of ISGs, mock-infected cells were treated with 2,000 U of recombinant IFN-α A/D per ml. Cells were harvested 24 and 48 h postinfection as indicated, and equal amounts of cell extract were subjected to SDS-PAGE followed by detection of MxA, STAT-2, BRSV N protein, and PCNA as a loading control.
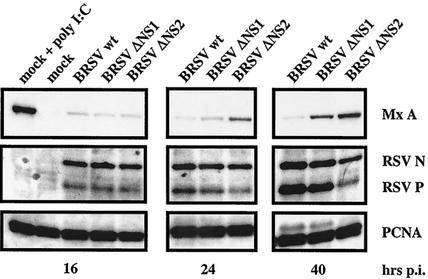
To further assess the contribution of individual NS proteins to successful blocking of ISG induction by wt BRSV, single NS deletion mutants were included in the experiments. Extracts from cell cultures infected with wt BRSV, BRSV ΔNS1, and BRSV ΔNS2 were harvested at 16, 24, and 40 h postinfection. Both deletion mutants induced MxA expression, although a delayed induction was always observed for the BRSV ΔNS1 deletion mutant (Fig. 2). Typically, the induction of MxA as a marker for antiviral proteins correlated with inhibition of virus propagation. In conclusion, it appears that both NS proteins are required for complete blocking of ISG expression, but the NS2 protein on its own appears to have some activity in preventing or delaying ISG expression.
FIG. 2.
Induction of MxA is delayed in BRSV ΔNS1-infected cells. MDBK cells were mock infected or infected at an MOI of 0.2 with BRSV wt, BRSV ΔΝS1, or BRSV ΔΝS2. When indicated, 2,000 U of IFN-α A/D per ml was added immediately after infection. Cells were harvested 16, 24, and 40 h postinfection, and equal amounts of cell extract were subjected to SDS-PAGE followed by detection of MxA, BRSV N and P protein, and PCNA.
BRSV does not affect IFN JAK/STAT signaling.
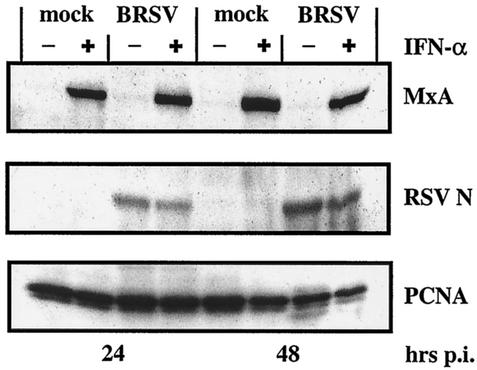
To examine how BRSV manages to prevent ISG expression, we first checked the integrity of the IFN JAK/STAT signaling, as this pathway is an established target for paramyxovirus V and C proteins (for reviews see references 18 and 19). MDBK cells were mock infected or infected with BRSV at an MOI of 0.2 and subsequently treated with 2,000 U of recombinant IFN-α/ml. Differences in the amounts of upregulated ISGs should then have disclosed possible interference in the IFN pathway. Immunoblot analyses, however, revealed equal MxA protein levels in mock-infected and BRSV-infected cells treated with exogenous IFN-α (Fig. 3). Thus, the lack of ISG expression in BRSV-infected cells is due not to defects in IFN signaling but to a more upstream event in the IFN defense pathway. In addition, RSV protein levels were not significantly reduced in IFN-treated infected cells compared to untreated control cells, showing the high IFN resistance capacity of BRSV.
FIG. 3.
BRSV wt does not interfere with IFN signaling and induction of ISGs by IFN. MDBK cells were mock infected or infected at an MOI of 0.2 with BRSV wt. When indicated, 2,000 U of IFN-α A/D per ml was added immediately after infection. Cells were harvested 24 or 48 h postinfection, and equal amounts of cell extract were subjected to SDS-PAGE followed by detection of MxA, BRSV N protein, and PCNA.
BRSV prevents production of IFN.
