Abstract
During the early stages of herpes simplex virus type 1 (HSV-1) infection, viral immediate-early regulatory protein ICP0 localizes to and disrupts cellular nuclear structures known as PML nuclear bodies or ND10. These activities correlate with the functions of ICP0 in stimulating lytic infection and reactivating quiescent HSV-1. The disruption of ND10 occurs because ICP0 induces the loss of the SUMO-1-modified forms of PML and the subsequent proteasome-mediated degradation of the PML protein. The functions of ICP0 are largely dependent on the integrity of its zinc-binding RING finger domain. Many RING finger proteins have been found to act as ubiquitin E3 ligase enzymes, stimulating the production of conjugated polyubiquitin chains in the presence of ubiquitin, the ubiquitin-activating enzyme E1, and the appropriate E2 ubiquitin-conjugating enzyme. Substrate proteins that become polyubiquitinated are then subject to degradation by proteasomes. We have previously shown that purified full-length ICP0 acts as an efficient E3 ligase in vitro, producing high-molecular-weight polyubiquitin chains in a RING finger-dependent but substrate-independent manner. In this paper we report on investigations into the factors governing the degradation of PML induced by ICP0 in a variety of in vivo and in vitro assays. We found that ICP0 expression increases the levels of ubiquitinated PML in transfected cells. However, ICP0 does not interact with or directly ubiquitinate either unmodified PML or SUMO-1-modified PML in vitro, suggesting either that additional factors are required for the ICP0-mediated ubiquitination of PML in vivo or that PML degradation is an indirect consequence of some other activity of ICP0 at ND10. Using a transfection-based approach and a family of deletion and point mutations of PML, we found that efficient ICP0-induced PML degradation requires sequences within the C-terminal part of PML and lysine residue 160, one of the principal targets for SUMO-1 modification of the protein.
Herpes simplex virus type 1 (HSV-1) immediate-early protein ICP0 is an important regulator of both lytic and latent HSV-1 infection (for reviews, see references 7 and 23). ICP0 has been found to interact with a number of cellular proteins and to have profound effects on cells, both during infection and in cells expressing ICP0 from a transfected plasmid. One of the most obvious and widely studied effects of ICP0 is its ability to localize to and disrupt specific nuclear structures known as PML (promyelocytic leukemia) nuclear bodies or ND10 (9, 19). Since mutations in ICP0 that eliminate or reduce its effects on ND10 cause concomitant defects in the ability of ICP0 to stimulate lytic infection and to induce reactivation of quiescent viral genomes, the mechanism by which ICP0 brings about the disruption of ND10 and related effects is of obvious importance.
In recent years it has become clear that ICP0 functions via the ubiquitin proteasome pathway (12). ICP0 induces the proteasome-dependent degradation of the PML protein, a major and essential component of ND10, and also of modified isoforms of Sp100, another ND10 component (4, 8, 20, 21). Both PML and Sp100 are efficient substrates for modification by conjugation to the ubiquitin-like protein SUMO-1, and it is the SUMO-1-modified forms of these proteins that are most sensitive to ICP0-induced degradation (8, 22). These effects require the RING finger domain of ICP0, a zinc-stabilized structure near the N-terminal end of the protein that is related to sequence motifs present in a wide variety of cellular and viral proteins. Many RING finger proteins have been found to have E3 ubiquitin ligase activity, performing the final step of the ubiquitin conjugation reaction in which activated ubiquitin molecules, covalently bound by a thiolester linkage to a member of the E2 family of ubiquitin-conjugating enzymes, are serially linked to form a polyubiquitin chain anchored to a lysine residue on a substrate that is targeted for degradation (reviewed in reference 14).
Consistent with these observations, full-length ICP0 induces the formation of colocalizing conjugated ubiquitin at ND10 in infected cells, and the purified full-length protein has E3 ubiquitin ligase activity in vitro; both of these activities are RING finger dependent (3, 6). The in vitro assay used in the previous studies monitors the formation of high-molecular-weight polyubiquitin chains in assay mixtures containing full-length ICP0, ubiquitin, the E1 ubiquitin-activating enzyme, and the appropriate E2 ubiquitin-conjugating enzyme (UbcH5a or UbcH6) (3). These assays are substrate independent, and the results left open the question whether ICP0 could ubiquitinate PML directly. Since the most obvious initial effect of ICP0 expression is the loss of the SUMO-1-modified forms of PML, it has also been questioned whether ICP0 has any direct effect on the formation or stability of the PML-SUMO-1 conjugates. Finally, there has been no previous investigation into the characteristics of PML itself that contribute to degradation by ICP0.
To address these questions, we have used in vitro assays to test whether ICP0 can interact with PML or affect the formation or stability of its SUMO-1-modified forms and whether the minimal components of the ICP0 E3 ligase mixture can ubiquitinate PML directly. Although we found that ICP0 increases the accumulation of ubiquitinated forms of PML in transfected cells, biochemical assays using purified components indicated that ICP0 does not interact directly with or ubiquitinate PML in vitro. Similar in vitro assays demonstrated that ICP0 by itself does not affect the formation or stability of SUMO-1-modified PML. These data suggest that the degradation of PML induced by ICP0 in vivo is either indirect or requires other, so-far-unidentified factors in addition to the basic components of the ubiquitination pathway. We have also constructed a series of truncation and point mutations of PML to test the characteristics of the protein that contribute to the efficiency of its degradation induced by ICP0 in transfected cells. We found that PML mutants that do not localize to ND10, that have lost C-terminal sequences, or that lack the SUMO-1 modification site lysine residue 160 are degraded less efficiently in vivo by ICP0 expressed from a cotransfected plasmid.
