Abstract
We have previously reported the construction and characterization of an inducible recombinant virus in which expression of the vaccinia virus membrane protein A14 is experimentally regulated using the tetracycline operator-repressor system. Repression of A14, which results in a 1,000-fold reduction in viral yield, leads to an early block in viral morphogenesis characterized by the accumulation of large virosomes, empty “crescents” that fail to contact these virosomes, and, most strikingly, large numbers of aberrant 25-nm vesicles. Here we report the establishment of a transient-complementation system for the structure-function analysis of A14. We have constructed numerous mutant alleles of A14 designed to identify and test the importance of key structural and sequence motifs within A14, including sites of posttranslational modification, such as glycosylation, phosphorylation, and dimerization. From these studies we have determined that robust complementation ability requires an intact N terminus and two regions flanking the first membrane-spanning domain of A14. We show that A14 is modified by N-linked glycosylation both in vitro and in vivo. However, only a minority of A14 molecules are glycosylated in vivo and these are not encapsidated. In this report we also identify the sole phosphorylated serine residue of A14 as lying within the NHS85 motif that undergoes glycosylation. Additionally, we show that the Cys71 residue is required for intermolecular disulfide bond formation and describe the properties of a virus expressing an allele of A14 that cannot form disulfide-linked dimers.
Vaccinia virus (VV) morphogenesis can be divided into several visually distinct stages (6, 20). Within a cytoplasmic zone from which cellular organelles have been largely excluded, the viral proteins to be encapsidated coalesce into an electron-dense virosome. Crescent-shaped precursors of the virion membrane then develop adjacent to the periphery of the virosome. These viral crescents are thought to envelop a portion of the viroplasm as they enlarge and circularize, giving rise to the first intracellular form of VV, immature virions (IV). IV then acquire an electron-dense, eccentric nucleoid whose appearance has been associated with the encapsidation of the virion genome, a process that remains poorly understood (4; O. Grubisha and P. Traktman, submitted for publication). These immature, nucleoid-containing particles undergo several structural changes leading to their maturation into infectious, intracellular mature virions (IMV), the brick-shaped particles that contain a biconcave core (9, 13). This maturation is accompanied by a characteristic set of proteolytic processing events (3, 38, 43). A small minority of IMV transit to the Golgi apparatus on microtubules (14, 22, 41), where they become wrapped by a Golgi-derived double membrane, generating intracellular enveloped virions (IEV) (33). IEV move to the cellular periphery on microtubules and undergo exocytosis, retaining a single Golgi-derived envelope. Upon egress, some of the virions remain tethered to the plasma membrane (cell-associated enveloped virus) and are propelled outward on actin tails in a manner that facilitates infection of adjacent cells (12, 22, 28, 41, 42). Other virions are shed from the cell surface; these extracellular enveloped virions (EEV) mediate distal spread throughout the infected host (32, 33).
Previous work has placed the F10 kinase at the top of the hierarchy of proteins that play essential roles in the earliest stage of morphogenesis (35, 40). Temperature-sensitive (ts) lesions in the F10 protein block VV morphogenesis prior to virosome condensation or crescent formation, implying that protein kinase activity may be required for the diversion of cellular membranes as well as the maturation of viral proteins (1, 8). A ts lesion in the H5 protein, a substrate of F10, also results in the arrest of morphogenesis prior to crescent formation (7). The phenotype seen during nonpermissive infections with tsH5 is much like that seen with tsF10, except that nonfunctional virosomes with a “curdled” appearance are seen.
Several other phosphoproteins have been shown to play essential roles in virion morphogenesis (1, 7, 46). Among them is the A14 protein, which was identified as a membrane component that becomes hyperphosphorylated when the H1 phosphatase is repressed (18, 36). A14 was also identified as a binding partner of the A17 protein, another phosphorylated component of the virion membrane (26). When expression of either A14 or A17 is repressed, a dramatic inhibition of virion morphogenesis is seen. Electron-dense virosomes are observed, but no IV, immature, nucleoid-containing particles, or IMV are seen; this phenotype suggests that the block to A14- and A17-deficient infections occurs just downstream of the F10 and H5 defects. In the absence of A17, the virosomes are surrounded by a large number of membrane vesicles and/or tubules, but no crescents are seen (24, 25, 44). In the absence of A14, a few crescents that fail to make normal contacts with the viroplasm are seen, but the most dramatic phenotype is the accumulation of large clusters of 20- to 30-nm-diameter vesicles (and/or tubulovesicular membranes) (27, 36).
We envision the earliest stage of viral membrane biogenesis as involving F10 kinase-mediated diversion of membranous components of the cellular secretory pathway, which contain the viral proteins A14 and A17 within their lipid bilayer. We envision that, in the absence of A14 or A17, vesicles cannot undergo further maturation, either initially into crescents or later into the spherical membranes that surround IV. We also envision that, in the absence of A14, the membrane vesicles cannot move to the periphery of the virosome and/or make specific contacts with the viroplasm.
Inducible recombinants in which A14 expression can be manipulated experimentally have been previously generated (27, 36). With the vindA14 virus that has been previously described, expression of A14 is tightly repressed unless tetracycline (TET) is added to the culture medium. In the absence of inducer and hence the absence of A14, plaque formation is abolished and the 24-h yield from cultures infected at a multiplicity of infection (MOI) of 2 is 3 orders of magnitude less than that obtained during permissive infections. This aborted infection reflects the morphogenesis defect described above.
The goal of the work described here was to elucidate which features and properties of A14 are required for its biological function; to do so, we exploited the phenotype of the inducible vindA14 recombinant and developed a transient-complementation assay. We describe herein the construction of a variety of altered alleles of A14 that were designed to uncover key sequence elements within the protein, define the sites of posttranslational modification, and test the importance of these modifications on A14's function in vivo.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, T4 DNA ligase, calf intestinal phosphatase, pancreatic RNase, Taq polymerase, and DNA molecular weight standards were purchased from New England Biolabs (Beverly, Mass.) or Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and were used according to the instructions provided by the manufacturers. [32P]orthophosphate and [35S]methionine were obtained from Dupont/New England Nuclear Corp. (Boston, Mass.). Lipofectamine Plus Reagent was purchased from Gibco BRL (Gaithersburg, Md.), and Endofree Maxi Kits came from Qiagen (Valencia, Calif.). Tunicamycin was acquired from ICN (Aurora, Ohio), and endoglycosidase H (EndoH) came from Roche (Indianapolis, Ind.). DNA oligonucleotides were synthesized by IDT (Coralville, Iowa).
Cells and viruses.
Monolayers of BSC40 primate cells were maintained in Dulbecco modified Eagle medium (DMEM; Gibco BRL) containing 5% fetal bovine serum (FBS) at 37°C. Wild-type (wt) VV (strain WR), vtetR (36), vindA14 (36), and vindA17 (kindly provided by Bernard Moss, National Institutes of Health, Bethesda, Md.) (44) were used where indicated. All viral stocks were prepared from cytoplasmic lysates by ultracentrifugation through 36% sucrose and in some instances were further purified by banding on 25 to 40% sucrose gradients (18). During permissive infections with vindA14, TET was included in the culture medium at 1 μg/ml. Where indicated, tunicamycin was added to infected cultures at a final concentration of 1 μg/ml.
Construction of A14 mutants.
Mutant alleles of the A14 gene were generated by using overlap PCR; viral genomic DNA served as the template. Table 1 shows the primers used to generate the various A14 mutants.
TABLE 1.
