Abstract
Newcastle disease virus (NDV) edits its P gene by inserting one or two G residues at the conserved editing site (UUUUUCCC, genome sense) and transcribes the P mRNA (unedited), the V mRNA (with a +1 frameshift), and the W mRNA (with a +2 frameshift). All three proteins are amino coterminal but vary at their carboxyl terminus in length and amino acid composition. Little is known about the role of the V and W proteins in NDV replication and pathogenesis. We have constructed and recovered two recombinant viruses in which the expression of the V or both the V and W proteins has been abolished. Compared to the parental virus, the mutant viruses showed impaired growth in cell cultures, except in Vero cells. However, transient expression of the carboxyl-terminal portion of the V protein enhanced the growth of the mutant viruses. In embryonated chicken eggs, the parental virus grew to high titers in embryos of different gestational ages, whereas the mutant viruses showed an age-dependent phenomenon, growing to lower titer in more-developed embryos. An interferon (IFN) sensitivity assay showed that the parental virus was more resistant to the antiviral effect of IFN than the mutant viruses. Moreover, infection with the parental virus resulted in STAT1 protein degradation, but not with the mutant viruses. These findings indicate that the V protein of NDV possesses the ability to inhibit alpha IFN and that the IFN inhibitory function lies in the carboxyl-terminal domain. Pathogenicity studies showed that the V protein of NDV significantly contributes to the virus virulence.
Newcastle disease virus (NDV) causes a highly contagious respiratory, neurological, or enteric disease in chickens. The disease is prevalent worldwide and causes severe economic losses in the poultry industry. Naturally occurring NDV isolates display a wide range of virulence for chickens, which vary from a fatal to an inapparent infection (2). Strains of NDV are classified into three main pathotypes depending on the severity of disease produced in chickens. Lentogenic strains do not cause disease and are considered avirulent. Viruses of intermediate virulence are termed mesogenic, while virulent strains that cause high mortality are termed velogenic (2). The virulence determinants of NDV are not completely understood. The amino acid sequence at the fusion (F) protein cleavage site has been postulated as a major determinant of NDV virulence (2, 41). However, the role of other viral proteins in NDV pathogenicity remains unknown.
NDV is a member of the newly formed genus Avulavirus in the family Paramyxoviridae (30). The genome of NDV is a single-stranded negative-sense RNA consisting of 15,186 nucleotides (8, 23). The genomic RNA contains six genes in the order of 3′-NP-P-M-F-HN-L-5′. In common with other paramyxoviruses, NDV produces two additional proteins, V and W, from the P gene by alternative mRNAs that are generated by RNA editing (16, 20, 31, 46, 48). In NDV, insertion of one nontemplate G residue gives rise to a V-encoding mRNA, while insertion of two nontemplate G residues generates a W-encoding mRNA. Analysis of mRNAs produced from the P gene showed that 68% were P-encoding mRNA, 29% were V-encoding mRNA, and 2% were W-encoding mRNA (32). All three P gene-derived proteins are amino coterminal but vary at their carboxyl terminus in length and amino acid composition. The V protein of NDV, in common with its counterparts in other paramyxoviruses, is cysteine rich within its unique carboxyl-terminal region and binds to zinc (19, 39, 49). The V protein of NDV is found to be incorporated in virions, as are simian virus 5 (SV5) and mumps virus (25, 32, 39), and unlike the V proteins of Sendai virus and measles virus (MV), which are not incorporated (18, 52). Of the three NDV P gene products, the P protein, together with the L protein, is known to form part of the virus RNA polymerase complex (26). However, very little is known about the biological functions of the V and W proteins.
The interferon (IFN) system is the first line of host defense against virus infection. Interferons induce an antiviral state that may inhibit virus replication and control virus spread. The effectiveness of IFN responses has prompted many viruses to adopt strategies to evade the IFN-induced antiviral responses (13-15, 42). Recent studies using reverse genetics systems have gained insights into the functions of several paramyxovirus accessory proteins (4, 17, 21, 32). It was shown that the V proteins of many paramyxoviruses are responsible for blocking the antiviral action of IFN (3, 9, 36, 38, 45). Subsequently, it was demonstrated that paramyxoviruses achieved this goal by distinct molecular mechanisms (55). The V protein of SV5 and mumps virus target STAT1 for proteasome-mediated degradation and thus block IFN signaling (9, 25), whereas human parainfluenza virus type 2 (hPIV2) blocked the IFN signaling by degrading STAT2 protein (3, 36). The role of the NDV V protein has been investigated by creating recombinant viruses in which the expression of V protein has been abolished (32). It was demonstrated that the absence of the V protein retarded the growth of the recombinant virus in cell cultures and in 6-day-old embryonated chicken eggs. In contrast, no virus growth was detected in 9- to 11-day-old embryonated eggs, indicating that the V protein probably plays an important role in antagonizing the host's innate response. Recently, using an IFN-sensitive recombinant NDV-based assay, it was demonstrated that the V protein of NDV is an IFN antagonist (38). However, the molecular mechanism of IFN antagonism and the specific role of the NDV V protein in viral pathogenicity have not been investigated. Furthermore, in the previous studies, the role of the V protein in NDV virulence could not be determined, as the mutant viruses were generated from an infectious cDNA clone of avirulent NDV strain LaSota (32).
Therefore, the purpose of this study was to determine the role of the V protein in IFN antagonism and virulence using a virulent NDV strain. The reverse genetics system was used to introduce mutations into the editing site of the P gene of virulent NDV strain Beaudette C. Mutant viruses were recovered in which the expression of the V protein or both V and W proteins was inhibited. The mutant viruses were highly attenuated in 1-day-old and 6-week-old chickens, indicating an important role of the V protein in NDV pathogenicity. Our results also demonstrated that one of the mechanisms of the editing-defective viruses was an increased sensitivity to IFNs. We further demonstrated that the carboxyl-terminal portion of the V protein was responsible for blocking the antiviral functions of alpha IFN (IFN-α) by targeting STAT1 for degradation.
MATERIALS AND METHODS
Cells and viruses.
DF1 (chicken embryo fibroblast cell line), Vero, HEp-2, and human 2fTGH cells (kindly provided by George Stark, Cleveland Clinic Research Foundation, Cleveland, Ohio) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Virus-infected cells were maintained in DMEM containing 5% fetal calf serum. The recovered recombinant NDVs were propagated in 9-day-old embryonated specific-pathogen-free (SPF) chicken eggs. The allantoic fluids were harvested 60 h postinfection and clarified by centrifugation. The clarified allantoic fluids were stored as single-use aliquots at −80°C for all experiments. Virus titers were determined by standard plaque assay on DF1 cells.
Mutagenesis of full-length NDV cDNA and recovery of mutant viruses.
