Abstract
A recombinant infectious bronchitis virus (IBV), BeauR-M41(S), was generated using our reverse genetics system (R. Casais, V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton, J. Virol. 75:12359-12369, 2001), in which the ectodomain region of the spike gene from IBV M41-CK replaced the corresponding region of the IBV Beaudette genome. BeauR-M41(S) acquired the same cell tropism phenotype as IBV M41-CK in four different cell types, demonstrating that the IBV spike glycoprotein is a determinant of cell tropism.
Avian infectious bronchitis virus (IBV), a member of the Coronaviridae (order Nidovirales, genus Coronavirus), is a highly infectious pathogen of domestic fowl that replicates primarily in the respiratory tract but also in epithelial cells of the gut, kidney, and oviduct (3, 6, 7). Genetically very similar coronaviruses cause disease in turkeys and pheasants (4, 5). Coronaviruses are enveloped viruses that replicate in the cell cytoplasm and contain an unsegmented, single-stranded, positive-sense RNA genome of 27 to 32 kb (11, 16, 24).
All coronavirus lipid envelopes contain at least three membrane proteins: the spike glycoprotein (S), integral membrane protein (M), and small membrane protein (E). The coronavirus S protein is a type I glycoprotein which oligomerizes in the endoplasmic reticulum and is assembled into virion membranes through noncovalent interactions with the membrane protein (12). Following incorporation into coronavirus particles, the S glycoprotein is responsible for binding to the target cell receptor and fusion of the viral and cellular membranes. The S glycoprotein consists of four domains: a signal sequence that is cleaved during synthesis; the ectodomain, which is present on the outside of the virion particle; the transmembrane region responsible for anchoring the S protein into the lipid bilayer of the virion particle; and the cytoplasmic tail. The IBV S glycoprotein (1,162 amino acids) is cleaved into two subunits, S1 (535 amino acids; 90 kDa), comprising the N-terminal half of the S protein, and S2 (627 amino acids; 84 kDa), comprising the C-terminal half of the S protein. The S2 subunit associates noncovalently with the S1 subunit and contains the transmembrane and C-terminal cytoplasmic tail domains. The S1 subunit contains the receptor-binding activity of the S protein (14, 23). The ectodomain region of the S2 subunit contains a fusion peptide-like region (18) and two heptad repeat regions involved in oligomerization of the S protein (10). The shorter heptad repeat is adjacent to the transmembrane region and consists of a leucine zipper motif (1). Modification of the murine hepatitis coronavirus (MHV) S protein leucine zipper motif affected oligomerization and loss of cell-to-cell fusion (17).
Targeted recombination has been used for modifying the MHV S gene (9, 19, 21). Similarly, a recombinant porcine coronavirus transmissible gastroenteritis virus (TGEV) containing the S gene derived from an enteric TGEV within a respiratory TGEV genome was isolated in vivo and was shown to have acquired an enteric tropism (22). In addition, the S glycoprotein ectodomain of MHV has been replaced with the corresponding sequence from the coronavirus feline infectious peritonitis virus, and the resulting virus acquired the ability to infect feline cells with the concomitant loss of ability to infect murine cells in tissue culture (15). These results demonstrated that the S glycoprotein for a group 2 coronavirus (MHV) or a group 1 coronavirus (TGEV) was involved in the tropism of these coronaviruses. However, due to the lack of a suitable modification system, the role of the IBV S glycoprotein, a group 3 coronavirus, in cell tropism had yet to be confirmed. We have used our IBV reverse genetics system to investigate the role of the IBV S glycoprotein in cell tropism, demonstrating, for the first time, that the IBV reverse genetics system could be used to generate a recombinant coronavirus with a precisely modified genome to study the role of a coronavirus gene. Analysis of recombinant IBV (rIBV) showed that exchange of the S gene resulted in a virus that had the growth characteristics of the S gene donor strain, M41-CK, confirming that the S glycoprotein of a group 3 coronavirus (IBV) is a determinant of cell tropism of the virus in vitro.
Strategy for the construction of a chimeric S gene.
