Abstract
Initiation of retrovirus reverse transcription requires the selection of a tRNA primer from the intracellular milieu. To investigate the features of primer selection, a human immunodeficiency virus type 1 (HIV-1) and a murine leukemia virus (MuLV) were created that require yeast tRNAPhe to be supplied in trans for infectivity. Wild-type yeast tRNAPhe expressed in mammalian cells was transported to the cytoplasm and aminoacylated. In contrast, tRNAPhe without the D loop (tRNAPheD−) was retained within the nucleus and did not complement infectivity of either HIV-1 or MuLV; however, infectivity was restored when tRNAPheD− was directly transfected into the cytoplasm of cells. A tRNAPhe mutant (tRNAPheUUA) that did not have the capacity to be aminoacylated was transported to the cytoplasm and did complement infectivity of both HIV-1 and MuLV, albeit at a level less than that with wild-type tRNAPhe. Collectively, our results demonstrate that the tRNA primer captured by HIV-1 and MuLV occurs after nuclear export of tRNA and supports a model in which primer selection for retroviruses is coordinated with tRNA biogenesis at the intracellular site of protein synthesis.
All retroviruses use a host cell tRNA as a primer to initiate reverse transcription at a site in the viral genome designated the primer binding site (PBS) (6, 25). Elongation from the 3′-terminal nucleotides of the tRNA results in the conversion of the viral RNA genome to double-stranded DNA prior to integration into the host DNA (6). While the process of reverse transcription is conserved among all retroviruses, the specific tRNA captured varies for different retroviruses (16, 17). For example, the primer for all known wild-type human immunodeficiency viruses (HIVs) is tRNALys,3 (16, 22), while murine leukemia viruses (MuLVs) use tRNAPro (21) and avian leukosis viruses (ALVs) use tRNATrp (8).
Very little is known about how the tRNA primer is selected by retroviruses. Previous studies have shown that alteration of the PBS of HIV type 1 (HIV-1), MuLV, or ALV to correspond to several different tRNAs allows the virus to transiently use that tRNA for replication, suggesting that retroviruses have evolved a selection process that can access the majority of cellular tRNAs (4, 5, 13, 15, 26, 27). The biogenesis of tRNA is a complex process that requires processing of a tRNA precursor within the nucleus, subsequent transport to the cytoplasm, and incorporation into protein synthesis. The tRNA interacts with no fewer than 30 proteins during biogenesis and translation (28). Indeed, previous studies have shown that tRNAs are rarely free in the cytoplasm and are found most frequently in complexes with cellular proteins (24, 28). A tRNA cycle has been proposed by which the aminoacylated tRNA is channeled into the translational machinery and immediately reacquired by cellular proteins at the E site of the ribosome for reaminoacylation (18, 24).
Understanding how retroviruses access this tRNA cycle to select the primer has been difficult due to the inability to manipulate endogenous levels of the tRNA primer. Recent studies have shown that for both HIV-1 and MuLV it is possible to alter the tRNA specificity by changing the PBS (15, 29-31). In the case of HIV-1, the primer specificity can be altered to allow selection of many different tRNAs, including Saccharomyces cerevisiae tRNAPhe, while for MuLV, the PBS has been altered so that even synthetic tRNAs can be selected and used for replication (15). The fact that the primer preference for HIV-1 or MuLV can be changed by alteration of the PBS suggests that the selection must occur at an intracellular site where the virus has access to a variety of tRNAs. Two such subintracellular locations are the nucleus during tRNA biosynthesis and the cytoplasm during the tRNA translation cycle. In the present study, we have addressed this aspect of retroviral primer selection by constructing tRNA mutants that are defective in different steps of tRNA biogenesis and are localized to the nucleus or cytoplasm. A yeast tRNA with a deletion of the D loop was designed to be retained in the nucleus after transcription (tRNAPheD−). A second mutant, yeast tRNAPheUUA, is defective for aminoacylation but is known to be transported from the nucleus to the cytoplasm (1). Both tRNA mutants were tested for the capacity to rescue HIV-1 or MuLVs that require yeast tRNAPhe for replication. The results of our studies demonstrate for the first time that both HIV-1 and MuLV require the tRNA primer to be transported from the nucleus to the cytoplasm and suggest that the selection of the tRNA for use in reverse transcription occurs at or near the intracellular site of translation.
MATERIALS AND METHODS
Tissue culture.
293T, HeLa H1, EcoPack2-293, and XC cells were maintained in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal calf serum and 1% antibiotic-antimycotic (Gibco/BRL, Gaithersburg, Md.).
Plasmid construction.
