Abstract
Canine parvovirus (CPV) and feline panleukopenia virus (FPV) capsids bind to the transferrin receptors (TfRs) of their hosts and use these receptors to infect cells. The binding is partially host specific, as FPV binds only to the feline TfR, while CPV binds to both the canine and feline TfRs. The host-specific binding is controlled by a combination of residues within a raised region of the capsid. To define the TfR structures that interact with the virus, we altered the apical domain of the feline or canine TfR or prepared chimeras of these receptors and tested the altered receptors for binding to FPV or CPV capsids. Most changes in the apical domain of the feline TfR did not affect binding, but replacing Leu221 with Ser or Asp prevented receptor binding to either FPV or CPV capsids, while replacing Leu221 with Lys resulted in a receptor that bound only to CPV but not to FPV. Analysis of recombinants of the feline and canine TfRs showed that sequences controlling CPV-specific binding were within the apical domain and that more than one difference between these receptors determined the CPV-specific binding of the canine TfR. Single changes within the canine TfR which removed a single amino acid insertion or which eliminated a glycosylation site gave that receptor the expanded ability to bind to FPV and CPV. In some cases, binding of capsids to mutant receptors did not result in infection, suggesting a structural role for the receptor in cell infection by the viruses.
Canine parvovirus (CPV) and feline panleukopenia virus (FPV) infect a variety of different carnivore hosts. CPV is a host range variant of FPV that appears to have gained the ability to infect dogs and dog cells through the acquisition of a small number of mutations in the capsid protein gene which altered residues on the surface of the capsid (1, 9, 20, 26). CPV and FPV both bind to the feline transferrin receptor (TfR) and use that receptor for cell infection (25). The canine TfR binds to the capsids of CPV but does not bind to the capsids of FPV or of certain CPV mutants (16). The specificity of virus binding to the TfR determines the virus susceptibility of the host cells. When the feline or canine TfRs are expressed in otherwise nonsusceptible hamster (CHO) cells, they show the same binding specificities that are seen for the feline or canine cells (16). Viral capsids that bind to the TfR enter the cell rapidly and then slowly traffic within the endosomal system of the cell before entering the cytoplasm and the nucleus, where the viral DNA replicates (24, 36, 37). Although TfR binding is necessary for virus infection, it is not always sufficient, and some specific structural interactions or coreceptors may also be required (15a).
The TfR is a type 2 transmembrane protein that is expressed on the surface of cells as a homodimer, and the human TfR has a 67-residue cytoplasmic tail, a 21-residue transmembrane sequence, and a 672-residue extracellular domain (ectodomain) (19). TfR binds iron-loaded Tf, and then the cytoplasmic tail of the receptor associates with the μ2 subunit of the adapter protein 2 (AP2), leading to association with other protein and lipid cofactors and to uptake through clathrin-mediated endocytosis (4, 6, 34). In the low pH of the endosome, the iron is released from the Tf, and the iron-free Tf and TfR remain together and recycle to the cell surface (23, 31). The structures of the dimeric human TfR ectodomain determined alone or in complex with HFE, the protein altered in human hereditary hemachromatosis, show that each TfR monomer is composed of three domains, including a domain with homology to carboxypeptidases, an apical domain, and a helical domain which forms the interface between the TfR monomers (5, 19). There are three N-linked glycosylation sites at positions 251, 317, and 727 in the human TfR that can affect Tf binding (14, 17, 41), a fourth N-linked glycosylation site is found in the helical domain of the TfRs of many species but not humans, and the canine TfR has a fifth predicted site for N-linked glycosylation within its apical domain (16). The human TfR also has O-linked oligosaccharides that differ when receptors are expressed in different human cell lines (10). The Tf binding site on the human TfR involves mostly residues in the helical domain, and it partially overlaps the site of HFE binding, resulting in competition between those ligands (5, 7, 39).
Here we show that CPV and FPV binding to the feline TfR involves the apical domain of the receptor and that substitutions of one residue in that domain of the feline TfR can prevent it from binding to both CPV and FPV or can specifically prevent FPV binding while allowing CPV binding. The CPV-specific binding of the canine TfR was affected by several residues in the apical domain, indicating that multiple structural interactions determine the specificity of the receptors for the different viral capsids.
MATERIALS AND METHODS
Cells and ligands.
Feline CRFK and NLFK cells or canine A72, Cf2th, and Walter Reed cells were grown in a 50% mixture of McCoy's 5A and Leibovitz L15 media with 5% fetal calf serum. Chinese hamster ovary (CHO) cell-derived TRVb cells which do not express type 1 TfR (22) were grown in Ham's F-12 medium with 10% fetal calf serum. CPV type 2, FPV-b, and CPV type 2b were isolated from infectious plasmid clones by transfection into NLFK cells (16, 26, 27). Viruses for use in binding studies were grown in NLFK cells and concentrated using polyethylene glycol precipitation, and full and empty capsids were purified by banding twice on sucrose gradients, dialyzed against either phosphate-buffered saline (PBS) or 20 mM Tris-HCl (pH 7.5), and stored at 4°C (33).
Viral stocks were titrated using a 50% tissue culture infectious dose (TCID50) assay in NLFK cells. Cells in 96-well cultures were inoculated with dilutions of virus and then incubated for 2 days. To detect infected cells, cultures were fixed with 2.5% paraformaldehyde (PFA) in PBS for 10 min and then stained for the presence of viral capsid protein using a rabbit anti-CPV serum or stained with Texas red- or Cy2 (Amersham Biosciences, Piscataway, N.J.)-conjugated antibody CE10 against the viral NS1 protein (42).
TfR clones and mutagenesis.