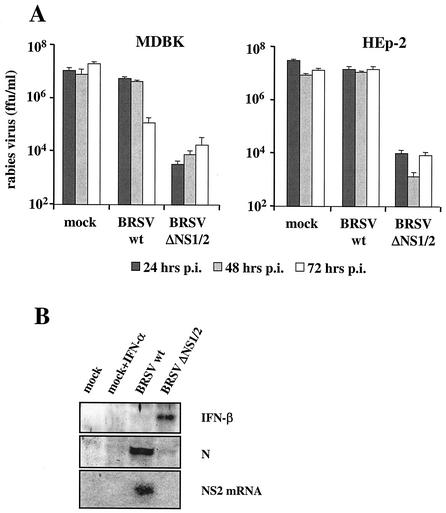
We were then interested to see whether the lack of ISG upregulation in BRSV-infected cells was caused by a lack of IFN production. To investigate whether biologically active IFN was present in the supernatants of MDBK and HEp-2 cells infected with wt BRSV or BRSV ΔNS1/2, we used an assay system based on RV, which is IFN sensitive in MDBK cells (36). Supernatants of BRSV- and BRSV ΔNS1/2-infected MDBK and HEp-2 cells were harvested 24, 48, and 72 h postinfection and incubated at 56°C for 30 min to inactivate replication-competent virus. RV-infected MDBK cells were then incubated with the inactivated supernatants, and RV titers were determined 2 days postinfection. All supernatants of BRSV ΔNS1/2-infected MDBK and HEp-2 cells were able to suppress RV replication about 3 log10 units, demonstrating the presence of active IFN (Fig. 4). However, supernatants taken from wt BRSV-infected cells did not influence RV titers significantly compared to control supernatants, indicating that in cells infected with wt BRSV production of IFN is prevented.
FIG. 4.
(A) IFN activity in supernatants of infected MDBK and HEp-2 cells. Cells were mock infected or infected at an MOI of 0.2 with BRSV wt or BRSV ΔNS1/2. Supernatants were harvested 24, 48, and 72 h postinfection and transferred onto RV-infected MDBK cells. RV titers were determined 2 days postinfection. Error bars indicate standard deviations. (B) IFN-β is expressed in BRSV ΔNS1/2-infected but not in wt BRSV-infected HEp-2 cells. Supernatant proteins were precipitated with TCA, and IFN-β was identified in a Western blot made with a polyclonal anti-human IFN-β serum. RSV N protein in cell extracts and NS2 mRNA in total RNA were demonstrated by Western blotting with an anti-RSV serum and an NS2-specific cDNA hybridization probe, respectively, at 37 h postinfection.
The presence of IFN-β in supernatants of HEp-2 cells infected with wt BRSV or BRSV ΔNS1/2 was shown after TCA precipitation of supernatant protein and Western blot analysis with polyclonal antibody to human IFN-β. At 37 h postinfection, a prominent band was present in the material from cells infected with BRSV ΔNS1/2 (Fig. 4B). In contrast, and in spite of heavy virus replication, a clear signal for IFN-β was not observed for wt BRSV. This demonstrated the induction of endogenous IFN-β expression by BRSV ΔNS1/2 and the ability of wt BRSV to interfere with induction.
Infection by BRSV prevents transcription from the IFN-β gene promoter.