Yeast tRNAPhe mutant genes were constructed via PCR extension. Yeast tRNAPhe wild type was created by PCR with oligonucleotides U6Phe5′ oligo (5′-AAACCCTCGAGGTCCGCGGATTTAGCTCAGTTGGGAGAGCGCCAGACTGAAGATCTGGAGG-3′) and U6Phe3′ oligo (5′-CTCCCAAGCTTCCAAAAAATGCGATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCT-3′). The following oligonucleotides were used for PCR extension of the yeast tRNAPheD− mutant: U6PheDLOOP-5′ oligo (5′-AAACCCTCGAGGTCCGCGGATTTAGCGCCAGACTGAAGATCTGGAGG-3′) and U6Phe3′ oligo. For construction of the yeast tRNAPheUUA mutant, the following oligonucleotides were used for PCR: U6PheAC = UUA5′ oligo (5′-CTCCCAAGCTTCCAAAAAATGCGAATTCTGTGGATCGAACACAGGACCTC CAGATCTAAAGTCT-3′) and U6PheAC = UUA3′ oligo (5′-AAACCCTCGAGGTCCGCGGATTTAG CTCAGTTGGGAGAGCGCCAGACTTTAGAT CTGGAGG-3′). PCR extension was also done with oligonucleotides designed for the construction of tRNAPro: U6Pro-For (5′-AAACCCTCGAGGTCCGGCTCGTTGGTCTAGGGGTATGATTCTCGCTTAGGGTGCGAGAGG-3′) and tRNAPro-REV (5′-CTCCCAAGCTTCCAAAAAAGGGCTCGTCCGGGATTTGAACCCGGGACCTCTCGCACCCTAAGC-3′). The PCR products were ligated into the pGEM-T Easy vector (Promega, Madison, Wis.). The resultant plasmids (pTAPheWt, pTAPheD−, pTAPheUUA, and pTAPro) were digested with HindIII and XhoI, generating fragments of approximately 100 bp containing the transcriptional units for the yeast tRNAPhe mutants. These fragments were then cloned into pLS9 that had been digested with HindIII and XhoI, resulting in four plasmids identified as pU6PheWt, pU6PheD−, pU6PheUUA, and pU6Pro (14). The correct sequences of the yeast tRNAPhe mutants and tRNAPro transcriptional units contained in pU6PheWt, pU6PheD−, pU6PheUUA, and pU6Pro were verified by DNA sequencing.
The pMuLV-Phe vector was made via mutagenesis of the PBS in pLEGFP-C1 (Clontech Laboratories, Palo Alto, Calif.) with the GeneEditor in vitro site-directed mutagenesis system (Promega). The mutagenic oligonucleotide used in the system was LEGFPC1mut oligo (5′-GGCAGGGGTCTCCAATCCACAGAATTCGCACCAAATGAAAGACCCC-3′). The resultant plasmid was identified as pMuLV-Phe, and the PBS sequence was verified by DNA sequencing.
In vitro transcription.
For in vitro transcription, the following two oligonucleotides that contain the T7 promoter followed immediately by the full-length yeast tRNAPhe were used: T7PheWT3′ oligo (5′-TGGTGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCTGGCGCTCTCCCAACTGAGCTAAATCCGCTATAGTGAGTCGTATTACTGCAG-3′) and T7PheD−3′ oligo (5′-TGGTGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCTGGCGCTAAATCCGCTATAGTGAGTCGTATTACTGCAG-3′). PCR extension of the three oligonucleotides was completed in separate reactions with T7P-5′ oligo (5′-CTGCAGTAATACGACTCACTATA-3′). The resulting PCR products contained the T7 promoter followed by the sequence for the full-length yeast tRNAPhe mutants (Wt and D−), including CCA nucleotides at the 3′ end. In vitro transcription reactions were carried out by use of the MEGAshortscript T7 kit (Ambion, Austin, Tex.).
Transfection and selection of drug-resistant colonies.
The procedure used for complementation of the defective HIV provirus psHIV-Phe has been previously described (29-31). Complementation of pMuLV-Phe was accomplished bypolyethylenimine (PEI)-mediated cotransfection of pMuLV-Phe and either pU6PheWt, pU6PheD−, or pU6PheUUA into EcoPack2-293 packaging cells (Clontech Laboratories). PEI (Sigma, St. Louis, Mo.) was diluted to 0.045% (wt/vol), and the pH was adjusted to 6.5 with HCl. For transfection of 3 μg of plasmid DNA, 13 μl of the PEI solution was further diluted in 100 μl of serum-free DMEM. The DNA-PEI transfection mixture was then added dropwise to one well of a six-well plate containing 60% confluent EcoPack2-293 cells. Approximately 24 h posttransfection, cells were washed with phosphate-buffered saline and fresh medium was added. At 48 h posttransfection, the supernatants were collected and filtered through a 0.45-μm-pore-size low-affinity protein binding filter (Pall Gelman Laboratory, Ann Arbor, Mich.). Serial dilutions of the virus-containing supernatant were used to transduce XC cells. At 24 h after infection, the cells were washed with phosphate-buffered saline and placed under selection with DMEM-G418 medium (DMEM containing 10% fetal calf serum, 1% antibiotics, and 0.55 mg of active G418 per ml) for 7 days for the formation of drug-resistant colonies. Cell colonies were fixed with 5% trichloroacetic acid (TCA) and stained with 2% Coomassie blue, and the number of drug-resistant colonies was counted.
RNA isolation and analysis.