Construction of A14 mutantsa
| Primer | Sequence |
|---|---|
| O1 | 5′ CCATCGATGGACATGATGCTTAT 3′ |
| O4 | 5′ CGGGATCCTTAGTTCATGGAAATAT 3′ |
| S12A-B | 5′ CACGCCGGCAAAATAATTTCCAATCA 3′ |
| S12A-C | 5′ ATTATTTTGCCGGCGTGCTAATCGCT 3′ |
| S25A-B | 5′ GATACACGCAAGAATCAAAAGAATGA 3′ |
| S25A-C | 5′ TGATTCTTGCGTGTATCTTCGCCTTT 3′ |
| S34A-B | 5′ GTAGACTTAGCAAAGTCAATAAAGGCGA 3′ |
| S34A-C | 5′ TTGACTTTGCTAAGTCTACCAGTCCCA 3′ |
| S36A-B | 5′ ACTGGTAGCCTTACTAAAGTCAATAA 3′ |
| S36A-C | 5′ TTAGTAAGGCTACCAGTCCCACTCGT 3′ |
| S38A-B | 5′ GAGTGGGAGCGGTAGACTTACTAAAGT 3′ |
| S38A-C | 5′ AGTCTACCGCTCCCACTCGTACATGGA 3′ |
| S34,36,38A-B | 5′ GAGTGGGAGCGGTAGCCTTAGCCAAAGTCAATAAAGGCG 3′ |
| S34,36,38A-C | 5′ TTGACTTTGCTAAGGCTACCGCTCCCACTCGTACATGGA 3′ |
| S47A-B | 5′ GCCATAATAGCCAATACTTTCCATGTA 3′ |
| S47A-C | 5′ AAGTATTGGCTATTATGGCGTTTATAC 3′ |
| S65A-B | 5′ CCACATAGCATAAATTAGCATTCCG 3′ |
| S65A-C | 5′ TAATTTATGCTATGTGGGGAAAGCA 3′ |
| S77A-B | 5′ AATGACTCCGGCAACTCTGTGGGGTGCG 3′ |
| S77A-C | 5′ CCCACAGAGTTGCCGGAGTCATTCATACC 3′ |
| S88A | 5′ GGGATCCTTAGTTCATGGCAATATCGCTATGATTG 3′ |
| N′ deletion | 5′ CCATCGATGATTGGAAATTATTTTTCCGGC 3′ |
| C′ deletion | 5′ CGGGATCCTTACATAGAATAAATTAG 3′ |
| PTRTWK-B | 5′ TGCCGCTGCAGCAGCGGCACTGGTAGACTTACTA 3′ |
| PTRTWK-C | 5′ GCCGCTGCTGCAGCGGCAGTATTGAGTATTATG 3′ |
| P39A-B | 5′ ACGAGTGGCACTGGTAGACTTACTA 3′ |
| P39A-C | 5′ CTACCAGTGCCACTCGTACATGGAAAG 3′ |
| R41K44-B | 5′ TACTGCCCATGTAGCAGTGGGACTGGTAGA 3′ |
| R41K44-C | 5′ ACTGCTACATGGGCAGTATTGAGTATTATG 3′ |
| W43A-B | 5′ ATACTTTCGCTGTACGAGTGGGACTG 3′ |
| W43A-C | 5′ CTCGTACAGCGAAAGTATTGAGTATT 3′ |
| W43K44-B | 5′ TCAATACTGCCGCTGTACGAGTGGGACTG 3′ |
| W43K44-C | 5′ CTCGTACAGCGGCAGTATTGAGTATTATG 3′ |
| NYF-B | 5′ AGCAGCAGCTCCAATCATAAGCATC 3′ |
| NYF-C | 5′ GCTGCTGCTTCCGGCGTGCTAATC 3′ |
| CIFAF-B | 5′ AGCGGCGGCGGCAGCCGAAAGAATCAAAAG 3′ |
| CIFAF-C | 5′ GCTGCCGCCGCCGCTATTGACTTTAGTAAG 3′ |
| DFSK-B | 5′ CGCAGCAGCGGCAATAAAGGCGAAGATA 3′ |
| DFSK-C | 5′ GCCGCTGCTGCGTCTACCAGTCCCACT 3′ |
| MWG-B | 5′ TGCCGCCGCAGAATAAATTAGCATT 3′ |
| MWG-C | 5′ GCGGCGGCAAAGCACTGCGCACCC 3′ |
| C71S-B | 5′ TGGGGTGCGCTGTGCTTTCCCCACAT 3′ |
| C71S-C | 5′ GAAAGCACAGCGCACCCCACAGAGTT 3′ |
| N83Q | 5′ CGGGATCCTTAGTTTACGGAAATATCGCTATGCAGGGTATGAATGACTCCA 3′ |
| S85A | 5′ CGGGATCCTTAGTTCAGTGAAATATCGGCATGATTGGTATGAATG 3′ |
| vS85A-B | 5′ GAAATATCCCGATGATTGGTATGAAT 3′ |
| vS85A-C | 5′ CCAATCATGCCGATATTTCCATGAAC 3′ |
Primer O1 inserts a ClaI site (in bold) at the 5′ terminus, overlapping the translational start site (underlined) of A14. Primer O4 inserts a BamHI site (in bold) at the 3′ terminus, immediately downstream of the translational stop site (underlined) of the A14 open reading frame. The B and C primers serve to mutate the various residues indicated in the table. Construction of most of these mutants involved two sequential rounds of PCR. In the first round, two reactions utilizing primers O1 plus B or O4 plus C were performed to generate subgenic fragments of the A14 gene containing the desired mutations within a 23-bp region of overlap. These products were purified on glass beads (39), with aliquots of each serving as the template for a second round of PCR using primers O1 and O4. Mutants of A14 not generated by overlap PCR include N′ deletion, C′ deletion, S85A, S88A, and N83Q. These mutants were generated in one round of PCR by substituting the N′ deletion primer for O1 or the C′ deletion S85A, S88A, or N83Q primer for O4. In all cases, the final products were purified on glass beads and were then subjected to digestion with BamHI and ClaI. These inserts were then ligated into pUC1246 plasmid DNA that had been previously digested with BamHI and ClaI and treated with calf intestinal alkaline phosphatase. pUC1246 places the A14 alleles under the regulation of a strong, late promoter derived from the cowpox ATI gene (23, 36). E. coli transformants carrying the pUC1246-A14 plasmids were generated, and the plasmids were purified by using Qiagen Endofree Maxi kits (Valencia, Calif.). All A14 alleles were verified by restriction enzyme digestion and automated DNA sequencing.
Infection-transfection assay.
Sixty-millimeter dishes of confluent BSC40 cell monolayers were infected with vindA14 at an MOI of 5 in the presence or absence of TET as indicated. The inoculum was removed, and fresh medium containing 5% FBS was applied after a 30-min adsorption period. Three hours postinfection (hpi), 10 μg of undigested pUC1246 vector or of the indicated pUC1246-A14 constructs was applied to cells by using Lipofectamine Plus Reagent (Gibco BRL) according to the manufacturer's instructions. Cells were harvested at 30 hpi; lysates were examined by immunoblot analysis and were also subjected to titration to determine the viral yield (number of PFU per milliliter). All experiments were performed in triplicate, and the results were averaged.
Immunoblot analysis.
Extracts of cells and virions were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and were transferred to nitrocellulose (Schleicher & Schuell, Keene, N.H.) filters in CAPS transfer buffer (10 mM CAPS [3-{cyclohexylamino-1-propanesulfonic acid}] in 10% methanol, pH 11.3). Filters were probed with the previously described anti-A14 serum (36) and then with horseradish peroxidase-conjugated secondary antibody (Bio-Rad, Richmond, Calif.). Immunoreactive species were visualized after development with enhanced chemiluminescence reagents (Pierce, Rockford, Ill.).
Metabolic labeling and immunoprecipitation. (i) Metabolic labeling with [35S]methionine.
Monolayers of BSC40 cells were infected at an MOI of 10 with either wt virus or vindA17 (without isopropyl-β-d-thiogalactopyranoside [IPTG] inducer) in the presence or absence of tunicamycin. At 6 hpi cultures were rinsed with methionine-free DMEM (ICN/FLOW, Costa Mesa, Calif.) supplemented with l-glutamine and were fed with the same medium containing 100 μCi of [35S]methionine (EXPRESS label; NEN-Dupont) per ml. Cells were labeled for a total of 3 h and were prepared for immunoprecipitation analysis. Cells were rinsed with phosphate-buffered saline (PBS) and were lysed in 1× 0.1 M NaPO4 (pH 7.4), 0.1 M NaCl, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate. Lysates were clarified by centrifugation and were incubated with primary serum for 4 h followed by incubation with protein A-Sepharose (Sigma) for 1.5 h; immunoprecipitates were then retrieved by centrifugation, washed thoroughly, and analyzed by SDS-PAGE and autoradiography or fluorography.
(ii) Labeling with [32P]orthophosphate.
Infected monolayers were incubated with phosphate-free DMEM (ICN-Flow, Costa Mesa, Calif.) supplemented with phosphate-free 5% FBS (prepared by dialysis against Tris-buffered saline [25 mM Tris-HCl, pH 7.4, 136 mM NaCl, and 2.7mM KCl]), and 100 μCi of 32PPi per ml. Labeling was performed from 6 to 24 hpi. Cells were then harvested as described above for immunoprecipitation analyses.
Construction of pTM1-A14 mutants.
All pTM1-A14 mutant constructs were generated by PCR by using the aforementioned pUC1246-A14 constructs as template DNA. The 5′ primer 5′ GGATCCCCATGGACATGATGCTTAT 3′ inserts an NcoI site (boldface) that overlaps the translational start site (underlined) of A14. Primer O4 (Table 1) inserts a BamHI site (boldface) downstream of the A14 open reading frame. Upon purification and digestion, final PCR products were ligated into pTM1 plasmid DNA (11) that had been previously digested with NcoI and BamHI and treated with calf intestinal alkaline phosphatase (CIP). Plasmids were purified from Escherichia coli transformants and were verified by restriction enzyme digestion and automated DNA sequencing.
In vitro transcription and translation of A14.
In vitro transcription and translation (IVTT) of all pTM1-A14 constructs was performed by using the TNT T7-Coupled Reticulocyte Lysate System purchased from Promega (Madison, Wis.). Canine pancreatic microsomal membranes (Promega) were included in all IVTT reactions. Proteins were transcribed and translated in 50-μl reactions containing Redivue l-[35S]methionine (Amersham Pharmacia, Piscataway, N.J.) per the manufacturer's protocol. Microsomal membranes were purified from completed reactions by centrifugation at 16,000 × g for 20 min at 4°C, resuspended in 200 μl of PBS, and sedimented for an additional 20 min. The supernatant was aspirated, and the membranes were resuspended in 50 μl of PBS, which was then layered on 75 μl of 0.5 M sucrose cushion and was sedimented for an additional 20 min. The supernatant was again removed, and the membranes were resuspended in 50 μl of PBS (5). Membranes containing the A14 protein were then analyzed by SDS-PAGE and autoradiography.
Construction of VVs containing A14 mutant alleles.
Overlap PCR was used to generate an 800-bp DNA fragment containing the appropriate 273-bp A14 allele as well as 200 bp of upstream (towards A15) and 300 bp of downstream (towards A13) sequence. The primers used to generate this fragment were U, 5′ CGGGATCCGTAAACCCGTCGTCTATA 3′, and D, 5′ CGGGATCCTGGTATTTTTTGGTTCTG 3′, as well as primers C71S-B and -C, N83Q-B and -C, or vS85A-B and -C (Table 1). Overlap PCR was performed as described in “Construction of A14 mutants.” Primers 1 and 4 were used to insert BamHI sites at the termini of the 800-bp fragment (boldface), while the B and C primers inserted the desired mutations. The final 800-bp fragments were purified on glass beads, digested with BamHI, and ligated to pUC19 DNA that had been previously digested with BamHI and treated with CIP. Appropriate plasmids were purified after selection in E. coli. Three micrograms of linearized DNA was used to transfect BSC40 cells infected with vindA14 at an MOI of 0.03 in the absence of inducer. Infected cells were harvested at 48 hpi, and TET-independent viruses were selected by plaque purification in the absence of inducer. To confirm that these TET-independent viruses had lost the tet operator by homologous recombination with the transfected plasmid, plaques were screened by PCR with primers that flanked the transcriptional start site of A14. These primers led to the amplification of 188- or 213-bp products from viruses lacking or containing the tet operator, respectively. Plaques that generated PCR products of 188 bp were further screened for the acquisition of the desired mutation by automated DNA sequencing. Plaques fulfilling both criteria were purified by four rounds of plaque purification prior to the preparation of high-titer viral stocks. In some cases, virions were also banded on sucrose gradients for the purpose of further characterization.
Determination of 24-h yield.