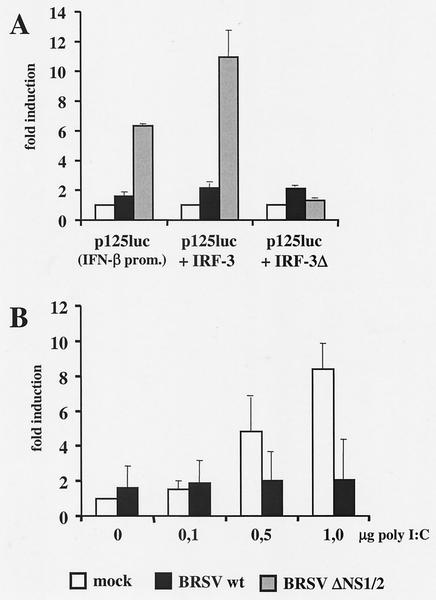
To address the mechanisms of BRSV involved in preventing IFN-β induction, we made use of a reporter construct harboring the entire IFN-β gene promoter-enhancer. Cultures of HEK 293 cells were transiently transfected with p125luc (52) followed by infection with wt BRSV or BRSV ΔNS1/2. Whereas in wt BRSV-infected cells only a slight twofold induction of the IFN-β enhancer was observed in comparison to mock-infected cells, in BRSV ΔNS1/2-infected cells a strong sevenfold induction was noted (Fig. 5A). The specificity of IFN-β promoter induction by BRSV ΔNS1/2 was further suggested by inclusion of plasmids encoding functional IRF-3 or a truncated, dominant-negative mutant of IRF-3 (ΔIRF-3). Whereas the IRF-3-encoding plasmid increased luciferase expression in BRSV ΔNS1/2-infected cells by approximately another twofold, the dominant-negative IRF-3 reduced luciferase levels to those observed with mock- or BRSV wt-infected cells (Fig. 5A).
FIG. 5.
Virus-induced activation of the IFN-β enhancer. (A) HEK 293 cells were transfected with a luciferase construct driven by the IFN-β promoter-enhancer (p125luc) and, as indicated, cotransfected with plasmids encoding IRF-3 or a dominant-negative IRF-3 mutant (ΔIRF-3). Ten hours posttransfection cells were infected at an MOI of 0.2 with BRSV wt or BRSV ΔNS1/2. Cell lysates were harvested 14 h postinfection and assayed for luciferase activity. Relative light units are given as fold induction and represent the mean values of four independent experiments with error bars indicating standard deviations. (B) BRSV wt inhibits IFN induction triggered by poly(rI):poly(rC). HEK 293 cells were mock infected or infected with BRSV wt at an MOI of 0.25. Twelve hours postinfection, cells were transfected with p125luc and the indicated amounts of poly(rI):poly(rC). Luciferase assays were done 24 h posttransfection. Relative light units are given as fold induction and represent the mean values of three independent experiments. Error bars indicate standard deviations.
In another experiment, the capacity of BRSV to block IFN-β promoter activity in the presence of an additional trigger was challenged. Mock- or BRSV-infected cells were transfected with increasing amounts of poly(rI):poly(rC), a dsRNA analog known to induce IFN production. Poly(rI):poly(rC) was not able to stimulate the IFN-β promoter in BRSV-infected cells, whereas a dose-dependent increase in luciferase activity was observed in mock-infected cells (Fig. 5B).
Infection by BRSV specifically blocks IRF-3 activation.
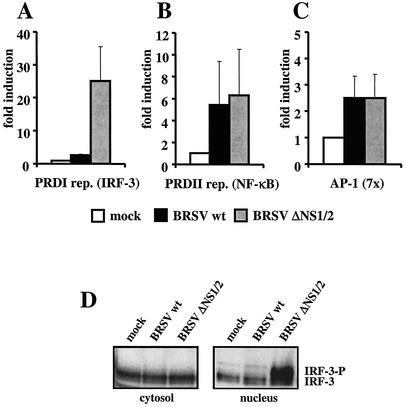
Induction of the IFN-β promoter is known to require the binding of transcription factors NF-κB, IRF-3, and AP-1 (dimers of ATF-2 or heterodimers of ATF-2/c-Jun), which are activated in response to virus infection. To assess the activation state of these three transcription factors, transient reporter gene expression assays were carried out in HEK 293 cells that had been mock infected or infected with wt BRSV or BRSV ΔNS1/2. Cells were transfected with plasmids encoding luciferase under the control of binding sites for NF-κB (PRD II; p55A2Luc) or IRF-3 (PRD I; p55CIBLuc) (52) or binding sites for AP-1 (pAP-1Luc; Stratagene). As predicted, in BRSV ΔNS1/2-infected cells all three transcription factors were found to be active. However, in cells infected with wt BRSV, activation of luciferase from the IRF-3-driven plasmid was clearly impaired, whereas NF-κB- or AP-1-driven expression was not affected (Fig. 6A to C). Thus, the IRF-3 pathway is the target of NS protein in preventing IFN-β induction.