Feline and canine TfR DNA clones in the plasmid pcDNA3.1(−) were used for all studies (16, 25). As it appeared from steric considerations that the apical domain would interact with the capsid, we initially altered sequences within that structure. There are many differences in sequence between the feline and canine TfRs within the apical domain (Fig. 1A), and by comparison with the human TfR structure, it is likely that many of the differences would be exposed on the surface (Fig. 1B). Most changes made within the apical domain were designed to alter the surface of the receptor (Fig. 1B). Mutant receptors prepared are listed in Tables 1 and 2 and were prepared directly in the plasmids using the GeneEditor in vitro site-directed mutagenesis system (Promega, Madison, Wis.) according to the manufacturer's directions. The mutations were confirmed by sequencing the plasmid, and for mutants for which virus binding was affected, the entire TfR gene was sequenced.
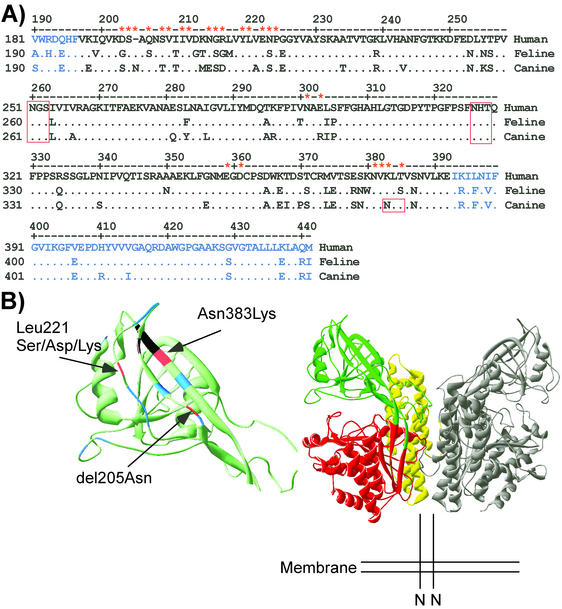
FIG. 1.
(A) The aligned sequences of the human, feline, and canine TfRs containing residues between positions 190 and 441 in the feline TfR sequence which transferred the CPV-specific binding to the feline TfR. Residues that are identical to those in the human TfR sequence are indicated by periods, while residues that differ between any of the three receptors are indicated. Residues that were mutated in the feline or canine TfR in these studies are marked with asterisks. The sequence corresponding to the apical domain of the human TfR is indicated in black, while protease-like domain sequences are shown in blue. Potential N-linked glycosylation sites in the sequences are boxed in red. (B) (Right) A ribbon diagram of the human TfR dimer structure, with the protease-like domain (red), the apical domain (green), and the helical domain (yellow) shown on one copy of the dimer. (Left) An enlarged view of the apical domain shows the positions of sequences analyzed by mutation in the feline or canine TfRs. Substitutions of positions indicated in blue had no clear effect on virus binding. Changes of residue 221 shown in red prevented CPV and FPV binding (feline TfR-L221S or feline TfR-L221D) or specifically prevented FPV binding (feline TfR-L221K). Other residues shown in red altered the specificity of virus binding of the canine TfR either when an Asn was deleted from position 205 of the canine TfR (delN205) or when Asn383 was changed to Lys, which altered a unique glycosylation site in the canine TfR. Positions marked in black affected receptor transport to the cell surface when mutated.
TABLE 1.
Mutations prepared within the feline TfR sequence and their effects on virus and canine Tf bindinga
| Feline TfR mutant | Human TfR | Bindingb to:
|
||
|---|---|---|---|---|
| FPV capsid | CPV capsid | Canine Tf | ||
| G203S/S206Q insertion N205 | D194/Q197+N195 | + | + | + |
| G203S/S206Q insertion N205 K383N/S385T | D194/Q197+N195 K374/S376 | + | + | + |
| S204A | S195 | + | + | + |
| S208A | S199 | + | + | + |
| V209E/I211G/V212D | V200/I202/V203 | NA | NA | NA |
| V209E/I211G | V200/I202 | NA | NA | NA |
| I211G/V212D | I202/V203 | NA | NA | NA |
| N215A/S216A/G217A | N206/G207/R208 | + | + | + |
| Y220T/L221S | Y211/L212 | − | − | + |
| Y220T | Y211 | + | + | + |
| L221S | L212 | − | + | + |
| L221D | L212 | − | + | + |
| L221K | L212 | − | + | + |
| E223A/P225A | E214/P216 | + | + | + |
| S224A | N215 | + | + | + |
| N310A | N292 | + | + | + |
| E303R | E294 | + | + | + |
| E303A | E294 | + | + | + |
| E359K/D361K | E350/D352 | + | + | + |
| Δ358-377 | Δ349-368 | NA | NA | NA |
| N381D/V382T/K383D | N372/V373/K374 | NA | NA | NA |
| K383N | K374 | + | + | + |
| K383N/S385T | K374/S376 | + | + | + |
| S385A | T376 | + | + | + |
The sequence positions for the feline TfR and the aligned sequence positions of the human TfR are given to allow comparison with the human TfR structure. Asn205 was inserted into the position after residue 204 (insertion N205) in the feline TfR to match the equivalent change in the canine TfR.
Capsid binding was measured by fluorescence microscopy or flow cytometry with Cy2-labeled CPV and FPV capsids, while canine Tf was labeled with Texas red. The lack of cell surface expression of some mutants was defined using capsid and Tf binding and with an antibody against a peptide from the ectodomain of the TfR or Cy5. Symbols: +, binding; −, no binding. NA, not applicable (not expressed on the surface).
TABLE 2.