Confluent 60-mm dishes of BSC40 cells were infected with wt virus in the presence or absence of brefeldin A (BFA) or vA14(N83Q) or vA14(C71S) at an MOI of 2 or 15 and were maintained at 37°C. At 24 hpi, the culture medium was collected and clarified by low-speed sedimentation; the extracellular virus (ECV) was retrieved by subjecting the clarified medium to a second high-speed sedimentation (16,000 × g, 30 min, 4°C). The cells were harvested by scraping in 1 ml of PBS and were collected by low-speed sedimentation. Both the cellular and ECV pellets were resuspended in 1 mM Tris (pH 9.0). Cells were disrupted by three rounds of freeze-thawing and two 15-s pulses of sonication. Yields of cell-associated virus (CAV) and ECV were determined by titration at 37°C on BSC40 cell monolayers.
Virion fractionation.
wt or vA14(C71S) virions were purified from cytoplasmic lysates of infected cells by sedimentation through a 36% sucrose cushion followed by banding on a 25 to 40% sucrose gradient. Five micrograms of banded virions was incubated at 37°C for 30 min in one of various reaction mixtures: 100 mM Tris (pH 9.0), 100 mM Tris (pH 9.0) supplemented with 0.1% or 1% NP-40 (vol/vol), or 100 mM Tris (pH 9.0) supplemented with 0.1% or 1% NP-40 (vol/vol) and 50 mM dithiothreitol (DTT). After the permeabilization reaction, solubilized and particulate fractions representing the membrane and core components of the virion, respectively, were separated by sedimentation (16,000 × g, 30 min, room temperature).
Virion detergent sensitivity assay.
A total of 2.0 × 108 wt or vA14[C71S] virions (purified as described above) were incubated at 37°C for 30 min in a reaction mixture containing 100 mM Tris (pH 9.0) and various percentages of NP-40 (vol/vol): 0, 0.01, 0.05, and 0.1%. Samples were then subjected to titration on BSC40 cells to determine the viral titer (number of PFU/milliliter). All titers were performed in duplicate, and the results were averaged.
Sequence analysis and preparation of figures.
Sequences were retrieved from the Poxvirus Bioinformatics Resource Center (http://www.poxvirus.org), and alignments were performed by using the Clustal V method and Lasergene software (DNASTAR, Inc., Madison, Wis.). Original data were scanned on a SAPHIR scanner (Linotype-Hell Co., Hauppauge, N.Y.) and were adjusted with Adobe Photoshop software (Adobe Systems, Inc., San Jose, Calif.). Graphs were prepared by using Sigma Plot software (SPSS, Chicago, Ill.). Final figures were prepared with Canvas software (Deneba Systems, Inc., Miami, Fla.).
RESULTS
In order to identify which functional and/or structural features of A14 were necessary for its role in morphogenesis, we undertook a mutational analysis of this essential protein. Our strategy was to construct various mutant alleles of A14L, analyze some of the resultant proteins after coupled transcription and translation in vitro, and in all cases test the ability of the altered alleles to substitute for wt A14 during nonpermissive infections with vindA14. The mutants that we generated can be divided into two main groups: those generated to define key structural regions and motifs within A14 and those generated to investigate posttranslational modification of A14 by glycosylation, phosphorylation, and intermolecular disulfide bond formation.
Identification of key structural regions of A14.
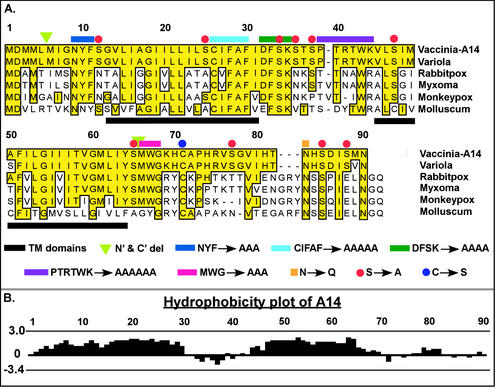
Figure 1A presents an alignment of VV A14 with the homologs encoded by the closely related poxviruses variola virus, rabbitpox virus, and monkeypox virus, as well as the more distantly related myxoma virus and molluscum contagiosum. VV A14 shares 97% identity with variola virus A14 and ≥42% identity with the other homologs shown; the highly conserved residues are highlighted in yellow. The A14 protein contains a methionine-rich N terminus, two predicted transmembrane regions (comprising residues 13 to 31 and 45 to 64, underlined with black bars) separated by a small loop region, and a hydrophilic C-terminal tail. Mutant alleles containing truncations at one or both termini (ΔN′, ΔC′, and ΔN′C′) were constructed, as shown by the green triangles. Several highly conserved regions of A14 that may contribute to its function can easily be seen in this alignment (yellow shading) and were chosen for analysis. Mutants were constructed in which the most highly conserved motifs, i.e., PTRTWK (purple line), NYF (dark blue), CIFAF (cyan), DFSK (dark green), and MWG (magenta), were changed to alanine residues.
FIG. 1.
VV A14 is highly conserved among the Poxviridae family. (A) Alignment of VV A14 with several of its orthopox homologs. Putative membrane-spanning regions are underlined in black. The A14 mutants constructed for this study are depicted as follows: green triangles indicate the new initiation and termination sites engineered for alleles containing N′ and/or C′ truncations. Several sequences chosen for their high level of conservation among the Poxviridae homologs were changed to an equivalent number of Ala residues: N9YF (dark blue overline), C26IFAF (cyan), D32FSK (dark green), P37TRTWK (purple), and M66WG (magenta). Asn83 (orange square), part of a highly conserved N-linked glycosylation motif (NXS) at the C terminus of A14, was changed to Gln. Cys71 (blue circle), likely to direct the covalent dimerization of A14 dimer, was changed to Ser. Red circles highlight Ser residues at positions 12, 25, 34, 36, 38, 47, 65, 77, 85, and 88 that were mutated to Ala, individually and in various combinations (see text). (B) Hydrophobicity plot of VV A14 as determined by the Kyte-Doolittle method. The highly hydrophobic A14 protein is thought to span the membrane twice; the two transmembrane domains comprise residues 13 to 31 and 45 to 64, which are predicted to adopt inside-to-outside and outside-to-inside orientations, respectively (http://www.ch.embnet.org/software/TMPRED_form.html). The hydrophobic domains are separated by a 13-aa hydrophilic loop region.
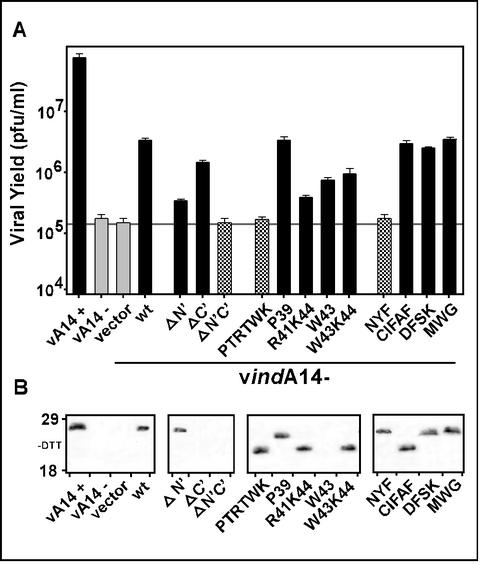
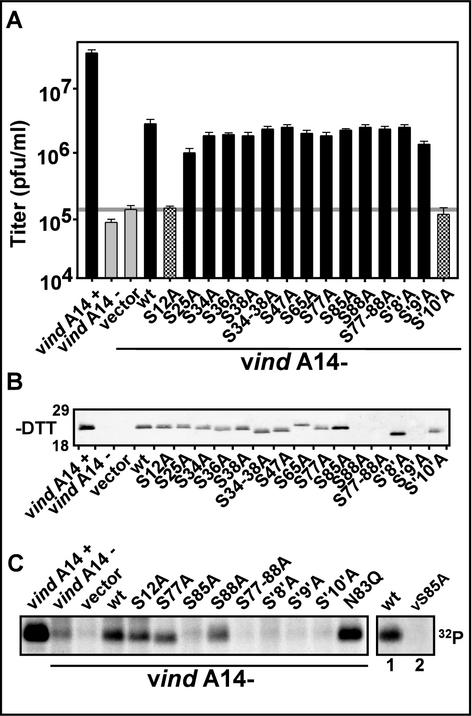
These mutant alleles were then tested in our transient-complementation assay in order to assess their ability to substitute for endogenous A14. The data shown in Fig. 2A illustrate the comparative ability of these mutants to augment virus production during nonpermissive vindA14 infections, in which the endogenous A14 gene is repressed. In the absence of inducer, ∼1,000-fold less virus was produced than in the presence of inducer. Transfection of a plasmid expressing wt A14 restored a significant level of virus production, approximately 1.5 logs greater than that obtained without transfection or upon transfection of empty vector. Expression of the ΔN′ construct was 10-fold less effective than that of wt, while the ΔC′ construct complemented nearly as well as the wt A14. When both the N′ and C′ termini were trimmed (ΔN′C′), no complementation was seen and virus production was comparable to that obtained upon transfection of empty vector.
FIG. 2.
Transient rescue of vindA14 performed with structural mutants of A14. (A) Determination of complementation competency by titration of viral yield from infections and transfections. BSC40 cells were infected at an MOI of 5 with vindA14 virus. Infections were performed in the presence (+) or absence (−) of TET, followed by transfection of 10 μg of empty vector or vector encoding the indicated alleles of A14. Extracts were harvested at 24 hpi and were titrated in order to assess the ability of the various A14 alleles to substitute for endogenous virus A14 in a transient-rescue assay. Cells infected in the presence (+) and absence (−) of TET served as positive and negative controls, respectively; cells infected in the absence of inducer and transfected with empty vector or with the plasmid encoding wt A14 served as the benchmarks for transient rescue. The horizontal grey line shows the titer obtained upon transfection of empty vector. The data shown represent the average of three independent experiments. (B) Immunoblot analysis of A14 expression. Aliquots of the extracts described above were resolved by nonreducing SDS-17% PAGE and were subjected to immunoblot analysis with anti-A14 serum to monitor A14 expression from the endogenous and transfected alleles. The positions of the 18,000- and 29,000-Mr standards are shown at the left.
