Abstract
By using a purified dengue virus RNA-dependent RNA polymerase and a subgenomic 770-nucleotide RNA template, it was shown previously that the ratio of the de novo synthesis product to hairpin product formed was inversely proportional to increments of assay temperatures (20 to 40°C). In this study, the components of the de novo preinitiation complex are defined as ATP, a high concentration of GTP (500 μM), the polymerase, and the template RNA. Even when the 3′-terminal sequence of template RNA was mutated from -GGUUCU-3′ to -GGUUUU-3′, a high GTP concentration was required for de novo initiation, suggesting that high GTP concentration plays a conformational role. Furthermore, utilization of synthetic primers by the polymerase indicated that AGAA is the optimal primer whereas AG, AGA, and AGAACC were inefficient primers. Moreover, mutational analysis of the highly conserved 3′-terminal dinucleotide CU of the template RNA indicated that change of the 3′-terminal nucleotide from U to C reduced the efficiency about fivefold. The order of preference for the 3′-terminal nucleotide, from highest to lowest, is U, A∼G, and C. However, change of the penultimate nucleotide from C to U did not affect the template activity. A model consistent with these results is that the active site of the polymerase switches from a “closed” form, catalyzing de novo initiation through synthesis of short primers, to an “open” form for elongation of a double-stranded template-primer.
Dengue viruses are members of the Flavivirus family of positive-strand RNA viruses. These viruses infect as many as 100 million individuals per year and are the causative agents of dengue fever, dengue hemorrhagic fever, and dengue shock syndrome (24, 35, 53). Approximately 500,000 cases of dengue hemorrhagic fever are reported worldwide, with as many as 25,000 deaths annually (23). The dengue virus, the most prevalent tropical infectious agent after malaria, is spread by the mosquito Aedes Aegypti, which is a common day-biting mosquito in tropical climates (23, 24). Epidemiological studies have demonstrated that dengue virus type 2 (DEN2) is the most prevalent of the four serotypes (DEN1 to -4).
The genome of the New Guinea-C strain of DEN2 is 10,723 nucleotides long and contains a type 1 cap structure at the 5′ terminus but lacks a poly(A) tail at the 3′ end (30; for a review see reference 12). The genome encodes a single poly-protein, NH2-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-COOH, which is processed within the endoplasmic reticulum (ER), producing the capsid (C), precursor membrane (prM) protein, and envelope protein (E) through cotranslational processing by the ER-resident signal peptidase (49, 55, 66). The C-terminal portion of the polyprotein, NS1 to NS5, is processed into at least seven nonstructural proteins in the ER by both the host protease(s) and the virally encoded serine protease, NS2B/NS3, of the trypsin family.
NS3 is the second largest protein encoded by the DEN2 virus and contains at least three enzymatic properties. Contained within the N-terminal 180 amino acids is a serine protease domain which, in the presence of the NS2B cofactor, recognizes and cleaves at dibasic amino acid motifs (3, 11, 13, 14, 19, 21, 60, 69, 77). NS3 also contains conserved motifs found in the DEXH family of RNA helicases (22, 41; for a review, see reference 31). Purified NS3 has been reported to contain both nucleoside triphosphatase (NTPase) and RNA helicase activities (6, 17, 37, 43, 44, 68, 70) as well as 5′-RNA triphosphatase activity (2, 71), the first enzyme required in 5′-cap synthesis.
NS5, the largest of the DEN2 structural proteins, contains conserved motifs consistent with those of RNA-dependent RNA polymerases (RdRP) encoded by several positive-strand RNA viruses (57, 59). This enzymatic activity has been demonstrated for purified NS5 from DEN1, DEN2, and Kunjin virus and for hepatitis C virus (HCV) NS5B (1, 25, 45, 48, 67, 72). NS5 also contains conserved motifs found in several 5′-RNA methyltransferases (40, 63). The crystal structure of the N-terminal region of DEN2 NS5 containing the guanylyltransferase/methyltransferase domain was recently reported (18). The facts that NS3 and NS5 exist in a stable complex in infected cells (34) and that NS3 has the NTPase/RNA helicase and the 5′-RNA triphosphatase activity suggest that replication and capping could occur in a closely coupled pathway.