The plasmid pNDVfl expressing the full-length antigenome of NDV Beaudette C has been described previously (24) and was used to construct P gene editing mutants. The AscI-SacII fragment containing the P gene editing site from pNDVf1 was subcloned into pGEM-7Z(+) (Promega, Madison, Wis.) between XbaI and HindIII by using a specific primer pair with XbaI and HindIII site overhangs. Mutations were introduced into the P gene editing site by PCR using a phosphorylated primer pair to amplify the plasmid pGEM-7Z(+) containing the AscI-SacII insert as described previously (6). Primers P1 (5′-2296GACCATAGGCCCTTTTTAGCATTGG2272-3′) and P2 (5′-2297tAGCCCCCAAGAGGGGAACCACC2319-3′; lowercase letter represents mutation) were used to introduce a stop codon in the V reading frame after the editing site to generate a C-terminus-truncated version of the V protein. Similarly, primer pair P3 (5′-2282gAAaGGCCTATGGTCGAGCCCCCAAG2307-3′) and P4 (5′-2281TTAGCATTGGACGATTTATTGCTGAGC2255-3′) was used in the same way to introduce two nucleotide changes to disrupt the P gene editing site. The mutations introduced were silent with regard to the P reading frame. The mutated AscI and SacII fragments were excised to replace the corresponding counterpart in pNDVfl. The presence of the introduced mutations was confirmed by sequencing the respective full-length clones. Mutant viruses were recovered from these full-length plasmids as described previously (24). Briefly, HEp-2 cells at 80 to 90% confluence in a six-well plate were infected with MVA-T7 at 1 focus-forming unit per cell. The cells were then transfected with the three expression plasmids encoding the NP, P, and L proteins of NDV strain Beaudette C and a fourth plasmid containing the mutated NDV cDNA. Three days after transfection, the cell culture supernatant was harvested, briefly clarified, and then used to infect fresh HEp-2 cells. Three days later, the supernatant was harvested and passed on to the DF1 cells until virus-specific cytopathic effect (CPE) appeared. The recovered viruses were plaque purified on DF1 cells and propagated in 9-day-old embryonated SPF chicken eggs for use as virus stocks.
Sequence analysis of recovered viruses.
Virus stocks were propagated twice in DF1 cells before sequence analysis. Total RNA from infected cells was prepared using Trizol (Invitrogen, Carlsbad, Calif.). The nucleotide sequence of the region of the P gene that contains the editing site was analyzed by sequencing a reverse transcriptase PCR (RT-PCR) fragment obtained using primer P5 (5′-1200GGAGACTTGGAGTAGAGTACGCT1222-3′) for reverse transcription and forward primer P6 (5′-1854GCTCTCTCCTCTACCTGATAGA1875-3′) and reverse primer P7 (5′-2550CTACAGGTGGCTGGACATGATCC2528-3′) for PCR. The resulting PCR product was purified by using a PCR purification kit (Qiagen, Hilden, Germany) and sequenced by using an automated sequencer.
Western blot analysis.
To examine the expression of the V protein in mutant viruses, Western blot analysis of purified viruses was performed using a standard protocol. The viruses were purified from infective allantoic fluids by centrifugation for 90 min through a 20% sucrose cushion in a Beckman SW28 rotor at 21,000 rpm. The virus pellet was resuspended, mixed with sodium dodecyl sulfate (SDS) gel-loading buffer, and boiled for 3 min before electrophoresis. Equal amounts of viral proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel, and the resolved proteins were transferred to polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The membranes were incubated with a cocktail of monoclonal antibodies against hemagglutinin-neuraminidase (HN) or rabbit antipeptide serum specific for the carboxyl-terminal 18 amino acids of the V protein, washed, and subsequently incubated with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit immunoglobulin G antibody (IgG; Kirkegaard & Perry Laboratories [KPL], Gaithersburg, Md.). Proteins were visualized after incubation with tetramethylbenzidine peroxidase substrate (KPL).
For detection of STAT protein degradation, cells were infected with respective viruses at a multiplicity of infection (MOI) of 3 or transfected with 3 μg of expression plasmid. Cells were harvested for analysis 20 h later. Cells were washed two times with phosphate-buffered saline (PBS) and lysed with lysis buffer (6.25 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 6 M urea, 0.01% bromophenol blue, 0.01% phenol red). Equal amounts of cell extracts were separated by SDS-PAGE on a 7.5% gel, transferred to nitrocellulose membrane, and incubated with a commercial antiserum (Santa Cruz Biotechnology, Santa Cruz, Calif.) specific for STAT1α-p91 (C24), STAT2 (C20). Protein-antibody interactions were detected using goat anti-rabbit IgG conjugate (KPL).
Virus growth in cells and embryonated eggs.
DF1 or Vero cells were infected with virus at an MOI of 0.01 and incubated at 37°C in DMEM with 5% fetal calf serum. Supernatants were harvested at 8-h intervals for multiple cycle growth condition. Virus titers were determined by plaque assay on DF1 cells. Briefly, serial 10-fold dilutions of the virus samples were made in serum-free medium, and 100 μl of each serial dilution was added to each well of a 12-well plate. After 60 min of adsorption, the cells were overlaid with DMEM containing 5% fetal calf serum, 0.9% methylcellulose, and incubated at 37°C. On the third day after infection, the cells were fixed with methanol and stained with crystal violet for examination of plaques. To compare virus growth in embryonated SPF chicken eggs, 103 PFU of the virus in a volume of 100 μl was inoculated into the allantoic cavity of eggs of different gestational ages. Allantoic fluid was harvested and pooled 60 h postinoculation for virus titration by plaque assay.
IFN sensitivity assay.
The relative sensitivity of the parental and mutant viruses to exogenously added recombinant chicken IFN-α (chIFN-α) was measured on DF1 cells. Briefly, cells in six-well culture dishes at 70 to 80% confluence were incubated with chIFN-α (kindly provided by Philip I. Marcus, University of Connecticut, Storrs) at various concentrations. After 24 h of incubation at 37°C, the cells were infected with rBC, rBC/V-Stop, or rBC/Edit at an MOI of 0.01 in a volume of 100 μl. Vesicular stomatitis virus (VSV) strain Indiana, a known IFN-sensitive virus, was included as a positive control. Cells were adsorbed with virus for 2 h, the residual virus in the inoculum was removed, and then cells were incubated for 48 h in medium containing 5% fetal calf serum. The virus yields in the culture supernatants were determined by plaque assay in DF1 cells.
Complementation assay.