To investigate the role of the IBV S protein in cell tropism, we produced rIBV in which the Beaudette S glycoprotein gene in Beau-R (2) was replaced with the corresponding sequence from IBV M41-CK (8), using the IBV reverse genetics system described by Casais et al. (2). In order to achieve our aim of generating the rIBV, several factors were taken into account. (i) IBV Beaudette can be propagated in CK, CEF, BHK-21, and Vero cells. In contrast, IBV M41-CK can only be propagated in CK cells, providing a mechanism for studying IBV cell tropism. (ii) The C-terminal end of the IBV replicase gene overlaps the N-terminal end of the S gene by 50 nucleotides (nt). (iii) The region of the S gene corresponding to the S2 subunit encodes part of the ectodomain, the transmembrane, and cytoplasmic tail regions, of which the latter are responsible for incorporation of the S glycoprotein into coronavirus particles (12).
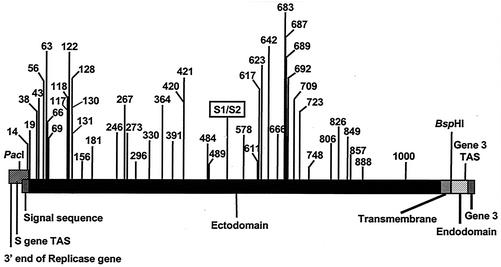
Sequence analysis of the M41-CK S gene identified 72 nt differences from the Beau-R S gene sequence, of which 50 represented nonsynonymous substitutions and 22 represented synonymous substitutions, resulting in a total of 47 amino acid differences between the two S glycoproteins. The last nonsynonymous substitution results in a premature stop codon within the M41 S gene, so that the M41-CK S glycoprotein is nine amino acids shorter than the Beaudette protein. Apart from the loss of the nine amino acids, there were no other amino acid differences between the cytoplasmic domains of the two viruses. Overall, the primary translation products of the two S genes are 1,153 and 1,162 amino acids in size for M41-CK and Beau-R, respectively, representing an identity of 95.2% between the two S proteins. Comparison of the replicase sequence that overlaps the S gene sequence showed that there is only one synonymous mutation, with no mutations between the S-gene transcription-associated sequence (TAS) and the initiation codon of the S gene (Fig. 1).
FIG. 1.
Schematic diagram of the IBV S gene. The 5′ end of the S gene overlaps the 3′ end of the replicase gene. The four domains of the S protein, the position of the S1/S2 cleavage point, and the positions of the PacI and BspHI restriction sites, gene 3 TAS, and the start of gene 3 are shown. The numbers refer to the positions of the amino acid differences resulting from nonsynonymous substitutions following exchange of the two S gene sequences.
A chimeric S gene, consisting of the signal sequence, ectodomain, and transmembrane regions derived from M41-CK and the cytoplasmic tail domain from Beau-R, was produced. The last 137 nt of the Beaudette S gene, comprising the cytoplasmic domain, were retained to maintain any interaction of the S protein C-terminal domain with the other Beaudette-derived proteins. The transmembrane regions are identical between the two S glycoproteins; therefore, the mature form of the chimeric S glycoprotein only differed in the ectodomain, derived from M41-CK, compared to the Beaudette S glycoprotein. A plasmid, pFRAG3-M41S, containing the chimeric S gene with the above characteristics was generated.
Recovery of a rIBV expressing a chimeric S protein.
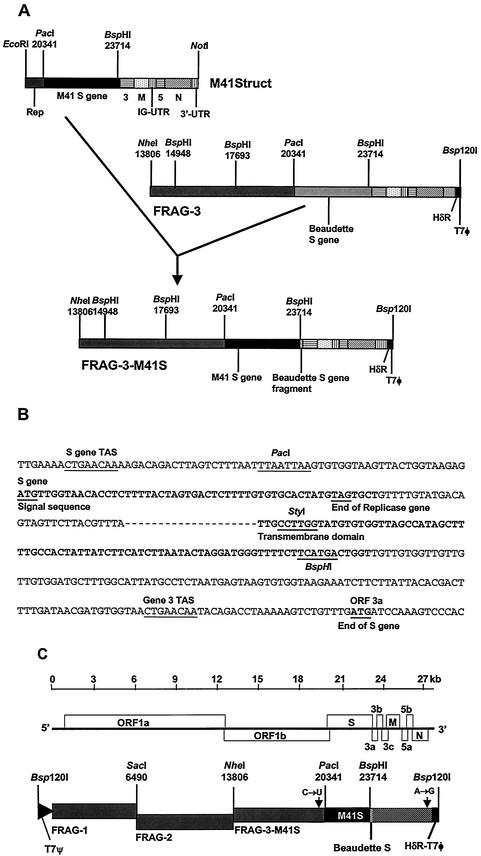
Plasmid pM41Struct (a gift from K. Tibbles, University of Cambridge) contained a cDNA, from nucleotide 19493 in the replicase gene to the poly(A) tail, derived from M41-CK genomic RNA, in pBluescript SK(+). The chimeric S gene was generated using two restriction sites, PacI and BspHI, present at the same respective positions in both virus sequences (Fig. 2A and B). The PacI site at nt 20337 is 30 nt proximal to the S gene initiation codon and 21 nt distal to the S gene TAS, and the BspHI site at nt 23714 is at the end of the transmembrane domain. The M41-CK-derived 3,378-bp PacI-BspHI fragment was used to replace the corresponding Beaudette-CK sequence in pFRAG-3 (2), generating pFRAG3-M41S. A full-length IBV cDNA containing the chimeric S gene was assembled in vitro and directly inserted into the vaccinia virus genome, following a procedure described previously (2), resulting in vNotI/IBVFL-M41S (Fig. 2C). PCR analysis of the IBV cDNA within vNotI/IBVFL-M41S confirmed that the cDNA represented a full-length IBV genome with the chimeric S gene.