An in vitro viral RNA replicase assay system which utilized cytoplasmic lysates prepared from mosquito (C6/36) cells infected with DEN2 and exogenous subgenomic 770-nucleotide RNA templates containing both 5′- and 3′-terminal regions was described previously. Two RNA products were formed. One was the same size as the input template (1×) and was resistant to RNase A treatment and therefore understood to be double-stranded RNA. The other product was shown to be a hairpin RNA twice the size of the template (2×), formed by 3′-end elongation of the template RNA by a “copy-back” mechanism. RNase A treatment of the hairpin RNA also gave rise to an RNase A-resistant, template-sized product upon digestion of the single-stranded loop formed by elongation of the “fold-back” region. The newly synthesized RNA was shown by RNase H mapping to be of minus-strand polarity (74, 75). This in vitro system revealed that both 5′- and 3′-terminal regions that include a 5′ untranslated region (5′ UTR), a 3′ stem-loop structure, 5′ and 3′ cyclization motifs, and a predictive pseudoknot tertiary structure are structural determinants involved in the long-range interactions between the two terminal regions regulating initiation of minus-strand RNA synthesis (74, 75).
In a subsequent study, DEN2 NS5 with an N-terminal His tag was expressed in Escherichia coli and purified to near-homogeneity. The purified polymerase also gave rise to the same two products, 1× and 2×, in the absence of any other viral or host proteins. Furthermore, we showed that the 1× product was formed by de novo initiation of RNA synthesis on the subgenomic 770-nucleotide RNA template, as opposed to the hairpin product, which was formed by 3′-end elongation of a fold-back structure of the template RNA. The ratio between the de novo product and the hairpin product was dependent on the incubation temperature (1). Using a heparin trap protocol (1, 65), we showed that lower temperatures promoted de novo synthesis of RNA at the initiation rather than the elongation stage. Once the de novo initiation complex was formed at lower temperatures (20 to 24°C) in the presence of the template RNA, NS5, ATP, GTP, and CTP, addition of heparin followed by UTP allowed the de novo product to be formed in preference to the hairpin product even when the elongation reaction was carried out at 30°C. If the same components in the initial partial system were incubated at higher temperatures (35 to 40°C), this preferred formation of the de novo product was not observed after the addition of heparin and the missing components, even when the second incubation was carried out at low temperatures (1).
The influence of temperature on the nature of the products formed could be explained by a proposed model in which the DEN2 polymerase exists in two conformational states, “closed” and “open,” that are in equilibrium. At lower temperatures, at which the equilibrium is in favor of a closed conformation, the polymerase is unable to initiate RNA synthesis at the 3′ end of the RNA primer either annealed to the template RNA or in the form of a fold-back structure. Thus, the polymerase is forced to synthesize short primers complementary to the 3′-terminal sequences of the template RNA. Once these primers are synthesized, the polymerase is in an open conformation, due to a conformational change mediated by the primer synthesis, and it can carry out an elongation reaction from the primer to produce the 1× product. At higher temperatures, the polymerase in an open conformation could bind to the 3′ fold-back structure of the template RNA and preferentially produce the hairpin product.
In the present study, using the heparin trap protocol, we further defined the temperature-dependent parameters for de novo initiation of RNA synthesis, such as the minimum nucleotide components and their concentrations required for de novo preinitiation complex formation versus the 3′-end elongation reaction. Our results show that the template RNA, ATP, and a high concentration of GTP, along with NS5 polymerase, are the minimum components that are required and sufficient for formation of the de novo preinitiation complex; omitting any of these components and adding it later, after the addition of heparin, did not result in the formation of the de novo product. The same components, including the high concentration of GTP, are required even if a mutant template that does not require a guanylate residue for incorporation within the first 6 nucleotides from the 3′-terminal end is used. Furthermore, as our model would predict that the polymerase is in an open conformation, the enzyme utilizes the AGAA primer efficiently to yield a template-sized product at 35°C. Mutational analysis of the highly conserved 3′-terminal dinucleotide sequence CU of the template RNA indicates that a change of the 3′-terminal U to C reduces the activity about threefold and that the order of preference for the 3′-terminal nucleotide (from highest to lowest) is U, A∼G, and C. However, mutation of the penultimate C to U did not have any significant effect. These results suggest that the 3′-terminal nucleotide is important for recognition and binding by the polymerase for initiation of RNA synthesis.
MATERIALS AND METHODS
RNA primers.
RNA primers 5′-AG, 5′-AGA, 5′-AGAA, and 5′-AGAACC were synthesized by Dharmacon Research, Inc. (Lafayette, Colo.). Radiolabeled [α-32P]CTP (800Ci/mmol) was obtained from Perkin-Elmer Corp. (Boston, Mass.).