Mutations prepared within the canine TfR sequence and their effects on virus and canine Tf bindinga
| Canine TfR mutant | Human TfR | Bindingb to
|
||
|---|---|---|---|---|
| FPV capsid | CPV capsid | Canine Tf | ||
| S203G/Q206S & ΔN205 | D194/Q197 | + | + | + |
| N383K/T385S | K374/S376 | + | + | + |
| S203G/Q206S & ΔN205 & N383K/T385S | D194/Q197 and K374/S376 | + | + | + |
The sequences shown in the canine TfR column are the aligned positions of the feline TfR, and the aligned sequence positions of the human TfR are also given. Asn205was deleted from the position after the residue equivalent to position 204 (ΔN205) in the feline TfR.
Capsid and Tf binding and TfR expression analyses were performed as described in Table 1, footnote b.
Chimeric receptor preparation.
Chimeras of the feline and canine TfRs were prepared as diagrammed in Fig. 2A. Some chimeras were prepared using restriction sites in common between the canine and feline TfR sequences, while other unique sites were engineered into the sequences by PCR with primers that altered the appropriate nucleotides using the Deep Vent polymerase (New England Biolabs, Beverly, Mass.). After assembly into the expression plasmid, all chimeric receptor genes were completely sequenced to confirm the changes introduced.
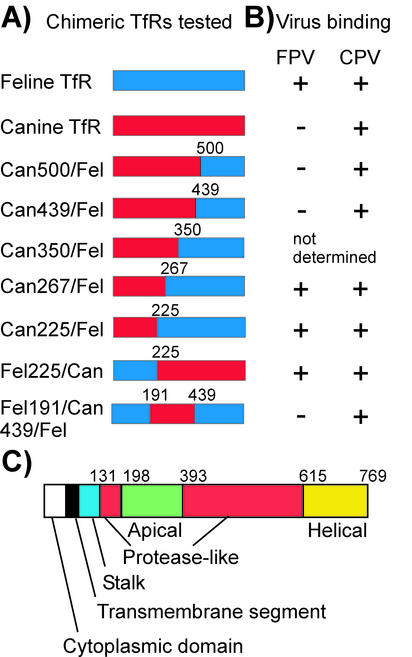
FIG. 2.
Chimeras of the feline and canine TfRs that were prepared and their binding to CPV or FPV capsids. (A) Feline-canine TfR chimeras tested. Clones of the feline (blue) and canine (red) TfRs were joined using either existing common restriction enzyme sites or sites engineered into the two sequences. Numbers indicate the residue positions (from the feline TfR sequence) at the recombination points. Can, canine; Fel, feline. (B) The binding of FPV or CPV capsids to the canine or feline TfRs or to chimeras of those receptors. The receptors were expressed in TRVb cells, and the virus capsid and Tf binding were measured by flow cytometry. Binding results were not obtained for the feline/canine chimera at position 350, as that receptor was not expressed on the cell surface. Symbols: +, binding; −, no binding. (C) A cartoon showing the TfR sequence, indicating the locations of the different domains of the proteins defined for the human TfR structure. Numbers indicate the residue positions in the feline TfR sequence at the boundaries of the structural domains.
Virus and transferrin labeling.
Purified CPV capsids, FPV capsids, and canine Tf were labeled with Texas red, Cy2, or Cy5 (Amersham) as previously described (16). Canine Tf (Sigma, St. Louis, Mo.) was iron loaded prior to labeling as previously described (2, 3).
TfR expression and virus and Tf binding assays.
Plasmids (2-μg amounts) were mixed with 6 μl of Lipofectamine (Life Technologies, Inc., Gaithersburg, Md.) and added to TRVb cells seeded at a density of 1.9 × 105 per cm2 in 9-cm2 dishes. The dishes were incubated for 2 h, growth medium was added, and the cells were incubated for 24 h. Cells were then incubated with Texas red- or Cy5-labeled canine Tf and Cy2-labeled CPV or FPV capsids for 1 h at 37°C, rinsed, fixed with 4% PFA for 10 min, and examined by fluorescence microscopy. For flow cytometric analysis, the cells were detached with 10 mM EDTA, stained as described above, and analyzed in a FACScalibur flow cytometer (Becton-Dickinson, San Jose, Calif.)
Antibody preparation and analysis of TfR expression.
A peptide from a conserved sequence of the TfR sequence (residues 558 and 571 of the human TfR) was conjugated to keyhole limpet hemocyanin and used to immunize rabbits. This antibody was used to detect TfR expression on the cell surface. Transfected cells were fixed with 4% PFA. The cells were then incubated with the rabbit antipeptide serum in PBS or with mouse antibody H65.4 against the TfR cytoplasmic tail (Zymed, South San Francisco, Calif.) (40) in PBS with 0.5% Triton X-100. The bound antibodies were detected with goat anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG conjugated to Cy2. For Western blot analysis of protein expression, feline cells, canine cells, or TRVb cells expressing the wild-type or mutant canine or feline TfRs were lysed in sodium dodecyl sulfate (SDS) sample buffer (2% SDS, 60 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol). Proteins separated on SDS-10% polyacrylamide gels were transferred to nitrocellulose membranes and incubated with antibody H65.4. Filters were incubated with goat anti-mouse IgG conjugated to horseradish peroxidase and then visualized with Supersignal substrate (Pierce Chemicals, Rockford, Ill.).
To examine the relative N-linked glycosylation of the TfRs, canine or feline cells or transfected TRVb cells were lysed in SDS sample buffer, boiled, and then incubated for 2 h at 37°C with peptide N-glycosidase F (PNGase F; New England Biolabs) in 50 mM sodium phosphate (pH 7.5) with 1% Nonidet P-40 (NP-40), before electrophoresis and Western blotting as described above.
Infection assays.
Cells expressing the wild-type or mutant feline TfR from plasmids or transfected with empty plasmid were inoculated with 10 TCID50 of CPV type 2 or 2b or FPV per cell for 1 h at 37°C, washed, and incubated for 48 h at 37°C. Cells were incubated in medium without serum for 30 min and then with Texas red-labeled canine Tf for 60 min at 37°C. After the cells were washed and incubated with 4% PFA, they were stained with a Cy2-labeled anti-NS1 antibody. The percentage of infected cells was determined and compared to the proportion of cells that became transfected and bound to canine Tf.