The structural feature that we chose to analyze next was the short hydrophilic loop between the transmembrane regions of A14 (Fig. 1B). Replacement of P39TRTWK with an equivalent number of Ala residues completely abolished the complementation ability of the transfected allele. To further investigate why this mutant was unable to restore virus production, additional alleles containing substitutions within PTRTWK were prepared and were then tested in our infection and transfection assay. (i) Since this region contains a Pro residue and since the PTRTWK→AAAAAA protein exhibited an altered mobility on SDS-PAGE (Fig. 2B), we wanted to know whether the structural perturbation induced by removing the proline might inhibit A14 function. We therefore generated an A14 allele in which only Pro39 was changed to Ala. As seen in Fig. 2A, this allele was capable of restoring viral yields as well as wt A14. (ii) This region of the hydrophilic loop region contains two charged residues, Arg41 and Lys44, and one aromatic residue, Trp43, which together might represent an interface for protein-protein interactions (2). To determine the importance of these residues to A14 function, we mutated them to Ala in the following combinations: R41K44 (PTATWA), W43 (PTRTAK), and W43K44 (PTRTAA). None of these mutants retained wt levels of complementation ability (1.5 logs > vector). Mutation of R41K44 caused the most dramatic diminution (0.5 logs > vector), while mutation of W43 or W43K44 decreased the complementation ability to 0.75 logs > vector. Together, these data confirm that several amino acids within the P39TRTWK region are important for the biological function of A14.
We next focused on the most highly conserved regions of the A14 protein, several of which flank the predicted transmembrane domains and one of which comprises the C-terminal portion of the first transmembrane domain (Fig. 1A). We constructed mutant alleles of A14 in which the amino acid clusters N9YF, C26IFAF, D32FSK, and M66WG were changed to an equivalent number of Ala residues. Ala substitution of CIFAF, DFSK, or MWG had no impact on the ability of these alleles to function at wt efficiency in our transient-complementation assay. In contrast, the NYF→AAA mutant was unable to complement, indicating that these residues lying just upstream of the first transmembrane domain of A14 are essential to its function.
Immunoblot analyses of lysates prepared from the infection-transfection experiments were performed to confirm that all of the mutant alleles of A14 were expressed (Fig. 2B). With the exception of the ΔC′, ΔN′C′, and W43A constructs, all of the A14 proteins expressed from the transfected alleles were detected at equivalent levels. Moreover, the levels obtained upon transfection were comparable to those seen for endogenous virus A14 expressed during permissive infections with vindA14(+). We believe that our inability to detect certain mutant forms of A14 by immunoblot analysis is most likely the result of their aggregation due to the intrinsic amino acid changes, altered intracellular localization, or loss of key protein-protein interactions. There is no correlation between the absence of a signal on the immunoblot and a loss of complementation activity, and immunoprecipitation analyses (not shown) confirm that the mutant proteins can indeed be recognized by our polyclonal anti-A14 serum.
A14 protein undergoes glycosylation in vitro and in vivo, but this form of A14 is not encapsidated.
VV proteins are subject to a plethora of posttranslational modifications, including phosphorylation, acylation, proteolytic processing, and glycosylation. Thirteen glycosylated VV proteins have been found; these proteins have been shown to be secreted, targeted to the plasma membrane of virally infected cells, or incorporated into the Golgi-derived membrane of EEV (31). None, to date, has been shown to be components of the IMV membrane, although some IMV proteins, like A14, contain a consensus site for N-linked glycosylation (N83HS).
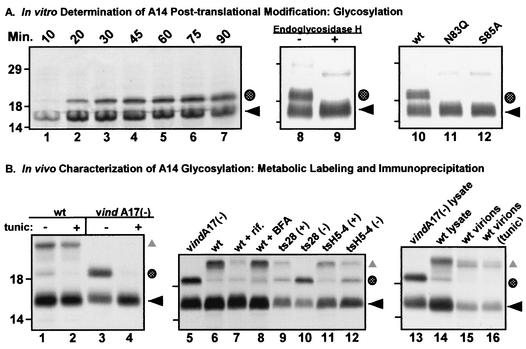
IVTT reactions programmed with the A14 gene in the presence of microsomal membranes generated the predicted primary translation product (apparent Mr, 16,000 [Fig. 3A, lanes 8 and 10, ◂) and an additional, abundant species with an Mr of ∼18,000 (Fig. 3A, lanes 8 and 10, shaded circle). Although glycosylation of A14 had previously been discounted (26), we considered it likely that the addition of N-linked sugars accounted for the 18-kDa form of A14. Because the addition of an N-linked carbohydrate is a posttranslational event, we performed an IVTT time course to assess whether the 18-kDa species of A14 represented a posttranslationally modified form of the 16-kDa primary translation product (Fig. 3A, lanes 1 to 7). Whereas the 16-kDa species of A14 (◂) accumulated rapidly, appearing within 10 min after initiation of the reaction (lane 1), the 18-kDa species of A14 (dotted circle) was not seen until 20 min and continued to accumulate over time. These results are consistent with the 16- and 18-kDa species having a precursor-product relationship. To elucidate the nature of the processing, we treated the reaction products with EndoH, which removes N-linked sugar moieties. As seen in Fig. 3A, lane 9, treatment with EndoH completely converted the 18-kDa form of A14 to the 16-kDa form, demonstrating that indeed the 18-kDa species of A14 represents an N-linked glycosylated form of the protein. Furthermore, mutation of either Asn83 (N83Q) or Ser85 (S85A), the two residues that define the functional N-linked glycosylation motif abolished the ability of A14 to be glycosylated in vitro (lanes 10 to 12).
FIG. 3.
A highly conserved N-linked glycosylation motif found at the carboxy terminus of A14 is functional both in vitro and in vivo. (A) In vitro characterization of posttranslational modification of A14 by N-linked glycosylation. Lanes 1 to 7, IVTT time course of A14. A 50-μl IVTT reaction was programmed with pTM1-A14 and was performed in the presence of microsomal membranes according to the manufacturer's instructions. Five-microliter aliquots were removed at the times indicated (10 to 90 min) and were analyzed by SDS-PAGE (under reducing conditions) and autoradiography (lanes 1 to 7). The black arrow indicates the primary translation product; the shaded circle indicates a species of lower mobility that is predicted to represent a glycosylated form of A14. Lanes 8 and 9, removal of N-linked carbohydrate moiety from IVTT-synthesized A14 by EndoH. wt A14 synthesized in an IVTT reaction in the presence of microsomal membranes was subsequently incubated at 37°C for 24 h in the absence (lane 8) or presence (lane 9) of EndoH. EndoH treatment led to the disappearance of the slowly migrating form of A14 (shaded circle). Lanes 10 to 12, IVTT reactions programmed with alleles of A14 containing a disrupted N-linked glycosylation motif. Parallel IVTT reactions performed in the presence of microsomal membranes were programmed with plasmids encoding wt A14 (lane 10) or mutant alleles of A14 containing Asn83-to-Gln (lane 11, N83Q) or Ser85-to-Ala (lane 12, S85A) substitutions. Both of these amino acid substitutions prevent the appearance of the slowly migrating form of A14 (shaded circle). For all three panels, reactions were analyzed by SDS-PAGE and autoradiography; the positions of the 14,000-, 18,000-, and 29,000-Mr standards are shown at the left. (B) Determination of N-linked glycosylation status of A14 expressed in vivo. Lanes 1 to 4, low levels of glycosylated A14 are seen during wt virus infections. BSC40 monolayers were infected with either wt (lanes 1 and 2) or vindA17(−) virus (lanes 3 and 4) at an MOI of 10 in the presence (+) or absence (−) of tunicamycin (tunic). Cultures were metabolically labeled with [35S]methionine from 6 to 9 hpi; cell lysates were prepared and were subjected to immunoprecipitation with anti-A14 serum. Immunoprecipitates were resolved by SDS-PAGE and were visualized by autoradiography. ◂ indicates unmodified A14; the shaded circle indicates the putative glycosylated form of A14; ▴ indicates coprecipitated A17 protein. Lanes 5 to 12, early blocks to virion morphogenesis increase the levels of glycosylated A14. Cells were infected with either vindA17(−) (lane 5), wt virus (lane 6), wt virus plus rifampin (rif, lane 7), wt virus plus BFA (lane 8), ts28 at permissive (+) and nonpermissive (−) temperatures (lanes 9 and 10), or tsH5-4 at permissive (+) and nonpermissive (−) temperatures (lanes 11 and 12). Cells were metabolically labeled with [35S]methionine from 6 to 9 hpi; cell lysates were subjected to immunoprecipitation with anti-A14 serum. Immunoprecipitates were resolved by SDS-PAGE and were visualized by autoradiography. The symbols indicate the unmodified (◂) and glycosylated (shaded circle) forms of A14 as well as A14's binding partner, A17 (▴). Lanes 13 to 16, the glycosylated form of A14 is not present within virions. BSC40 cells were infected at an MOI of 2 with wt virus in the absence (lane 15) or presence (lane 16) of tunicamycin. Virions were harvested 48 hpi and were purified by ultracentrifugation through a 36% sucrose cushion followed by banding on a 25 to 40% sucrose gradient. Virions were disrupted and subjected to immunoprecipitation with anti-A14 serum. Immunoprecipitates were resolved by SDS-PAGE and were visualized by fluorography. Immunoprecipitates from vindA17(−)- and wt virus-infected cell lysates (lanes 13 and 14, respectively) served as controls for high and low levels of glycosylated A14 (shaded circle) as well as the coprecipitation of the A17 protein (▴). For all three panels, the positions of the 14,000-, 18,000-, and 29,000-Mr standards are shown at the left.