RdRP assay.
DEN2 NS5 protein with an N-terminal His tag was expressed in E. coli and purified as previously described (1). Purified NS5 was stored in a 40% glycerol-containing buffer at −20°C before use in RdRP assays. The construction of the plasmid encoding the DEN2 subgenomic RNA has been described elsewhere (75). The RNA template was prepared by in vitro transcription using an Ampliscribe T7 Transcription kit (Epicentre Technologies). The standard reaction mixture (50 μl) contained 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, template RNA (0.1 μg; 0.1 pmol), 500 μM (each) ATP, GTP, and UTP, 10 μM unlabeled CTP, and 10 μCi of [α-32P]CTP along with 200 ng (2 pmol) of purified NS5 except when otherwise indicated. The reaction was carried out by incubation at 24 to 38°C for 1 h by using a gradient thermocycler (T-gradient; Biometra, Göttingen, Germany). The reaction was terminated by acid phenol-chloroform extraction, followed by purification with a Bio-Rad P-30 column to remove the unincorporated nucleotides. Radioactive RNA products were analyzed by formaldehyde-agarose gel electrophoresis and visualized by autoradiography. The labeled bands were excised from dried gels and quantified by liquid scintillation counting.
Heparin trap protocol.
To determine the factors necessary for de novo initiation of negative-strand synthesis, we utilized a modified heparin trap protocol (1, 65). In the original protocol, the subgenomic 770-nucleotide RNA was incubated with 200 ng of purified NS5, 500 μM (each) ATP, GTP, and CTP, and 10 μCi of [α-32P]CTP for 10 min at various temperatures (20 to 40°C) to allow partial synthesis of the negative-strand RNA (1). Under these conditions, the NS5 polymerase pauses in a stable, heparin-resistant initiation complex. After the addition of heparin (2 μl of 25 ng/μl) to inactivate polymerase that was not in a stable RNA-protein complex, followed by UTP (500 μM), the elongation reaction was allowed to proceed (1). In a slightly modified protocol, the template RNA, NS5, and either various combinations of two nucleoside triphosphates (NTPs) or a single NTP were incubated at 30°C before the addition of heparin and the remaining NTPs to make up the complete system, and the elongation was continued at 30°C. At this incubation temperature, when all components were added at the same time, equimolar amounts of the template-sized and hairpin products were formed (1). Therefore, we surmised that only if the right combination of components that could form a functional de novo initiation complex was present would we be able to detect the template-sized product; otherwise, only the hairpin product would be detectable.
RESULTS
Components required for de novo initiation of RNA synthesis by DEN2 RdRP.
In previous studies, it was reported that the DEN2 NS5 RdRP exhibits a temperature-dependent shift toward de novo initiation from 3′-end elongation in negative-strand RNA synthesis from the subgenomic positive-strand RNA template (1). If the RdRP assay was carried out at lower temperatures (i.e., 24°C), a predominance of de novo initiation occurred to produce the template-sized product (1×), whereas at higher temperatures (∼38°C), the hairpin product was predominantly formed (2×). We used a heparin trap protocol to isolate the initiation events as opposed to 3′-end elongation events and to identify the components of the preinitiation complex that are required to form the de novo product. A partial RdRP assay system containing ATP, GTP, and CTP, NS5, and the template RNA produced the template-sized de novo product after the addition of heparin and UTP (1). According to this protocol, if the components that are required for the formation of the de novo initiation complex are present in the partial RdRP assay system, then addition of heparin followed by the missing components would produce the 1× product through heparin-resistant elongation events. On the other hand, if the initial incubation conditions with selected components are favorable only for 3′-end elongation, then addition of heparin and complete reconstitution of the assay would predominantly produce the hairpin product.