RESULTS
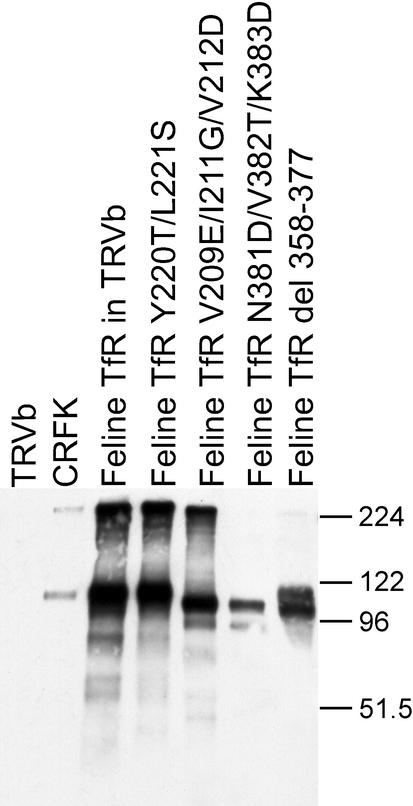
We prepared a variety of single, double, and triple mutations which changed conserved residues in the feline TfR that were exposed on the surface of the structure (Fig. 1 and Table 1). We also prepared chimeras of the feline and canine TfR sequences or changed residues in the canine TfR which differed from those in the feline TfR (Fig. 1 and 2; Tables 1 and 2). Several receptors showed altered virus binding properties, and those were all full-length, bound canine Tf, and were expressed at high levels after transfection of TRVb cells (Fig. 3). Some mutant TfR proteins examined showed slight differences in migration on SDS-polyacrylamide gels, most likely due to the deletion of 20 residues from the Δ349-368 mutant or to differences in the protein glycosylation or charge (Fig. 3).
FIG. 3.
Western blot showing the size and level of expression of various TfRs. Wild-type TfR and examples of mutant feline TfRs recovered from transfected TRVb cells compared to proteins from feline CRFK cells or from nontransfected TRVb cells are shown. TRVb cells were transfected with plasmids expressing either the wild-type feline TfR or mutants of that receptor. Cells were lysed in SDS sample buffer, electrophoresed on a 10% acrylamide gel, and transferred to a membrane. The TfR was detected with an antibody against the cytoplasmic tail. The positions of molecular size markers (in kilodaltons) are indicated to the right of the gel. del, deletion.
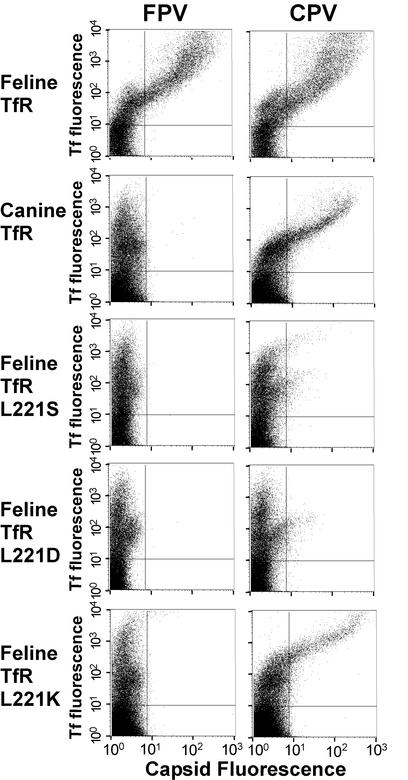
Of the feline TfR with substitutions of conserved residues that were expressed on the cell surface, only the substitutions of Leu221 affected virus binding (Table 1). The feline TfR-L221S and feline TfR-L221D did not bind to either CPV or FPV capsids to a significant level, while feline TfR-L221K bound CPV, but not FPV, capsids (Fig. 4).
FIG. 4.
Binding of canine Tf (y axis) or CPV or FPV capsids (x axis) to wild-type or mutant canine or feline TfRs expressed in TRVb cells. Receptors were expressed in TRVb cells by transfection, and the cells were then incubated with Cy5-labeled canine Tf and CPV or FPV capsids for 1 h at 37°C. Capsids were detected by antibody staining, and the cell-associated capsids and Tf were quantified by flow cytometry.
The residues controlling the specific binding of the canine TfR to CPV capsids were mapped by analysis of feline/canine TfR chimeras and also by the preparation of single or double substitutions with sequences of the alternate receptor of the feline or canine TfR sequences (Fig. 2; Tables 1 and 2). Transferring the specific CPV binding properties of the canine TfR to the feline receptor required at least two regions from the apical domain. The double chimera of the feline TfR containing the sequence from residues 191 to 439 from the canine TfR (Fig. 2 and 5) showed that the virus binding specificity mapped within that region, as it bound CPV but not FPV. However, two single chimeras which contained reciprocal portions of the canine TfR apical domain (TfR-feline225canine and TfR-canine225feline) both bound FPV (Fig. 2 and 5). Within the apical domain, several residues were involved in controlling the binding specificity. Various combinations of residues in the canine TfR were changed to the feline sequences (Table 2), and in each case, that receptor gained the ability to bind FPV (Table 2). In addition, changing single or multiple combinations of positions 203, 206, 383, or 385 in the feline TfR to the canine TfR sequence or inserting Asn after residue 204 did not prevent that receptor from binding to FPV (Tables 1 and 2). These results show that the specific virus binding of the canine TfR required the combined effects of more than one sequence.
FIG. 5.