A Mammalian Transfection kit (Stratagene, San Diego, Calif.) was used to transfect HeLa H1 cells with either pU6PheWt, pU6PheD−, or pU6PheUUA. Tri-Reagent (Sigma) was used for the collection of total cellular RNA from transfected HeLa H1 cells 48 h after transfection. For extraction of cytoplasmic RNA, 107 transfected cells were washed twice with phosphate-buffered saline 48 h after transfection. The cells were then lysed on the plate on ice with 0.4 ml of lysis buffer (0.03% Triton X-100, 0.15 M NaCl, 0.01 M Tris [pH 7.8], 0.0015 M MgCl2) for exactly 1 min. The solution was then collected and centrifuged at 1,000 × g for 3 min at 4°C. The supernatant was removed from the tube, immediately added to 0.2 ml of urea-sodium dodecyl sulfate buffer (7 M urea, 0.35 M NaCl, 0.01 M Tris [pH 7.4], 0.01 M EDTA, 1.0% sodium dodecyl sulfate), and mixed. RNA was then extracted twice with phenol-chloroform. Prior to Northern blot analysis, residual plasmid DNA was removed by digestion with DNA Free (Ambion) according to the manufacturer's instructions. For Northern blot analysis, oligonucleotide probes were designed to be complementary to yeast tRNAPhe, mammalian tRNALys, and U6 snRNA as follows: tRNALys Probe oligo (5′-CGCCCGAACAGGGACTTGAACCCTGGACCCTCAGATTAAAAGTCTGATGCTCTACCGACTGAGCTATCC3′), tRNAPhe Probe oligo (5′-TGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCTGGCGCTCTCCCAACTGAGCTAAATCC3′), and U6 Probe oligo (5′-CGCTTCACGAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAATCTTCTCTGTATCGT-3′). The probes were 5′ end labeled with [γ-32P]ATP by use of Ready-To-Go T4 polynucleotide kinase (Amersham Pharmacia Biotech, Piscataway, N.J.). Free nucleotides were removed by centrifugation through ProbeQuant G-50 Micro columns (Amersham Pharmacia); equal amounts (15 pmol) of probes were used for each blot (106 cpm/pmol). The NortherMax-Gly kit (Ambion) was used for Northern blotting, and BrightStar-Plus (Ambion) was used as the positively charged nylon membrane. Blots were exposed on a phosphor screen and analyzed with a PhosphorImager. Isolation and blotting of aminoacylated tRNAs collected from HeLa H1 cells at 48 h posttransfection with pU6PheWt or pU6PheUUA were carried out as previously described (9).
RESULTS
Design of yeast tRNAPhe mutants.
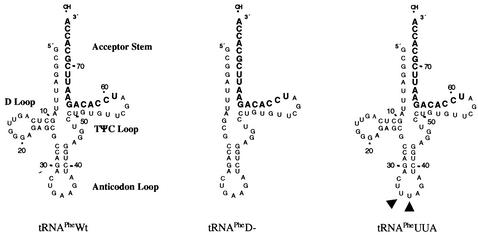
Two yeast tRNAPhe mutants were designed such that they would be defective with respect to different aspects of mammalian tRNA biogenesis. The first mutant, tRNAPheD−, contains a deletion of the D loop such that the tertiary structure of the tRNA is disrupted (Fig. 1). Consequently, tRNAPheD− should be sequestered within the nucleus since intact tertiary structure is required for binding to exportin-t and for subsequent transport from the nucleus to the cytoplasm (1, 7). Another mutant, tRNAPheUUA, contains two nucleotide substitutions within the anticodon (G34→U and A35→U). Nucleotides 34 and 35 represent several of the yeast tRNAPhe identity elements responsible for tRNA recognition by its cognate aminoacyl-tRNA synthetase. The G34→U and A35→U substitutions have been previously reported to render the tRNA unable to be aminoacylated when injected into Xenopus oocytes although the tRNA can be transported from the nucleus to the cytoplasm (1).
FIG. 1.
Engineered yeast tRNAPhe mutants. A schematic of the two engineered yeast tRNAPhe mutants and wild-type yeast tRNAPhe is shown. The yeast tRNAPheD− mutant contains a deletion of nucleotides 11 through 24 relative to wild-type yeast tRNAPhe. The yeast tRNAPheUUA mutant contains two mutations within the anticodon (G34→U and A35→U [closed triangles]) relative to wild-type yeast tRNAPhe.
Wild-type tRNAPhe, tRNAPheD−, and tRNAPheUUA were constructed via PCR extension with oligonucleotides designed to contain at the 3′ terminus thymidine nucleotides necessary for termination of polymerase III (Pol III) transcription. After PCR, the products were cloned into an expression vector immediately downstream of a U6 snRNA Pol III promoter. The three plasmids were designated pU6PheWt, pU6PheD−, and pU6PheUUA. tRNA transcription normally occurs via an intragenic Pol III promoter consisting of an A and B box generally located within the D loop and TΨC stem and loop of the tRNA. The capacity of the U6 snRNA promoter to drive tRNA transcription in the presence or absence of the native intragenic promoter has been previously described (14, 20).