MATERIALS AND METHODS
Plasmids.
Plasmid pPML(F) expresses PML isoform IV (13, 16). Plasmid pSV5-PML560 expresses PML isoform VI with an N-terminal SV5 tag, and pSV5-PML560(SUMO-) is a derivative that contains lysine-to-arginine mutations at PML codons 160 and 490, the two major SUMO-1 modification sites on PML. These two plasmids were kind gifts from Dan Bailey and Peter O'Hare (Marie Curie Research Institute, Oxted, Surrey, United Kingdom). Plasmid pSV5-PML560-K160R includes the K160R mutation but not that at lysine residue 490. Plasmid pPML(F)2KR is a derivative of pPML(F) containing both the K160R and K490R mutations. Plasmid pCImycPML-fl contains the complete PML isoform IV-coding region with an N-terminal Myc tag inserted into vector pCIneo (Promega). Derivatives pCImycPML-NE, -NK, -NKnls, -NX, -NXnls, -NS2, and -NM contain 3′ truncations of the PML isoform IV open reading frame, ending at the EagI, KpnI, XmaI, SacII, and MluI restriction sites, thus expressing PML proteins consisting of residues 1 to 135, 1 to 304, 1 to 447, 1 to 502, and 1 to 556, respectively (see Fig. 5). The nls versions contain additional sequences encoding the simian virus 40 large-T-antigen nuclear localization signal. Plasmid pCImycPML-ΔEag contains an in-frame deletion removing PML codons 136 to 263. Plasmids pCI-110 and pCI-FXE express full-length ICP0 and RING finger deletion mutant ICP0, respectively (10). Plasmid pCW7 expresses ubiquitin with N-terminal polyhistidine and Myc tags, and pCW8 is a derivative containing an arginine substitution at lysine residue K48 of ubiquitin, causing the termination of K48-linked polyubiquitin chains (27).
FIG. 5.
Map of PML isoform IV (633 amino acid residues [aa]), showing the RING finger (R), the two B boxes (B1 and B2), the coiled-coil region (CC), the nuclear localization signal (nls), and the two major characterized sites of SUMO-1 modification, lysine 160 and lysine 490. At the top are shown the positions of restriction sites in the cDNA that were used for the generation of the truncated derivatives depicted below. The addition of nls to the name of a truncated plasmid indicates that the simian virus 40 large-T-antigen nuclear localization signal had been added to the C-terminal end of the open reading frame.
In vivo ubiquitination of PML.
HEp-2 cells were propagated in Dulbecco's modified Eagle's medium containing 100 U of penicillin per ml and 100 μg of streptomycin per ml and supplemented with 10% fetal calf serum. Cells were seeded into 35-mm-diameter dishes at a density of 1.5 × 105 cells/dish and transfected by using Lipofectamine Plus reagent (GibcoBRL) according to the manufacturer's instructions. Cells were cotransfected with 10 ng of pPML(F), 50 ng of pCW7, and 100 ng of either pCI-110 or pCI-FXE. Promoter competition was balanced by including the appropriate amounts of pCI-neo vector, and pUC9 was used to bring the total DNA amount to 500 ng. The cells were treated with MG132 (final concentration, 10 μM) 16 h after transfection and incubated for an additional 8 h. Alternatively, instead of pCW7, plasmid pCW8 was used to express K48R mutant ubiquitin, which terminates the extension of polyubiquitin chains and thus stabilizes the ubiquitinated forms of PML in the absence of MG132. The two methods gave equivalent results. Cell monolayers were subsequently washed twice in ice-cold phosphate-buffered saline (PBS) before being harvested either in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) boiling mix buffer containing 3 M urea and 25 mM dithiothreitol or in 1 ml of guanidinium chloride (GdnCl) buffer A (PBS with 6 M GdnCl, 0.1% NP-40, and 5% glycerol [pH 8.0]). Polyhistidine-tagged ubiquitinated proteins were isolated by nickel affinity chromatography with 35 μl of equilibrated Ni-nitrilotriacetic acid beads (Qiagen) per sample. Samples were incubated end-over-end at 4°C for 4 h, and then the beads were washed three times with 1 ml of GdnCl buffer A, three times with 1 ml of a 1:2 mixture of GdnCl buffer A and buffer B (PBS, 0.1% NP-40, 5% glycerol, 20 mM imidazole), three times with 1 ml of a 1:3 mixture of GdnCl buffer A and buffer B, and finally three times with 1 ml of buffer B. This procedure removes weakly bound proteins and sequentially dilutes the GdnCl so that the samples can be applied to a gel. The beads were resuspended in 30 μl of 1× SDS-PAGE boiling mix buffer and boiled for 5 min before analysis. Samples were resolved by SDS-10% PAGE and transferred to nitrocellulose membranes, and PML was detected with an antibody that recognizes the F tag (16).
In vitro ubiquitination assays.
Radiolabeled PML was produced by in vitro transcription-translation with pPML(F) as a template and the TnT rabbit reticulocyte lysate system (Promega) in the presence of [35S]methionine, according to the manufacturer's instructions. In vitro ubiquitination assays were performed with 0.5 μl of [35S]methionine-labeled substrate in 50 mM Tris-HCl (pH 7.5)-50 mM NaCl-5 mM ATP containing an ATP-regenerating system (10 mM creatine phosphate, 3.5 U of creatine kinase per ml, 0.6 U of inorganic pyrophosphatase per ml), 20 ng of E1, 50 ng of E2, 5 μg of ubiquitin, and 50 ng of purified full-length ICP0. Reactions were carried out in a final volume of 10 μl for 3 h at 28°C and were terminated by the addition of 5 μl of 3× SDS-PAGE boiling mix buffer containing 9 M urea and 100 mM dithiothreitol. Samples were resolved either by SDS-7.5% PAGE or with 4 to 12% NuPAGE gels (Invitrogen). Gels were either Western blotted and probed with an antiubiquitin monoclonal antibody (either FK2 from Affiniti or P4D1 from Santa Cruz Biotechnology) or stained with Coomassie brilliant blue, destained, dried, and analyzed with a Bio-Rad phosphorimager. The ubiquitin-activating enzyme E1, the E2 ubiquitin-conjugating enzymes, and full-length ICP0 were expressed and purified as described previously (3).
