Abstract
The 65 human enterovirus serotypes are currently classified into five species: Poliovirus (3 serotypes), Human enterovirus A (HEV-A) (12 serotypes), HEV-B (37 serotypes), HEV-C (11 serotypes), and HEV-D (2 serotypes). Coxsackie A virus (CAV) serotypes 1, 11, 13, 15, 17, 18, 19, 20, 21, 22, and 24 constitute HEV-C. We have determined the complete genome sequences for the remaining nine HEV-C serotypes and compared them with the complete sequences of CAV21, CAV24, and the polioviruses. The viruses were most diverse in the capsid region (4 to 36% amino acid difference). A high degree of capsid sequence conservation (96% amino acid identity) suggests that CAV15 and CAV18 should be classified as strains of CAV11 and CAV13, respectively. In the 3CD region, CAV1, CAV19, and CAV22 differed from one another by only 1.2 to 1.4% and CAV11, CAV13, CAV17, CAV20, CAV21, CAV24, and the polioviruses differed from one another by only 1.2 to 3.6%. The two groups, however, differed from one another by 14.6 to 16.2%. The polioviruses as a group were monophyletic only in the capsid region. Only one group of serotypes (CAV1, CAV19, and CAV22) was consistently monophyletic in multiple genome regions. Incongruities among phylogenetic trees based on different genome regions strongly suggest that recombination has occurred between the polioviruses, CAV11, CAV13, CAV17, and CAV20. The close relationship among the polioviruses and CAV11, CAV13, CAV17, CAV20, CAV21, and CAV24 and the uniqueness of CAV1, CAV19, and CAV22 suggest that revisions should be made to the classification of these viruses.
The human enteroviruses were originally classified into four categories on the basis of human disease and virulence and pathogenesis in intracranially inoculated suckling mice, as follows: (i) polioviruses (PV; agents of human poliomyelitis; generally not pathogenic in mice), (ii) coxsackie A viruses (CAV; associated with human central nervous system disease, exanthems, and herpangina; cause flaccid paralysis in mice), (iii) coxsackie B viruses (CBV; associated with human central nervous system and cardiac disease; cause spastic paralysis in mice), and (iv) echoviruses (at the time, not known to cause human disease; not pathogenic in mice) (7, 26). It quickly became apparent, however, that this scheme was insufficient to describe the universe of human enteroviruses, because viruses that were pathogenic to mice and antigenically identical to known echoviruses were isolated, and the echoviruses were shown to be associated with a wide range of human diseases (6, 7). Thereafter, new human enterovirus serotypes were simply named “enterovirus” (EV) and numbered sequentially, starting with EV68 (27, 51). A total of 64 serotypes are currently recognized (19), and at least 6 additional serotypes have been proposed (30, 35) (S. Michele, M. S. Oberste, and M. A. Pallansch, unpublished data). In the current classification scheme, which takes into account both biological and molecular properties of the viruses, the human enteroviruses are divided among five species: (i) Poliovirus (PV1-3), (ii) Human enterovirus A (HEV-A) (CAV2 to CAV8, CAV10, CAV12, CAV14, CAV16, and EV71), (iii) HEV-B (CAV9, CBV1 to CBV6, E1 to E7, E9, E11 to E21, E24 to E27, E29 to E33, EV69, and EV73 [proposed]), (iv) HEV-C (CAV1, CAV11, CAV13, CAV15, CAV17 to CAV22, and CAV24), and (v) HEV-D (EV68 and EV70) (19).
The enterovirus genome is a 7.4- to 7.5-kb single-stranded, polyadenylated RNA of positive sense, with a 22-amino-acid virus-encoded protein (VPg) covalently linked to the 5′-end. Flanked by 5′- and 3′-nontranslated regions (NTRs), the single, long open reading frame encodes a polyprotein of approximately 2,200 amino acids that is processed by viral proteinases to yield the mature viral polypeptides. The P1 region encodes the capsid proteins 1A to 1D (VP1 to VP4). The P2 region encodes a protease, 2Apro, and two proteins involved in RNA replication, 2B and 2C. VPg (3B) and its precursor (3AB), the major viral protease (3Cpro), and the RNA-dependent RNA polymerase (3Dpol) are encoded in the P3 region. Complete genome sequences are available for each of the poliovirus serotypes and for CAV21 and CAV24, but only partial sequences (partial 5′-NTR, partial VP2, complete VP1, and partial 3D) are available for the remaining members of HEV-C. Phylogenetic analyses of available sequences have shown that the polioviruses and HEV-C viruses are very closely related to one another in multiple regions of the genome (18, 33, 34, 38), but details of the relationship remain unknown because of missing data from all the genomic regions and from the other serotypes within HEV-C.
We present here the first comprehensive analysis of genetic variation at the complete genome level from an entire enterovirus species and compare the polioviruses with the members of HEV-C. The data described here show that the polioviruses are distinct from most members of HEV-C only in the capsid-coding region and strongly suggest that recombination has played a major role in their evolution. Furthermore, CAV1, CAV19, and CAV22 are similar to one another but distinct from other HEV-C viruses and from the polioviruses in all genome regions. These results suggest that modifications to the current classification of several of these serotypes, including the polioviruses, should be considered.
MATERIALS AND METHODS
Viruses.
The prototype strains of CAV1, CAV11, CAV13, CAV15, CAV17, CAV18, CAV19, CAV20, CAV21, and CAV22 were obtained from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (Table 1) and propagated for at least nine serial passages by intracranial inoculation of newborn mice (28).
TABLE 1.
Complete genome sequences analyzed
| Serotype | Strain | Yr | Location | GenBank accession no. (reference) |
|---|---|---|---|---|
| CAV1 | Tomkins | 1947 | New York | AF499635 (this work) |
| CAV11 | Belgium-1 | 1951 | Belgium | AF499636 (this work) |
| CAV13 | Flores | 1952 | Mexico | AF499637 (this work) |
| CAV15 | G-9 | 1950 | South Africa | AF499638 (this work) |
| CAV17 | G-12 | 1951 | South Africa | AF499639 (this work) |
| CAV18 | G-13 | 1950 | South Africa | AF499640 (this work) |
| CAV19 | NIH-8663 | 1952? | Japan | AF499641 (this work) |
| CAV20 | IH-35 | 1955 | New York | AF499642 (this work) |
| CAV21a | Kuykendall | 1952 | California | AF546702 (this work) |
| CAV22 | Chulman | 1955 | New York | AF499643 (this work) |
| CAV24 | EH24/70 | 1970 | Singapore | D090457 (49) |
| PV1 | Mahoney | 1942 | Ohio | J02281 (40) |
| PV2 | MEF-1 | 1942 | Egypt | M12197 (20) |
| PV3 | Leon | 1937 | California | K01392 (47) |
A complete genome sequence is also available for CAV21-Coe (accession no. D00538).