FIG. 2.
Schematic diagram for the construction of the chimeric S gene and production of a full-length IBV cDNA. (A) Replacement of the ectodomain region of the Beaudette S gene by the corresponding sequence from IBV M41 for construction of FRAG-3-M41S. (B) Junction of the S gene sequences at the PacI and BspHI restriction sites with the intervening sequence derived from M41. The nucleotides in bold correspond to the signal and transmembrane sequences, respectively. (C) Schematic diagram of the BeauR-M41(S) full-length cDNA composed of FRAG-1, FRAG-2, and FRAG-3-M41S.
Infectious rIBV was recovered (2) from vNotI/IBVFL-M41S using CK cells, previously infected with rFPV/T7, to provide T7 RNA polymerase, and cotransfected with AscI-restricted vNotI/IBVFL-M41S DNA and pCi-Nuc (13). A rIBV, BeauR-M41(S), containing the chimeric S gene was passaged five times on CK cells and used for further characterization. Sequence analysis of the complete genome of BeauR-M41(S) confirmed that the sequence was as expected.
Characterization of BeauR-M41(S).
IBV strains Beau-CK and M41-CK have different cell tropisms and both viruses replicate to similar titers in CK cells, but only Beau-CK produces infectious virus on Vero cells. Therefore, by using the recombinant isogenic viruses, Beau-R and BeauR-M41(S), which differ only in the ectodomain of the S protein, we sought to determine whether the IBV S glycoprotein was responsible for the observed differences in the abilities of different IBV strains to infect and replicate in different cell lines.
Beau-R, M41-CK, and BeauR-M41(S) were used to infect four different cell types, and the titer of progeny virus was determined over a 96-h period. All three viruses displayed similar growth profiles on CK cells (Fig. 3A). Progeny virus was detectable 8 h postinfection, with peak titers of 107 PFU/ml at 18 to 24 h postinfection, showing that the three viruses replicated to a similar extent in CK cells irrespective of the origin of the S gene. The growth profiles of the three viruses were then analyzed in CEF cells, previously shown to support the growth of 10 different IBV strains, including M41, which produced 1000-fold less virus than Beaudette (20), and on two mammalian cell lines, Vero and BHK-21. Only two strains of IBV, including Beaudette, had been demonstrated to grow on BHK-21 cells (20).
FIG. 3.
Growth profiles of the three IBVs on four cell types. Cells were infected with 1.5 × 106 PFU of each IBV, and cell medium was analyzed for progeny virus by plaque titration assay on CK cells 0 to 96 h postinfection. The panels show the growth patterns of Beau-R (solid line with triangle), M41-CK (dashed line with diamond), and BeauR-M41(S) (dotted line with square) on CK cells (A), Vero cells (B), CEF cells (C), and BHK-21 cells (D).
Analysis of the growth profiles of the three viruses in Vero, CEF, and BHK-21 (Fig. 3B to D) showed that only Beau-R replicated to any significant extent in the different cells, usually with maximum titer by 24 h postinfection. In Vero cells, Beau-R replicated to a titer of 107 PFU/ml at 24 h postinfection, while BeauR-M41(S) and M41-CK showed 104-fold-lower titers (Fig. 3B). Both M41-CK and BeauR-M41(S) barely increased in titer above the titer observed at time zero and after 30 h postinfection showed little evidence for production of new infectious virus.