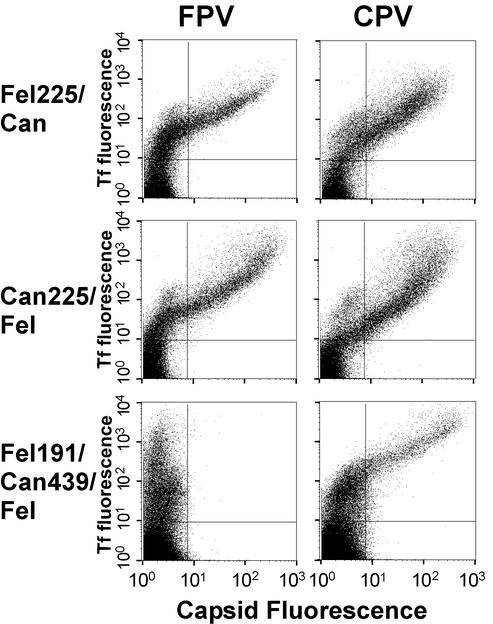
Binding of canine Tf and CPV or FPV capsids to TfRs that were chimeras of the feline and canine receptors (Fig. 2). Feline225canine (Fel225/Can) and canine225feline (Can225/Fel) were reciprocal chimeras recombined around residue 225 in the feline TfR sequence, while feline191canine439feline (Fel191/Can439/Fel) was a double recombinant with residues 192 to 439 of the feline TfR replaced by the equivalent sequence of the canine TfR. Capsid and Tf binding to the wild-type canine and feline TfRs are depicted as shown in Fig. 4. Receptors were expressed in TRVb cells by transfection, and the cells were then incubated with canine Tf and CPV or FPV capsids and examined as described in the legend to Fig. 4.
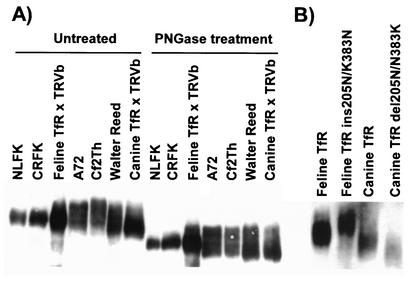
The specific changes in the canine and feline TfRs that were tested included an insertion of Asn at position 205, and addition or removal of a potential glycosylation site involving Asn383 (Tables 1 and 2). To determine whether glycosylation occurred at Asn383, we treated the receptor with PNGase F to remove N-linked oligosaccharides and then determined the sizes of the proteins. Before treatment, all the feline and canine TfRs from cat or dog cells or expressed from plasmids in TRVb cells were glycosylated as expected, and some forms of the canine TfRs were slightly larger than those from the feline cells (Fig. 6). After PNGase F treatment, the sizes of all receptors were reduced, but those from canine cells still showed broad bands, suggesting that they were still glycosylated, perhaps due to the presence of O-linked glycans (Fig. 6A). The mutants that differed at residue 383 showed differences in size when expressed in TRVb cells, with an increase in the mutant form of the feline TfR and a decrease in the mutant form of the canine TfR (Fig. 6B).
FIG. 6.
Expression and N-linked glycosylation of TfRs from feline (NLFK and CRFK) and canine (A72, Cf2Th, and Walter Reed) cells or of wild-type or mutant TfRs expressed from plasmids by transfection of TRVb cells. Proteins were collected by lysis of the cells and then were either not treated or incubated with PNGase before electrophoresis on 10% acrylamide gels and detection of the TfR by Western blotting. (A) The feline or canine TfR recovered from cell lines of feline or canine origin or after expression from cDNA clones in hamster TRVb cells. Samples were either not treated or were treated with PNGase before electrophoresis. (B) The wild-type feline and canine TfRs and mutants containing an insertion (ins) or deletion (del) of Asn at position 205 and with an Asn or Lys at position 383 to alter those positions to the sequence of the alternative host receptor. All plasmids were expressed in TRVb cells.
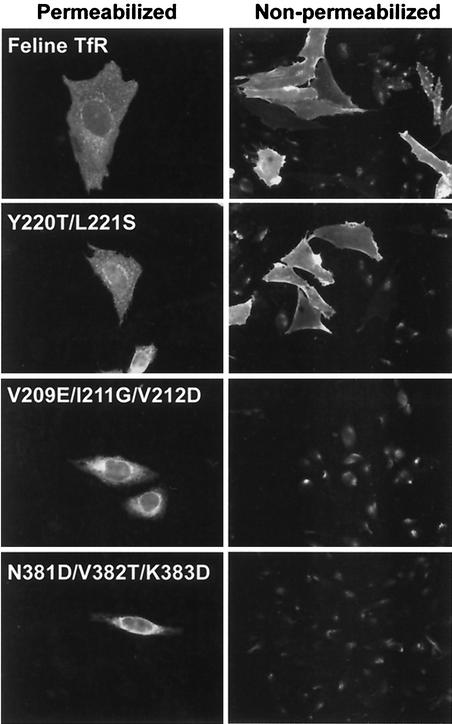
Most feline TfR mutants besides those with mutations at position 221 were not distinguished from the wild type in the binding assays used here (Table 1). Some mutant receptors expressed in TRVb cells did not bind to capsids or Tf, but in those cases, the TfR was not detected at the cell surface when permeabilized and nonpermeabilized cells expressing those receptors were compared by antibody staining (Table 1 and Fig. 7), showing that those mutations prevent transport to the plasma membrane.
FIG. 7.
Microscopic analysis of the feline TfR expressed in TRVb cells, showing the cellular localization of wild-type receptor (Feline TfR) compared to feline TfRs containing mutations which prevented virus binding (Y220T/L221S) or which affected cell surface transport (V209E/I211G/V212D and N381D/V382T/K383D). Cells were fixed with PFA. The cells were then either permeabilized with 0.5% Triton X-100 and stained with an antibody against the cytoplasmic domain of the receptor or were not permeabilized and stained with a rabbit anti-TfR antipeptide antibody that recognizes the ectodomain of the receptor.