In vitro SUMO-1 modification assays.
Recombinant SAE1/SAE2, SUMO-1GG, and Ubc9 were expressed and purified from bacteria as previously described (25). Radiolabeled PML was produced in vitro by TnT wheat germ transcription-translation (Promega) (to avoid the presence of trace amounts of mammalian SUMO-1 isopeptidases) with pPML(F) as a template in the presence of [35S]methionine, according to the manufacturer's guidelines. Aliquots (0.5 μl) of radiolabeled PML were incubated in the presence or absence (as indicated in the appropriate figure) of 10 μg of SUMO-1GG, 10 μg Ubc9, and 1 μg of SAE1/SAE2, in the presence of an ATP-regenerating system (as described above), in a final volume of 10 μl for 3 h (unless otherwise indicated in the figure) at 37°C.
Western blotting.
Proteins in cell extracts or other samples were separated by electrophoresis on SDS-7.5% or 10% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The filters were blocked overnight in PBS containing 0.1% Tween 20 and 5% dry milk and then incubated with primary antibodies for 2 h in the same buffer. After extensive washing, the filters were incubated with horseradish peroxidase-conjugated secondary antibody in PBS-0.1% Tween 20-2% dry milk for 1 h and then washed again extensively before detection of bound antibody by the enhanced chemiluminescence method (NEN) and exposure to film.
Transfections and immunofluorescence.
HEp-2 cells on glass coverslips were transfected with PML expression plasmids by using Lipofectamine Plus reagent. On the following day, the cells were washed with PBS, fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose), and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The coverslips were incubated with for 1 h at room temperature with monoclonal antibody 9E10 (diluted in PBS containing 1% newborn calf serum), which recognizes the Myc tag on the expressed protein, and then washed several times before treatment with fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma) in the same manner. The coverslips were mounted with Citifluor AF1 and examined with a Zeiss LSM 510 confocal microscope at an excitation wavelength of 488 nm. Data were collected with fourfold averaging at a resolution of 1,024 by 1,024 pixels. The image files were prepared for printing with Adobe Photoshop.
PML cotransfection degradation assays.
The effect of ICP0 on epitope-tagged PML expressed from a cotransfected plasmid in HEp-2 cells, using a total of 1 μg of DNA for 5 × 104 cells in 35-mm-diameter dishes, was analyzed as described previously (21). Given the competing effects of ICP0 transactivation of promoters on cotransfected plasmids and ICP0-induced degradation of PML, these assays are sensitive to the relative amounts of all of the plasmids in the transfection mix. Care was taken to equalize the total input of cytomegalovirus promoter equivalents in the mixtures by adding appropriate amounts of pCIneo vector and to equalize total DNA concentrations by using pUC9. Amounts of the PML expression plasmids were kept to a minimum (10 ng) to avoid saturating the degradation ability of ICP0. Amounts of the ICP0 expression plasmid were also kept to a minimum (50 ng) to avoid the possibility of promoter competition effects.
RESULTS
ICP0 increases the amount of ubiquitinated PML in transfected cells.
Previous studies have shown that HSV-1 infection leads to the rapid loss of the presumed SUMO-1-modified isoforms of endogenous PML and to a subsequent reduction in total PML levels in an ICP0-dependent manner (8, 21). Addition of the proteasome inhibitor MG132 demonstrated that these processes were proteasome dependent, and cotransfection experiments indicated that, in the absence of other viral proteins, ICP0 reduced the half-life of exogenous PML and its modified isoforms (8, 21). Despite the ample evidence that PML was being degraded by the proteasome, proof that ICP0 caused ubiquitination of PML was lacking. This question has assumed a greater importance since it was demonstrated that ICP0 possesses ubiquitin E3 ligase activity in vitro (3, 26). These experiments were initiated to explore whether the ubiquitin E3 ligase activity of ICP0 could target PML in vivo or in vitro.
Substrates that are ubiquitinated in response to a relevant E3 ligase activity should be stabilized and therefore accumulate in the presence of the proteasome inhibitor MG132. The previous failure to detect such forms on PML in vivo in cells expressing ICP0 could be due to several factors, such as low abundance, limiting ubiquitin pools, difficulty in resolving the high-molecular-weight branched species, and band patterns complicated by the stabilized SUMO-1-modified forms of PML. An alternative approach is to cotransfect plasmids expressing the proteins of interest with a plasmid expressing ubiquitin with an N-terminal polyhistidine tag and the K48R mutation, which acts as a chain terminator in polyubiquitin chain growth, thereby stabilizing the ubiquitinated protein. Total ubiquitinated cell proteins can be purified and concentrated by nickel affinity column chromatography under denaturing conditions, thus both stabilizing and allowing the detection of low levels of ubiquitinated proteins. Application of these methods to HEp-2 cells cotransfected with plasmids expressing epitope-tagged PML and His(K48R)-ubiquitin revealed low levels of ubiquitinated PML, indicating a degree of turnover of PML through the proteasome pathway by endogenous enzymes. Inclusion of a plasmid expressing ICP0 in the transfection mixture resulted in a clear increase in the levels of the ubiquitinated PML species (Fig. 1), but this increase did not occur in cotransfections with the ICP0 RING finger deletion mutant FXE (data not shown). These data demonstrate that ICP0 does indeed induce the formation of ubiquitinated PML species in vivo and that this process requires the ICP0 RING finger E3 ligase domain.