Because no glycoproteins have been found associated with the IMV membrane to date, we wanted to determine if A14 was glycosylated in vivo and, if so, whether or not this form of A14 was encapsidated. [35S]methionine-labeled A14 was immunoprecipitated from lysates of cells infected with either wt virus (Fig. 3B, lanes 1 and 2) or with vindA17 in the absence of IPTG, which results in the repression of A17 synthesis and hence an early block in virion morphogenesis (lanes 3 and 4) (24, 44). In each case, infections were performed both in the presence or absence of tunicamycin, a drug that inhibits GlcNAc transferase, thereby preventing N-linked glycosylation (10). The glycosylated form of A14 (shaded circle) was retrieved at detectable but low levels from cells infected with wt VV (lane 1). Significantly increased levels of glycosylated A14 (shaded circle) were seen when A17 expression was repressed (compare lanes 1 and 3; the coimmunoprecipitated A17 protein [▴] was evident in lanes 1 and 2 but was absent in lanes 3 and 4). In either case glycosylation was abolished in the presence of tunicamycin; a concomitant increase in the nonglycosylated form of A14 was seen in the vindA17(−) infection (lane 4). These results indicate that in vivo, in the context of a wt virus infection, a minor portion of A14 is posttranslationally modified by N-linked glycosylation. In the absence of expression of the A17 protein, which leads to a disruption in A14-A17 interaction and causes an early arrest in virion morphogenesis, there is a dramatic increase in the fraction of A14 molecules that become glycosylated.
This result led us to examine the levels of A14 glycosylation under a variety of conditions that affect virion morphogenesis (Fig. 3B, lanes 5 to 12). Cells were infected with wt virus in the presence of drugs that inhibit morphogenesis (lanes 5 to 8) or with ts mutants defective in one of several genes required for the early stages of VV morphogenesis (lanes 9 to 12). [35S]methionine-labeled A14 immunoprecipitates were then analyzed by SDS-PAGE and autoradiography, and the levels of the unmodified and glycosylated forms were monitored. Cells infected with vindA17(−) (lane 5) served as a positive control for increased A14 glycosylation levels, while wt virus-infected cells (lane 6) represented the basal level of A14 glycosylation in vivo. Neither rifampin (rif; lane 7), which blocks VV morphogenesis at the stage of crescent formation (21), or BFA (lane 8) which spares the formation of IMV but inhibits their subsequent wrapping with Golgi membranes to form IEV (37), affected the levels of A14 glycosylation. Conversely, cells infected with either ts28 or tsH5-4, viruses that contain lesions in the F10 kinase (35, 40) or the H5 protein (7), respectively, showed elevated levels of A14 glycosylation at the nonpermissive but not the permissive temperature (lanes 9 to 12, shaded circle). These results suggest that an early block in VV morphogenesis, prior to the diversion of cellular membranes to the region surrounding the virosome, can lead to increased levels of A14 glycosylation.
Upon determination that a minority of A14 molecules were glycosylated during the course of a wt virus infection, we wanted to investigate whether these modified molecules were encapsidated. BSC40 cells were infected with wt virus (MOI of 2) in the presence of [35S]methionine and the presence or absence of tunicamycin; at 24 hpi, cytoplasmic lysates were prepared and virions were further purified by ultacentrifugation through a 36% sucrose cushion followed by banding on a 25 to 40% sucrose gradient. The various species of A14 within the virions were retrieved by immunoprecipitation and were analyzed by SDS-PAGE and fluorography (Fig. 3B, lanes 13 to 16). Lysates prepared from vindA17(−) or wt virus infections (lane 13 or 14), which contain the unmodified (◂) and glycosylated (shaded circle) forms of A14, served as controls. wt virions prepared from infections performed in either the absence or presence of tunicamycin did not appear to contain glycosylated A14 (lane 15 or 16, respectively). Thus, although A14 is readily glycosylated in vitro on Asp83 and although a minority of molecules sustain the same modification in vivo, the glycosylated form of A14 does not appear to become associated with mature virions.
A recombinant virus harboring the N83Q allele of A14 shows no defect in IMV or EEV production.
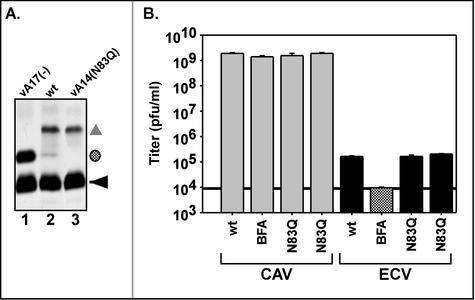
In our transient-complementation assay, the A14 allele containing the N83Q substitution, which encodes a protein that cannot undergo glycosylation, showed no defect in its ability to restore virus production (data not shown). To enable a more careful analysis of any subtle role that A14 glycosylation might play in the virus life cycle, we constructed a recombinant virus containing the N83Q allele in place of the endogenous gene (vA14[N83Q]). Metabolic labeling of cells infected with vA14(N83Q) demonstrated that, as expected, none of the A14 protein produced by this virus was modified by glycosylation (Fig. 4A, lane 3). Infections with vindA17(−) and wt virus (lanes 1 and 2) served as the controls for glycosylation of the A14 protein. We also monitored the size of plaques generated on BSC40 cells in 48 h (not shown) and the 24-h yield of both CAV and ECV (Fig. 4B). In each of these respects, the behavior of vA14(N83Q) was indistinguishable from that of wt virus. Thus, at least in tissue culture, glycosylation of A14 does not appear to modulate virus production.
FIG. 4.
Characterization of vA14(N83Q). (A) The A14 protein encoded by vA14(N83Q) is not glycosylated in vivo. Cells were infected with vindA17 (without IPTG) (lane 1), wt virus (lane 2), or vA14(N83Q) (lane 3) and were metabolically labeled with [35S]methionine from 6 to 9 hpi. Cell lysates were subjected to immunoprecipitation with anti-A14 serum; immunoprecipitates were resolved by SDS-PAGE and were visualized by fluorography. As above, the symbols indicate unmodified A14 (◂), glycosylated A14 (shaded circle), and coprecipitated A17 protein (▴). (B) Determination of CAV and ECV yields. Cells were infected at an MOI of 2 with wt virus in the absence or presence of BFA or with two isolates of vA14(N83Q). At 24 hpi cells and culture media were harvested and viral yield was determined by plaque assay. CAV is largely IMV but also scores low levels of IEV and cell-associated enveloped virus (see introduction). The black horizontal line, set at the titer of virus found in the supernatant fluid of cultures infected in the presence of BFA, represents the background level of IMV that had leaked into the supernatant fluid. Titers above this threshold are interpreted as representing bona fide EEV. No significant differences are seen in the levels of CAV or ECV produced by vA14(N83Q) versus those produced by wt virus.
VV A14 is phosphorylated exclusively on serine 85, but this modification is not essential for A14's role in VV morphogenesis.
It has been previously shown that A14 is phosphorylated during viral infections (1, 26, 36); phosphorylation occurs solely on Ser residues and is modulated both by the F10 kinase and the H1 phosphatase (1, 36). We sought to identify which Ser residue(s) undergoes phosphorylation and to determine whether this modification is necessary for A14's role in viral morphogenesis. Therefore, we constructed multiple alleles of A14 in which the 10 Ser residues (S12, S25, S34, S36, S38, S47, S65, S77, S85, and S88) were changed to Ala, both individually and in clusters (S34 -38, S77-88, S34-88 [S'8′A], S25-88 [S'9′A], and S12-88 [S'10′A]). Cells infected with vindA14 under nonpermissive conditions were transfected with the various Ser-to-Ala alleles in order to monitor the loss or retention of complementation activity. Figure 5A presents a graphic representation of the ability of these alleles to restore virus production. Two alleles, S12A and S′10′A, showed a complete loss of ability to substitute for wt A14; the other alleles retained full complementation activity. Immunoblot analysis was performed on the lysates prepared from the infection-transfection experiments in order to confirm that the transfected A14 alleles had been expressed (Fig. 5B). Indeed, both the S12A and S′10′A alleles were expressed at wt levels, indicating that their inability to complement the vindA14 infection was not due to trivial reasons but reflected instead a key role for the Ser12 residue in A14 function. The immunoblot analyses showed wt levels of expression for most of the other alleles. The proteins encoded by the S88A, S77-88A, and S′9′A alleles could not be detected by immunoblotting, although they possessed full complementation activity.
FIG. 5.
Transient rescue of vindA14 performed with Ser-to-Ala mutants of A14. (A) Determination of complementation competence by titration of viral yield from infections and transfections. Infection-transfection assays were performed as described above. Cells infected with vindA14 in the absence of inducer were transfected with empty vector, plasmid encoding wt A14, or plasmids encoding the various Ser-to-Ala mutants in order to assess their ability to substitute for endogenous A14. All experiments were performed in triplicate, and the titers of viral yield were averaged. The horizontal grey line illustrates the titer of virus obtained upon transfection of empty vector. (B) Immunoblot analysis of A14 expression. Aliquots of the extracts described above were resolved by nonreducing SDS-17% PAGE and were subjected to immunoblot analysis to monitor A14 expression from the endogenous and transfected alleles. The positions of the 18,000- and 29,000-Mr markers are shown at the left. (C) 32PPi labeling of A14 Ser-to-Ala mutants. Infections and transfections were performed as described earlier. Cells were metabolically labeled with 32PPi from 6 to 24 hpi; cell lysates were subjected to immunoprecipitation analysis with anti-A14 serum in order to determine the phosphorylation status of the various A14 proteins. In the case of wt and vA14(S85A) virus infections (rightmost lanes 1 and 2), cells were labeled with [35S]methionine from 5 to 6 hpi and with 32PPi from 6 to 9 hpi prior to being harvested at 9 hpi and subjected to immunoprecipitation with anti-A14 serum as described above. In all cases, immunoprecipitates were resolved by SDS-PAGE and were visualized by autoradiography.
Having shown that the Ser12-to-Ala mutation abolished complementation activity, we wanted to determine whether this loss of function reflected the requirement for Ser12 phosphorylation or an unrelated contribution of Ser12 to A14's function. We therefore transfected vindA14(−)-infected cells with each of our mutants in the presence of 32PPi and monitored the phosphorylation of the A14 proteins by immunoprecipitation. All of the individual Ser-to-Ala proteins were phosphorylated with the exception of S85A (Fig. 5C and data not shown). Of the alleles containing clustered Ser-to-Ala mutations, S85-88, S′8′A, S′9′A, and S′10′A also failed to show phosphorylation, consistent with the identification of S85 as the sole site of A14 phosphorylation. This assignment was further confirmed upon generation of a recombinant virus containing the A14 Ser-to-Ala allele at the endogenous locus [vA14(S85A)]. When cells were infected with either wt virus or vA14(S85A) in the presence of [35S]methionine and 32PPi, immunoprecipitation analyses clearly showed that phosphorylation of A14 was abolished by the Ser85-to-Ala substitution (Fig. 5C, compare rightmost lanes 1 and 2), although [35S]-A14 was retrieved at similar levels from both samples (not shown).