Nucleotide sequencing.
Complete genomic sequences were determined for each of the nine strains (Table 1). Overlapping fragments representing each complete viral genome were amplified by reverse-transcription-PCR by using degenerate, inosine-containing primers designed to anneal to sites encoding amino acid motifs that are highly conserved among enteroviruses. Specific primers were designed from preliminary sequences to close gaps between the original PCR products. The PCR products were purified for sequencing by using the High-Pure PCR product purification kit (Roche Molecular Biochemicals, Indianapolis, Ind.). Both strands were sequenced by automated methods using fluorescent dideoxy-chain terminators (Applied Biosystems, Foster City, Calif.). The complete genome sequences for PV1, PV2, PV3, and CAV24 were obtained from GenBank (Table 1).
Sequence analysis.
The pairwise sequence identities among the nucleotide and deduced amino acid sequences of all of the Poliovirus and HEV-C serotypes were calculated by using the programs Gap and Distances (Wisconsin Sequence Analysis Package, version 10.2; Genetics Computer Group, Inc., Madison, Wis.). Nucleotide sequences were aligned by using the Pileup program (Wisconsin Sequence Analysis Package) and adjusted manually to conform to the optimized alignment of deduced amino acid sequences. Similarity plots depicting the relationships among the aligned amino acid sequences were generated by using SimPlot, version 3.2 (24). To simplify visualization of the analysis results, sequences were grouped on the basis of pairwise identities in the P1 region as described below. The identity of the query sequence (PV1-PV2-PV3 consensus) with each target sequence was plotted in the center of a window of 300 amino acids that was successively advanced by 30 residues. Phylogenetic relationships were inferred from the aligned nucleic acid sequences by the maximum-likelihood method implemented in the program DNAML (PHYLIP: Phylogeny Inference Package, version 3.57; University of Washington, Seattle), using a transition-transversion ratio of 10. Support for specific tree topologies was estimated by bootstrap analysis with 100 pseudoreplicate data sets. Branch lengths in consensus maximum-likelihood trees were calculated by the maximum-likelihood quartet-puzzling method, using the nucleotide substitution model of Tamura and Nei (50), as implemented in Tree-Puzzle 5.0 (48). To assess potential recombinational relationships, nucleotide sequences were analyzed by using the bootscanning method implemented in SimPlot (24). Functional amino acid motifs were identified by comparison with motifs of known function in other picornavirus proteins and by searching the PROSITE dictionary of protein sites and patterns, version 16.45, using the program Motifs (Wisconsin Sequence Analysis Package).
Nucleotide sequence accession numbers.
The sequences reported here were deposited in the GenBank sequence database under accession no. AF499635 to AF499643 and AF546702.
RESULTS
The nine genomes sequenced varied in length from 7,397 nucleotides (CAV1 and CAV22) to 7,458 nucleotides (CAV13 and CAV18), which is similar to the length range among the previously sequenced genomes of PV1, PV2, PV3, CAV21, and CAV24 (7,401 to 7,461 nucleotides) (Table 2). The genomes of CAV1, CAV19, and CAV22 were composed of 43.0 to 43.7% G and C residues, whereas those of the other HEV-C viruses contained 44.7 to 46.0% G+C, compared to the 46.3 to 46.9% G+C content of the poliovirus genomes (Table 2). For all viruses, the G+C content was higher in the 5′-NTR than in the coding region and differed between the capsid and noncapsid regions of the genome.
TABLE 2.
General properties of the poliovirus and HEV-C genomes
| Serotype | Length | Total G+C (%) | 5′NTR G+C (%) | P1 G+C (%) | P2-P3-3′NTR G+C (%) | 5′NTR length (nt) | ORF locationa | Polyprotein length (aa) | 3′NTR length (nt)b |
|---|---|---|---|---|---|---|---|---|---|
| CAV1 | 7397 | 43.1 | 49.4 | 43.5 | 41.7 | 711 | 712-7323 | 2204 | 74 |
| CAV11 | 7454 | 45.3 | 50.4 | 46.6 | 43.7 | 747 | 747-7382 | 2212 | 72 |
| CAV13 | 7458 | 44.7 | 49.0 | 45.4 | 43.6 | 745 | 746-7387 | 2214 | 71 |
| CAV15 | 7441 | 46.0 | 50.9 | 47.0 | 44.4 | 734 | 735-7370 | 2212 | 71 |
| CAV17 | 7457 | 45.5 | 49.3 | 46.6 | 44.1 | 747 | 748-7386 | 2213 | 71 |
| CAV18 | 7458 | 44.9 | 47.8 | 45.8 | 43.8 | 745 | 745-7386 | 2214 | 72 |
| CAV19 | 7410 | 43.0 | 49.9 | 42.7 | 41.9 | 715 | 716-7336 | 2207 | 74 |
| CAV20 | 7436 | 45.2 | 48.7 | 46.2 | 43.9 | 744 | 745-7365 | 2207 | 71 |
| CAV21 | 7401 | 45.1 | 50.0 | 45.8 | 43.8 | 711 | 712-7329 | 2206 | 72 |
| CAV22 | 7406 | 43.7 | 49.0 | 45.0 | 41.8 | 715 | 716-7333 | 2206 | 73 |
| CAV24 | 7461 | 45.9 | 50.4 | 46.6 | 44.6 | 750 | 751-7392 | 2214 | 69 |
| PV1 | 7440 | 46.3 | 49.9 | 48.5 | 44.3 | 742 | 743-7369 | 2209 | 71 |
| PV2 | 7440 | 46.7 | 50.3 | 48.6 | 44.8 | 747 | 748-7368 | 2207 | 72 |
| PV3 | 7431 | 46.9 | 53.1 | 48.0 | 45.1 | 742 | 743-7360 | 2207 | 71 |
Nucleotide coordinates, starting with the first “open” AUG and ending with the last base before the stop codon. ORF, open reading frame.