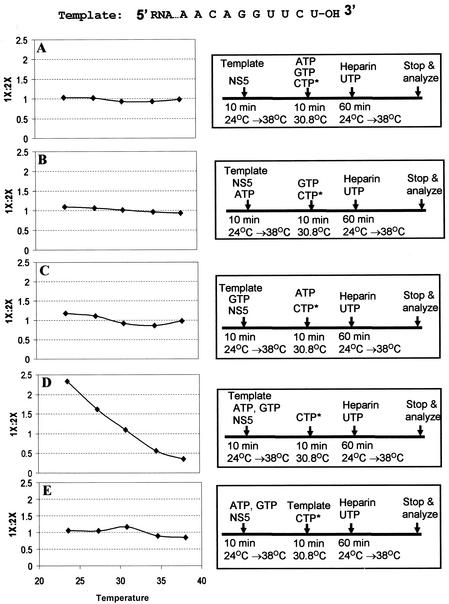
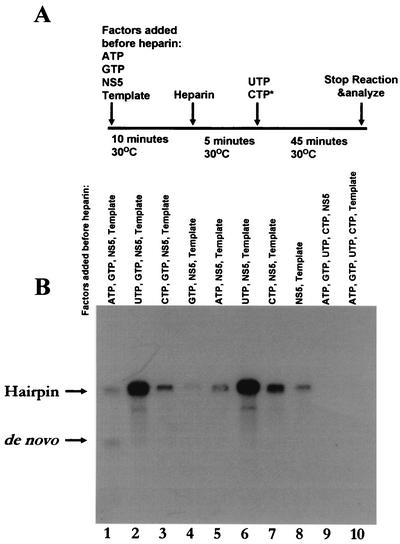
In this study, we further defined the minimum components that are required for the formation of the de novo initiation complex. By using the heparin trap protocol, it was first determined whether NS5 binding to RNA alone at different temperatures was sufficient to elicit the de novo-to-hairpin switch (Fig. 1). If NS5 could stably bind to different RNA conformations, and would stay bound to the template until nucleotides were supplied after heparin addition, then one would expect a temperature-dependent shift from formation of the de novo (1×) product at low temperatures to formation of the hairpin (2×) product at higher temperatures. As shown in Fig. 1, five tubes containing various components in the preinitiation complex were incubated at five different temperatures for 10 min. This step was then followed by addition of the remaining components and [α-32P]CTP, and incubation was continued for 10 min at a constant temperature of 30.8°C. After the addition of heparin and UTP, a final incubation for elongation was carried out at the same temperatures used in the first step for 60 min. The RNA products formed at the end of final incubation were analyzed by formaldehyde-agarose gel electrophoresis, visualized by autoradiography, and quantified by scintillation counting as described previously (1). The ratio of de novo to hairpin product formation (1×:2×) was plotted. As shown in Fig. 1A to C, incubation of the template and the polymerase alone or in the presence of ATP or GTP was insufficient to cause a shift in the ratio of 1× to 2× toward the de novo product at lower temperatures; the ratio remained close to 1 at all five temperatures of incubation. However, when the template, polymerase, ATP, and GTP were preincubated at various temperatures, this ratio shifted, and the de novo product was present in about 2.2-fold-larger amounts than the hairpin product at 24°C (Fig. 1D). A similar ratio was observed when the template, NS5, ATP, GTP, and CTP were incubated at 24°C and UTP was added subsequent to heparin (1). Furthermore, if the template RNA was omitted during the preinitiation step but added later, before the second incubation, this temperature-dependent shift from the 1× to the 2× product was not observed (Fig. 1E). These experiments were repeated twice with similar results, which indicated that the minimum components that are required for a shift in favor of the de novo product at 24°C are the polymerase, template RNA, ATP, and GTP (Fig. 1D). Even when the polymerase assay was carried out at a constant temperature of 30°C, the de novo product was still formed at the same level as the hairpin product instead of at the 3:1 ratio observed at 24°C (Fig. 2, lane 1). However, the de novo product could not be detected under any of the other assay conditions, although the hairpin (2×) product was readily formed (Fig. 2, lanes 2 to 8). If the template or the enzyme was omitted in the assays, no products could be detected; these conditions served as negative controls (Fig. 2, lanes 9 and 10).
FIG. 1.
Components of the de novo preinitiation complex. The protocol for determining the components for the de novo preinitiation complex is outlined in each of the panels on the right. The conditions under which de novo initiation occurs in preference to 3′-end elongation (1×/2× ratio, ≥2) define the components for formation of the de novo preinitiation complex. It was previously established that one of the determinants for preferential de novo initiation is incubation at a low temperature (1). In experiments A to E, if components in an initial preincubation mixture are favorable for de novo initiation, then subsequent additions of missing components should not affect the preferential formation of the de novo product. RdRP assays were performed, and the products were fractionated on formaldehyde-agarose gels and subjected to autoradiography. The template-sized and hairpin products were cut out and quantitated by scintillation counting. The 1×/2× ratio on the y axis is plotted against the temperature of the RdRP assay on the x axis.