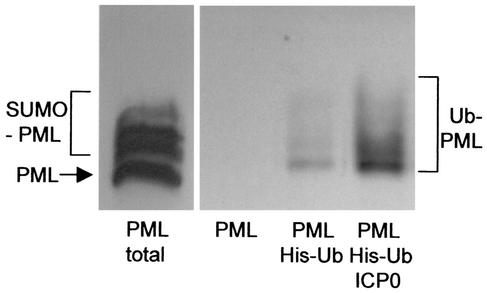
FIG. 1.
ICP0 induces the formation of ubiquitinated PML in transfected cells. HEp-2 cells were transfected with combinations of plasmids expressing epitope-tagged PML, polyhistidine-tagged ubiquitin with the lysine 48-to-arginine (K48R) mutation (His-Ub), and ICP0 (as indicated below the panels). The left panel shows a sample of a whole-cell extract of cells transfected with the PML expression plasmid alone (with the unmodified and SUMO-1-modified forms of PML indicated). The right panel shows a Western blot of ubiquitinated total cell proteins that had been purified by metal chelate affinity chromatography as described in Materials and Methods and probed for PML. PML was not detected in the left lane of this panel, which is a control with proteins purified from cells transfected with the PML plasmid in the absence of the tagged K48R ubiquitin plasmid. Inclusion of this plasmid results in the isolation of ubiquitin-modified PML bands (center lane). The quantity of these bands was greatly increased in the presence of coexpressed ICP0 (right lane).
ICP0 does not induce the ubiquitination of unmodified PML in vitro.
Having established that PML can indeed be ubiquitinated in response to ICP0 in vivo, we tested whether ICP0 could ubiquitinate PML directly in vitro with purified components. The E1 ubiquitin-activating enzyme and full-length ICP0 were expressed and purified as described previously (3), and the E2 conjugating enzymes UbcH5a, UbcH6, UbcH10, and Cdc34 were expressed in bacteria and purified by virtue of their polyhistidine tags. UbcH7 was similarly expressed and then purified by ion-exchange chromatography. The E1 enzyme and all of the E2 enzymes were shown to be catalytically active by formation of a reducing agent-sensitive thiolester linkage with ubiquitin in the presence of ATP (data not shown). UbcH5a and UbcH6 were chosen for these experiments since we had previously shown that ICP0 stimulates the formation of polyubiquitin chains in the presence of these two E2 enzymes (3), and Cdc34 was chosen because a glutathione S-transferase fusion protein containing a segment of ICP0 near its C terminus has been reported to stimulate autoubiquitination of Cdc34 in vitro (26). As previously reported, full-length ICP0 acted as a highly active E3 ubiquitin ligase in a substrate-independent manner with both UbcH5a and UbcH6 but not with the other E2 enzymes (Fig. 2A).
FIG. 2.
ICP0 does not ubiquitinate unmodified PML in vitro. (A) The ubiquitin-activating enzyme E1, ICP0, and various E2 ubiquitin-conjugating enzymes were expressed, purified, and mixed in a buffer containing ATP and ubiquitin (Ub). The products of these substrate-independent reactions were analyzed by Western blotting, with probing for the formation of high-molecular-weight polyubiquitin chains by using monoclonal antibody FK2. Such chains were readily produced in the presence of UbcH5a and UbcH6 but not with the other E2 enzymes tested. (B) PML protein was produced by coupled in vitro transcription-translation and added to reaction mixtures containing ICP0, UbcH5a, and the other basic components of the ubiquitin conjugation machinery. There was no evidence of the formation of ubiquitinated PML species with lower gel mobility.
To test whether this ubiquitination activity could be directed towards PML, isoform IV of PML (which is known to be subject to ICP0-induced degradation in transfected cells [21]) was expressed by coupled in vitro transcription-translation and then mixed with the components of the ICP0-activated in vitro ubiquitination assay. Neither UbcH5a (Fig. 2B) nor UbcH6 (data not shown) had any detectable affect on PML in the presence of ICP0. To test whether an E2 conjugation enzyme other than UbcH5a or UbcH6 was required for PML ubiquitination, the experiment was repeated with a commercial fraction from a rabbit reticulocyte extract that contains a mixture of ubiquitin pathway enzymes. As expected, ICP0 stimulated the basal ubiquitination activity of this extract (Fig. 3, lower panel), but the activity could not be directed onto PML (Fig. 3, upper panel).
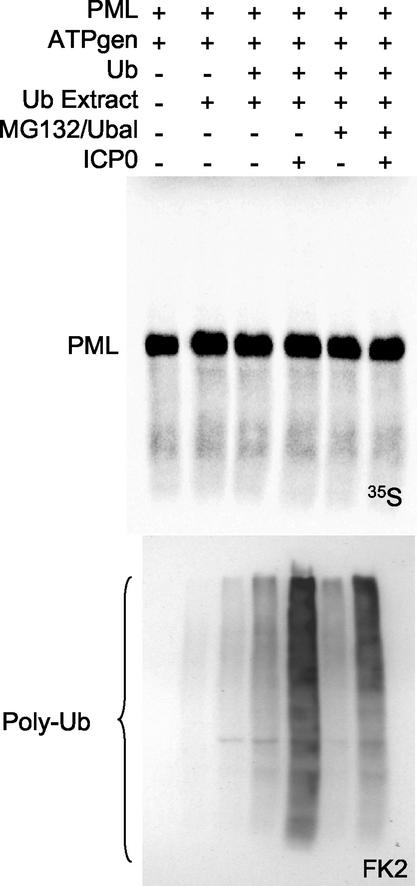
FIG. 3.