Two expression plasmids which encode the carboxyl terminus of the V and W proteins were constructed for the complementation assay. The expression plasmid pV was made using forward primer P8 (5′-atcgctagcgtaccatgGGGCCTATGGTCGAGCCCCCAAGA-3′ [uppercase letters are for the NDV-specific sequence, bold is for the cloning site, and underlined portion indicates translation start or stop codon) and reverse primer P9 (5′-cgtctgcagTTACTTACTCTCTGTGATATATTGCC-3′) to amplify the carboxyl-terminal portion of the V protein after the editing site. The insert contained a Kozak sequence (CGTACC) and an initiation codon (ATG) for optimal translation (22). The PCR product was digested and cloned into pVAX1 (Invitrogen) between NheI and PstI. Similarly, the expression plasmid pW was generated by PCR using forward primer P10 (5′-atcgctagcgtaccatgGGGGCCTATGGTCGAGCCCCCAA-3′) and reverse primer P11 (5′-cgtctgcagCTACAGGTGGCTGGACATGATCCG-3′). The resulting clones were confirmed by DNA sequencing. The complementation assays were performed as described previously, with slight modification (5). Transient transfection was performed on DF1 cells by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Briefly, DF1 cells at 80 to 90% confluence were washed two times with serum-free Opti-MEM I and used for transfection. The expression plasmids pV (3 μg) and pW (1 μg), either alone or in combination, were added to a tube containing 250 μl of Opti-MEM I serum-free medium (Invitrogen). The plasmid pVAX1 backbone was used as a negative control. Four microliters of Lipofectamine 2000 reagent diluted in 250 μl of Opti-MEM was added to the tube containing the DNA, and the tube was gently agitated. After 15 min of incubation at room temperature, 600 μl of serum-free medium was added to the tube and gently mixed by flicking the tube. The DNA-Lipofectamine mix was overlaid onto the washed DF1 cells. After 6 h of incubation at 37°C, the cells were washed with Opti-MEM and refed with DMEM supplemented with 5% fetal calf serum. The cells were infected with parental or mutant viruses at an MOI of 0.01 after 24 h of transfection. The infected cells were incubated for 48 h at 37°C. The virus titers in the culture supernatants were determined by plaque assay.
Pathogenicity studies.
The mean death time (MDT) was determined to examine the pathogenicity of the mutant viruses in chicken embryonated eggs as described previously (1). Fresh infective allantoic fluid was diluted in PBS to give a 10-fold dilution series. For each dilution, 100 μl was inoculated into the allantoic cavity of each of five 9-day-old embryonated SPF chicken eggs. The eggs were incubated at 37°C and examined three times daily for 7 days. The MDT was calculated as the mean time in hours for the minimum lethal dose to kill the embryos.
To examine the pathogenicity of mutant viruses in vivo, the intracerebral pathogenicity index (ICPI) and the intravenous pathogenicity index (IVPI) tests were performed as described elsewhere, with modifications (1). For ICPI, 103 PFU of each virus/bird was inoculated into groups of 10 1-day-old SPF chicks, via the intracerebral route. The inoculation was performed using a 27-gauge needle attached to a 1-ml stepper syringe dispenser that was set to dispense 0.05 ml of inoculum per inoculation. The birds were inoculated by inserting the needle to the hub into the right or left rear quadrant of the cranium. For IVPI, 103 PFU of each virus/chicken was inoculated intravenously into groups of 10 6-week-old SPF chickens. The dispenser was set to dispense 0.1 ml of inoculum per inoculation to each nostril. In both studies, the birds were observed for clinical symptoms and mortality at 12-h intervals for a period of 8 days for the ICPI test and 10 days for the IVPI test. Each experiment had mock-inoculated controls that received a similar volume of sterile PBS by the respective route. The ICPI and IVPI values were calculated as described by Alexander (1).
To study the virus growth kinetics and tissue distribution in vivo, groups of 3-week-old SPF chickens were inoculated with 103 PFU of each virus/bird by the intranasal route. Tissues, such as lung, trachea, spleen, and brain, were collected from inoculated chickens on days 1, 3, 5, 7, and 10 postinfection and snap-frozen immediately. Blood was also collected from these birds on the above-indicated days and assayed on the same day for viremia. The tissues were homogenized for determination of virus content by plaque assay on DF1 cells. All animal experiments were performed in our U.S. Department of Agriculture-approved biosafety level 3 animal facility.
RESULTS
Generation and characterization of mutant NDVs from cDNA.
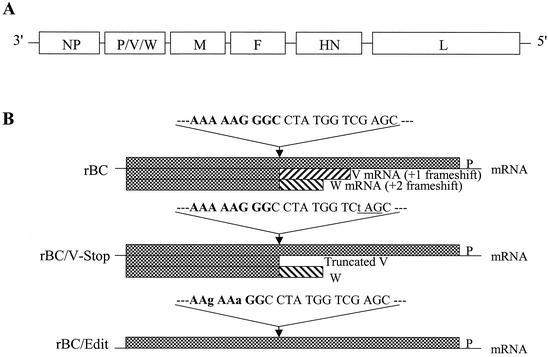
We have previously reported the recovery of recombinant NDV, rBC, from an infectious cDNA clone (pNDVfl) derived from virulent NDV strain Beaudette C (24). For the purpose of this study, we generated two P gene-editing mutant viruses, rBC/V-Stop and rBC/Edit. The recovered viruses were purified from infective allantoic fluids, and viral genomic RNA was extracted using Trizol (Invitrogen). RT-PCR was performed to amplify the fragment spanning the conserved P gene RNA editing region. Nucleotide sequencing of the RT-PCR products confirmed that the introduced mutations were retained in the recovered viruses. rBC/Edit virus contained a P gene whose editing site had been disrupted, and rBC/V-Stop virus contained a P gene in which a stop codon had been introduced after the editing site (Fig. 1).
FIG. 1.
Construction and generation of the recombinant NDV mutants. (A) Schematic diagram of the genomic organization of the parental rBC NDV. NDV contains six genes. All genes are monocistronic except the P gene, which allows for potential expression of three proteins, P, V, and W, by RNA editing. (B) Nucleotide sequences around the editing site and patterns of P gene expression. Sequences are shown in positive sense, and mutated nucleotides are in lower case. Expression of the carboxyl-terminus-truncated V protein was obtained by introducing a stop codon TAG (underlined) in the V frame, which was silent to the P frame. Disruption of the P gene mRNA editing by introducing two silent nucleotide mutations (A to G and G to A) resulted in the inhibition of both V and W protein expression.
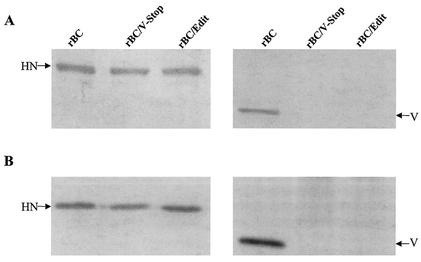
Since V protein is a structural component of NDV, we used Western blot analysis of purified viruses to assess the levels of the V protein expression. Equal amounts of purified virions were loaded onto the gel in duplicate to compare the amount of V protein incorporated into the virions between parental and mutant viruses. Our results showed that only the parental virus reacted with anti-V peptide serum, whereas the mutant viruses did not show the presence of any protein on the blot (Fig. 2A). The rBC, rBC/V-Stop, and rBC/Edit viruses reacted with equal sensitivity and intensity to a cocktail of monoclonal antibodies to the HN protein, indicating that comparable amounts of viral proteins were subjected to Western blot analysis. In order to differentiate V protein synthesis and packaging, lysates prepared from infected Vero cells at the time of maximum CPE were subjected to Western blot analysis. The results showed that the mutant viruses did not express detectable amounts of V protein in infected cells (Fig. 2B).