Analysis of yeast tRNAPhe mutant expression and intracellular localization.
To assess the level of tRNAPheD− and tRNAPheUUA expression relative to that of wild-type yeast tRNAPhe within mammalian cells, we transfected HeLa H1 cells with pU6PheWt, pU6PheD−, and pU6PheUUA. Total RNA was collected at 48 h posttransfection and subjected to Northern blot analysis with an oligonucleotide probe designed to be complementary to yeast tRNAPhe (Fig. 2). In preliminary studies, we determined that the oligonucleotide probe recognized both mutant tRNAs as well as wild-type yeast tRNAPhe. Furthermore, we have shown the probe to be specific for yeast tRNAPhe, in part due to 17 nucleotide differences relative to mammalian tRNAPhe (12). Northern blot analysis of the two mutant tRNAs and wild-type yeast tRNAPhe revealed that the three tRNAs were expressed at the same levels when equal amounts of plasmids were transfected.
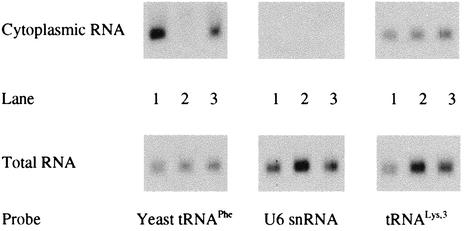
FIG. 2.
Intracellular distribution of yeast tRNAPhe mutants following transfection. Northern blots of 5 μg of cytoplasmic RNA (top) or total RNA (bottom) collected from HeLa H1 cells after transfection with pU6PheWt, pU6PheD−, or pU6PheUUA are shown. Lanes 1, RNA from HeLa cells transfected with pU6PheWt; lanes 2, RNA from HeLa cells transfected with pU6PheD−; lanes 3, RNA from HeLa cells transfected with pU6PheUUA. The Northern blots were probed for yeast tRNAPhe and mammalian tRNALys as depicted; the U6 snRNA probe served as a control to ensure that no nuclear RNA was present in the samples.
We designed tRNAPheD− so that it would be sequestered within the nucleus due to its inability to interact with exportin-t for subsequent transport to the cytoplasm. To confirm that tRNAPheD− was deficient with regard to transport, we collected cytoplasmic RNA from HeLa H1 cells transfected with pU6PheWt, pU6PheD−, and pU6PheUUA. Northern blot analysis of the cytoplasmic RNA revealed that tRNAPheD− was not present in the cytoplasm and was sequestered in the nucleus. As expected, both tRNAPheWt and tRNAPheUUA were present in the cytoplasm in nearly equal amounts. An oligonucleotide probe complementary to the U6 snRNA was used to ensure that the cytoplasmic fraction was not contaminated with nuclear RNA. Analysis with an oligonucleotide probe specific for tRNALys,3 confirmed that equal amounts of RNA were used for Northern blots of the three samples.
Analysis of mutant yeast tRNAPhe aminoacylation.
In a previous study we found that the overwhelming majority of the cytoplasmic yeast tRNAPhe detected following transfection of the cDNA was aminoacylated, similar to other mammalian tRNAs (12). The fact that wild-type yeast tRNAPhe is aminoacylated when expressed in mammalian cells suggests that the tRNA accesses the normal tRNA biogenesis pathways. The yeast tRNAPheUUA mutant was not aminoacylated following injection into the nuclei of Xenopus oocytes (1). To determine if tRNAPheUUA is capable of aminoacylation within mammalian cells, HeLa H1 cells were transfected with either pU6PheWt or pU6PheUUA and RNA was collected under acidic conditions at 48 h posttransfection. The RNA samples were then analyzed by Northern blotting with probes specific for yeast tRNAPhe and tRNALys (Fig. 3). Similar to the results previously obtained in Xenopus oocytes (1), we found that tRNAPheUUA in mammalian cells was not aminoacylated. The tRNALys within the same sample was shown to be aminoacylated, ensuring that the result was not due to deacylation at some point during RNA isolation or Northern blotting.
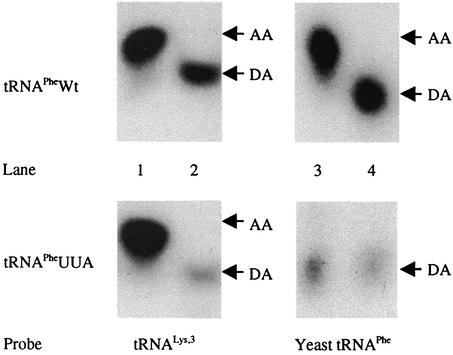
FIG. 3.