ICP0 does not ubiquitinate unmodified PML in the presence of a mixture of endogenous cellular ubiquitination enzymes. PML was produced by coupled in vitro transcription-translation and mixed with ICP0, an ATP generating system (ATPgen), the proteasome inhibitor MG132 and ubiquitin aldehyde (to inhibit proteasomes and ubiquitin-specific proteases, respectively), and an extract containing a mixture of endogenous ubiquitin conjugating enzymes (Ub Extract). No ubiquitination of PML was observed (upper panel) despite ICP0 inducing increased amounts of conjugated ubiquitin (lower panels).
ICP0 does not affect the in vitro SUMO-1 modification of PML or ubiquitinate SUMO-1-modified PML in vitro.
The failure of ICP0 to ubiquitinate PML in our in vitro assays could be due to a variety of reasons. As described above, the available data indicate that the SUMO-1-modified forms of PML are particularly susceptible to degradation in both infection and cotransfection assays. To test whether ICP0 affects the process of SUMO-1 modification of PML or the intrinsic stability of the SUMO-1-modified forms, the heterodimeric Sae1/Sae2 SUMO-1 activating enzyme, the SUMO-1-conjugating enzyme Ubc9, and SUMO-1 were expressed in bacteria and purified. A thiolester assay showed that the purified proteins were active in vitro (data not shown) and that they could modify in vitro-translated PML to give multiple modified bands similar to those seen in vivo (Fig. 4A). Addition of purified ICP0 alone either during or after the SUMO-1 modification reaction had no effect on the efficiency of formation or the stability of SUMO-1-modified PML, showing that ICP0 by itself does not target these processes (Fig. 4B). Further addition of the components of the in vitro ubiquitination machinery to SUMO-1-modified PML did not result in ubiquitination of any of the forms of PML (Fig. 4C, left), despite ICP0 producing polyubiquitin chains in this reaction (Fig. 4C, right). Similar results were obtained with UbcH5a as the E2 in the reaction mixture (data not shown).
FIG. 4.
ICP0 does not hinder the conjugation of SUMO-1 to PML, affect the stability of SUMO-1-modified PML, or ubiquitinate SUMO-1-modified PML in vitro. (A) In vitro-expressed PML was mixed with purified components of the SUMO-1 modification pathway, i.e., Sae1/Sae2, Ubc9, and SUMO-1. SUMO-1-modified PML was readily observed, as indicated by the bracketed bands above the labeled major unmodified PML band. The modified and unmodified forms of PML are similarly marked in panels B and C. (B) SUMO-1-modified PML was produced in the absence (lane 3) or presence (lanes 4 to 6) of ICP0. The reaction mixtures in lanes 1 to 6 were incubated for 180 min. In lanes 7 to 10, similar reaction mixtures were incubated for 180 min in the absence of ICP0 and then ICP0 was added to the reaction mixtures in lanes 8 to 10 and the incubations were continued for a further 180 min. ICP0 affected neither the efficiency of formation nor the stability of SUMO-1-modified PML. (C) ICP0 does not ubiquitinate SUMO-1-modified PML in vitro. PML was expressed in vitro and then mixed with the components of the SUMO-1 modification pathway, ICP0, UbcH6, and the other components of the ubiquitination pathway, as indicated. There was no evidence for the formation of additional ubiquitinated PML species (left panel), despite ICP0 producing polyubiquitin chains in these reactions, as detected by probing of parallel reactions for conjugated ubiquitin by using monoclonal antibody FK2 (right panel). Mock TnT in the right panel refers to the use of an amount of the TnT wheat germ transcription-translation mixture equivalent to that in the lanes in the left panel in the absence of added substrate plasmid and labeled methionine.
From these and other similar experiments, it appears that ICP0 does not act as a substrate-specific E3 ubiquitin ligase on either unmodified or SUMO-1-modified PML in vitro. While some RING finger E3 ligases can interact directly with and ubiquitinate their substrates in vitro, others require additional factors (14). This may be the case for the ability of ICP0 to induce degradation of PML; it is worth noting that PML could not be coimmunoprecipitated with ICP0 from our in vitro mixtures (data not shown). In analogous experiments we also found that ICP0 neither interacted directly with nor induced the ubiquitination of two other ND10 components, Sp100 and hDaxx (data not shown). Therefore, we decided to investigate the characteristics of PML that contribute to ICP0-induced degradation in vivo.
Mutants of PML that are cytoplasmic or not recruited into ND10 are not efficiently degraded by ICP0.