FIG. 2.
Western blot analysis performed with either purified viruses (A) or infected Vero cell extracts (B). Viral proteins were resolved in an SDS-12% polyacrylamide gel and transferred to polyvinylidene difluoride membranes in duplicate. The membranes were incubated with a cocktail of monoclonal HN antibodies or antipeptide serum specific for the carboxyl-terminal 18 amino acids of the V protein. The membrane was subsequently incubated with horseradish peroxidase-conjugated goat anti-mouse antibody or goat anti-rabbit IgG antibody, respectively. Reactive proteins were visualized in a visualization buffer.
Growth of the mutant viruses in cultured cells.
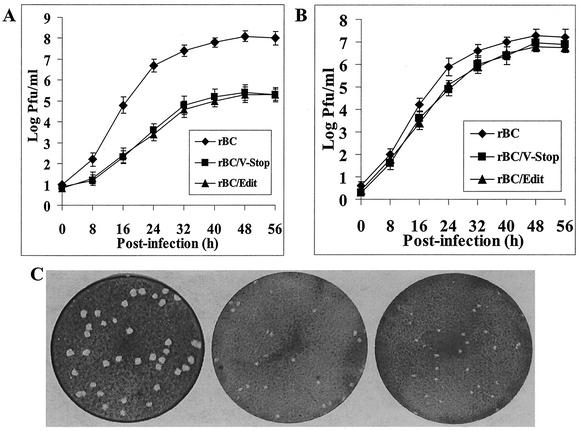
The growth characteristics of rBC, rBC/V-Stop, and rBC/Edit viruses were compared under multistep growth conditions using the chicken embryo fibroblast-derived DF1 cell line or the monkey kidney-derived Vero cell line. In DF1 cells, we found the growth rate of viruses lacking the V protein (rBC/V-Stop or rBC/Edit) was significantly lower than that of the parental virus (Fig. 3A). After 2 days of infection, rBC virus grew to titers of 8 × 107 PFU/ml, whereas the titers of rBC/V-Stop and rBC/Edit reached only up to 8 × 105 PFU/ml (Fig. 3A). The plaques produced on DF1 cells by the mutant viruses were much smaller (1 to 2 mm in diameter) with fuzzy edges, whereas the plaques produced by rBC were bigger (3 to 5 mm in diameter) with sharp edges (Fig. 3C). In contrast, in Vero cells, a cell line that lacks an intact IFN-α/β system (9), the growth rates of the mutant viruses were comparable to that of rBC virus (Fig. 3B). Both the mutant and parental viruses reached similar titers of 107 PFU/ml after 2 days of infection in Vero cells. The growth rates of mutant and parental viruses were further examined in primary chicken embryo fibroblast cells and in BHK21 cells. In these cells, the production of the mutant viruses relative to the parental virus was also delayed and achieved a maximum titer which was more than 100-fold lower than that of the parental virus (data not shown). These results suggested that the absence of the V protein is deleterious to virus replication in cells harboring an intact IFN system.
FIG. 3.
Growth kinetics and plaque formation in tissue culture. Graphs show multiple-step growth curves of the parental rBC and mutant viruses on DF1 (A) or Vero (B) cells. Values are from two independent experiments, each performed in triplicate. Bars show standard deviations. (C) Plaque morphology on DF1 cells by the parental rBC and mutant viruses at 3 days postinfection.
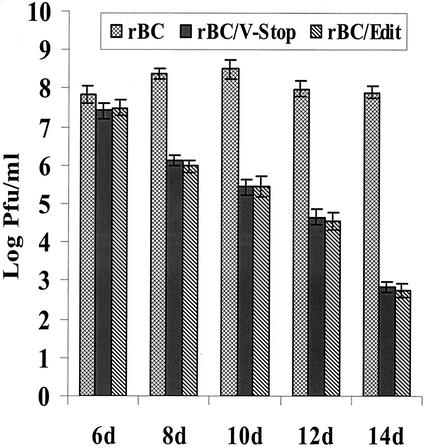
Virus growth in embryonated SPF chicken eggs of different gestational ages.
The growth characteristics of rBC, rBC/V-Stop, and rBC/Edit viruses were examined in SPF chicken eggs of different gestational ages. Briefly, embryonated chicken eggs ranging in gestational age from 6 to 14 days were inoculated by the allantoic route at a dose of 103 PFU/egg, with five eggs for each gestational age for each virus. Sixty hours postinoculation, the allantoic fluids were harvested, clarified, and titrated on DF1 cells. Our results showed that the parental virus was able to grow to high titers in embryos of all gestational ages, whereas the titers of the mutant viruses decreased with the gestational age of the embryos (Fig. 4). The yields of the mutant viruses were comparable to that of rBC in 6-day-old eggs but were approximately 105-fold lower in 14-day-old eggs, indicating an age-dependent resistance of chicken embryos to mutant viruses.
FIG. 4.
Growth properties of rBC, rBC/V-Stop, and rBC/Edit viruses in SPF chicken embryonated eggs. Aliquots of 2 × 103 PFU of parental rBC or mutant viruses in a volume of 100 μl were injected into the allantoic cavity of different gestational ages of embryonated eggs. Sixty hours after incubation at 37°C, allantoic fluids were harvested for virus titration on DF1 cells by plaque assay. Titers were determined from three independent experiments, each with pooled allantoic fluids from six inoculated eggs, with bars indicating standard deviations.
Viruses with an editing-site mutation are more sensitive to exogenous IFN-α/β.
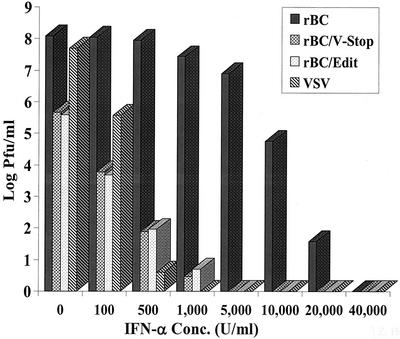
The mutant viruses were severely impaired in replication in IFN-competent cells and in chicken embryos with a more developed innate immune system. These results indicated that the altered growth characteristics of the mutant viruses might be, in part, due to an increased sensitivity to the antiviral effects of IFN-α/β. To examine this possibility, DF1 cells were pretreated with increasing amounts of recombinant chIFN-α to stimulate the host cell antiviral pathways. After 24 h of IFN priming, cells were infected with rBC, rBC/V-Stop, or rBC/Edit virus at an MOI of 0.01. VSV, a known IFN-sensitive virus, was used as the positive control. At 48 h postinfection, supernatants were harvested and virus yields were determined by plaque assay.