In CEF cells, M41-CK and BeauR-M41(S) initially grew, but to a lower level than observed for Beau-R. The Beau-R titer increased approximately 1,800-fold by 30 h postinfection, whereas the titers for M41-CK and BeauR-M41(S) only increased approximately 15-fold over the same time period (Fig. 3C). However, after 30 h postinfection, only the titer of Beau-R continued to increase, while the titers of M41-CK and BeauR-M41(S) decreased.
Similarly, Beau-R replicated to a titer of 105 PFU/ml at 24 h postinfection on BHK-21 cells, while BeauR-M41(S) and M41-CK showed more than 102-fold-lower titers with little to no growth compared to the titers at time zero (Fig. 3D). This observation corroborates the results of Otsuki et al. (20), who demonstrated that Beaudette but not M41 replicated on BHK-21 cells. Our results demonstrated that BeauR-M41(S) had the same tropism as M41-CK on all four cell types.
The growth experiments represented the measurement of progeny virus, following a single infection, over a time period. Therefore, we decided to investigate the possibility that serial passage might result in amplification following adaptation or selection of progeny virus in the different cell types. The four cell types were infected with the three viruses at 2 × 107 PFU (P1), and after 24 h postinfection, progeny virus was serially passaged twice (P2 and P3). Total cellular RNA was extracted from the cytoplasm of P1 to P3 cells and analyzed by reverse transcription (RT)-PCR using oligonucleotides located in the N gene and 3′ untranslated region. Detection of an RT-PCR product of 666 bp for Beau-R and BeauR-M41(S) or 481 bp for M41-CK was indicative of IBV replication.
Analysis of the RT-PCR products from RNA isolated from P1 to P3 CK cells infected with Beau-R, M41-CK, and BeauR-M41(S) confirmed that all three viruses were serially passaged on CK cells (Fig. 4A). RT-PCR analysis of RNA derived from Vero cells showed that only Beau-R was passaged on Vero cells (Fig. 4B). Although IBV RNA was detected in P1 cells infected with M41-CK and BeauR-M41(S), there was no evidence of IBV-derived RNA following passage of the viruses in P2 and P3 Vero cells (Fig. 4B). Detection of M41-CK and BeauR-M41(S)-derived RNA in P1 Vero cells may have resulted from (i) the virus inoculum, indicating that neither virus replicated in Vero cells, or (ii) low levels of replication in P1 Vero cells, resulting in low titers of progeny virus, so that resultant amounts of RNA in P2 and P3 cells were below the threshold of detection by RT-PCR.
FIG. 4.
Detection of IBV by RT-PCR following passage of Beau-R, M41-CK, or BeauR-M41(S) on CK, Vero, CEF, or BHK-21 cells. Cells (P1) were infected with 2 × 107 PFU of each virus, and 24 h postinfection any progeny viruses were passaged twice (P1 to P3). Total cellular RNA was isolated from the IBV-infected cells and used for RT-PCR analysis; an RT-PCR product of 666 bp corresponded to detection of Beau-R- and BeauR-M41(S)- derived RNA, and a product of 481 bp corresponded to detection of M41-CK-derived RNA. The RT-PCR products detected following passage (P1 to P3) of Beau-R, M41-CK, or BeauR-M41(S) on CK (A), Vero (B), CEF (C), or BHK-21 (D) cells were analyzed on 1% agarose gels.
Analysis of RNA isolated from CEF cells showed that all three viruses were detected in P1-P2 cells, indicating that the viruses grew in this cell type (Fig. 4C). This confirmed the results obtained from the growth experiment (Fig. 3C), with the RT-PCR analysis detecting growth of the three viruses in the initial 24-h period. The reason for the decline in growth of M41-CK and BeauR-M41(S) after 30 h postinfection is not known (Fig. 3C).
RT-PCR analysis of RNA isolated from BHK-21 cells infected with the three viruses showed that only Beau-R could be passaged on this cell type (Fig. 4D). The analysis showed the presence of IBV-derived RNA in P1 BHK-21 cells infected with all three viruses. There was no evidence that M41-CK was passaged beyond P1 on BHK-21 cells. Although BeauR-M41(S) was detected in P1 BHK-21 cells, the amount detected at P2 was lower, and no IBV RNA was detected in P3 cells. Overall, these results demonstrated that exchange of the ectodomain of the S protein resulted in the loss of the ability of Beau-R to establish a successful infectious cycle in Vero, CEF, and BHK-21 cells.