Two additional conclusions can be drawn from our analysis of the Ser-to-Ala mutants. First, since the protein encoded by the S12A allele retained phosphorylation, the loss of complementation activity caused by this mutation is due to perturbation of a process other than phosphorylation. It is worth noting here that Ser12 lies adjacent to the N9YF motif shown above to be essential for A14 function. Second, since the S85A allele retained full complementation activity, phosphorylation of A14 is apparently dispensable for its role in IMV morphogenesis.
As shown above, S85 is the sole site of A14 phosphorylation. Intriguingly, Ser85 is part of the functional N-linked glycosylation motif discussed above and documented in Fig. 3 and 4. To investigate a potential dynamic tension between phosphorylation and glycosylation of A14, we examined the level of A14 phosphorylation seen upon transfection with the N83Q allele (described above). The A14 encoded by this allele retains S85 but no longer has a functional glycosylation site (Fig. 5C). Phosphorylation of A14N83Q was clearly elevated compared to that of wt A14 in both infection-transfection assays (Fig. 5C) and upon infection with the stable vA14(N83Q) recombinant (not shown). These data suggest that that glycosylation and phosphorylation of A14 may be mutually exclusive; moreover, the overlap of a functional glycosylation site with the phosphorylation site implies that phosphorylation of A14 may take place within the lumen of the endoplasmic reticulum (ER).
Cysteine 71 is necessary and sufficient for the formation of disulfide-linked A14 dimers both in vitro and in vivo.
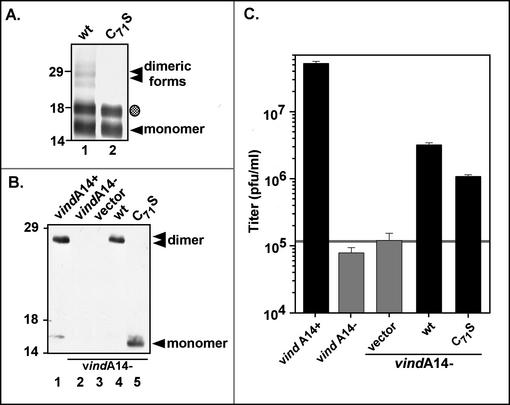
The A14 protein has been shown previously to exist in two forms: a monomer with an apparent Mr of 16,000 and a dimer with an Mr of 25,000 (15, 26, 27, 34). The dimeric form predominates within the virion membrane. When treated with both β-mercaptoethanol and DTT, the 25,000-Mr form can be completely reduced to the 16,000-Mr monomer, indicating that A14 forms an intermolecular disulfide bond(s). The A14 protein contains only two cysteine residues: Cys26, which lies within the first predicted membrane-spanning region, and Cys71. We focused on Cys71 and generated an A14 allele in which this residue was changed to serine (C71S). Initially, we compared the wt virus and C71S proteins generated by IVTT in the presence of [35S]methionine and microsomal membranes. As shown in Fig. 6A, lane 1, multiple species of wt A14 were seen after electrophoresis under nonreducing conditions, corresponding to unglycosylated (◂) and glycosylated (shaded circle) monomers and unglycosylated (lowest band), hemiglycosylated (middle band), and fully glycosylated (upper band) (stacked triangles) dimers. In contrast, when the C71S form was analyzed in parallel, no dimeric forms of A14 were seen (Fig. 6A, lane 2). These results indicated that, in vitro, Cys71 is responsible for intermolecular disulfide bond formation.
FIG. 6.
Cys71 is necessary and sufficient for intermolecular disulfide bond formation by A14, both in vitro and in vivo. (A) Synthesis of wt A14 (lane 1) and the C71S A14 mutant (lane 2) by using IVTT. [35S]methionine-labeled IVTT products were subjected to SDS-PAGE in the absence of reducing agents in order to assess the ability of the A14 proteins to undergo covalent dimerization; proteins were visualized by autoradiography. The unmodified (◂) and glycosylated (shaded circle) monomers of A14 were seen, as were several more slowly migrating forms that represent dimeric forms of wt A14 (stacked triangles). The positions of the 14,000-, 18,000-, and 29,000-Mr standards are shown at the left. (B) Immunoblot analysis of the wt and C71S A14 proteins expressed in vivo. Transient-complementation analysis was performed by using the infection-transfection protocol described above. Lysates were resolved by nonreducing SDS-PAGE; A14 species were visualized by immunoblot analysis. Infections with vindA14 were performed in the presence (lane 1) or absence (lanes 2 to 5) of TET. The latter were transfected with vector alone (lane 3) or plasmids encoding wt A14 (lane 4) or the C71S allele of A14 (lane 5). The A14 monomer ([◂]) and dimer (stacked triangles) are indicated; no dimer is seen in cells expressing the C71S allele of A14 (lane 5). The positions of the 14,000-, 18,000-, and 29,000-Mr standards are shown at the left. (C) Transient-complementation assay using A14 C71S. BSC40 monolayers were infected with vindA14 in the presence (+) or absence (−) of TET and were transfected with empty vector or plasmids expressing wt A14 or the C71S allele. Twenty-four-hour viral yield was determined by plaque assay; the horizontal grey line represents the titer obtained upon transfection of empty vector.
To investigate covalent dimer formation in vivo, we analyzed the C71S allele in our transient-rescue assay. Lysates prepared after infection-transfection were subjected to nonreducing SDS-PAGE and immunoblot analysis to determine whether A14 C71S could form disulfide-linked dimers in vivo (Fig. 6B) and to virus titration to determine whether it could support virus production (Fig. 6C). When wt A14 protein was expressed, either during permissive infections with vindA14 (lane 1) or upon transfection into cells infected nonpermissively with vindA14 (lane 4), >95% of the protein migrated as a disulfide-linked dimer in nonreducing SDS-PAGE, as expected. On the contrary, when cells were infected with vindA14(−) and were transfected with a plasmid encoding the C71S allele (lane 5), only the 16,000-Mr monomeric form of A14 was seen. These results indicate that Cys71 is required for the disulfide-linked dimerization of A14 in vivo. Figure 6C demonstrates that preventing disulfide-linked dimerization has a reproducible but modest effect on the ability of the A14 protein to complement vindA14(−) infections, with viral yield being only threefold less than that seen after transfection of the plasmid encoding wt A14. These results suggest that covalent dimerization of A14 is not essential for A14 function during ongoing morphogenesis.
Generation of a recombinant virus carrying the C71S allele: disulfide-linked dimers of A14 do not form, and viral yield is reduced fourfold.
Given the observation by our laboratory and others that the vast majority of the A14 protein encapsidated into virion membranes is present as disulfide-linked dimers, we were surprised that prevention of covalent dimerization appeared to have only a minor impact on the viral life cycle. In order to further characterize the impact of A14 dimerization on virion morphogenesis, stability, or infectivity, we constructed a virus in which the C71S allele of A14 was inserted in place of the endogenous allele. Our strategy was to first infect cells with the TET-dependent virus vindA14 and then transfect them with a plasmid containing the C71S allele of A14 flanked by the normal upstream and downstream sequences derived from wt virus genomic DNA. We then selected for TET-independent viral progeny generated as a result of recombination between the viral genome and the plasmid. Plaques that formed in the absence of TET were screened by PCR to confirm the loss (rather than mutagenic inactivation) of the TET operator and by DNA sequence analysis to identify those plaques in which recombination had resulted in the simultaneous acquisition of the Cys-to-Ser mutation. Two plaques satisfying both criteria were subjected to four subsequent rounds of plaque purification and DNA sequencing. The viruses generated were designated vA14(C71S). Lysates prepared from vA14(C71S) virus-infected cells were analyzed by reducing and nonreducing SDS-PAGE; immunoblot analysis confirmed that the A14 protein was found exclusively in the monomeric form (Fig. 7A, lanes 2 and 3). As a preliminary comparison of the infectivity of vA14(C71S) and wt virus, we determined the size of the plaques formed at 48 hpi: no difference was apparent (data not shown). We also quantitated the yield of intracellular virus and ECV at 24 hpi (MOI = 2) with either vA14(C71S) or wt virus. Parallel infections performed in the presence of BFA allowed us to define the subset of ECV that was indeed EEV and not IMV that had been artifactually released into the culture medium. The yield of CAV from cells infected with vA14(C71S) was consistently fourfold lower than that obtained from cells infected with wt virus (Fig. 7B). There was an ∼2-fold reduction in EEV production. A comparison of gradient-purified vA14(C71S) and wt virions showed no impact of the C71S substitution on specific infectivity, as assessed by determinations of particle-to-PFU ratios (data not shown). Finally, preventing covalent dimerization of A14 did not appear to affect virion ultrastructure, as determined by electron microscopy examination of embedded and sectioned samples of wt and vA14(C71S) virions (data not shown). We have therefore determined that lack of covalent dimerization of the A14 protein has no effect on virion infectivity or overall viral morphology, while having a modest but consistent impact (fourfold reduction) on virion production.
FIG. 7.