FIG. 2.
Preincubation of either ATP or GTP alone with NS5 and template is not sufficient for de novo initiation. (A) RdRP assays were carried out at 30°C with limiting components before the addition of heparin, and then the complete system was reconstituted after its addition. At 30°C, if all components were added at the same time, de novo initiation and 3′-end elongation occurred on the template RNA, and equal amounts of 1× and 2× products were formed (1). (B) The reaction products were analyzed by formaldehyde-agarose gel electrophoresis and detected by autoradiography.
Effects of nucleotide concentrations on 3′-end elongation versus de novo synthesis of RNA.
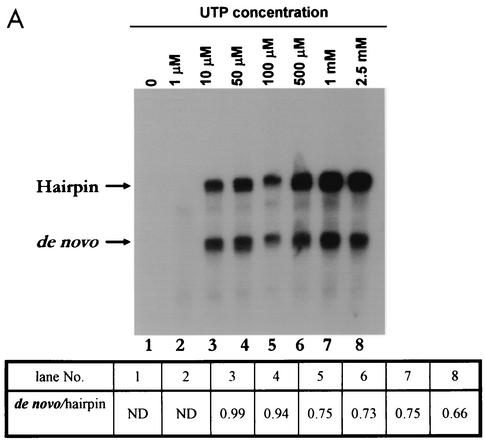
It has been reported that high concentrations of GTP stimulate de novo initiation of RNA synthesis by brome mosaic virus (BMV) RdRP (33) as well as by HCV NS5B RdRP (46, 48, 56, 79). The results described above indicated that de novo initiation requires preincubation of the template RNA and the polymerase along with ATP and GTP; CTP and UTP could be added later and are required only for the elongation reaction. Therefore, we sought to determine the concentrations of ATP, GTP, and UTP that are required for de novo initiation by DEN2 polymerase. In the RdRP assay, all nucleotide concentrations were kept at standard levels (500 μM) except for the one that was varied. The results shown in Fig. 3A indicate that at 1 μM UTP no RNA synthesis occurred, whereas at 10 μM UTP both de novo synthesis and elongation from the 3′ end of the template RNA occurred. Moreover, the concentration of UTP in the range between 10 μM and 1 mM did not affect the ratio of the de novo to the hairpin product, suggesting that UTP is required for elongation only.
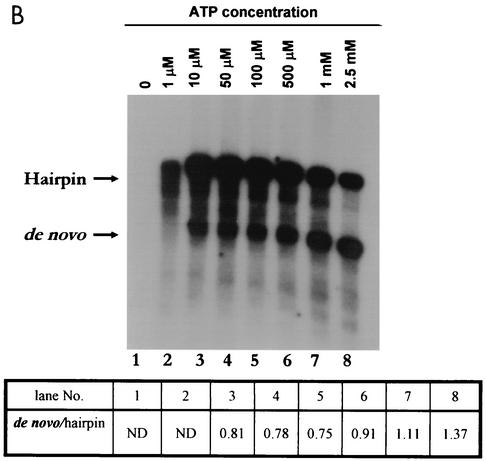
FIG. 3.
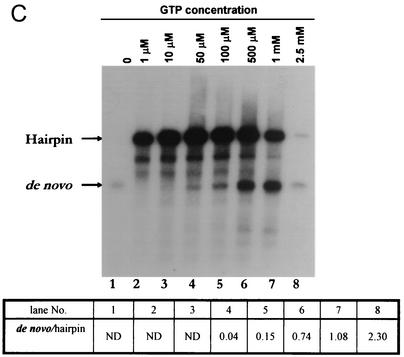
Analysis of NTP requirement for de novo initiation or 3′-end elongation. For RdRPs, the initiating nucleotide is required at high concentrations for de novo synthesis whereas lower concentrations are sufficient for elongation (see the text). Standard RdRp assays in which all components were held at a fixed concentration except for the NTP that was varied (UTP [A], ATP [B], or GTP[C]) were performed at 30°C. Products were analyzed by formaldehyde-agarose gel electrophoresis followed by autoradiography. The 1× and 2× products were cut out and quantified by either scintillation counting (A and B) or density analysis using the imagej program, a free software product available at the National Institutes of Health website (http://rsb.info.nih.gov/ij/). ND, not done.