FIG. 6.
BRSV wt selectively blocks the activation of IRF-3. (A to C) HEK 293 cells were transfected with plasmids harboring the luciferase gene under the control of promoters containing either IRF-3 (A), NF-κB (B), or AP-1 (C) binding sites. Ten hours posttransfection, cells were infected with the indicated viruses at an MOI of 0.2. Fourteen hours postinfection cells were harvested and luciferase activity was determined. Relative light units are given as fold induction. Results show the mean values of four independent experiments with error bars showing standard deviations. (D) IRF-3 is not phosphorylated in BRSV wt-infected cells. HEK 293 cells were mock infected or infected with the indicated viruses at an MOI of 0.25. Ten hours postinfection, cells were harvested, and nuclear extracts were prepared as described in Materials and Methods and subjected to SDS-PAGE. Nonphosphorylated and phosphorylated forms of IRF-3 were detected with an anti-IRF-3 antibody (Dianova).
C-terminal phosphorylation of IRF-3 is a prerequisite for its nuclear localization, association with CBP/p300 coactivators, and activation of the IFN-β gene transcription. The phosphorylation status of IRF-3 is readily detectable in virus-infected cells since phosphorylated IRF-3 migrates slower in SDS-PAGE than nonphosphorylated IRF-3 does. To investigate whether phosphorylated IRF-3 is present in BRSV-infected cells, HEK 293 cells were mock infected or infected with BRSV or BRSV ΔNS1/2 and harvested 8 h postinfection. Nuclei were prepared as described in Materials and Methods, and equal amounts of nuclear extract, or cytosolic extract as a control, were subjected to SDS-PAGE. In cells infected with BRSV ΔNS1/2 substantial amounts of phosphorylated IRF-3 were detectable in nuclear extracts (Fig. 6D). However, in nuclear extracts of wt BRSV-infected cells no slower-migrating form of IRF-3 was present, indicating that in the presence of the NS proteins NS1 and NS2 phosphorylation of IRF-3 is not possible.
DISCUSSION
In the present study we provide evidence that BRSV has the capacity to block the early stages of the IFN response to virus infection by interfering with the activation of IRF-3, thereby preventing induction of IFN-β and the establishment of an antiviral state. This novel function is correlated with the presence of the NS proteins NS1 and NS2. Recombinant BRSV lacking either NS1 or NS2 or both NS proteins efficiently induces IFN and ISGs in infected cells. Though not able to completely prevent IFN induction, the NS2 protein present in BRSV ΔNS1 virus is able to at least inhibit IFN induction.
The first indication of an impaired IFN response in BRSV-infected cells was observed in the bovine MDBK cell line. Infection with the NS deletion mutant BRSV ΔNS1/2 led to the successful establishment of an antiviral state indicated by increased expression levels of ISGs, including STAT-1, p48, dsRNA-activated protein kinase (data not shown), STAT-2, and, in particular, MxA, the expression of which is tightly regulated and induced specifically by IFN (1, 21). However, in cells infected with wt BRSV, this ISG upregulation was not observed, suggesting the involvement of the NS proteins in blocking one or several steps in the cellular IFN pathway. Similar results were also obtained in a human cell line harboring an intact IFN system, HEp-2 cells, confirming the finding of an obstructed cellular IFN pathway in BRSV-infected MDBK cells.
As several members of the subfamily Paramyxovirinae, such as Simian virus 5, Human parainfluenzavirus 2, or Sendai virus, are able to interfere with IFN signaling and thus prevent upregulation of ISGs (11, 15, 51), we first investigated whether BRSV was able to inhibit IFN JAK/STAT signaling. This pathway, however, was not affected in BRSV-infected cells, since small amounts of IFN added to infected and to noninfected cells caused a similar and strong induction of MxA and other ISGs. Therefore, an impaired production and/or secretion of IFN was suggested instead.