Addition of chIFN-α significantly inhibited yields of rBC/V-Stop and rBC/Edit viruses. Both the mutant viruses showed a dose-dependent sensitivity to chIFN-α, with only 100 U of chIFN-α/ml resulting in a 104-fold reduction in virus yield and 500 U of chIFN-α/ml resulting in a 106-fold reduction in virus yield (Fig. 5). No virus replication was detected when the cells were pretreated with chIFN-α at a concentration of 5,000 U/ml. In contrast, the parental virus, rBC, was relatively unaffected by similar doses of chIFN-α, with 5,000 U of chIFN-α/ml inhibiting virus replication to a titer approximately 10-fold lower than that of the untreated control sample. As expected, addition of chIFN-α significantly inhibited the yield of VSV. These results suggest that both the mutant viruses are more sensitive than the parental virus to the antiviral effects of exogenous chIFN-α.
FIG. 5.
Viruses with editing-site mutation are more sensitive to IFN-α. DF1 cells were pretreated with an increasing concentration of chIFN-α for 24 h and were infected with rBC, rBC/V-Stop, or rBC/Edit at an MOI of 0.01. VSV was included as a control. Forty-eight hours postinfection, supernatant was harvested and used for virus titration. Results are mean values of three independent experiments, and bars show standard deviations.
The carboxyl terminus of the V protein enhanced the growth of the mutant viruses in vitro.
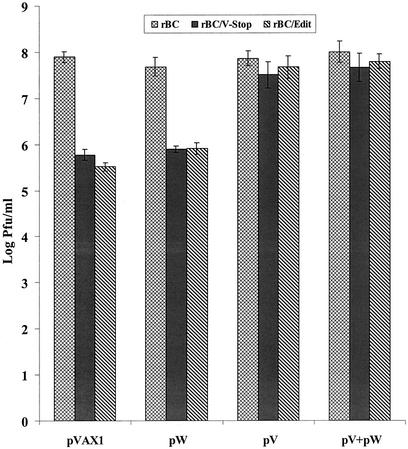
Both the mutant viruses showed the impaired growth properties in cell cultures except in Vero cells, suggesting that the impaired growth resulted from the elimination of the carboxyl-terminal portion of the V protein. Therefore, we performed a complementation assay to examine whether the transient expression of the carboxyl-terminus portion of the V and/or W protein would enhance the growth of the mutant viruses in cell culture. DF1 cells were transfected with the expression plasmids pV and pW, either alone or in combination. The vector backbone without insert was used as a control. The cells were infected with the respective viruses 24 h after transfection. Supernatants were harvested for plaque assay to determine the virus titer after 48 h of infection. The results showed that the expression of the carboxyl-terminal portion of the V protein alone was found to enhance the growth of the mutant viruses, whereas the expression of plasmid pW had no effect on virus growth. The expression of both pV and pW had a similar effect as that of pV alone (Fig. 6). The complementation assay clearly demonstrated that the carboxyl terminus of the V protein enhanced the growth of the mutant viruses in vitro and that the W protein probably plays very little or no role in NDV growth.
FIG. 6.
Transient expression of the carboxyl-terminal portion of the NDV V protein enhanced the growth of editing-mutant viruses. DF1 cells were transfected with expression plasmid and 3 μg of pV, 1 μg of pW, or pV and pW combined. Blank plasmid pVAX1 was used as a negative control. Twenty-four hours posttransfection, the cells were infected with rBC, rBC/V-Stop, or rBC/Edit at an MOI of 0.01. Supernatant was collected 48 h after infection for assessment of the virus titers by plaque assay. Titers reflect two independent experiments, with bars indicating standard deviations.
Infection with the parental but not with the mutant viruses and expression of the C terminus of the NDV V protein preferentially degraded STAT1.
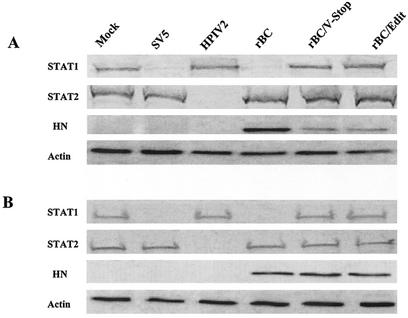
It has been demonstrated that the V protein of SV5 and mumps virus blocks IFN signaling by degrading the STAT1 protein, whereas the V protein of hPIV2 degrades STAT2 rather than STAT1 (3, 36). NDV grew to high titers and induced typical CPE in human 2fTGH cells. Furthermore, the growth of mutant viruses in this cell line was severely impaired compared to that of parental virus (data not shown). Thus, it was of interest to determine the effect of NDV on the levels of STAT1 and STAT2 in 2fTGH cells. Briefly, cells were infected with parental and mutant viruses at a high MOI. Cell lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with either STAT1- or STAT2-specific antiserum. Lysates prepared from 2fTGH cells infected with SV5 and hPIV2 were included as controls to show degradation of STAT1 and STAT2, respectively. Our results demonstrated that NDV infection resulted in preferential degradation of STAT1 protein, but not STAT2 protein (Fig. 7A). Both the mutant viruses, which lacked expression of the V protein, failed to degrade the STAT1 protein.
FIG. 7.
NDV infection preferentially degraded STAT1 protein. 2fTGH cells (A) or Vero cells (B) were mock infected or infected with rBC, rBC/V-stop, rBC/Edit, SV5, or hPIV2. At 20 h postinfection, whole-cell extracts were subjected to electrophoresis and probed for STAT1 and STAT2 as well as viral protein HN by immunoblotting. Actin was used as a loading control.
The mutant viruses grew to a lower level in 2fTGH cells and expressed less viral proteins than the parental virus (Fig. 7A). However, both parental and mutant viruses grew to comparable levels in Vero cells (Fig. 3B). In order to rule out the possibility that other viral proteins might mediate the STAT1 degradation, we compared the degradation pattern of both STAT1 and STAT2 in Vero cells following virus infection. The results were consistent with those obtained with 2fTGH cells, suggestive of the role of the NDV V protein in degrading STAT1 protein (Fig. 7B). The Vero cells are known to be defective in IFN synthesis; therefore, our data here indicate that the C terminus of the NDV V protein functions as an IFN-α antagonist in part by degrading the STAT1 protein.