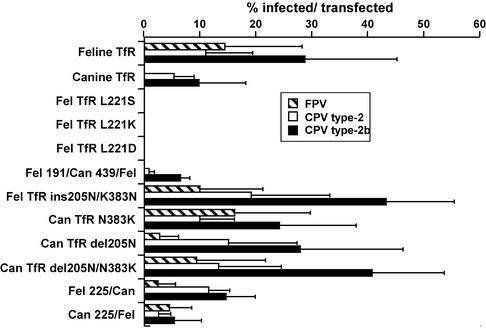
Virus infection of cells expressing the mutant TfRs generally paralleled the capsid binding results, and the canine TfRs that gained the ability to bind FPV also allowed that virus to infect TRVb cells (Fig. 8). An exception was feline TfR-L221K, which bound CPV capsids to TRVb cells but did not mediate infection of those cells. Mutant receptors that did not bind virus did not mediate infection of the TRVb cells (Fig. 8).
FIG. 8.
Virus susceptibility of TRVb cells expressing the wild-type feline or canine TfR or mutant TfRs that were altered within the apical domain. The cells were transfected with the plasmids expressing the receptors, incubated for 2 days at 37°C, and then inoculated with 10 TCID50 per cell of CPV type 2 or 2b or FPV. The cells were fixed 2 days later and stained for the presence of infected cells using an antibody against the NS1 protein. The data are the mean percentages of infected cells among the cells that became transfected in the experiment ± 1 standard deviation (error bars) of the data from at least three separate infection experiments for all the receptors tested. Fel, feline; Can, canine; ins, insertion; del, deletion.
DISCUSSION
Infection of feline cells by CPV and FPV involves binding and endocytosis of the capsid by the feline TfR, and infection of canine cells by CPV requires specific binding to the canine TfR (16, 25). Here we demonstrate that the feline TfR binds FPV and CPV capsids through structures in its apical domain and that the specific CPV interaction with the canine TfR is also controlled by differences of multiple residues in the apical domain.
How does the TfR interact with the capsid? The feline TfR binds to CPV, FPV, and to many different mutants of those capsids, and must therefore make sufficient interactions with conserved structures of the capsid surface so that most changes in the virus do not disrupt that binding (9, 26, 33). The virus binding by the feline TfR apical domain also proved quite tolerant of changes, and many substitutions introduced did not affect its binding to either CPV or FPV capsids (Table 1). However, the feline TfR binding must involve a structure controlled by Leu221, since changing that residue to either Ser or Asp prevented it from binding to FPV capsids and those receptors also showed greatly reduced binding to CPV. An interesting finding was that another substitution at that site (feline TfR-L221K) was CPV specific in binding (Fig. 4), showing that the structure around residue 221 must interact with the capsid in a region that differs between CPV and FPV. That mutant was therefore functionally similar to the canine TfR in capsid binding.
The specific binding of the canine TfR to CPV was also controlled by sequences within or near the apical domain, since the double recombinant (TfR-feline191canine439feline) was CPV specific in binding (Fig. 2 and 5). However, that CPV-specific binding required differences of more than one position in the feline TfR sequence, since the reciprocal TfR chimeras TfR-feline229canine and TfR-canine229feline both bound to FPV (Fig. 2B and 5). This control of specificity was also confirmed by the finding that single changes within the apical domain of the canine TfR gave it the ability to bind to both CPV and FPV and mediate FPV infection of the TRVb cells. Changes tested in the canine TfR included the deletion of Asn after position 204 (feline TfR numbering) and the Asn-to-Lys change of residue 383 (feline TfR numbering) which removed an Asn-linked glycosylation site (Table 2 and Fig. 1). However, although those sites allowed the canine TfR to bind FPV, the reciprocal changes in the feline TfR did not introduce the CPV-specific binding (Table 1). This indicates that the canine TfR has a very specific structure that makes it bind and allow cell infection by CPV but not by FPV.
The canine and feline TfRs differ in sequence by 11%, and both differ by about 22% from the human TfR (16). The differences from the known human TfR structure cannot be modeled precisely, but the proteins likely show similar folded structures. There are many differences in primary amino acid sequence of the apical domains of those receptors which affect surface-exposed residues (Fig. 1), and one of those differences introduces a new N-linked glycosylation site into the canine TfR which is functional when the receptors are expressed in TRVb cells (Fig. 6B). In addition, O-linked glycosylation may occur, particularly when the receptor is expressed in canine cells, since after extensive PNGase F treatment, the TfRs from three different dog cells still showed broad bands after electrophoresis on SDS-polyacrylamide gels, while the native feline TfR from NLFK or CRFK cells or the canine or feline TfRs expressed in TRVb cells formed more tightly resolved bands on SDS-polyacrylamide gels (Fig. 6A). In the human TfR, at least one O-linked glycan is attached, and the oligosaccharide added differs in different human cell lines (10). Analysis of the human, feline, and canine TfRs for potential O-glycosylation sites using the NetOGlyc program version 2.0 (13) showed the following numbers of predicted sites for TfR sequences from different species: one for the human TfR, two for the feline TfR, and six for the canine TfR. These results indicate that additional glycans may also influence virus-specific binding. These various results implicate a combination of primary surface changes in the control of the altered receptor interactions with FPV and CPV capsids, and the various carbohydrates may also influence the binding when the receptors are expressed in cells from different hosts or tissues. Differential glycosylation of the receptors for various other viruses have been shown to influence their ability to bind to virus; examples are poliovirus and the human and mouse poliovirus receptors (CD155), human coronavirus and CD13 (aminopeptidase N), human immunodeficiency virus and CXCR4, and various retroviruses and Na+-dependent neutral amino acid transporter type 2 (8, 12, 21, 38).