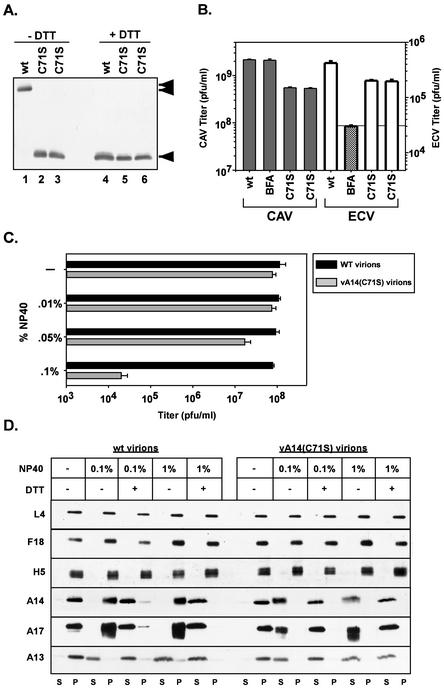
Characterization of vA14(C71S). (A) Immunoblot analysis of A14 encapsidated within purified wt or vA14(C71S) virions. One microgram of virions purified from cells infected with wt virus (lanes 1 and 4) or with two isolates of vA14(C71S) (lanes 2 and 5 and 3 and 6) was resolved by SDS-PAGE under nonreducing (lanes 1 to 3) or reducing (lanes 4 to 6) conditions and was subjected to immunoblot analysis with anti-A14 serum. The monomeric and dimeric forms of A14 are indicated (◂ and stacked triangles, respectively). (B) Determination of CAV and ECV yields. Cells were infected at an MOI of 2 with wt virus in the absence or presence of BFA or with vA14(C71S). Harvesting and titration of virus were performed as described for Fig. 4B and in Materials and Methods. Note the different scales used to plot the yields of CAV (left axis) and ECV (right axis). (C) Biological sensitivity of wt and vA14(C71S) virions to detergent treatment. A total of 2.0 × 108 wt or vA14(C71S) virions were incubated at 37°C for 30 min in various concentrations of NP-40 (0, 0.01, 0.05, or 0.1%) and were then titrated to determine the impact on viral titer (number of PFU/milliliter). (D) Permeabilization of wt and vA14(C71S) virions and fractionation of membrane and core components. wt and vA14(C71S) virions were subjected to detergent permeabilization and centrifugation to separate membrane and core components. Virions were incubated at 37°C for 30 min in 100 mM Tris (pH 9.0) containing NP-40 (0.1 or 1%) in the absence or presence of 50 mM DTT (− and +, respectively). After permeabilization, solubilized (S) and particulate (P) fractions were separated by sedimentation. Samples were resolved by SDS-PAGE, and the partitioning of various viral proteins was catalogued by immunoblot analysis by using sera directed against core (L4, F18, and H5) and membrane (A14, A17, and A13) proteins.
The infectivity of vA14(C71S) virions shows a dramatic increase in detergent sensitivity.
We analyzed the biological sensitivity of both wt and vA14(C71S) virion preparations to incubation at 45°C for 0 to 90 min, to incubation at pH 4 to 11 for 30 min at 37°C, and to incubation with various concentrations of nonionic detergent (data not shown and Fig. 7C). We found no differences in the infectivity profiles of heat- or pH-treated virions. Detergent sensitivity assays were performed by incubating 2.0 × 108 wt or vA14(C71S) virions at 37°C for 30 min in buffer containing 100 mM Tris and concentrations of NP-40 ranging from 0 to 0.1%. As Fig. 7C illustrates, treatment of wt virions with even the highest concentration of NP-40, 0.1%, did not diminish their infectivity, as measured by titration in a plaque assay. In contrast, vA14(C71S) virions showed a striking increase in detergent sensitivity. Treatment with 0.05% NP-40 caused an eightfold reduction in the infectious titer of the sample, and treatment with 0.1% NP-40 reduced the infectious titer by nearly 3.5 orders of magnitude (99.97% reduction). In the absence of covalent dimerization of the A14 protein, the virion membrane responds to mild detergent treatment in a manner that profoundly reduces virion infectivity.
vA14(C71S) virions show an altered fractionation pattern upon NP-40 permeabilization.
Having determined that the infectivity of vA14(C71S) virions was highly sensitive to treatment with a nonionic detergent in the absence of reducing agents, we wanted to investigate what physical changes might be responsible for this sensitivity. In order to determine if the structural integrity of the vA14(C71S) virions differed from that observed for wt virions, we subjected both virion preparations to permeabilization with either 0.1 or 1% NP-40 in the absence or presence of 50 mM DTT (Fig. 7D). Permeabilized virions were sedimented at 16,000 × g for 30 min to partition the solubilized membrane proteins from the particulate core fraction. Both samples were resolved by SDS-PAGE; the fractionation of several core and membrane proteins was monitored by immunoblot analysis. In both the wt and C71S virions, the core proteins L4 (45), F18 (17), and H5 (7; U. Sankar and P. Traktman, unpublished data) remained associated with the particulate core fraction under all fractionation conditions. These data indicated that the absence of disulfide-linked dimers of A14 in the vA14(C71S) virions had no apparent impact on the integrity of the VV cores or the associated core proteins.
In contrast, the fractionation pattern of some but not all membrane components was indeed shown to be altered in the vA14(C71S) virions. Partitioning of the A13 protein (16, 29; B. Unger and P. Traktman, submitted for publication) but not of the A14 or A17 proteins was equivalent for the two virion preparations. In wt virions treated with 0.1 or 1% NP-40, the A14 and A17 proteins remained in the particulate fraction; only upon the addition of 50 mM DTT were these proteins released into the solubilized fraction (S). In contrast, in vA14(C71S) virions treated with 0.1% NP-40 alone, both the A14 and A17 proteins were released into the solubilized fraction (S). We conclude that the documented requirement for DTT in the solubilization of the virion membrane can be overcome by preventing the formation of disulfide-linked dimers of A14. As a consequence of maintaining the monomeric form of A14, both A14 and its binding partner, A17, can be released by treatment with mild detergent alone. Release of these membrane proteins is likely to explain the dramatic loss of infectivity seen upon treatment of vA14(C71S) virions with 0.1% NP-40.
DISCUSSION
This structure-function analysis of A14 was initiated because of the essential role that the A14 phosphoprotein plays in the morphogenesis of the vaccinia virion membrane. A14 is essential for the remodeling of vesicles and/or tubular membranes into the characteristic crescents that form during the process of virion formation. It is known to interact stably with the A17 protein and to be a substrate for both the F10 kinase and the H1 phosphatase (1, 26, 36). A mere 90 amino acids (aa) must confer all of these properties and capabilities. In order to perform a structure-function analysis of this protein, we established a transient-rescue assay that utilized a previously characterized TET-inducible recombinant, vindA14 (36). In the absence of inducer, A14 is not detectable by immunoblot analysis and vindA14 fails to form plaques. The viral yield is reduced by ∼1,000-fold compared to that obtained in the presence of inducer. We show here that transient expression of wt A14 can restore a significant amount of virus production (50-fold greater than that obtained upon transfection of empty vector). This assay was sufficiently robust to permit us to test the complementation competence of a large number of mutant alleles of A14. Our emerging view of the important sequences and sites of posttranslational modification is illustrated schematically in Fig. 8.
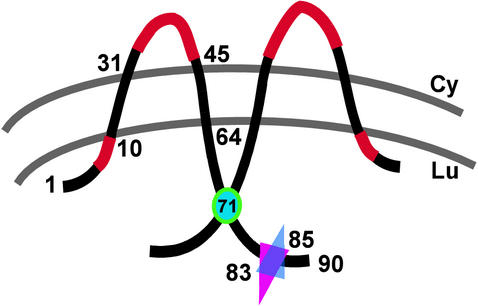
FIG. 8.
Predicted topology and functional motifs of VV A14. A model of membrane-spanning, disulfide-linked dimers of A14 is depicted. The N′ and C′ termini are predicted to be luminal (Lu) upon synthesis in the ER; the hydrophilic loop region is predicted to be cytosolic (Cy). Regions highlighted in red represent essential sequence elements within A14 (N9YFS and P39TRTWK). The position of Cys71, the residue required for disulfide formation, is indicated by a cyan circle. The overlapping triangles indicate the sites of N-linked glycosylation (Asn83) and phosphorylation (Ser85).
Taking advantage of computer predictions of A14 topology and sequence alignments of poxviral A14 homologs, we identified several motifs as being of potential functional or structural importance. As presented in Fig. 1 and illustrated in Fig. 8, the A14 protein has two predicted transmembrane domains (residues 13 to 31 and 45 to 64) that are separated by a 13-aa loop. The first transmembrane domain is preceded by a short (12-aa) hydrophobic region, and the second transmembrane domain is followed by a 26-aa hydrophilic tail. Deletion of amino acids 1 to 5 or 65 to 90 led to a 10- or 5-fold decrease in complementation activity, respectively, whereas deletion of both termini abolished complementation activity. The extreme N terminus is likely to be important for facilitating membrane insertion of the A14 protein. It was surprising that deletion of the C′-terminal tail had so little impact on complementation activity, since this region represents the majority of the exposed portion of the molecule and contains several well-conserved residues and motifs that will be discussed below. Since deletion of the C′ terminus was well tolerated, it was understandable that mutation of the well-conserved M66WG motif to A66AA was not detrimental. In contrast, mutation of the well-conserved N9YFS sequence that lies just before the first transmembrane domain abolished complementation activity. It will be of interest to determine whether this region affects the topology of A14, its interactions with A17 or other viral proteins, and/or its ability to remodel membrane vesicles into crescents. This allele did retain the ability to associate cotranslationally with microsomal membranes when expressed in IVTT reactions (not shown). Mutation of the well-conserved C26IFAF cluster within the first transmembrane region did not impair complementation activity, perhaps because the alanine residues preserve the hydrophobic character of this region.
With respect to the hydrophilic loop separating the two transmembrane domains, alteration of the D32FSK motif had no impact, but a profound impairment was seen upon mutation of the P39TRTWK region. Further mutational analyses of this region implicate the highly conserved T40, W43, and K44 as being most necessary for biological function; indeed, we have recently constructed an allele in which the original sequence has been changed to P39ARTAA and have shown that it lacks any complementation activity (not shown). We believe that this loop will be cytosolic (see below) and propose that the hydrophobic and basic residues participate in key protein-protein interactions. Our future plans include a determination of whether this loop region mediates interactions between the A14 and A17 proteins.
The 10 Ser residues within the A14 protein were mutated to Ala, individually and in clusters. We determined that the protein is phosphorylated on Ser85 in vivo but that an alteration to Ala at this position does not compromise the ability of the protein to substitute for endogenous A14. Because A14 is dephosophorylated by the H1 phosphatase and because the latter enzyme is essential for the production of infectious virions, we also constructed an allele in which Ser85 was changed to Glu85, in order to mimic constitutive phosphorylation. This allele also exhibited wt levels of complementation ability (not shown). Cumulatively, these data suggest that, at least in the context of our transient-rescue assay, the reversible phosphorylation of Ser85 is irrelevant.