To test for IFN activity in the supernatant of BRSV-infected cells, we used an assay based on the IFN sensitivity of RV in MDBK cells (36). Whereas IFN antiviral activity was readily detectable in supernatants from cells infected with BRSV ΔNS1/2, we could not detect IFN activity in supernatants from cells infected with wt BRSV up to 3 days postinfection. IFN-β was not present in the supernatants of wt BRSV-infected cells, in contrast to those of BRSV ΔNS1/2-infected cells, pointing toward a function for the NS proteins in interfering with the pathway leading to induction of IFN gene expression. Therefore, we directly looked at IFN-β gene promoter activity by using established reporter gene assays. While cells infected with the double deletion mutant BRSV ΔNS1/2 displayed high levels of IFN induction, only a minor increase in IFN-β gene promoter activity could be detected in BRSV wt-infected cells, indicating that BRSV is able to block the induction of IFN-β during the course of viral infection or, as a potential alternative, does not produce a suitable trigger for induction of IFN, such as dsRNA or protein. Transfection of poly(rI):poly(rC) led to a dose-dependent stimulation of IFN-β gene promoter activity in noninfected cells. However, in BRSV-infected cells the dsRNA-triggered IFN induction was completely suppressed by BRSV, suggesting an active block of the IFN induction pathway. In addition, superinfection experiments have indicated that wt BRSV is also able to diminish IFN induction by BRSV ΔNS1/2 (46a), which is consistent with the ability to block IFN induction by dsRNA.
Induction of IFN-β expression is controlled by an enhanceosome that binds three distinct transcription factor complexes in the context of chromatin-organizing proteins (12, 17, 32, 44-46, 48). Each of these complexes, IRF-3, NF-κB, and ATF-2/c-Jun, becomes activated following protein phosphorylation events induced in response to viral infection. To reveal the activity of these transcription factors in BRSV-infected cells, assays were performed in which the reporter gene, luciferase, was under the transcriptional control of the binding site for the individual transcription factors. As could be expected from the successful induction of IFN by the BRSV ΔNS1/2 gene deletion virus, all three transcription factors were activated in cells infected with this mutant. Interestingly, NF-κB- and AP-1-driven expression of luciferase was equal in BRSV ΔNS1/2- and wt BRSV-infected cells, suggesting that activation of these factors is not affected by BRSV. However, a striking reduction of activity was observed for IRF-3-controlled expression, suggesting the IRF-3 activation pathway as a specific target of the BRSV NS proteins.
The C-terminal phosphorylation of IRF-3 following virus infection or dsRNA stimulation is mediated by a unique VAK activity and leads to conformational changes in the IRF-3 protein, triggering its association with the CBP/p300 coactivator and stimulation of transcriptional activity (30, 37, 40, 52). We therefore investigated whether this activity was impaired in BRSV wt-infected cells by appraising the phosphorylation state of IRF-3 in these cells. While phosphorylated IRF-3 was present in the nuclei of BRSV ΔNS1/2-infected cells, we were not able to detect phosphorylated IRF-3 in cytoplasmic or nuclear extracts of BRSV wt-infected cells. This provides an explanation for the complete inhibition of IFN-β induction after BRSV infection and defines activation of IRF-3 by the VAK or the upstream activation of VAK by a virus-specific pattern as a target of BRSV NS proteins.
How cells sense viral entry or replication is only poorly understood. It is generally accepted that dsRNA that might be generated during virus infection serves as a major recognition pattern (for a review see reference 24). The mammalian Toll-like receptor 3 senses dsRNA (3) and leads to IRF-3 activation and IFN-β induction via specific adaptor molecules (TICAM-1 [31] or TRIF [50]). Recently, tenOever et al. showed that IRF-3 is not only associated with but also directly activated by a viral protein, the measles virus nucleocapsid protein (43). RSV NS proteins are assumed to be associated with RSV nucleocapsid structures, as NS1 is able to bind viral M protein (13) and NS1 and NS2 affect replication of RSV model genomes (5). The ability of BRSV NS proteins to prevent virus-independent, poly(rI):poly(rC)-triggered IFN induction demonstrates active interference with a cellular pathway, rather than a possible function in just hiding a virus pattern that might lead to activation of the IFN alert. Further studies aimed at elucidating the mechanism by which NS-deficient BRSV induces IFN and the NS proteins interfere with the pathway(s) leading to IFN gene expression may help us to gain more insight into the intricate mechanisms by which cells are able to recognize and respond to virus infection.