Includes the termination codon.
The 5′-NTRs of CAV1, CAV19, CAV21, and CAV22 were 711 to 715 nucleotides long. Those of the other viruses varied in length from 734 to 750 nucleotides, with all except CAV15 (734 nucleotides) clustering in the range of 742 to 750 nucleotides (Table 2). The 5′-NTR sequences differed from one another by 12 to 24% (Fig. 1), .with CAV21 and CAV24 the most distant from one another. Fifty-six percent of the 5′-NTR residues were invariant among all of the viruses; almost one-third of the variable sites were concentrated in the hypervariable region, the 80 to 110 residues immediately upstream of the initiation codon. Most of the differences in length of the 5′-NTR between the serotypes were also located in this region. Structural elements that are important for the function of the poliovirus internal ribosome entry site were well conserved among the HEV-C strains (data not shown). The 3′-NTRs of all viruses were similar in length, 69 to 74 nucleotides (Table 2), but the 3′-NTRs of CAV1, CAV19, and CAV22 differed from those of the other viruses by 11 to 14%, whereas the remaining serotypes differed from one another by no more than 4% (Fig. 1). The 3′-NTR sequences of the polioviruses and CAV18 were identical to one another, as were those of CAV13 and CAV21 and those of CAV17 and CAV20. For all of the viruses, the predicted 3′-NTR structures were similar to that of the PV1-Mahoney 3′-NTR, consisting of two stem-loops (data not shown).
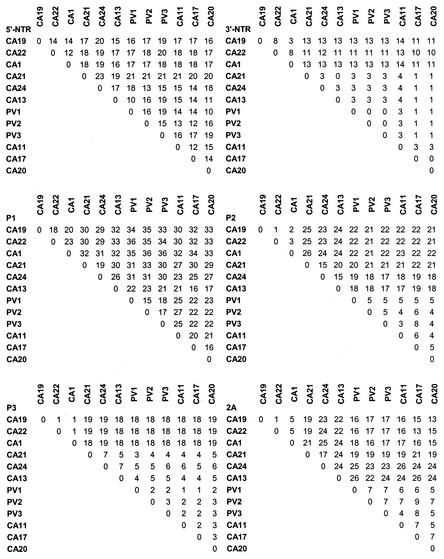
FIG. 1.
Nucleotide and deduced amino acid sequence relationships (percent identity). For noncoding regions (5′NTR and 3′-NTR), nucleotide comparisons are shown. Deduced amino acid sequence comparisons are shown for all other regions.
The predicted proteolytic cleavage sites within the deduced polyproteins of the newly sequenced viruses were consistent with those predicted or experimentally determined for the polioviruses, CAV21, and CAV24 (data not shown). Pairwise sequence comparisons, multiple alignments, and similarity plots were used to examine the relationships among the complete amino acid sequences of the polioviruses and species C enteroviruses. The greatest sequence variation occurred in the capsid region, whereas the 3D gene was the most highly conserved. CAV11 and CAV15 differed from one another by only 4% in their capsid sequences, consistent with their belonging to a single serotype as previously suggested from their antigenic cross-reactivities (6, 46) and by their sequence similarities in the amino-terminal part of VP2 (34) and in VP1 (33). Similarly, CAV13 and CAV18 differed from one another by only 4% and probably also represent variants of a single serotype, as previously proposed (33). Because of their close relationship to other established strains, we propose that CAV15 and CAV18 should no longer be considered distinct serotypes. For that reason, CAV15 and CAV18 have been omitted from further analyses.
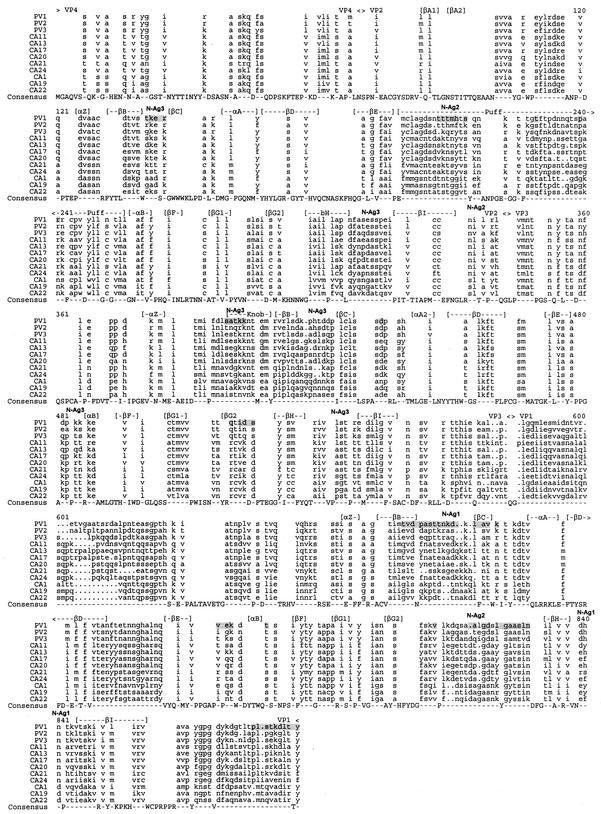
The complete capsid polypeptide sequences differed by 15 to 18% among the polioviruses, by 16 to 34% within HEV-C, and by 21 to 36% between the two groups (Fig. 1). The close relationship among the PVs in the capsid region may reflect their shared use of the poliovirus receptor, CD155, or it may simply reflect the limited number of isolates represented by these three prototypes. The poliovirus and HEV-C capsid sequences were aligned and compared to determine the distribution of amino acid differences and alignment gaps relative to known structural motifs, antigenic sites, and receptor contacts (Fig. 2). As expected, sequences that are predicted to participate in important structural motifs, such as the beta strands that form the eight-stranded beta barrels of VP1, VP2, and VP3, were highly conserved. In contrast, the “loop” regions between the beta strands, as well as the VP2 “puff” and VP3 “knob,” were poorly conserved. Gaps in the sequence alignment also occurred in these regions of peptide backbone flexibility, such as the VP2 puff, the VP3 B-C loop, the VP3-VP1 junction, the VP1 B-C and G-H loops, and the amino- and carboxyl-terminal domains of VP1 (Fig. 2). In general, these variable regions correlated with PV1 residues that have been shown to form the neutralization antigenic sites, N-Ag1, N-Ag2, and N-Ag3 (9). Similarly, poliovirus residues that have been implicated in receptor interaction were also not conserved among HEV-C viruses.