Ser85 lies within a predicted site of N glycosylation (N83HS) that is highly conserved within diverse poxviral genomes. In vitro, A14 is readily glycosylated at this site, but in vivo, only a fraction of the A14 protein undergoes modification. Interestingly, the level of glycosylation increases dramatically when morphogenesis is blocked at an early stage by inactivation of the F10 or H5 protein or repression of the A17 protein. We suspect that this phenomenon is due to a prolonged residency in the ER when viral morphogenesis is blocked. Although this is the first demonstration of an IMV membrane protein that is modified by N-linked glycosylation, we have been unable to detect glycosylated A14 within the virion. It may be that A14 destined for crescent and IMV formation leaves the ER too rapidly to be glycosylated or that the glycosylated form is in fact excluded from the developing virion membrane. We do not support a model in which only a fraction of the A14 molecules are positioned such that their C′ terminus extends into the ER lumen, where glycosylation enzymes are found (19), because essentially all of the A14 undergoes covalent dimerization (see below) through the Cys71 residue, which must therefore be positioned within the ER lumen. Disruption of the glycosylation motif had no impact on the ability of the A14 protein to substitute for endogenous A14 in our transient-rescue assay; likewise, a stable recombinant virus encoding only the glycosylation-deficient A14 showed no defect in either IMV or EEV production. Interestingly, abolition of the glycosylation motif led to an increased level of A14 phosphorylation, suggesting that these two modification pathways may be in competition. Since N′ glycosylation occurs only within the lumen of the ER, these data suggest that phosphorylation of A14 also occurs within this compartment. This conclusion presents a dilemma, since the F10 kinase and the H1 phosphatase have been implicated in the regulation of A14 phosphorylation and since neither of these enzymes has been shown to associate with the ER. Further analysis of how and when F10 and H1 gain access to A14 will be of interest in the future.
It has been known for some time that the predominant form of A14 within mature virions is a disulfide-linked dimer. Of the two Cys residues present within the A14 open reading frame, we focused on Cys71, since Cys26 is present within a predicted transmembrane region. Indeed, A14 containing a Cys71-to-Ser substitution was unable to form covalent dimers and was present exclusively in the monomeric form. These data indicate that the C′-terminal tail of the vast majority of the A14 molecules must lie within the lumen of the ER, which supports disulfide bond formation. Although VV does encode a novel protein complex that directs disulfide bond formation within the cytoplasm, dimerization of the A14 protein appears to be independent of this system (30) (B. Moss, personal communication).
The A14 Cys71-to-Ser allele could substitute for wt A14, but the viral yield produced from infections with a stable A14 Cys71-to-Ser recombinant, vA14(C71S), was consistently fourfold less than that obtained with wt virus. Although it was initially surprising that a more severe deficiency was not seen, it is likely that the A14 protein can still dimerize via noncovalent interactions. The reproducible reduction in the viral titer obtained after infection with vA14(C71S) probably occurs at the level of virus production rather than infectivity, since careful study of purified virions showed that the particle-to-PFU ratio was similar to that of wt virions. Comparative immunoblot analysis of wt and vA14(C71S) virions indicates that A14 Cys71-to-Ser is encapsidated at wt levels (Fig. 7A). The modest reduction in viral yield during vA14(C71S) infections, therefore, is unlikely to be due to instability of the mutant form of A14 but rather to an alteration of protein structure or function. Whether the biological impact is a direct reflection of the inability of A14 to undergo covalent dimerization or to other subtle changes in the structure of the protein remains to be determined.
To see whether the virions would show a greater sensitivity to environmental perturbations when covalent dimers of A14 were not present, we investigated the impact of exposure to a range of pH, temperature, and detergent treatments. While exposure to various conditions of pH and temperature revealed no defect, the vA14(C71S) virions showed a greatly increased sensitivity to treatment with nonionic detergent. Whereas the preparation of wt virions showed no loss in titer when treated with 0.1% NP-40, the vA14(C71S) preparation showed a 4,000-fold loss of titer. To investigate the cause of this fragility, both preparations of virions were permeabilized with either 0.1 or 1.0% NP-40 in the absence or presence of 50 mM DTT. The solubilized (representing membrane proteins liberated from the virion) and particulate (representing the core) fractions were separated by sedimentation, and the partitioning of various viral proteins was determined. In the wt virions, neither the A14 or A17 protein was released in the absence of exposure to DTT. In the vA14(C71S) virions, however, both were released in the absence of DTT. Clearly, the major requirement for DTT in membrane permeabilization pertains to the disruption of covalent A14 dimers, a role that is rendered moot by the incorporation of the Cys71-to-Ser substitution. In wt virions, the interaction of dimeric A14 with A17 must generate a stable lattice that is resistant to disruption by NP-40, allowing the virions to retain full integrity and infectivity. In the absence of covalent dimers of A14, the architecture of these two abundant membrane proteins must be far more fluid, enabling NP-40 treatment to disrupt virion integrity and abolish virion infectivity. It seems likely that, within an infected individual, poxvirus virions encounter environmental conditions that may mimic NP-40 in terms of membrane perturbation. If so, then covalent dimerization of A14 is likely to play a significant role in maintaining virion infectivity in vivo, although this scenario was not recapitulated in tissue culture. Determination of whether the virions containing the A14 Cys71-to-Ser substitution show diminished pathogenicity in a mouse model would be of interest and will be undertaken in the future. If these virions are attenuated, then a vaccinia virus derivative containing the A14 Cys71-to-Ser allele might be a good vaccine candidate, since it would retain full immunogenicity while showing reduced pathogenicity.
In sum, these studies have allowed us to define several regions of the A14 membrane protein whose perturbation eliminates the biological competency of the protein. Further examination of how these regions, as well as the sites of phosphorylation, glycosylation, and dimerization, govern interactions of the A14 protein with the A17 protein and/or with membrane lipids will be informative in establishing a more in-depth appreciation of the biogenesis of the vaccinia virus membrane.
Acknowledgments
This work was supported by a grant awarded to P.T. by the NIH (2 R01 GM 53601).
We thank Beth Unger for sharing the anti-A13 antiserum prior to publication and all the members of the Traktman laboratory for helpful discussions.
REFERENCES
- 1.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. [DOI] [PubMed] [Google Scholar]
- 3.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2002. The vaccinia virus I7L gene product is the core protein proteinase. J. Virol. 76:8973-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassetti, M. C., M. Merchlinsky, E. J. Wolffe, A. S. Weisberg, and B. Moss. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dales, S., and E. H. Mosbach. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564-583. [DOI] [PubMed] [Google Scholar]
- 7.DeMasi, J., and P. Traktman. 2000. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J. Virol. 74:2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrien, M., A. Punjabi, M. Khanna, O. Grubisha, and P. Traktman. 1999. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J. Virol. 73:7287-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubochet, J., M. Adrian, K. Richter, J. Garces, and R. Wittek. 1994. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J. Virol. 68:1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbein, A. D. 1987. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 56:497-534. [DOI] [PubMed] [Google Scholar]
- 11.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths, G., N. Roos, S. Schleich, and J. K. Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analyses. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollinshead, M., G. Rodger, H. van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichihashi, Y., M. Oie, and T. Tsuruhara. 1984. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J. Virol. 50:929-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and L. J. Krijnse. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, S. Y., and W. R. Bauer. 1987. Biosynthesis and phosphorylation of vaccinia virus structural protein VP11. Virology 159:399-407. [DOI] [PubMed] [Google Scholar]
- 18.Liu, K., B. Lemon, and P. Traktman. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69:7823-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Locker, J. K., and G. Griffiths. 1999. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J. Cell Biol. 144:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2884. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 21.Moss, B., E. N. Rosenblum, E. Katz, and P. M. Grimley. 1969. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature 224:1280-1284. [DOI] [PubMed] [Google Scholar]
- 22.Moss, B., and B. M. Ward. 2001. High-speed mass transit for poxviruses on microtubules. Nat. Cell Biol. 3:E245-E246. [DOI] [PubMed] [Google Scholar]
- 23.Patel, D. D., C. A. Ray, R. P. Drucker, and D. J. Pickup. 1988. A poxvirus-derived vector that directs high levels of expression of cloned genes in mammalian cells. Proc. Natl. Acad. Sci. USA 85:9431-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez, D., M. Esteban, and J. R. Rodríguez. 1995. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 69:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodríguez, D., C. Risco, J. R. Rodríguez, J. L. Carrascosa, and M. Esteban. 1996. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J. Virol. 70:7641-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodríguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Röttger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. K. Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 99:6667-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, G. L. 1993. Vaccinia virus glycoproteins and immune evasion. The sixteenth Fleming Lecture. J. Gen. Virol. 74:1725-1740. [DOI] [PubMed] [Google Scholar]
- 32.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440:395-414. [PubMed] [Google Scholar]
- 33.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, T., M. Oie, and Y. Ichihashi. 1994. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology 202:844-852. [DOI] [PubMed] [Google Scholar]
- 35.Traktman, P., A. Caligiuri, S. A. Jesty, K. Liu, and U. Sankar. 1995. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1995. Brefeldin A inhibits vaccinia virus envelopment but does not prevent normal processing and localization of the putative envelopment receptor P37. J. Gen. Virol. 76:103-111. [DOI] [PubMed] [Google Scholar]
- 38.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 39.Vogelstein, B., and D. Gillespie. 1979. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. USA 76:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S., and S. Shuman. 1995. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J. Virol. 69:6376-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehead, S. S., and D. E. Hruby. 1994. Differential utilization of a conserved motif for the proteolytic maturation of vaccinia virus proteins. Virology 200:154-161. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, W. P., and W. R. Bauer. 1988. Purification and characterization of vaccinia virus structural protein VP8. Virology 167:578-584. [PubMed] [Google Scholar]
- 46.Zhang, Y., and B. Moss. 1991. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J. Virol. 65:6101-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]