It is becoming increasingly clear that many viruses have acquired functions that allow them to simultaneously target different steps of the cellular innate response to virus infection. In particular, it appears necessary to prevent the action of IFN or of IFN-induced antiviral proteins, as the IFN alert may be triggered by companion, opportunistic, or competitor pathogens. IFN resistance appears important for a virus like BRSV, which is a major agent of the multifactorial enzootic bronchopneumonia. In addition, it appears worthwhile to prevent IFN induction, as observed here for BRSV. Intriguingly, other paramyxoviruses as well are able to concurrently interfere with both IFN induction and IFN action. Reminiscent of the situation with BRSV is that the same proteins are involved in the two functions. The V proteins of simian virus 5 and human parainfluenzavirus 2 trigger STAT-1 and STAT-2, respectively, to degrade, thus preventing IFN signaling and the induction of ISGs (4). As revealed recently, they also prevent the induction of IFN-β, however, and in contrast to BRSV NS proteins, interfere with the activation of both NF-κB and IRF-3 (22, 33). Also the Sendai virus V protein was found to be active in preventing IFN induction (33), although in this case another protein, the C protein, rather than V, interfered with JAK/STAT signaling (16, 27).
The ability of viruses such as BRSV to prevent IFN-β induction is certainly of great importance not only with respect to the immediate antiviral activities of some ISGs but also with respect to the outcome of adaptive immunity, as alpha/beta IFNs are known to be potent enhancers of immune responses (2, 26, 29, 34). Therefore, RSV NS deletion mutants appear to be promising candidates for attenuated live vaccines with improved immunogenic potential, as supported by recent work (46a).
Acknowledgments
This work was supported by the European Commission (EC 5th FP-RSV Vac QLK2-CT-1999-00443) and the Deutsche Forschungsgemeinschaft (SFB 455-A3 and SPP1089, Co260/1-1).
Reporter plasmids p125luc, p55CIBLuc, and p55A2Luc and the IRF-3-encoding plasmids were a generous gift from Takashi Fujita, Kyoto, Japan. Otto Haller and Georg Kochs, Freiburg, Germany, kindly provided the MxA monoclonal antibody, and Jose Antonio Melero, Madrid, Spain, provided RSV peptide serum. We thank Stefan Finke and Jörg Schlender for critical reading and help with infections and Friedemann Weber, Freiburg, for discussions of all sorts.