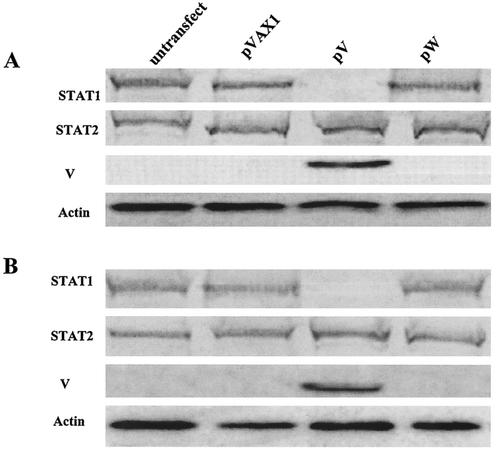
To test whether the expression of the V protein directly degrades the STAT1 protein, we carried out transfection experiments. Transfection of the pW and empty expression plasmid pVAX1 did not result in a decrease in either the STAT1 or STAT2 protein, whereas the expression of the C terminus of the NDV V protein by pV degraded the STAT1 protein in 2fTGH and Vero cells (Fig. 8). These results clearly showed that the C terminus of the NDV V protein suppressed IFN signaling by lowering the concentration of the STAT1 protein.
FIG. 8.
Expression of the C terminus of the NDV V protein degraded STAT1 protein. 2fTGH cells (A) or Vero cells (B) were mock transfected or transfected with 3 μg of empty plasmid pVAX1, pV, or pW. Cells were lysed 20 h posttransfection. Lysates were separated by SDS-PAGE, and polypeptides were transferred to a nitrocellulose membrane and immunoblotted using antibodies specific for V, STAT1, or STAT2 proteins. Actin was used as a loading control.
Pathogenicity studies.
NDV isolates show a continuous spectrum of virulence for chickens. The degree of virulence of a given NDV isolate can be assessed by the following in vivo tests: the MDT test in embryonated SPF chicken eggs, the ICPI test in 1-day-old chicks, and the IVPI test in 6-week-old chickens. We examined the virulence of the parental and mutant viruses by all these tests (Table 1). The MDT results showed that it took a much longer time for the rBC/V-Stop and rBC/Edit viruses to cause embryo mortality. The MDT values for rBC/V-Stop and rBC/Edit were 96 and 98 h, respectively, which were higher than that of the parental rBC (62 h). The MDT values of the mutant viruses were typical of lentogenic isolates (>90 h), and the MDT of the parental virus was typical of a mesogenic isolate (60 to 90 h).
TABLE 1.
Pathogenesis study of mutant viruses in embryos and chicks
MDT is the mean time in hours for the minimum lethal dose to kill the embryos when inoculated into the allantoic cavity of 9-day-old embryonated eggs (1).
ICPI was determined as described by Alexander (1) on 1-day-old chicks.
IVPI was determined as described by Alexander (1) on 6-week-old chickens.
The ICPI values of both mutant viruses were much lower compared to the parental virus (Table 1). The ICPIs of rBC/V-Stop and rBC/Edit viruses were 0.72 and 0.68, respectively, which is between lentogenic (0.2 to 0.5) and mesogenic (1.0 to 1.5). The ICPI value of rBC virus was 1.58 (out of a maximum of 2.0), falling between mesogenic (1.0 to 1.5) and velogenic (1.5 to 2.0).
The results obtained by IVPI tests in 6-week-old chickens for the mutant viruses were quite surprising (Table 1). The rBC virus had an IVPI value of 1.45 (out of a maximum of 3.0), which was higher than classical mesogenic (0.0 to 0.5) and lower than velogenic (2.0 to 3.0). But the IVPI values of both mutant viruses were 0.0, indicating complete attenuation in older chickens.
To study the pathogenesis of the mutant viruses, 3-week-old chickens were infected intranasally with 103 PFU of each virus. The virus distribution in blood, lung, spleen, and brain on days 1, 3, 5, 7, and 10 after infection was examined by plaque assay. The rBC virus first appeared by day 3 in the blood, lung, spleen, and brain and reached highest titers in the lung and spleen at day 5 and in the brain at day 7. The rBC virus was undetectable in the blood by day 5. Surprisingly, the mutant viruses could not be detected in any of these organs or blood. These results demonstrated that NDVs lacking the V protein are highly debilitated in growth in chickens and the attenuation of virulence may be due to the inability of the mutant viruses to counteract the host innate immune system.
DISCUSSION
In this report, we have used the reverse genetics approach to provide experimental evidence that the V protein of NDV functions as a virulence factor in its natural host, chickens. Furthermore, our results demonstrated that the V protein, specifically the cysteine-rich carboxyl terminus, functions as an IFN-α antagonist by selectively targeting the STAT1 protein for degradation.
In a previous study, it was shown that a recombinant NDV lacking V protein expression was highly impaired in growth in tissue culture and was unable to replicate in 10-day-old embryonated chicken eggs (32). In that study, an avirulent NDV strain was used as the parent for all the mutants. Since avirulent NDV strains do not cause an apparent disease in chickens, the actual role of the V protein in virulence could not be determined in chickens. In our study, a virulent NDV strain was used as the parent for all the mutants, thereby making it possible to infer the role of the V protein in pathogenesis in chickens.
The mutant rBC/Edit was constructed to determine the role of both the V and W proteins, whereas the mutant rBC/V-stop was constructed to distinguish the role played by the V protein from that of the W protein. Both the mutant viruses showed severe growth impairment in DF1 and BHK21 cells and grew to titers which were as much as 1,000-fold lower than that of the parental virus. Interestingly, the mutants and the parental viruses grew to similar titers in Vero cells, which lack an intact IFN system (10). These results suggested that the mutant viruses were probably unable to counteract the host-specific IFN responses. Differences in growth between the mutant and parental viruses were also observed in embryonated eggs. Both parental and mutant viruses grew to identical titers in 6-day-old embryonated chicken eggs, but not in 10-day-old embryonated chicken eggs. The mutant viruses grew to titers more than 1,000-fold lower in 10-day-old embryos than the parental virus did. This age-dependent growth phenomenon was also reported with a recombinant influenza A virus which lacks the IFN-antagonist NS1 gene (12, 50). We speculate that this reduction in growth of the mutant viruses in older embryonated eggs is due to maturation of the host innate immune system and, probably, the inability of the mutant viruses to counteract the IFN response, which is more developed in older embryos (47). The difference in the ability to counteract host-specific IFN responses between parental and mutant viruses was much more evident in IFN-treated cells. In DF1 cells, the parental virus was protected against high doses of exogenous chIFN-α, whereas the mutant viruses were severely attenuated and highly sensitive to low doses of exogenously added chIFN. Our experiment did not test the sensitivity to IFN signaling, because the viruses were applied after the IFN treatment. However, our results clearly showed that the V protein of NDV is able to mediate virus escape from the IFN-α-induced cellular antiviral mechanism.