FIG. 2.
Aligned poliovirus and HEV-C capsid precursor proteins. Cleavage sites are indicated above the alignment. Regions predicted to form structurally defined alpha helices and beta strands (16, 29, 36), as well as the location of neutralization antigenic sites N-Ag1, N-Ag2, and N-Ag3 in PV1 (9), are also indicated. Amino acids implicated by structural or genetic studies to be involved in PV1-poliovirus receptor interaction are shaded (2, 15).
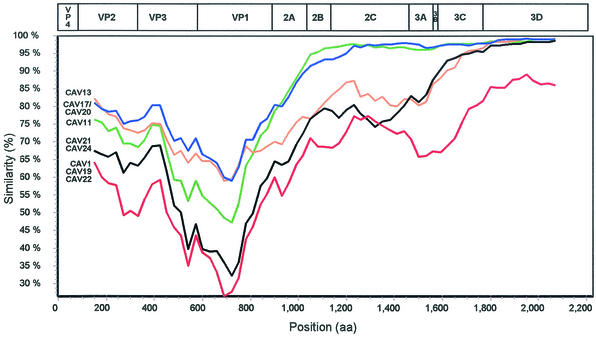
To analyze the patterns of amino acid sequence relationships across the polyprotein, viruses whose capsid sequences differed by less than 20% were considered a group in similarity plot comparisons. CAV13 was considered separately, despite its relationship to CAV17 and CAV20 in the capsid region, because of its higher degree of divergence from CAV17 and CAV20 in P2 (Fig. 1). CAV1, CAV19, and CAV22 were considered to be a single group, even though they differed from one another by up to 23% in the capsid, because they were very similar to one another in P2 and P3 but distinct from all other viruses throughout the polyprotein (Fig. 1). The resulting groups were (i) CAV1, CAV19, and CAV22; (ii) PV1, PV2, and PV3; (iii) CAV11; (iv) CAV21 and CAV24; (v) CAV13; and (vi) CAV17 and CAV20. Similar results were obtained by using each of the sequences individually and comparing them to all of the other sequences, in every combination (data not shown). The individual similarity plots were also confirmed by the region-by-region pairwise comparisons (Fig. 1). CAV11, CAV17, and CAV20 were the most closely related viruses to the PVs throughout the polyprotein (Fig. 1 and 3), whereas the CAV1-CAV19-CAV22 group was distinct from all of the other viruses throughout the polyprotein (Fig. 1 and 3; also data not shown). CAV13 was as similar to PV1-PV2-PV3 in the capsid region as were CAV11, CAV17, and CAV20 but sharply diverged near the carboxyl terminus of VP1 and remained at an intermediate distance from the polioviruses, relative to CAV11-CAV17-CAV20, and CAV1-CAV19-CAV22, from 2A through 3B and into 3C (Fig. 1 and 3). CAV21 and CAV24 were nearly as distant from the poliovirus group in the capsid region as were CAV1, CAV19, and CAV22, and both groups were distinct from the polioviruses from 2A through 2C (about the same distance as CAV13). CAV21-CAV24 was more similar to the polioviruses in P3 than was the CAV1-CAV19-CAV22 group. Although the CAV1-CAV19-CAV22 and CAV21-CAV24 groups were roughly equidistant from the PV consensus in P2, they were not similar to one another in this region (Fig. 1). All of the viruses except CAV1, CAV19, and CAV22 were at least 96% identical to the polioviruses in the 3CD region; CAV1, CAV19, and CAV22 were only 85% identical to the polioviruses in this region (Fig. 1). The noncapsid proteins were fully colinear among all of the polioviruses and HEV-C viruses, with the exception of a one-amino-acid deletion near the carboxyl terminus of 2A in CAV13 and a one-amino-acid insertion in the 3A protein of CAV1, CAV19, and CAV22. The polioviruses and HEV-C viruses differed in nucleotide sequence from members of other enterovirus species by 41 to 56% in P1, 39 to 44% in P2, and 28 to 39% in P3 (data not shown).
FIG. 3.
Similarity plots of poliovirus and HEV-C viruses deduced polyprotein sequences, calculated and plotted by SimPlot 3.2 (24), relative to the polyprotein consensus sequence of PV1, PV2, and PV3. To facilitate the analysis, CAV1, CAV19, and CAV22 were grouped together, as were CAV17 and CAV20 and also CAV21 and CAV24, as described in the text. Each point represents the percent identity to the poliovirus consensus, within a sliding window of 300 amino acids centered on the position plotted, with a step of 30 amino acids between points. Positions containing gaps were excluded from the analysis. The PV1 genetic map is shown at the top, approximately to scale.
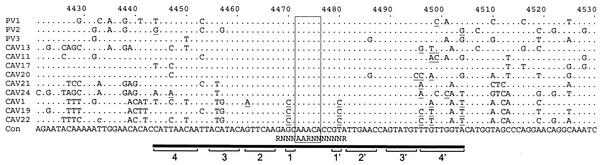
A distinct RNA structural element, the cis-acting replication element (cre), has been shown to be required for replication of PV1 and PV3 (12, 41). The poliovirus cre is a four-part stacked stem and conserved loop located in the region encoding 2C. Similar elements have been identified at different locations in the cardioviruses (23) and human rhinoviruses (11, 25). To determine whether members of HEV-C contain a similar sequence and/or structure, the analogous poliovirus and HEV-C 2C nucleotide sequences were aligned and compared (Fig. 4). The AAACA motif in the loop, which is required for cre function (12), was completely conserved among all of the viruses. The expanded and generalized version of this motif, RN3AARN6R, which models stem 1 (described by Goodfellow et al. [12]) as part of the loop (52), was also conserved. The structure of the predicted stem region was also well conserved, with a total of only 38 pair-interrupting changes in the 14 viruses. For example, while CAV22 was the most divergent from PV1, containing 13 differences in the 46 base-paired residues, only 6 of the differences would interrupt the predicted base-pairing interaction (either because of alternative G:U base pairs or compensating changes at a second site). In addition, stems 1 to 3 appear to be the most important, as 29 of the 38 pair-interrupting differences occurred in stem 4. PV1 contains a non-base-paired C residue in stem 4 (nucleotide 4499) that is shared with CAV11 and CAV24. Only one pair-breaking difference occurred in stem 2, and two occurred in stem 3. CAV1, CAV19, and CAV22 shared two noncompensated differences in complementary residues of stem 1.