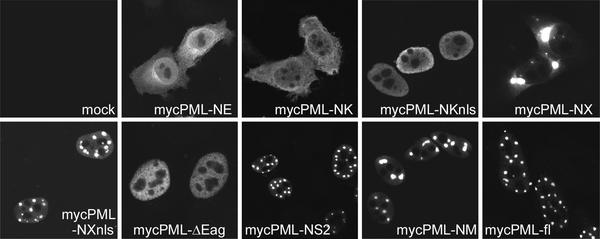
A number of truncation mutants of PML were constructed (Fig. 5), and their cellular localizations were investigated by immunofluorescence (Fig. 6). Only those mutants that showed a normal distribution in typical ND10 foci colocalized with ICP0, and the aberrantly localized PML mutant proteins did not affect the typical distribution of ICP0 in punctate foci (data not shown). These mutant PML proteins were used in transfection assays to test their sensitivity to degradation induced by ICP0. This assay involves cotransfection of plasmids expressing PML and ICP0, using an amount of the PML plasmid (10 ng per 35-mm-diameter plate) that gives detectable yet not excessive expression of exogenous PML. The plasmid-expressed PML resolves into a number of bands on a Western blot, of which the major band is taken to be the unmodified protein, while the ladder of more slowly migrating species has been presumed in a number of previous publications to represent exogenous PML modified by endogenous SUMO-1 (5, 15, 20, 21, 24). The precise pattern of these modified PML isoforms varies between different PML expression cassettes for reasons that are not clear.
FIG. 6.
Typical immunofluorescent localizations in transfected cells expressing the PML proteins depicted in Fig. 5.
Inclusion of an ICP0 expression plasmid, in a mixture carefully controlled to avoid promoter competition effects, resulted in greatly reduced recovery of both the presumed SUMO-1-modified and the unmodified isoforms of PML expressed by plasmid pCImycPML-fl (Fig. 7A). The reduced levels of PML reflect proteasome-mediated degradation induced by ICP0, since subsequent addition of the proteasome inhibitor MG132 in similar cotransfection experiments increased the levels of all forms of PML (21). The C-terminal truncation mutants PML-NM and PML-NS2 retain the SUMO-1 modification sites and present a pattern of modified bands in transfected cells that is consistent with SUMO-1 modification to a degree similar to that of the wild-type protein (Fig. 7A). Although these modified bands were sensitive to coexpressed ICP0, the major unmodified band was degraded to a lesser degree than that of the full-length protein (Fig. 7A). Similar data were obtained with mutant PML-NXnls, which has a larger deletion of C-terminal residues but retains the modification site at K160; the modified forms were sensitive to the effects of ICP0, but the major unmodified form was less sensitive than that of the full-length protein (data not shown). These data confirm that the SUMO-1-modified forms of PML are preferentially sensitive to the effects of ICP0 and indicate that the C-terminal sequences of PML isoform IV contribute to the efficiency of ICP0-induced degradation.
FIG. 7.
Degradation of SUMO-1-modified and unmodified forms of PML by cotransfection expression of ICP0. The extracts were prepared 20 h after transfection. (A) Top panel, Western blot detection of Myc-tagged PML and deletion mutant variants with monoclonal antibody 9E10. The left lane shows a control (cont) whole-cell extract from cells transfected with vector plasmids. The other lanes show pairs of extracts from cells transfected with the indicated PML plasmid (see Fig. 5 for their structures) either without or with an ICP0 expression plasmid (on the left and right of each pair, respectively). The arrows indicate the positions of the unmodified forms of full-length PML and the NM and NS2 deletions. The modified forms of these proteins are of lesser intensity and migrate more slowly. The SUMO-1-modified forms of full-length PML are marked by the bracket. The modified forms of the truncated proteins have correspondingly increased mobilities. Although the modified PML isoforms expressed by each construct are subject to ICP0-induced degradation, the unmodified forms of the deletion mutants are relatively stable compared to the full-length protein. Middle panel, a lower exposure of the same blot, showing that the unmodified forms of the truncation mutants PML-NM and PML-NS2 are more resistant to ICP0-induced degradation that the parent full-length protein. Bottom panel, reprobing of the blot filter for ICP0, demonstrating equivalent expression levels in the three experiments. (B) PML truncation mutant PML-NK is stable in the presence of ICP0, and its level of expression is increased. The left three lanes are equivalent to the left trio of panel A, showing the modified and unmodified forms of full-length PML in the presence and absence of ICP0 as a control. The right four lanes show PML-NK expression levels in the absence of ICP0 and with increasing amounts of the ICP0 expression plasmid in the transfection mixture (20, 50, and 100 ng, as indicated by the wedge).
The PML mutants that were either cytoplasmic or nuclear-diffuse, such as mycPML-NK, PML-NKnls, mycPML-NX, and mycPMLΔEag were not at all sensitive to ICP0; indeed, increased amounts of the ICP0 expression plasmid resulted in a transactivation of expression of the PML proteins (Fig. 7B and data not shown). That increased levels of these proteins were expressed in the presence of a cotransfected ICP0 plasmid confirms that the effects on the full-length protein are indeed due to degradation and suggests that the apparently stable levels of the C-terminal deletion mutants are not due to an absence of degradation but that this occurs at a reduced rate such that the degradation activity of ICP0 on the protein and its ability to transactivate expression from the cotransfected plasmid cancel each other out.
PML residue lysine 160, a major SUMO-1 modification site, is required for efficient ICP0-induced degradation.
One of the hallmark characteristics of PML is its conjugation to multiple SUMO-1 moieties at multiple lysine residues. The major PML sites of SUMO-1 modification have been mapped to lysine residues 160 and 490, and PML mutants with these lysine residues changed to arginine are poorly modified by SUMO-1 both in vitro and in transfected cells (5, 15). Mutations K160R and K490R were introduced into plasmid pPML(F), and the stabilities of the wild-type and double mutant proteins were compared in the ICP0-induced degradation cotransfection assay. As expected, the double mutant protein did not form the major, presumably SUMO-1-modified higher-molecular-weight species present in the normal protein, although two minor two bands migrating more slowly than the major unmodified species were detected (Fig. 8). Coexpression of ICP0 caused a much lesser degree of degradation of the mutant than of the wild-type protein (Fig. 8A), implying that either SUMO-1 modification itself or the lysine residues that are modified by SUMO-1 play an important role in the efficiency of ICP0-induced degradation of PML. Although it has been reported that PML that is not SUMO-1 modified is poorly localized into ND10, we found that in transfected HEp-2 cells the double lysine mutant efficiently colocalized with both endogenous ND10 proteins and cotransfected ICP0 (data not shown). Therefore, the reduced degradation of the mutant in these experiments could not be explained simply by failure of the proteins to colocalize in the overall ND10 structure.