The carboxyl-terminal domain of the V protein is conserved among all the paramyxoviruses except hPIV1 and hPIV3, which do not express V protein (11, 33). In common with its counterparts in other paramyxoviruses, the V protein of NDV possesses a carboxyl-terminal domain that is cysteine rich and binds to zinc (48, 49). The high level of amino acid sequence conservation suggests that the region of the V protein plays a critical role in paramyxovirus replication and pathogenesis. In this study, we have shown that the IFN-antagonist activity of the NDV V protein is located in the carboxyl-terminal domain. This result is similar to that of the mumps virus and hPIV2, where the carboxyl-terminal domain of the V protein was sufficient for IFN-antagonist activity (25, 35). For SV5, a single amino acid substitution (N to D) in the N terminus of the V protein was responsible for the difference in its ability to block IFN signaling in human and murine cells, which may explain, in part, the host constraints that prevented the virus from crossing species barriers (54). Moreover, two canine isolates of SV5 (CPI+ and CPI−) were isolated. While CPI+ targeted STAT1 for degradation and thus blocked IFN signaling, CPI− failed to do so (7, 53). Three amino acid differences in the P/V N coterminus of the V protein were responsible for the difference in the ability to block IFN signaling. However, expression of the SV5 P protein, which contains the N coterminal region did not inhibit activation of the IFN-α/β-responsive promoter, indicating that the unique cysteine-rich V domain is essential for targeting STAT1 for proteasome-mediated degradation (54). These data also indicated that the N-terminal domain of SV5 V protein could modulate the ability to target STAT1 for degradation (7, 53, 54). Whether the N coterminus contains a functional domain indispensable for blocking IFN signaling or only a domain modulating this function remains to be elucidated. Recently, it was demonstrated that Nipah virus V protein escapes IFN by a distinct mechanism involving direct inhibition of STAT protein function (45). Nipah virus V protein does not induce STAT degradation but instead efficiently prevents STAT1 and STAT2 activation and nuclear translocation in response to IFN, thus inhibiting cellular responses to both IFN-α and IFN-γ (45). It was identified that the N-terminal but not the C-terminal domain of the Nipah virus V protein was responsible for the IFN-antagonist activity (38). However, our data clearly demonstrated that the C terminus of the NDV V protein is sufficient to degrade the STAT1 protein. Our results are in agreement with the recent data obtained from a novel NDV-green fluorescent protein-based assay that showed that the cysteine-rich C-terminal domain of the NDV V protein was involved in IFN-antagonist activity (38).
Recently, the IFN-antagonist mechanisms used by some paramyxoviruses have been determined (3, 9, 36, 38). For example, SV5 was found to evade host IFN response by specifically targeting the STAT1 protein for proteolytic degradation. The degradation of STAT1 was mediated by the virus-encoded V protein (9). Similarly, the V protein of hPIV2 was found to block IFN signaling by preferentially inducing degradation of STAT2, but not STAT1 (36). However, both STAT1 and STAT2 were strictly required in the host cells to establish a degradation-permissive environment enabling both SV5 and hPIV2 to target the respective STAT protein (37). In this report, we showed that NDV blocks IFN signaling by specifically targeting the STAT1 protein. Clearly, there are many potential advantages for NDV to target STAT1, since STAT1 degradation can block both IFN-α/β and IFN-γ signaling. Our available evidence with NDV mutant viruses and from the transfection experiments indicates that the C terminus of the NDV V protein targets STAT1 for degradation and thus blocks IFN signaling. At present, the molecular mechanism of how the V protein of NDV degrades STAT1 remains to be elucidated.
In additional to blocking IFN signaling, paramyxoviruses have also evolved mechanisms to circumvent the IFN antiviral effects by inhibiting IFN synthesis. Studies with Sendai virus have shown that different strains of the virus can induce highly variable amounts of IFN-α/β (29). For MV, it was reported that laboratory-passaged strains and clinically isolated strains exhibited different capabilities of inducing IFN synthesis, though a gene responsible for the difference has not yet been identified (34). It was reported that SV5 infection inhibited IFN-β production, whereas recombinant SV5 lacking the V protein carboxyl-terminal domain (rSV5VΔC) permitted production of IFN-β (17, 43). Analysis of two naturally occurring related strains of SV5 revealed that these strains possessed different abilities to induce IFN-α/β synthesis and that substitution in the N-terminal region of the V protein was responsible for the observed difference (7, 53). For NDV, it was documented that different strains have different abilities to induce IFN production and to counteract the IFN effect (28). Whether the parental and mutant viruses we described here possess different abilities to induce IFN synthesis remains to be elucidated.
Our data clearly demonstrated that the NDV-editing mutants were highly attenuated in vivo. The first indication that the mutant viruses were attenuated in vivo came from the size of the plaques produced by the mutant viruses on cell culture (Fig. 3C). It has long been shown that the ability of NDV strains to produce large plaques is related to virulence for chickens (44). Results from in vivo pathogenicity tests (MDT, ICPI, and IVPI) provided convincing evidence that the mutants were highly attenuated in their natural host, the chicken (Table 1). The pathogenicity of the mutants was decreased to the level of lentogenic viruses by MDT test in embryonated chicken eggs and by ICPI test in 1-day-old chicks. Mutant viruses were recovered from the allantoic fluids of infected eggs and from the brains of infected 1-day-old chicks, indicating virus replication in vivo. However, the mutants were completely attenuated in an IPVI test in 6-week-old chickens. Moreover, intranasal infection of 3-week-old chickens with the mutant viruses showed that the mutants were avirulent and that the viruses could not be recovered from blood, trachea, lung, spleen, or brain, indicating rapid clearance by the host immune system. These results suggested that the mutant viruses were unable to antagonize the innate immune responses in older chickens when administered by the natural route. Our work is in agreement with studies of MV (40, 51, 52) and Sendai virus (19, 21) pathogenesis, whereby deletion of the V protein attenuates viral pathogenesis. Our results from the rBC/V-Stop mutant, which presumably encodes only the amino-terminal domain but not the V-specific carboxyl-terminal domain of the V protein, indicated that the pathogenicity determinant of the V protein lies in its carboxyl terminus. This hypothesis was supported by our transfection experiments, in which the expression of the C terminus of the NDV V protein degraded STAT1 (Fig. 8). The pathogenicity indices of the rBC/V-Stop and rBC/Edit mutants were similar, suggesting that the W protein of NDV probably plays very little, if any, role in pathogenesis. Considering the very low level of expression of the W protein, and even the absence of expression in some strains (27), this finding is not surprising. However, additional studies will be required to fully assess the role of the W protein in pathogenesis.
We have shown in this study that the V protein is one of the factors which determine the virulence of NDV. Sequence alignment of the V protein shows that differences exist among the NDV strains; therefore, we speculate that the broad range of virulence shown by natural NDV strains could be due to the ability of an altered V protein encoded by these viruses to act as an IFN antagonist. Further studies are needed to identify the amino acids in the V protein that determine the virulence and pathogenesis of NDV. The ability to adjust virulence will have important applications for the rational development of live NDV vaccines that are optimally attenuated and immunogenic.