FIG. 4.
Nucleotide sequence alignment of the region surrounding the poliovirus cis-acting replication element (cre). The majority rule consensus sequence is shown at the bottom (Con). Residues that are identical to those of the consensus are indicated by a dot. The bar below the sequence alignment indicates the location of the conserved stem-loop structure. The conserved AAACA motif in the loop is boxed (12, 41), and the location of the RN3AARN6R motif is indicated below the consensus (52). Base-paired residues in the stem are indicated by brackets and numbered 1-1′ to 4-4′, as previously described (12). Residues predicted to disrupt base pairing are underlined. Nucleotides are numbered according to the genome of PV1-Mahoney.
Amino acid sequence motifs that are important for the function of picornaviral nonstructural proteins were highly conserved among all of the polioviruses and the members of HEV-C. For example, the catalytic triads in the active sites of the viral proteases were fully conserved as H20-D38-C109 and H40-E71-C147, in 2Apro and 3Cpro, respectively. In both cases, the cysteine residue occurred in the context of a conserved GXCG motif. The 3Cpro sequences also contained the conserved putative RNA-binding domain, 82-KFRDI-86 (RFRDI in CAV17). The cysteine-rich motif, 269-CX2CX8CX4C-286, which has been shown to bind zinc and to play a role in the replication of PV1 (37), was identified in all of the 2C sequences, as was a putative NTP-binding motif, GX4GK(S/T). The latter motif was present as 109-GSPGTGKS-116 in all of the viruses except CAV1, CAV19, and CAV22, which contained the motif GTPGTGKS. A nucleotide-binding motif, 166-(S/T)KVEQGKS-173, and the RNA-dependent RNA polymerase signature motifs 159-KDELR-163, 283-GGMPSG-288, 325-YGDD-328, and 372-FLKR-375 were fully conserved in all of the 3Dpol sequences.
Alignments of amino acid sequences revealed the longest stretches of conserved amino acids to be in the 3D region, with 85% amino acid identity. Twelve percent of the sites in the 3D polymerase were unique in the CA1, CA19, CA22 group; of these distinct conserved sites (total of 55), almost half (47%) were in the palm region, which composes one-third of the polymerase.
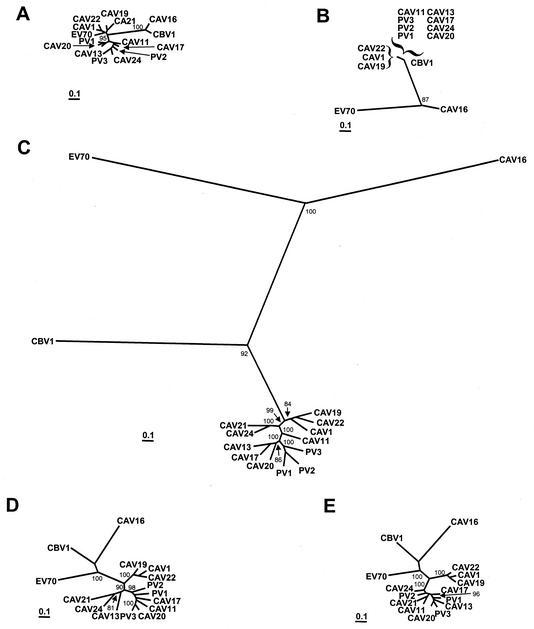
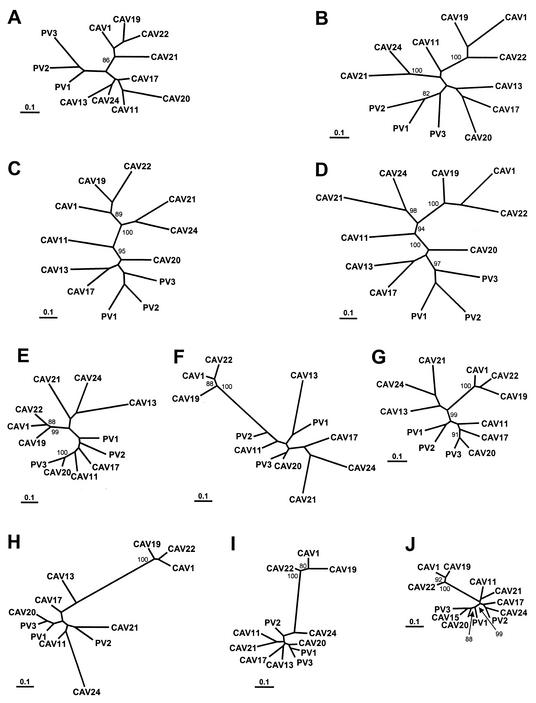
Maximum-likelihood nucleotide sequence phylogenies were constructed separately for the 5′- and 3′-NTR, the P1, P2, and P3 regions, and the regions encoding each of the mature viral proteins (Fig. 5 and 6). The branch lengths in the trees were consistent with the pairwise distances in Fig. 1; that is, the branch lengths were shorter in the P3 region trees than in the capsid region trees. In agreement with previously published phylogenies (18, 33, 34, 38), the poliovirus and HEV-C sequences were monophyletic relative to members of HEV-A, HEV-B, and HEV-D throughout the coding region and in the 3′-NTR (Fig. 5B to E), but the 5′-NTR sequences clustered with those of HEV-D viruses (Fig. 5A). Within the capsid region as a whole, the viruses clustered into five major monophyletic groups: (i) CAV1, CAV19, and CAV22; (ii) PV1, PV2, PV3; (iii) CAV11; (iv) CAV21 and CAV24v; and (v) CAV13, CAV17, and CAV20 (Fig. 5C). Two of these groups, CAV1-CAV19-CAV22 and PV1-PV2-PV3, were monophyletic in each of the regions encoding the individual capsid proteins, with strong bootstrap support (Fig. 6A to D). The VP3 and VP1 trees were congruent except for the relative positions of CAV1, CAV19, and CAV22 and of PV1, PV2, and PV3 within their respective clusters (Fig. 6C and D). The VP2 tree differed from the VP1 and VP3 trees only in the position of CAV20, but the difference may not be significant given the relatively low bootstrap support for the CAV20 branches (Fig. 6B). Outside the capsid region, CAV1, CAV19, and CAV22 formed the only consistently monophyletic group, with 99 to 100% bootstrap support (Fig. 5D and E and 6E to J). For example, CAV21 clustered with CAV24 in 2A, 2B, and 2C (Fig. 6E to G) but branched directly from an internal node in 3AB and 3C and clustered with CAV11 in 3D (Fig. 6J).