REFERENCES
- 1.Aebi, M., J. Fah, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 4.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atreya, P. L., M. E. Peeples, and P. L. Collins. 1998. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 72:1452-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossert, B., and K. K. Conzelmann. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1352. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 9.Conzelmann, K. K. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu. Rev. Genet. 32:123-162. [DOI] [PubMed] [Google Scholar]
- 10.Conzelmann, K. K., J. H. Cox, and H. J. Thiel. 1991. An L (polymerase)-deficient rabies virus defective interfering particle RNA is replicated and transcribed by heterologous helper virus L proteins. Virology 184:655-663. [DOI] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, W., D. Thanos, and T. Maniatis. 1993. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell 74:887-898. [DOI] [PubMed] [Google Scholar]
- 13.Evans, J. E., P. A. Cane, and C. R. Pringle. 1996. Expression and characterisation of the NS1 and NS2 proteins of respiratory syncytial virus. Virus Res. 43:155-161. [DOI] [PubMed] [Google Scholar]
- 14.Finke, S., and K. K. Conzelmann. 1999. Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a 3′ copy-back ambisense rabies virus. J. Virol. 73:3818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 17.Garoufalis, E., I. Kwan, R. Lin, A. Mustafa, N. Pepin, A. Roulston, J. Lacoste, and J. Hiscott. 1994. Viral induction of the human beta interferon promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J. Virol. 68:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 20.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 21.Haller, O., and G. Kochs. 2002. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 22.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 23.Iwamura, T., M. Yoneyama, N. Koizumi, Y. Okabe, H. Namiki, C. E. Samuel, and T. Fujita. 2001. PACT, a double-stranded RNA binding protein, acts as a positive regulator for type I interferon gene induced by Newcastle disease virus. Biochem. Biophys. Res. Commun. 282:515-523. [DOI] [PubMed] [Google Scholar]
- 24.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 25.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 26.Kadowaki, N., and Y. J. Liu. 2002. Natural type I interferon-producing cells as a link between innate and adaptive immunity. Hum. Immunol. 63:1126-1132. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2002. Sendai virus C protein impairs both phosphorylation and dephosphorylation processes of Stat1. FEBS Lett. 511:139-144. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14:432-436. [DOI] [PubMed] [Google Scholar]
- 30.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 32.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125-129. [DOI] [PubMed] [Google Scholar]
- 33.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 34.Proietti, E., L. Bracci, S. Puzelli, T. Di Pucchio, P. Sestili, E. De Vincenzi, M. Venditti, I. Capone, I. Seif, E. De Maeyer, D. Tough, I. Donatelli, and F. Belardelli. 2002. Type I IFN as a natural adjuvant for a protective immune response: lessons from the influenza vaccine model. J. Immunol. 169:375-383. [DOI] [PubMed] [Google Scholar]
- 35.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 38.Stott, E. J., G. Taylor, L. A. Ball, K. Anderson, K. K.-Y. Young, A. M. Q. King, and G. W. Wertz. 1987. Immune and histopathological responses in animals vaccinated with recombinant vaccinia viruses that express individual genes of human respiratory syncytial virus. J. Virol. 61:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhara, W., M. Yoneyama, T. Iwamura, S. Yoshimura, K. Tamura, H. Namiki, S. Aimoto, and T. Fujita. 2000. Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. J. Biochem. (Tokyo) 128:301-307. [DOI] [PubMed] [Google Scholar]
- 40.Suhara, W., M. Yoneyama, I. Kitabayashi, and T. Fujita. 2002. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 277:22304-22313. [DOI] [PubMed] [Google Scholar]
- 41.Teng, M. N., and P. L. Collins. 1999. Altered growth characteristics of recombinant respiratory syncytial viruses which do not produce NS2 protein. J. Virol. 73:466-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng, M. N., S. S. Whitehead, A. Bermingham, M. St. Claire, W. R. Elkins, B. R. Murphy, and P. L. Collins. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317-9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 71:777-789. [DOI] [PubMed] [Google Scholar]
- 45.Thanos, D., and T. Maniatis. 1995. Identification of the rel family members required for virus induction of the human beta interferon gene. Mol. Cell. Biol. 15:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 46a.Valarcher, J.-F., J. Furze, S. Wyld, R. Cook, K.-K. Conzelmann, and G. Taylor. 2003. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J. Virol. 77:8426-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Poel, W. H., A. Brand, J. A. Kramps, and J. T. van Oirschot. 1994. Respiratory syncytial virus infections in human beings and in cattle. J. Infect. 29:215-228. [DOI] [PubMed] [Google Scholar]
- 48.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 49.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St. Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto, M., S. Sato, K. Mori, K. Hoshino, O. Takeuchi, K. Takeda, and S. Akira. 2002. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J. Immunol. 169:6668-6672. [DOI] [PubMed] [Google Scholar]
- 51.Yokoo, J., B. Gotoh, T. Komatsu, K. Takeuchi, and T. Miyadai. 1999. Replication-incompetent Sendai virus can suppress the antiviral action of type I interferon. Arch. Virol. 144:1043-1055. [DOI] [PubMed] [Google Scholar]
- 52.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]