Aminoacylation status of yeast tRNAPhe expressed in mammalian cells. Northern blot analyses of cytoplasmic yeast tRNAPhe and tRNALys from cells transfected with either pU6PheWt or pU6PheUUA are shown. All lanes contain 5 μg of cytoplasmic RNA isolated under acidic conditions; the RNAs loaded in lanes 2 and 4 were adjusted to pH 9.0 and incubated at 37°C for 1 h to serve as the deacylated controls. The migration of aminoacylated tRNA and deacylated control tRNA are denoted AA and DA, respectively. Lanes 1 and 2 were probed for tRNALys, and lanes 3 and 4 were probed for yeast tRNAPhe. Phosphorimager analysis of the aminoacylated versus deacylated tRNA in lanes 1 and 3 revealed a ratio of greater than 99 to 1, indicating that the vast majority of the yeast tRNAPhe and tRNALys isolated from the cells was aminoacylated. However, for RNA collected from cells transfected with pU6PheUUA, no aminoacylation of tRNAPheUUA was detected, while the majority of mammalian tRNALys from the same sample was aminoacylated. Note that the blots were not performed simultaneously and autorad exposure times varied.
Complementation of HIV replication with mutant yeast tRNAPhe.
In previous studies, the construction and characterization of an HIV proviral genome in which the PBS was altered to correspond to the 18 3′-terminal nucleotides of yeast tRNAPhe were described (29-31). This defective proviral genome, designated psHIV-Phe, has an expression cassette consisting of the simian virus 40 early promoter and a gene encoding xanthine-guanosine phosphoribosyl transferase (gpt) in place of a deleted env gene. Pseudoviruses were derived from cotransfection of psHIV-Phe and pLGRNL, which encodes the vesicular stomatitis virus G protein.
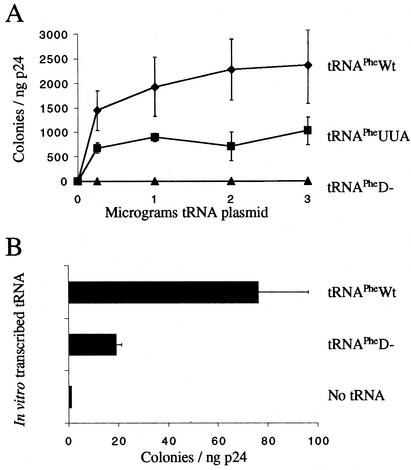
In our previous studies, we found that cotransfection of psHIV-Phe, pLGRNL, and in vitro-generated yeast tRNAPhe or a plasmid carrying the gene for yeast tRNAPhe resulted in infectious virus, as judged by the capacity to convert HeLa cells to carry resistance to mycophenolic acid (29-31). The number of CFU of infectious viruses generated following cotransfection was substantially higher than background levels seen in the absence of yeast tRNAPhe. To delineate the initial interaction between the tRNA and the HIV-1 primer selection process, we tested the mutants defective in different steps in tRNA biogenesis for the capacity to rescue psHIV-Phe. In the case of tRNAPheD−, we found that the mutant tRNA was not selected for use in reverse transcription when endogenously expressed from cDNA (Fig. 4A). In contrast, we show here, as previously reported, that when tRNAPheD− was transcribed in vitro and transfected directly into the cytoplasm, it was used by HIV, albeit at lower levels than those when in vitro-transcribed wild-type tRNAPhe was used for complementation (30) (Fig. 4B). For tRNAPheUUA, we observed that when expressed in vivo, the mutant was able to complement psHIV-Phe. However, the number of infectious viruses produced was approximately 50% of that seen when wild-type tRNAPhe was used to rescue psHIV-Phe.
FIG. 4.
Complementation of psHIV-Phe by either in vitro-transcribed or intracellularly expressed yeast tRNAPhe mutants or in vitro-transcribed tRNAPhe. (A) Numbers of CFU from infection of psHIV-Phe pseudoviruses complemented by either yeast tRNAPheWt (diamonds), tRNAPheUUA (squares), or tRNAPheD− (triangles) expressed from designated quantities of cotransfected pU6PheWt, pU6PheUUA, or pU6PheD−, respectively. The represented data are means ± standard deviations from three independent transfections. (B) Two micrograms of the indicated in vitro-transcribed yeast tRNAPhe mutants was added to cotransfections to complement the psHIV-Phe virus; previous studies have shown this to be the optimal amount for complementation (29). The data are means ± standard deviations from three independent transfections.
Intracellular location of MuLV primer tRNA selection.
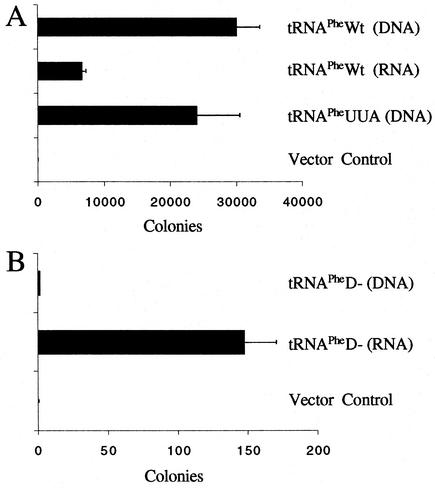
Previous studies have shown that replacement in the MuLV PBS of tRNAPro with alternative tRNAs results in the use of the alternative tRNA as a primer for replication during a single round of infection (15). Given that MuLV constitutively uses tRNAPro, while HIV uses tRNALys,3, we wanted to determine if these different retroviruses shared common characteristics for the intracellular locale from which the tRNA primer is selected. We constructed an MuLV vector, identified as pMuLV-Phe, in which the PBS was mutated to be complementary to yeast tRNAPhe. Cotransfection of the proviral genome and tRNA mutant plasmids into EcoPack2-293 cells resulted in the production of viruses that, upon infection of XC cells, conferred resistance to G418. We obtained the greatest number of infectious virus following cotransfection with the cDNA encoding tRNAPhe (Fig. 5A). Transfection of in vitro-transcribed tRNAPheWt also rescued virus infectivity, although the level of complementation was considerably lower than that found from transfection of the cDNA. The tRNAPheUUA mutant derived from cDNA was also able to rescue virus, albeit at a level lower than that for wild-type tRNAPhe.