FIG. 5.
Phylogenetic trees based on poliovirus and HEV-C virus nucleotide sequences. Each of the major functional regions of the genome was analyzed independently. Bootstrap values (percent of 100 pseudoreplicate data sets) of over 80% supporting each cluster are shown at the nodes. All trees are plotted to the same scale (see scale bar). (A) 5′-NTR; (B) 3′-NTR; (C) complete P1 region; (D) complete P2 region; (E) complete P3 region.
FIG. 6.
Phylogenetic trees based on poliovirus and HEV-C virus nucleotide sequences. The regions encoding each of the mature viral proteins were analyzed independently. Bootstrap values (percent of 100 pseudoreplicate data sets) of over 80% supporting each cluster are shown at the nodes. All trees are plotted to the same scale (see scale bar). (A) 1A (VP4); (B) 1B (VP2); (C) 1C (VP3); (D) 1D (VP1); (E) 2A; (F) 2B; (G) 2C; (H) 3AB; (I) 3C; (J) 3D.
DISCUSSION
This first comprehensive analysis of genetic variation of an entire enterovirus species at the complete genome level has demonstrated that the polioviruses and members of HEV-C are very similar to each other in genomic structure, genome composition, and sequence relationships. Outside the capsid region, the polioviruses are as similar to most of the HEV-C serotypes as they are to one another. In the noncapsid coding region, it is not readily apparent that there is any poliovirus-specific sequence, amino acid, or motif. Thus, the polioviruses and members of HEV-C are distinguished from one another only in the capsid region.
Intra- and intertypic recombination has been shown to occur among poliovirus vaccine strains (3, 10, 21) and between wild polioviruses and other, unidentified donor strains (14, 22). There is also evidence for intertypic recombination among certain nonpolio enteroviruses (1, 43). In our analysis of poliovirus and HEV-C complete genome sequences, the incongruities among phylogenetic trees constructed from different genome regions suggest that recombination has occurred between the polioviruses and nonpolio enteroviruses of HEV-C. The reference point for these comparisons appears to be arbitrary, in that it is impossible to distinguish whether a given noncapsid gene sequence originated with a poliovirus or with a nonpolio enterovirus of species C. One could explain the relatedness of poliovirus and HEV-C noncapsid sequences by saying that certain CAVs have acquired poliovirus noncapsid sequences, but the data are equally consistent with the view that the polioviruses have acquired CAV noncapsid sequences. In the absence of data to specifically support or refute either of these views, the most conservative explanation is that recombination effectively results in a lack of linkage between noncapsid sequences and the serotype of a virus within a species. That is, viruses with a poliovirus capsid (hence, recognized as polioviruses by all standard identification methods) may recombine with CAVs to acquire different noncapsid sequences. Likewise, a virus with a CAV capsid may acquire different noncapsid sequences by recombination with a poliovirus. In this respect, an enterovirus might be conceptually defined as a capsid sequence in search of noncapsid sequences of the highest fitness (i.e., the greatest replication capacity and, hence, a selective advantage). Recombination results in homogenization of P2 and P3 within a group of related serotypes, because the sequences are readily shuffled among viruses, and the regions conferring the greatest selective advantage tend to become the dominant lineage of every serotype. Clearly, however, it is possible to envision that restrictions on the compatibility of recombination partners exist. Cell tropism (probably due to receptor utilization) must play a role, as the two recombination partners must be simultaneously present in the same cell in order to recombine. CAV1, CAV19, and CAV22 are distinct from most other enteroviruses in that they are the only three EV serotypes that are seldom, if ever, isolated or propagated in cell culture (44), implying an inability to use the receptors commonly used by other enteroviruses. This might partly explain the apparent absence of recombination between the CAV1-CAV19-CAV22 group and the other viruses. In addition, the nonstructural proteins, particularly 3Cpro and 3Dpol, must interact with sites distant from their own coding region (the capsid precursor and the 3′-end of the minus strand, respectively), so these enzymes must be compatible with the potentially heterologous substrates. The observation that the 3C proteinases of human rhinovirus type 14 (Rhinovirus genus) and coxsackievirus B3 (HEV-B) can correctly process the PV1 nonstructural protein precursor but not its capsid protein precursor provides evidence for this compatibility requirement (8). Phylogenetic studies using partial genome sequences also argue that recombination between more distantly related serotypes is rare, if it occurs at all (17, 38, 39).
VP1 sequence analysis has been successfully implemented for enterovirus serotype identification (4, 5, 31-33, 35). The close genetic relationships between CAV11 and CAV15 and between CAV13 and CAV18 are consistent with VP1 sequence comparisons (33) and provide further evidence that the pairs CAV11-CAV15 and CAV13-CAV18 each comprise geographically distinct isolates of a single serotype. The genetic differences in the noncapsid regions within the two pairs are consistent with their isolation on different continents—CAV15 and CAV18 in Africa, CAV13 in North America, and CAV11 in Europe. Based on their similarities in complete capsid sequences, we propose that CAV15 and CAV18 be vacated as enterovirus serotype designations and that existing CAV15 and CAV18 isolates be considered isolates of CAV11 and CAV13, respectively. Additional clinical isolates of both serotypes should be analyzed to confirm these findings. Conversely, pairwise sequence comparisons and phylogenetic analysis suggest that CAV1, CAV19, and CAV22 are genetically distinct from the other serotypes and that the prototype strains show no evidence of recombination with any of the other members of HEV-C. It will be important to analyze additional clinical isolates of these serotypes to fully assess their ability to recombine with viruses of other serotypes within HEV-C.