The relative affinity or avidity of capsid binding to the different cells or tissues was not determined directly in these studies. In our binding assays, cell-associated virus or Tf was detected after incubation at 37°C using antibody staining and flow cytometry, which would show the combined effects of binding and uptake of those ligands. The level of cell-associated virus gives only an approximate estimate of the affinity of the receptor-capsid association (15a). The binding site for Tf on the human TfR is mostly in the helical domain (39), and we do not observe any competition between the canine Tf and capsids in our assays (K. Hueffer, unpublished). Some of the TfR mutants (e.g., feline TfR-L221K) bound virus only when expressed at very high levels as measured by Tf binding (Fig. 4). Since there are up to 60 potential receptor binding sites on each capsid, those mutant receptors likely have a lower affinity of binding to the capsids so that high densities are required to give sufficient virus-receptor contacts for virus binding and endocytosis. We will be examining the affinities and avidities of the different receptor-virus interactions in future studies using other assays.
In general, the susceptibility of TRVb cells expressing the different receptors was correlated with the levels of receptor binding for FPV and CPV type 2 capsids. The same trend was seen for CPV type 2 and 2b viruses, although in some receptors, CPV type 2b infected at higher levels than the same titer of CPV type 2, as has been seen in other studies of virus infection using the canine TfR in TRVb cells (16). As we have observed previously (15a, 16), binding of the capsids to receptors was not always sufficient for infection. In these studies, the feline TfR-L221K mutant bound CPV capsids when expressed in TRVb cells but did not mediate infection (Fig. 8). This suggests that the TfR acts as more than just a tether for endocytosis but that it may have specific structural effects on the capsid that enhance its ability to infect the cells. If this is correct, these viruses appear to differ in this regard from the adeno-associated viruses, which appear to be able to use a variety of alternative and glycan-based receptors to successfully infect host cells (11, 18, 32). Other explanations could involve the need for a specific binding affinity or control of trafficking into different endosomal pathways after uptake by the altered receptor-capsid interaction.
The sites on the capsids that affect canine TfR binding have been defined in a variety of studies, but mutations affecting feline TfR binding have not yet been determined. Differences in three regions on the top or side of the threefold spike can influence host range through their combined influence on canine TfR binding (9, 15, 26). Two changes between FPV and CPV (VP2 residues Lys93 to Asn and Asp323 to Asn) are required to be introduced together into FPV to allow that virus to bind to the canine TfR and to infect canine cells at only slightly lower levels than those of wild-type CPV (9, 15, 15a). Introducing the changes of VP2 residues Asn93 to Lys and Asn323 to Asp together into CPV gives a virus which was unable to infect canine cells, although it still bound to the canine TfR (15a). A third structural region affecting canine TfR binding is seen around VP2 residues 299 and 300 (16, 26). Residues 93, 299 or 300, and 323 are separated by 20 to 35 Å, and there are no direct structural interactions between them, indicating that those three sites are separately involved in adapting the CPV to bind to the canine TfR. The distances between the various different sites in the capsid and those that we have defined here in the TfR suggest that there is a broad structural complementarity between the receptor and its binding site on the capsid. In future studies, we will examine the interactions between the feline and canine TfRs and the capsids using cryoelectron microscopy, which should positively identify the contacts between the receptor and the capsid, as has been demonstrated for other virus-receptor complexes (29, 30).
Animal viruses are exquisitely adapted to the many interactions they have with their natural hosts, and there appear to be high biological and evolutionary barriers that prevent viruses from adapting to new hosts. This has been clearly shown for CPV, where the process of canine cell adaptation involved a series of structurally distinct capsid mutations that allowed the virus to accommodate several differences in the structure of the canine TfR, including an additional glycan, and allowed the virus to both bind to that receptor and to use it for cell infection. Those mutations were acquired in a step-wise series which allowed the intermediates to remain viable in the feline or canine hosts (28, 35), and after a period of rapid selection during the initial transfer into dogs, the CPV strains have continued to acquire small numbers of changes in the capsid regions that influence receptor binding and host range.
Acknowledgments
Wendy S. Weichert and Gail Sullivan provided expert technical assistance.
This work was supported in part by grants AI28385 and A33468 from the National Institutes of Health to C.R.P.