FIG. 5.
Complementation of MuLV with yeast tRNAPhe mutants. (A) Numbers of CFU (drug-resistant colonies) derived from infection of pMuLV-Phe complemented by yeast tRNAPheWt or yeast tRNAPheUUA expressed from 1.5 μg of cotransfected tRNAPhe mutant plasmids (tRNAPhe DNA) or in vitro-transcribed tRNAPhe (tRNAPhe RNA). A plasmid expressing tRNAPro from the U6 promoter was used for complementing MuLV-Phe as a negative control. The presented data were collected from three independent complementation experiments, and standard deviations are provided. (B) Complementation with tRNAPheD−. The plasmid expressing tRNAPheD− or in vitro-transcribed tRNAPheD− was cotransfected with pMuLV-Phe. The number of drug-resistant colonies was determined after infection. Under these conditions, the number of CFU for tRNAPheD− (DNA) was 1 or 2 colonies, which was the background level seen with a vector clone.
We next examined whether cellular transport from the nucleus to the cytoplasm was required for MuLV primer tRNA selection (Fig. 5B). Similar to our results with HIV-1, the expression of tRNAPheD− from the cDNA did not complement the replication of pMuLV-Phe. However, direct transfection of tRNAPheD− into the cytoplasm resulted in complementation that was approximately 100-fold more than that seen with transfection of cDNA encoding tRNAPheD−. The results of the studies with both HIV-1 and MuLV demonstrate that tRNA transport from the nucleus to the cytoplasm is an absolute prerequisite for selection of the tRNA primer for reverse transcription. Selection of the tRNA primer from the pool of aminoacylated tRNAs that has entered the channeled tRNA cycle is probably more efficient than selection of free tRNA (from transfection of in vitro-transcribed tRNA). Previous reports from our laboratory have indicated that the decreased efficiency of transfected in vitro-transcribed tRNAs is most probably a result of the intracellular locale in which they reside rather than of limiting concentrations of tRNA, since excess tRNA was readily detectable at the time of virus collection (29-31).
DISCUSSION
For the present study, we investigated the mechanism by which the tRNA primer used for retrovirus replication is selected for use in reverse transcription. For these studies, we constructed HIV-1 and MuLV mutants that require yeast tRNAPhe for infectivity; previous studies have demonstrated that both HIV-1 and MuLV can be engineered to use noncognate tRNA primers by altering the PBS (15, 29-31). We have shown that transfection of cDNA encoding yeast tRNAPhe results in the expression of yeast tRNAPhe that is almost entirely aminoacylated, indicating that it is fully functional in mammalian tRNA biogenesis and has undergone incorporation (i.e., channeling) into protein synthesis pathways (24, 28). To investigate the coordination of tRNA biogenesis with primer selection, we utilized a complementation system by which mutant tRNAs with PBSs complementary to yeast tRNAPhe are provided in trans to the defective retroviral genomes. One mutant was designed that eliminated the D loop of the tRNA (tRNAPheD−); consistent with previous studies, we have shown that this mutation results in nuclear sequestration of the tRNA (1). A second mutant was constructed that altered the identity elements required for aminoacylation without affecting tRNA transport from the nucleus to the cytoplasm. Analysis of the capacity of these tRNAs to function as primers for retrovirus replication gave strikingly different results. tRNAPheD− expressed from the plasmid was not capable of complementing replication due to sequestration in the nucleus; however, if this tRNA was provided to the cell cytoplasm via transfection, it was able to complement the replication of both retroviruses at levels well above the background. In contrast, the mutant tRNAPhe that was unable to undergo aminoacylation did complement the replication of either retrovirus, although the level was not equal to that for wild-type tRNAPhe.