While the genetic basis of complex phenotypes, such as transmissibility, host range, and receptor usage may not be clearly understood, all intrinsic properties of a picornavirus must ultimately derive from the viral genome. In response to the increase in viral sequence data and a greater understanding of the molecular details of enterovirus biology, explicit molecular criteria have been added recently to the taxonomic classification scheme as an adjunct to the existing physical and biological criteria (19). In the current system, members of an enterovirus species “(i) share greater than 70% amino acid identity in P1, (ii) share greater than 70% amino acid identity in the nonstructural proteins 2C and 3CD, (iii) share a limited range of host cell receptors, (iv) share a limited natural host range, (v) have a genome base composition (G+C) which varies by no more than 1%, and (vi) share a significant degree of compatibility in proteolytic processing, replication, encapsidation, and genetic recombination” (19). Previous molecular studies had compared poliovirus sequences only with those of CAV21 and CAV24 because they were the only HEV-C complete capsid or complete genome sequences available (18, 38, 42), and these comparisons contributed to the classification of Poliovirus and HEV-C as separate species (19). Our results show that CAV11, CAV17, and CAV20, and not CAV21 and CAV24, are the serotypes most closely related to the polioviruses. Furthermore, the polioviruses are distinct from CAV11, CAV17, and CAV20 only in the capsid region.
The current classification of HEV-C and polioviruses as separate species is not compatible with the application of the existing criteria, given the new sequence data presented here. HEV-C P1 amino acid identities range as low as 66%, yet the CAV11, CAV13, CAV17, and CAV20 capsid sequences are all at least 73% identical to those of the polioviruses (criterion i above). Similarly, the 2C polypeptides of all of the HEV-C viruses and the polioviruses are at least 75% identical to one another and those of CAV11, CAV17, and CAV20 are at least 97% identical to those of the polioviruses (criterion ii). All of the viruses except CAV1, CAV19, and CAV22 are at least 96% identical to one another in 3CD. While the base composition (G+C content) among PV1-Mahoney, PV2-MEF-1, and PV3-Leon differs by only 0.6%, the composition within the existing recognized HEV-C species varies by 3.0% (failing to meet criterion v). The differences in G+C content between polioviruses and HEV-C viruses (other than CAV1, CAV19, and CAV22) are largely restricted to P1, as the range of G+C content in P2-P3-3′-NTR varied by only 1.5%. The limited range of G+C content among the poliovirus prototype strains may simply be due to sampling—comparison of a large number of poliovirus complete genome sequences may help to clarify this point. In any case, this criterion may need to be reassessed because of these inconsistencies.
Among the viruses analyzed here, the host cell receptor is known only for the polioviruses and for CAV21, making it difficult to consistently apply criterion iii. The CAV21 receptor (intracellular adhesion molecule 1) (45) is also the host cell receptor for the major group of human rhinoviruses (13), further clouding the issue of receptor utilization as a practical taxonomic tool. Criterion v fails to discriminate among the five human enterovirus species, as they all share a primary host, namely humans. Lastly, there is no evidence to suggest that the human enteroviruses differ fundamentally from one another in proteolytic processing, replication, or encapsidation (criterion vi), but we have shown that recombination may occur between the polioviruses and members of HEV-C during natural transmission in the human population (22).
In summary, analysis of the complete molecular data for all members of these species and application of the existing classification criteria does not support the existence of a separate Poliovirus species. We propose that the polioviruses should be considered members of HEV-C and that Poliovirus should be dropped as an enterovirus species designation. We also propose that the two serotypes CAV15 and CAV18 be reclassified into existing serotypes. Furthermore, our data suggest that CAV1, CAV19, and CAV22 appear to form a separate group, distinct from the other HEV-C serotypes in terms of recombination and genetic differences, and should be further investigated. The proposed changes to HEV-C would increase the number of recognized serotypes within the species to 12. Overall, the data presented here suggest that modifications to the current classification of viruses in the HEV-C and Poliovirus species should be considered.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype. Evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Belnap, D. M., B. M. J. McDermott, D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cammack, N., A. Phillips, G. Dunn, V. Patel, and P. D. Minor. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167:507-514. [PubMed] [Google Scholar]
- 4.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ′serotyping' of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 5.Casas, I., G. F. Palacios, G. Trallero, D. Cisterna, M. C. Freire, and A. Tenorio. 2001. Molecular characterization of human enteroviruses in clinical samples: comparison between VP2, VP1, and RNA polymerase regions using RT nested PCR assays and direct sequencing of products. J. Med. Virol. 65:138-148. [PubMed] [Google Scholar]
- 6.Committee on Enteroviruses. 1962. Classification of human enteroviruses. Virology 16:501-504. [Google Scholar]
- 7.Committee on the Enteroviruses. 1957. The enteroviruses. Am. J. Public Health 47:1556-1566. [PMC free article] [PubMed] [Google Scholar]
- 8.Dewalt, P. G., M. A. Lawson, R. J. Colonno, and B. L. Semler. 1989. Chimeric picornavirus polyproteins demonstrate a common 3C proteinase substrate specificity. J. Virol. 63:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond, D. C., B. A. Jameson, J. Bonin, M. Kohara, S. Abe, H. Itoh, T. Komatsu, M. Arita, S. Kuge, A. Nomoto, A. D. M. E. Osterhaus, R. Crainic, and E. Wimmer. 1985. Antigenic variation and resistance to neutralization in poliovirus type 1. Science 229:1090-1093. [DOI] [PubMed] [Google Scholar]
- 10.Furione, M., S. Guillot, D. Otelea, J. Balanant, A. Candrea, and R. Crainic. 1993. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology 196:199-208. [DOI] [PubMed] [Google Scholar]