Acknowledgments
We thank Peter L. Collins for his invaluable suggestions for this work and Peter Savage and Daniel Rockemann for their excellent technical assistance.
This work was partially supported by U.S. Department of Agriculture grant no. 2002-35204-1601.
REFERENCES
- 1.Alexander, D. J. 1989. Newcastle disease, p. 114-120. In H. G. Purchase, L. H. Arp, C. H. Domermuth, and J. E. Pearson (ed.), A laboratory manual for the isolation and identification of avian pathogens, 3rd ed. American Association for Avian Pathologists, Inc., Kennett Square, Pa.
- 2.Alexander, D. J. 1997. Newcastle disease and other avian Paramyxoviridae infections, p. 541-569. In B. W. Calnek (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 3.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V protein of simian virus 5 and human parainfluenza virus type 2, respectively: consequence for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, M. D., and T. Barrett. 2000. Rinderpest viruses lacking the C and V proteins show specific defects in growth and transcription of viral RNAs. J. Virol. 74:2603-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., X. Wang, E. M'uhlberger, V. Volchkov, J. Paragas, H. Klenk, A. García-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 7.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 8.De Leeuw, O., and B. Peeters. 1999. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxoviridae. J. Gen. Virol. 80:131-136. [DOI] [PubMed] [Google Scholar]
- 9.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defective in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 11.Galinski, M. S., R. M. Troy, and A. K. Banerjee. 1992. RNA editing in the phosphoprotein gene of the human parainfluenza virus type 3. Virology 186:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Sastre, A., A. Egorov, D. Matassov, B. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient system. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 13.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signaling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 14.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2001. Paramyxovirus accessory proteins as interferon antagonist. Microbiol. Immunol. 45:787-800. [DOI] [PubMed] [Google Scholar]
- 15.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 16.Hausmann, S., D. Garcin, C. Delenda, and D. Kolakofsky. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, B., R. G. Paterson, N. Stocks, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V P protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-β induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 18.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genomic RNA replication. Virology 222:383-390. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C., K. Kiyotani, Y. Fujii, N. Fukuhara, A. Kato, Y. Nagai, T. Yoshida, and T. Sakaguchi. 2000. Involvement of the zinc-binding capacity of Sendai virus V protein in viral pathogenesis. J. Virol. 74:7834-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan, I. K., B. A. Sutter IV, and M. A. McClure. 2000. Molecular evolution of the Paramyxoviridae and Rhabdoviridae multiple-protein-encoding P gene. Mol. Biol. E. 17:75-86. [DOI] [PubMed] [Google Scholar]
- 21.Kito, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:125-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnamurthy, S., and S. K. Samal. 1998. Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J. Gen. Virol. 79:2419-2424. [DOI] [PubMed] [Google Scholar]
- 24.Krishnamurthy, S., Z. Huang, and S. K. Samal. 2000. Recovery of a virulent strain of Newcastle disease virus from cloned cDNA: expression of a foreign gene results in growth retardation and attenuation. Virology 278:168-182. [DOI] [PubMed] [Google Scholar]
- 25.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal Cys-rich region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 26.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 27.Locke, D. P., H. S. Sellers, J. M. Crawford, S. Schultz-Cherry, D. J. King, R. J. Meineramann, and B. S. Seal. 2000. Newcastle disease virus phosphoprotein gene analysis and transcriptional editing in avian cells. Virus Res. 69:55-68. [DOI] [PubMed] [Google Scholar]
- 28.Lomniczi, B. 1973. Studies on interferon production and interferon sensitivity of different strains of Newcastle disease virus. J. Gen. Virol. 21:305-313. [DOI] [PubMed] [Google Scholar]
- 29.Mattana, P., and G. C. Viscomi. 1998. Variations in the interferon-inducing capacity of Sendai virus subpopulations. J. Interferon Cytokine Res. 18:399-405. [DOI] [PubMed] [Google Scholar]
- 30.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 31.McGinnes, L., C. McQuain, and T. Morrison. 1988. The P protein and the nonstructural 38K and 29K proteins of Newcastle disease virus are derived from the same open reading frame. Virology 164:256-264. [DOI] [PubMed] [Google Scholar]
- 32.Mebatsion, T., S. Verstegen, L. T. C. de Vaan, A. R'omer-Oberd'orfer, and C. C. Schrier. 2001. A recombinant Newcastle disease virus with low-level V protein expression is immunogenic and lacks pathogenicity for chicken embryos. J. Virol. 75:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuoka, Y., J. Curran, T. Pelet, D. Kolakofsky, R. Ray, and R. W. Compans. 1991. The P gene of human parainfluenza virus type 1 encodes P and C proteins but not a cysteine-rich V protein. J. Virol. 65:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naniche, D., A. Yeh, D. K. Eto, M. Manchester, R. M. Friedman, and M. B. A. Oldstone. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferon: cysteine-rich V-specific domains is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 37.Parisien, J.-P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 76:4190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. García-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson, R. G., G. P. Leser, M. A. Haughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virion. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 40.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. A. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 41.Peeters, B. P. H., O. S. de Leeuw, G. Koch, and A. L. Gielkens. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 73:5001-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 43.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-β. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 44.Reeve, P., and G. Poste. 1971. Studies on the cytopathogenicity of Newcastle disease virus: relation between virulence, polykaryocytosis and plaque size. J. Gen. Virol. 11:17-24. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samson, A. C., I. Levesley, and P. H. Russell. 1991. The 36K polypeptide synthesized in Newcastle disease virus-infected cells possesses properties predicted for the hypothesized V protein. J. Gen. Virol. 72:1709-1713. [DOI] [PubMed] [Google Scholar]
- 47.Sekellick, M. J., W. J. Biggers, and P. I. Marcus. 1990. Development of the interferon system. I. In chicken cells development in ovo continues on time in vitro. In Vitro Cell Dev. Biol. 26:997-1003. [DOI] [PubMed] [Google Scholar]
- 48.Steward, M., I. B. Vipond, N. S. Millar, and P. T. Emmerson. 1993. RNA editing in Newcastle disease virus. J. Gen. Virol. 74:2539-2547. [DOI] [PubMed] [Google Scholar]
- 49.Steward, M., A. C. Samson, W. Errington, and P. T. Emmerson. 1995. The Newcastle disease virus V protein binds zinc. Arch. Virol. 140:1321-1328. [DOI] [PubMed] [Google Scholar]
- 50.Talon, J., M. Salvatore, R. E. O'Neil, Y. Nakaya, H. Zheng, T. Muster, A. García-Asatre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. D. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valsamakis, A., H. Schneider, P. G. Auwaerter, H. Kaneshima, M. A. Billeter, and D. E. Griffin. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wansley, E., and G. D. Parks. 2002. Naturally occurring substitutions in the P/V gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. Single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block the interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]