The results of our studies provide new insights into the complexity of the primer tRNA selection process for retroviruses. In previous studies, it was found that alteration of the PBS to correspond to tRNAs other than the wild type have resulted in either HIV-1, MuLV, or ALV that uses these alternative tRNAs for replication (4, 5, 13, 15, 26, 27). Therefore, it seems feasible that the tRNA used for retroviral reverse transcription may be selected prior to encapsidation, since many of the alternatively used tRNAs are not enriched into virions. Collectively, the results of these studies suggest that retroviruses select the tRNA primer used for reverse transcription from a location that contains a variety of tRNAs. Several intracellular locales exist that would meet these criteria. To gain further insight into the selection process, we utilized novel HIV-1 and MuLV that require a yeast tRNAPhe primer to be supplied in trans. Since the yeast tRNAPhe can be distinguished from endogenous mammalian tRNAPhe, we could track the intracellular fate of the wild-type and mutant tRNAs. We first established that the expressed yeast tRNAPhe could enter into the tRNA biosynthetic pathway within mammalian cells. The tRNA is initially produced as a precursor and subsequently processed within the nucleus of the cell (7, 28). During this process, the tRNA associates with a number of cellular proteins that facilitate its transport from the nucleus to the cytoplasm and its interaction with the translational machinery; it has been estimated that a minimum of 30 different proteins associate with the tRNA molecule during biogenesis and translation (28). The tRNA molecule is believed to be channeled into the protein synthesis machinery and is thus rarely free within the cytoplasm (24), consistent with the results presented in this study by which we found that the majority if not all of the yeast tRNAPhe expressed in mammalian cells was aminoacylated. Previous studies have established that tRNA molecules that have defects in conformation that preclude them from interaction with exportin-t are not transported from the nucleus to the cytoplasm (1). One possibility, then, is that retroviruses select the tRNA primer from a nuclear pool of tRNAs. If this were the case, retroviruses would not have to compete with cellular translation for tRNAs. Indeed, the fact that tRNAPheD− transfected into the cytoplasm could complement HIV-1 or MuLV with a PBS complementary to tRNAPhe supported the possibility that retroviruses might select the tRNA primer from a pool of nonfunctional tRNAs, of which the majority would be found sequestered in the nucleus. The tRNA might be selected by the viral genome in the nucleus and transported with the genome to the cytoplasm prior to incorporation into virions. The results of our study clearly show that tRNAPheD− was retained in the nucleus after expression and, most importantly, it was not selected by either HIV-1 or MuLV as a primer for reverse transcription. Since we know that tRNAPheD− can function as a primer, albeit at a low level, when transfected into the cytoplasm, we conclude that HIV-1 and MuLV have not evolved to select the primer tRNA from a nuclear pool of defective tRNAs.
The second intracellular locale in which a variety of tRNAs are present is the site of protein synthesis. A recent study reported that for HIV-1 the ability of mutant tRNALys,3 to be aminoacylated correlated with the capacity of the tRNA to be incorporated into the virion (10). However, due to the endogenous background levels of wild-type tRNALys,3, it was not possible to evaluate the contribution of these mutant tRNAs as primers for reverse transcription and production of infectious virus. Indeed, a correlation between the enrichment of certain tRNAs in the virion and the selection of the actual tRNA that is used to prime reverse transcription is uncertain. Alternative tRNAs that are not enriched in virions can be stably used by mutant HIV-1 in extended culture (11, 32). Using our complementation system, which relies on exogenously supplied tRNAPhe, we found that the nonaminoacylated tRNA (tRNAPheUUA) complemented HIV-1 and MuLV replication, although it did so at lower levels than did the wild-type tRNA. Although this result implies that aminoacylated tRNA is not an absolute requirement for selection by HIV-1 or MuLV, it is still possible that interaction with the synthetase is important for selection of the primer used for reverse transcription. Previous studies have shown that a variety of mutations in the tRNAPhe identity elements, although not the exact mutations as those in tRNAPheUUA, retained the capacity to interact with the synthetase, albeit with a lower affinity than that of wild-type tRNAPhe (23). While the decreased level of complementation with tRNAPheUUA may not be significant, if HIV-1 or MuLV preferred to select tRNAs that are not involved in translation, the level of complementation would have been significantly greater than that found using wild-type tRNAPhe. Additional studies will be needed to determine if this is the case.
Finally, the results of our studies suggest a common theme between HIV-1 and MuLV with respect to selection of the tRNA primer used for replication. Both viruses show similar features, such as the intracellular location for primer selection and the ability to alter tRNA preference by mutation of the PBS. Under normal physiological conditions in an infected cell, HIV-1 or MuLV has access to numerous tRNAs at the site of translation. The initial interaction between the PBS and tRNA might occur with the tRNA in complex with the synthetase or possibly with elongation factor 1A (α) (EF1A), since the majority of tRNA in the cytoplasm is in complex with EF1A-GTP. Most, if not all, of the tRNA is expected to be aminoacylated (9, 18, 28). Interestingly, both proteins have been found in the HIV-1 virions (2, 3, 19). An amino acid at the 3′ end of the tRNA would preclude the use of an aminoacylated tRNA as the primer for initiation of reverse transcription. However, the selection of the aminoacylated tRNA while in complex with either EF1A or its cognate synthetase might result in destabilization, causing the deacylation of the selected tRNA. Additional experiments will be needed to understand the interrelationship between the translation of the viral genome and selection of the tRNA primer required for retrovirus replication.
Acknowledgments
We thank members of the Morrow laboratory for helpful comments. We thank Susan Lobo-Rupert for the plasmid containing the U6 promoter. We thank Adrienne Ellis for help with preparation of the manuscript. C.D.M. acknowledges the help of M.A.R.