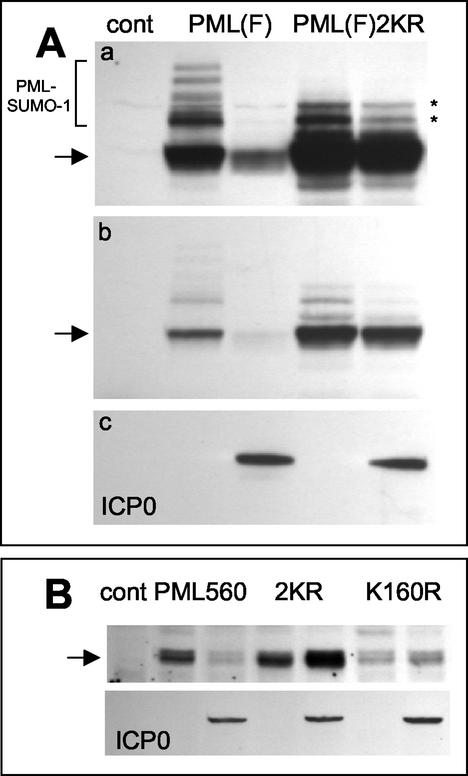
FIG. 8.
PML lysine residue 160, which is subject to conjugation to SUMO-1, is required for efficient ICP0-induced PML degradation. (A) Panel a, the left three lanes are equivalent to those in Fig. 7A, top panel, showing the modified (bracket) and unmodified (arrow) forms of full-length PML in the presence and absence of ICP0 as a control (cont). The right two lanes show the levels of expression of a PML derivative with lysine-to-arginine mutations at residues 160 and 490, the major characterized SUMO-1 modification sites of PML, in the presence and absence of coexpressed ICP0. The asterisks indicate uncharacterized modified forms of the mutant PML protein that migrate more rapidly than most of the SUMO-1-modified forms of the normal protein. Panel b, a shorter exposure of the same blot, showing the relative insensitivity of the double lysine mutant to ICP0 expression. Panel c, a reprobe of the same blot to detect ICP0 in the relevant lanes. (B) A similar experiment using the 560-residue PML isoform VI. The upper panel shows the control lane and then paired lanes of PML isoform VI (PML560), the double lysine mutant PML560SUMO- (2KR), and the single K160R mutant detected by Western blotting in extracts of cells cotransfected or not with an ICP0 expression plasmid (as detected in the lower panel by reprobing of the same filter). The amount of PML isoform VI was decreased by ICP0, whereas the amounts of the double and single lysine mutants were increased. The basal level of the single K160R mutant was lower than those of the other two proteins in this experiment.
Similar results were obtained when the mutations were present in the 560-residue isoform VI version of PML. This PML protein is not as efficiently modified by SUMO-1 in our transfection assays, with the modified forms visible only on prolonged exposures of the Western blot (data not shown). Despite this, the major unmodified band of PML VI was efficiently degraded in response to ICP0 expression (Fig. 8B). In confirmation of the results with isoform IV, isoform VI containing the K160R and K490R mutations was stable in the presence of ICP0, and indeed there was a reproducible effect of the ICP0 transactivation function increasing the levels of the mutant PML protein. The absence of transactivated expression of the double lysine mutant PML isoform IV in the presence of ICP0 (Fig. 8A) suggests that the 633-residue isoform IV of PML may be more sensitive to ICP0-mediated degradation than the 560-residue isoform VI, an implication that is consistent with the role of the C-terminal end of isoform IV being involved in fully efficient targeting by ICP0 (Fig. 7). Interestingly, PML isoform VI carrying only the K160R mutation was as resistant to the effects of ICP0 as the double mutant protein (Fig. 8B). This result identifies lysine residue 160 of PML as a critical determinant in the mechanism of ICP0-mediated PML degradation.
DISCUSSION
This paper demonstrates that ubiquitinated forms of PML can be detected in transfected cells in response to ICP0 expression. Previous work had shown that ICP0 works through the ubiquitin pathway during infection and in the reactivation of quiescent viral genomes (12), that full-length ICP0 has E3 ligase activity in vitro (3), and that ICP0 induces the proteasome-dependent degradation of PML in vivo (8, 21). Taken together, these data provide strong evidence that ICP0 functions as a RING finger-dependent E3 ligase that is able to target specific proteins for ubiquitination and degradation. However, we could not reproduce the ubiquitination of either unmodified or SUMO-1 unmodified PML in vitro. The most likely explanations for the negative in vitro data are either that a component in addition to PML, ICP0, and the E2 conjugation enzyme is required for the active reaction complex or that PML may not be a direct substrate of ICP0 but is degraded as an indirect consequence of ICP0's E3 ligase activity on some other substrate at ND10 in vivo. Despite the failure of ICP0 to ubiquitinate PML in vitro, we have evidence that purified full-length ICP0 is indeed capable of ubiquitinating other specific substrates in vitro, producing a ladder of multiply ubiquitinated forms of the target proteins in question (C. Boutell, M. Canning, and R. Everett, unpublished data).