We have used our IBV reverse genetics system to produce a rIBV, BeauR-M41(S), consisting of the Beaudette genome but with the ectodomain region of the S gene replaced with the corresponding sequence from M41-CK. Our results demonstrated that BeauR-M41(S) had the growth characteristics of M41-CK on four cell types and that replacement of only the ectodomain of the Beaudette S glycoprotein with the M41-CK homologue resulted in the altered growth characteristics of Beau-R. The results demonstrated that the IBV S protein plays a major role in the cell tropism of the virus.
Acknowledgments
This work was supported by the Commission of the European Communities specific RTD program Quality of Life and Management of Living Resources, QLK2-CT-1999-00002, the Department of Environment, Food and Rural Affairs, project code OD0712, and the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Britton, P. 1991. Coronavirus motif. Nature 353:394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh, D. 2001. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 30:109-115. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh, D., K. Mawditt, M. Sharma, S. E. Drury, H. L. Ainsworth, P. Britton, and R. E. Gough. 2001. Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. Avian Pathol. 30:365-378. [DOI] [PubMed] [Google Scholar]
- 5.Cavanagh, D., K. Mawditt, D. B. B. Welchman, P. Britton, and R. E. Gough. 2002. Coronaviruses from pheasants (Phasianus colchicus) are genetically closely related to coronaviruses of domestic fowl (infectious bronchitis virus) and turkeys. Avian Pathol. 31:81-93. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh, D., and S. Naqi. 2003. Infectious bronchitis, p. 101-119. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames.
- 7.Cook, J. K., J. Chesher, W. Baxendale, N. Greenwood, M. B. Huggins, and S. J. Orbell. 2001. Protection of chickens against renal damage caused by a nephropathogenic infectious bronchitis virus. Avian Pathol. 30:423-426. [DOI] [PubMed] [Google Scholar]
- 8.Darbyshire, J. H., J. G. Rowell, J. K. A. Cook, and R. W. Peters. 1979. Taxonomic studies on strains of avian infectious bronchitis virus using neutralisation tests in tracheal organ cultures. Arch. Virol. 61:227-238. [DOI] [PubMed] [Google Scholar]
- 9.Das Sarma, J., L. Fu, J. C. Tsai, S. R. Weiss, and E. Lavi. 2000. Demyelination determinants map to the spike glycoprotein gene of coronavirus mouse hepatitis virus. J. Virol. 74:9206-9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. M. Van der Zeijst, W. J. M. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196:963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organisation of the Nidovirales: similarities and differences between arteri-, toro- and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiscox, J. A., T. Wurm, L. Wilson, P. Britton, D. Cavanagh, and G. Brooks. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch, G., L. Hartog, A. Kant, and D. J. van Roozelaar. 1990. Antigenic domains of the peplomer protein of avian infectious bronchitis virus: correlation with biological function. J. Gen. Virol. 71:1929-1935. [DOI] [PubMed] [Google Scholar]
- 15.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, Z., A. M. Matthews, and S. R. Weiss. 1999. Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 73:8152-8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, Z. L., and S. R. Weiss. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navas, S., S. H. Seo, M. M. Chua, J. D. Sarma, E. Lavi, S. T. Hingley, and S. R. Weiss. 2001. Murine coronavirus spike protein determines the ability of the virus to replicate in the liver and cause hepatitis. J. Virol. 75:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otsuki, K., K. Noro, H. Yamamoto, and M. Tsubokura. 1979. Studies on avian infectious bronchitis virus (IBV). II. Propagation of IBV in several cultured cells. Arch. Virol. 60:115-122. [DOI] [PubMed] [Google Scholar]
- 21.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sánchez, C. M., A. Izeta, J. M. Sánchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Durán, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultze, B., D. Cavanagh, and G. Herrler. 1992. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependant on sialic acid containing receptors on erythrocytes. Virology 89:792-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddell, S. G. 1995. The Coronaviridae, p. 1-10. In S. G. Siddell (ed.), The Coronaviridae. Plenum, New York, N.Y.