- 11.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of the human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greve, J. M., G. Davis, A. M. Meyer, C. P. Forte, S. C. Yost, C. W. Marlor, M. E. Kamarck, and A. McClelland. 1989. The major human rhinovirus receptor is ICAM-1. Cell 56:839-847. [DOI] [PubMed] [Google Scholar]
- 14.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, Y., V. D. Bowman, S. Mueller, C. B. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 17.Huttunen, P., J. Santti, T. Pulli, and T. Hyypia. 1996. The major echovirus group is genetically coherent and related to coxsackie B viruses. J. Gen. Virol. 77:715-725. [DOI] [PubMed] [Google Scholar]
- 18.Hyypiä, T., T. Hovi, N. J. Knowles, and G. Stanway. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1-11. [DOI] [PubMed] [Google Scholar]
- 19.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-678. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 20.La Monica, N., C. Meriam, and V. R. Racaniello. 1986. Mapping of sequences required for mouse neurovirulence of poliovirus type 2 Lansing. J. Virol. 57:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipskaya, G. Y., A. R. Muzychenko, O. K. Kutitova, S. V. Maslova, M. Equestre, S. G. Drozdov, R. Perez Bercoff, and V. I. Agol. 1991. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J. Med. Virol. 35:290-296. [DOI] [PubMed] [Google Scholar]
- 22.Liu, H.-M., D.-P. Zheng, L.-B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobert, P.-E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Channock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 27.Melnick, J. L., V. I. Agol, H. L. Bachrach, F. Brown, P. D. Cooper, W. Fiers, S. Gard, J. H. Gear, Y. Ghendon, L. Kasza, M. LaPlaca, B. Mandel, S. McGregor, S. B. Mohanty, G. Plummer, R. R. Rueckert, F. L. Schaffer, I. Tagaya, D. A. Tyrrell, M. Voroshilova, and H. A. Wenner. 1974. Picornaviridae. Intervirology 4:303-316. [DOI] [PubMed] [Google Scholar]
- 28.Melnick, J. L., H. A. Wenner, and C. A. Phillips. 1979. Enteroviruses, p. 471-534. In E. H. Lennette and N. J. Schmidt (ed.), Diagnostic procedures for viral, rickettsial, and chlamydial infections, 5th ed. American Public Health Association, Washington, D.C.
- 29.Muckelbauer, J. K., and M. G. Rossmann. 1997. The structure of coxsackievirus B3. Curr. Topics Microbiol. Immunol. 223:191-208. [DOI] [PubMed] [Google Scholar]
- 30.Norder, H., L. Bjerregaard, L. Magnius, B. Lina, M. Aymard, and J.-J. Chomel. 2003. Sequencing of ′untypable' enteroviruses reveals two new types, EV-77 and EV-78, within human enterovirus type B and substitutions in the BC loop of the VP1 protein for known types. J. Gen. Virol. 84:827-836. [DOI] [PubMed] [Google Scholar]
- 31.Norder, H., L. Bjerregaard, and L. O. Magnius. 2001. Homotypic echoviruses share aminoterminal VP1 sequence homology applicable for typing. J. Med. Virol. 63:35-44. [PubMed] [Google Scholar]
- 32.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberste, M. S., K. Maher, D. R. Kilpatrick, and M. A. Pallansch. 1999. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 73:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Molecular phylogeny of all human enterovirus serotypes based on comparison of sequences at the 5′ end of the region encoding VP2. Virus Res. 58:35-43. [DOI] [PubMed] [Google Scholar]
- 35.Oberste, M. S., D. Schnurr, K. Maher, S. al-Busaidy, and M. A. Pallansch. 2001. Molecular identification of new picornaviruses and characterization of a proposed enterovirus 73 serotype. J. Gen. Virol. 82:409-416. [DOI] [PubMed] [Google Scholar]
- 36.Palmenberg, A. C. 1989. Sequence alignments of picornaviral capsid proteins, p. 215-230. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 37.Pfister, T., K. W. Jones, and E. Wimmer. 2000. A cysteine-rich motif in poliovirus protein 2CATPase is involved in RNA replication and binds zinc in vitro. J. Virol. 74:334-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pöyry, T., L. Kinnunen, T. Hyypia, B. Brown, C. Horsnell, T. Hovi, and G. Stanway. 1996. Genetic and phylogenetic clustering of enteroviruses. J. Gen. Virol. 77:1699-1717. [DOI] [PubMed] [Google Scholar]
- 39.Pulli, T., P. Koskimies, and T. Hyypia. 1995. Molecular comparison of coxsackie A virus serotypes. Virology 212:30-38. [DOI] [PubMed] [Google Scholar]
- 40.Racaniello, V. R., and D. Baltimore. 1981. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc. Natl. Acad. Sci. USA 78:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigo, M. J., and J. Dopazo. 1995. Evolutionary analysis of the picornavirus family. J. Mol. E. 40:362-371. [DOI] [PubMed] [Google Scholar]
- 43.Santti, J., H. Harvala, L. Kinnunen, and T. Hyypiä. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1361-1372. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, N. J., H. H. Ho, and E. H. Lennette. 1975. Propagation and isolation of group A coxsackieviruses in RD cells. J. Clin. Microbiol. 2:183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shafren, D. R., D. J. Dorahy, S. J. Greive, G. F. Burns, and R. D. Barry. 1997. Mouse cells expressing human intercellular adhesion molecule-1 are susceptible to infection by coxsackievirus A21. J. Virol. 71:785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sickles, G. M., M. Mutterer, P. Feorino, and H. Plager. 1955. Recently classified types of coxsackie virus, group A. Behavior in tissue culture. Proc. Soc. Exp. Biol. Med. 90:529-531. [DOI] [PubMed] [Google Scholar]
- 47.Stanway, G., P. J. Hughes, R. C. Mountford, P. Reeve, P. D. Minor, G. C. Schild, and J. W. Almond. 1984. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc. Natl. Acad. Sci. USA 81:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. E. 13:964-969. [Google Scholar]
- 49.Supanaranond, K., N. Takeda, and S. Yamazaki. 1992. The complete nucleotide sequence of a variant of Coxsackievirus A24, an agent causing acute hemorrhagic conjunctivitis. Virus Genes 6:149-158. [DOI] [PubMed] [Google Scholar]
- 50.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. E. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 51.Wildy, P. (ed.). 1971. Classification and nomenclature of viruses, vol. 5. S. Karger, Basel, Switzerland.
- 52.Yang, Y., R. Rijnbrand, K. L. McKnight, E. Wimmer, A. V. Paul, A. Martin, and S. M. Lemon. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]