The initial observations on the effect of ICP0 on PML identified the SUMO-1-modified forms of PML as being particularly sensitive to degradation (8). One explanation for this activity could be interference by ICP0 in the process of SUMO-1 modification of PML. The data presented here show that ICP0 neither affects the activity of the enzymes required for SUMO-1 modification of PML in vitro nor directly destabilizes the modified PML forms in vitro. Since ICP0 binds very strongly to the ubiquitin-specific protease USP7 (11), it had been thought possible that ICP0 might redirect the peptidase activity of USP7 onto SUMO-1 conjugated PML. However, addition of purified enzymatically active USP7 to our reaction mixes had no effect on SUMO-1-modified PML (data not shown). The negative in vitro data described in this paper are nonetheless significant because they answer a number of questions that have been raised in publications from several different laboratories in recent years. It is possible that ICP0 has additional effects on authentic SUMO-specific proteases. Indeed, it has recently been observed that SUMO-specific protease SENP-1, an enzyme capable of deconjugating SUMO-1 from PML, accumulates in ICP0-defined domains in both transfected and infected cells (2). Whether SENP-1 plays a role in the ICP0-induced loss of PML-SUMO-1 conjugates in infected cells remains to be determined.
Our transfection assays on the characteristics of PML that contribute to its ICP0-induced degradation provide a number of new insights. We found that the C-terminal region of PML is required for its efficient degradation and that when the two major SUMO-1 modification sites (K160 and K490) were mutated to arginine, PML became markedly less sensitive to the effects of ICP0. Use of the single K160R mutant form of PML in both the isoform IV and isoform VI backgrounds demonstrated that this residue is crucial for ICP0-mediated degradation. The involvement of the C-terminal sequences of PML and the essential role of residue lysine 160 in the efficiency of degradation induced by ICP0 are reminiscent of similar data on the degradation of PML that occurs in response to arsenic treatment (17). However, the situation appears to be complex. Examination of a number of PML isoforms with different C termini in the arsenic-induced degradation assay did not reveal a common sequence that was required for degradation (17). Similarly, we found that PML isoform VI (560 residues) retains sensitivity to ICP0 but differs from the relatively resistant mycPML-NM truncation mutant only in the N-terminal tag and by a limited number of C-terminal residues.
These data indicate that slight changes in PML structure can have a significant effect on sensitivity to ICP0. These changes may cause fine-structure alterations in the localization of PML within ND10 domains or changes in its interactions with other proteins. For example, although a PML mutant that cannot be modified by SUMO-1 forms punctate structures visible by fluorescence that are similar to normal ND10, at the level of electron microscopy these appear to be irregular aggregates rather then the highly structured entities seen in normal ND10 (17). Furthermore, the PML K160R mutant protein was unable to recruit 11S proteasome regulatory subunits to ND10 (17). Our data show that PML localization in ND10 and colocalization with ICP0 is necessary but not sufficient for maximally efficient PML degradation. Similarly, ICP0 mutants that do not localize efficiently to ND10 have reduced abilities to induce PML degradation (8, 22). Therefore it appears that the ability of PML and ICP0 to accumulate in the same nuclear structures is important but is not the only factor in the ICP0-induced degradation of PML; fine-structure considerations within the ND10 domain itself may also contribute.
Although the degradation of PML and disruption of ND10 induced by ICP0 are one of the hallmarks of HSV-1 infection in cultured cells, the significance of these events for the progress of the infection has remained elusive. PML and ND10 remain stable for relatively long periods during infection by ICP0-deficient viruses, yet these viruses replicate well in high-multiplicity infections. Although overexpression of PML isoform VI has been shown to slow human cytomegalovirus infection (1), high levels of PML expression appear to have no significant effect on HSV-1 infection (4, 18). The conclusions of the latter studies are supported by our own unpublished investigations. However, it is clear that the ubiquitin E3 ligase activity of the RING finger domain of ICP0 correlates extremely well with the principal biological functions of ICP0. It is possible that PML, and indeed the other targets of ICP0 so far identified, are merely innocent victims and that the biologically relevant target is another protein (perhaps with some shared biochemical characteristics, such as SUMO-1 modification). Alternatively, it could be that the important factor involves structural and environmental changes within ND10 but that these are too subtle to be observed at the present level of analysis and are not affected by simple overexpression of PML.
Acknowledgments
We are particularly grateful to Ron Hay for advice and reagents used in the in vitro SUMO-1 modification assays. Seth Sadis, Dan Bailey, Peter O'Hare, Christina Ward, and Pierre Chambon generously supplied several plasmids used in these studies. We acknowledge helpful discussions with several members of the MRC Virology Unit, including Duncan McGeoch and Chris Preston, and the contribution of some of the unpublished work of Wei-Li Hsu, Mary Canning, and Alexandros Zafiropoulos.
This work was supported by the Medical Research Council and by the award of a Human Frontiers Science Programme Fellowship to C.B.
REFERENCES
- 1.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. [DOI] [PubMed] [Google Scholar]
- 3.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 5.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 6.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. BioEssays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 8.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, K., C. Shiels, and P. S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223-7233. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 15.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 16.Kastner, P., A. Perez, Y. Lutz, C. Rochette-Egly, M. P. Gaub, B. Durand, M. Lanotte, R. Berger, and P. Chambon. 1992. Structure, localization and transcriptional properties of two classes of retinoic acid receptor alpha fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 11:629-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As(2)O(3)-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus type 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 20.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roizman, R., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams and Wilkins, Philadelphia, Pa.
- 24.Sternsdorf, T., K. Jensen, and H. Will. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139:1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 26.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121-127. [DOI] [PubMed] [Google Scholar]