REFERENCES
- 1.Agbandje, M., R. McKenna, M. G. Rossmann, M. L. Strassheim, and C. R. Parrish. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155-171. [DOI] [PubMed] [Google Scholar]
- 2.Bates, G. W., and M. R. Schlabach. 1973. The reaction of ferric salts with transferrin. J. Biol. Chem. 248:3228-3232. [PubMed] [Google Scholar]
- 3.Bates, G. W., and J. Wernicke. 1971. The kinetics and mechanism of iron(3) exchange between chelates and transferrin. IV. The reaction of transferrin with iron(3) nitrilotriacetate. J. Biol. Chem. 246:3679-3685. [PubMed] [Google Scholar]
- 4.Benmerah, A., C. Lamaze, B. Begue, S. L. Schmid, A. Dautry-Varsat, and N. Cerf-Bensussan. 1998. AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J. Cell Biol. 140:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, M. J., J. A. Lebron, and P. J. Bjorkman. 2000. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature 403:46-53. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky, F. M., C. Y. Chen, C. Knuehl, M. C. Towler, and D. E. Wakeham. 2001. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17:517-568. [DOI] [PubMed] [Google Scholar]
- 7.Buchegger, F., I. S. Trowbridge, L. F. Liu, S. White, and J. F. Collawn. 1996. Functional analysis of human/chicken transferrin receptor chimeras indicates that the carboxy-terminal region is important for ligand binding. Eur. J. Biochem. 235:9-17. [DOI] [PubMed] [Google Scholar]
- 8.Chabot, D. J., H. Chen, D. S. Dimitrov, and C. C. Broder. 2000. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J. Virol. 74:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do, S. I., C. Enns, and R. D. Cummings. 1990. Human transferrin receptor contains O-linked oligosaccharides. J. Biol. Chem. 265:114-125. [PubMed] [Google Scholar]
- 11.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052-1056. [DOI] [PubMed] [Google Scholar]
- 12.Gromeier, M., H. H. Lu, G. Bernhardt, J. J. Harber, J. A. Bibb, and E. Wimmer. 1995. The human poliovirus receptor. Receptor-virus interaction and parameters of disease specificity. Ann. N. Y. Acad. Sci. 753:19-36. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, J. E., O. Lund, N. Tolstrup, A. A. Gooley, K. L. Williams, and S. Brunak. 1998. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15:115-130. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, G. R., A. Williams, C. E. Costello, C. A. Enns, and J. J. Lucas. 1995. The critical glycosylation site of human transferrin receptor contains a high-mannose oligosaccharide. Glycobiology 5:227-232. [DOI] [PubMed] [Google Scholar]
- 15.Horiuchi, M., H. Goto, N. Ishiguro, and M. Shinagawa. 1994. Mapping of determinants of the host range for canine cells in the genome of canine parvovirus using canine parvovirus/mink enteritis virus chimeric viruses. J. Gen. Virol. 75:1319-1328. [DOI] [PubMed] [Google Scholar]
- 15a.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 16.Hueffer, K., J. S. Parker, W. S. Weichert, R. E. Geisel, J. Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt, R. C., R. Riegler, and A. A. Davis. 1989. Changes in glycosylation alter the affinity of the human transferrin receptor for its ligand. J. Biol. Chem. 264:9643-9648. [PubMed] [Google Scholar]
- 18.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence, C. M., S. Ray, M. Babyonyshev, R. Galluser, D. W. Borhani, and S. C. Harrison. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science 286:779-782. [DOI] [PubMed] [Google Scholar]
- 20.Llamas-Saiz, A. L., M. Agbandje-McKenna, J. S. L. Parker, A. T. M. Wahid, C. R. Parrish, and M. G. Rossmann. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65-71. [DOI] [PubMed] [Google Scholar]
- 21.Marin, M., D. Lavillette, S. M. Kelly, and D. Kabat. 2003. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J. Virol. 77:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGraw, T. E., L. Greenfield, and F. R. Maxfield. 1987. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J. Cell Biol. 105:207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellman, I. 1996. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12:575-625. [DOI] [PubMed] [Google Scholar]
- 24.Parker, J. S., and C. R. Parrish. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74:1919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker, J. S. L., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker, J. S. L., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish, C. R. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195-205. [DOI] [PubMed] [Google Scholar]
- 28.Parrish, C. R., C. Aquadro, M. L. Strassheim, J. F. Evermann, J.-Y. Sgro, and H. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossmann, M. G., J. Bella, P. R. Kolatkar, Y. He, E. Wimmer, R. J. Kuhn, and T. S. Baker. 2000. Cell recognition and entry by rhino- and enteroviruses. Virology 269:239-247. [DOI] [PubMed] [Google Scholar]
- 30.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with x-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 31.Sheff, D. R., E. A. Daro, M. Hull, and I. Mellman. 1999. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J. Cell Biol. 145:123-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, W., G. S. Arnold, and J. S. Bartlett. 2001. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum. Gene Ther. 12:1697-1711. [DOI] [PubMed] [Google Scholar]
- 33.Simpson, A. A., V. Chandrasekar, B. Hebert, G. M. Sullivan, M. G. Rossmann, and C. R. Parrish. 2000. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300:597-610. [DOI] [PubMed] [Google Scholar]
- 34.Takei, K., and V. Haucke. 2001. Clathrin-mediated endocytosis: membrane factors pull the trigger. Trends Cell Biol. 11:385-391. [DOI] [PubMed] [Google Scholar]
- 35.Truyen, U., A. Gruenberg, S. F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vihinen-Ranta, M., W. Yuan, and C. R. Parrish. 2000. Cytoplasmic trafficking of the canine parvovirus capsid and its role in infection and nuclear transport. J. Virol. 74:4853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wentworth, D. E., and K. V. Holmes. 2001. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 75:9741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, A. P., Jr., A. M. Giannetti, A. B. Herr, M. J. Bennett, J. S. Nangiana, J. R. Pierce, L. P. Weiner, P. M. Snow, and P. J. Bjorkman. 2001. Mutational analysis of the transferrin receptor reveals overlapping HFE and transferrin binding sites. J. Mol. Biol. 313:385-397. [DOI] [PubMed] [Google Scholar]
- 40.White, S., K. Miller, C. Hopkins, and I. S. Trowbridge. 1992. Monoclonal antibodies against defined epitopes of the human transferrin receptor cytoplasmic tail. Biochim. Biophys. Acta 1136:28-34. [DOI] [PubMed] [Google Scholar]
- 41.Yang, B., M. H. Hoe, P. Black, and R. C. Hunt. 1993. Role of oligosaccharides in the processing and function of human transferrin receptors. Effect of the loss of the three N-glycosyl oligosaccharides individually or together. J. Biol. Chem. 268:7435-7441. [PubMed] [Google Scholar]
- 42.Yeung, D. E., G. W. Brown, P. Tam, R. H. Russnak, G. Wilson, I. Clark-Lewis, and C. R. Astell. 1991. Monoclonal antibodies to major nonstructural nuclear protein of minute virus of mice. Virology 181:35-45. [DOI] [PubMed] [Google Scholar]