DNA sequencing was carried out at the CFAR Sequencing Core (AI-27767). N.J.K. was supported by a training grant (AI-07493). The research was supported by a grant from the NIH (AI-34745) to C.D.M.
REFERENCES
- 1.Arts, G.-J., S. Kuersten, P. Romby, B. Ehresmann, and I. W. Mattaj. 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17:7430-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colicelli, J., and S. P. Goff. 1987. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J. Virol. 57:37-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, A. T., B. Klaver, and B. Berkhout. 1995. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA3Lys. J. Virol. 69:3090-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilboa, E., S. W. Mitra, S. Goff, and D. Baltimore. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93-100. [DOI] [PubMed] [Google Scholar]
- 7.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 8.Harada, F., R. C. Sawyer, and J. E. Dahlberg. 1975. A primer ribonucleic acid for initiation of in vitro Rous sarcoma virus deoxyribonucleic acid synthesis. J. Biol. Chem. 250:3487-3497. [PubMed] [Google Scholar]
- 9.Ho, Y.-S., and Y. W. Kan. 1987. In vivo aminoacylation of human and Xenopus suppressor tRNAs constructed by site-specific mutagenesis. Proc. Natl. Acad. Sci. USA 84:2185-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javanbakht, H., S. Cen, K. Musier-Forsyth, and L. Kleiman. 2002. Correlation between tRNALys3 aminoacylation and its incorporation into HIV-1. J. Biol. Chem. 277:17389-17396. [DOI] [PubMed] [Google Scholar]
- 11.Kang, S.-M., Z. Zhang, and C. D. Morrow. 1997. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J. Virol. 71:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly, N. J., and C. D. Morrow. Yeast tRNAPhe expressed in human cells can be selected by HIV-1 for use as a reverse transcription primer. Virology, in press. [DOI] [PubMed]
- 13.Li, X., J. Mak, E. J. Arts, Z. Gu, L. Kleiman, M. A. Wainberg, and M. A. Parniak. 1994. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J. Virol. 68:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobo, S. M., and N. Hernandez. 1989. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell 58:55-67. [DOI] [PubMed] [Google Scholar]
- 15.Lund, A. H., M. Duch, J. Lovmand, P. Jorgensen, and F. S. Pedersen. 1997. Complementation of a primer binding site-impaired murine leukemia virus-derived retroviral vector by a genetically engineered tRNA-like primer. J. Virol. 71:1191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 71:8087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marquet, R., C. Isel, C. Ehresmann, and B. Ehresmann. 1995. tRNAs as primers of reverse transcriptases. Biochimie 77:113-124. [DOI] [PubMed] [Google Scholar]
- 18.Negrutskii, B. S., and M. P. Deutscher. 1991. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. USA 88:4991-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. I. Sowder, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 20.Parry, H. D., and I. W. Mattaj. 1990. Positive and negative functional interactions between promoter elements from different classes of RNA polymerase III-transcribed genes. EMBO J. 9:1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters, G., F. Harada, J. E. Dahlberg, A. Panet, W. A. Haseltine, and D. Baltimore. 1977. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J. Virol. 21:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhim, H., J. Park, and C. D. Morrow. 1991. Deletions in the tRNALys primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J. Virol. 65:4555-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampson, J. R., L. S. Behlen, A. B. DiRenzo, and O. C. Uhlenbeck. 1992. Recognition of yeast tRNAPhe by its cognate yeast phenylalanyl-tRNA synthetase: an analysis of specificity. Biochemistry 31:4161-4167. [DOI] [PubMed] [Google Scholar]
- 24.Stapulionis, R., and M. P. Deutscher. 1995. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 92:7158-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varmus, H. 1988. Retroviruses. Science 240:1427-1435. [DOI] [PubMed] [Google Scholar]
- 26.Wakefield, J. K., A. G. Wolf, and C. D. Morrow. 1995. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNALys,3. J. Virol. 69:6021-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitcomb, J. M., B. A. Ortiz-Conde, and S. H. Hughes. 1995. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J. Virol. 69:6228-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolin, S. L., and A. G. Matera. 1999. The trials and travels of tRNA. Genes Dev. 13:1-10. [DOI] [PubMed] [Google Scholar]
- 29.Yu, Q., and C. D. Morrow. 1999. Complementarity between 3′ terminal nucleotides of tRNA and primer binding site is a major determinant for selection of the tRNA primer used for initiation of HIV-1 reverse transcription. Virology 254:160-168. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Q., and C. D. Morrow. 2000. Essential regions of the tRNA primer required for HIV-1 infectivity. Nucleic Acids Res. 28:4783-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, Q., and C. D. Morrow. 2001. Identification of critical elements in the tRNA acceptor stem and TΨC loop necessary for human immunodeficiency virus type 1 infectivity. J. Virol. 75:4902-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, Z., S. M. Kang, A. LeBlanc, S. L. Hajduk, and C. D. Morrow. 1996. Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for a human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNAHis. Virology 226:306-317. [DOI] [PubMed] [Google Scholar]