Next, we examined the effect of varying the ATP concentration on the shift between the de novo and 3′-end elongation pathways of the polymerase. The 3′-terminal nucleotide of DEN2 RNA is U and the penultimate nucleotide is C, so ATP and GTP would be expected to be components of the de novo preinitiation complex (Fig. 1C and D). When ATP concentrations were varied (Fig. 3B), RNA synthesis mainly by 3′-end elongation readily occurred at 1 μM, but no de novo synthesis occurred at this low concentration. However, in the range of 10 to 500 μM ATP, de novo synthesis and 3′-end elongation reactions were observed to approximately similar extents. At 1 mM, this ratio did not change significantly, although at 2.5 mM ATP, there was a slight increase in the proportion of the de novo product compared to the hairpin product (Fig. 3B). From these results, we conclude that although ATP is a component of the de novo preinitiation complex, a high concentration of ATP is not required for a preferred de novo initiation pathway.
Next, when the GTP concentration was varied in the RdRP assay from 1 to 100 μM, 3′-end elongation occurred predominantly, giving rise to the hairpin product (Fig. 3C). A 500 μM concentration of GTP was required to shift the equilibrium toward equal levels of de novo and hairpin products. When the concentration of GTP was varied in the 100 to 500 μM range, there seemed to be a threshold for GTP concentration (∼150 μM) below which no appreciable de novo synthesis (1× product) was detectable (see below). On the other hand, both the de novo and hairpin reactions are inhibited at 2.5 mM GTP. This inhibition may be due to an imbalance of the ratio between GTP and Mg2+, as was previously shown for HCV NS5B (46). It was verified that the high GTP concentration requirement by the enzyme for de novo synthesis was not due to any phosphatase present in the enzyme by incubation of the enzyme with 4-nitrophenol phosphate as a substrate. No hydrolysis of the substrate occurred under the assay conditions, although calf intestinal alkaline phosphatase at 0.01 U, used as a positive control, could hydrolyze the substrate (data not shown). Thus, in this regard, DEN2 polymerase exhibits characteristics similar to those of the BMV and HCV polymerases reported earlier with regard to the requirement of high GTP concentrations for de novo initiation (33, 46, 48, 79). In the absence of GTP (Fig. 3C, lane 1), the polymerase presumably catalyzes a non-template-dependent addition of a labeled nucleotide(s) at the 3′ end of the template RNA, which is visualized as a 1× product. This property of the polymerase may be due to its intrinsic terminal nucleotidyltransferase activity, similar to that of HCV NS5B polymerase (61). It was previously shown by its sensitivity to RNase A digestion that this product is single stranded (1).
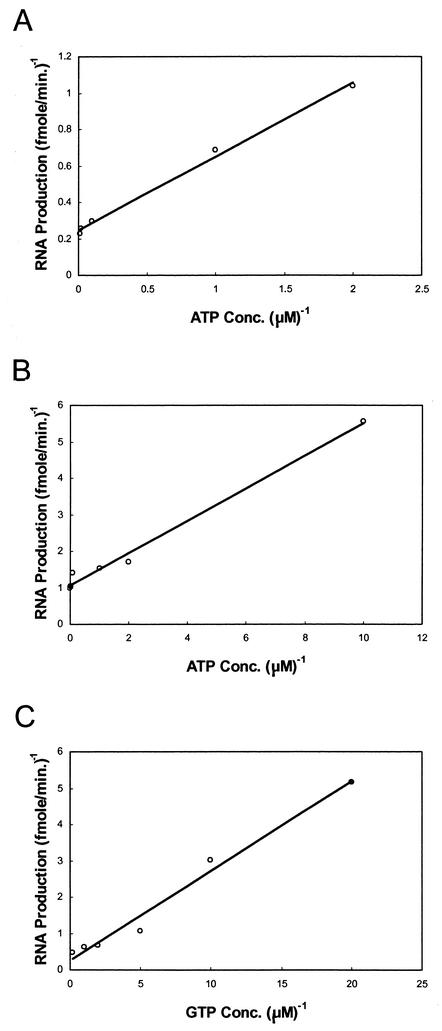
Since both ATP and GTP are required for formation of the de novo preinitiation complex under a set of concentrations different from those required for 3′-end elongation, we sought to determine the Km values of the polymerase for these two NTPs for the 3′ elongation reaction from the 3′ fold-back structure and for de novo synthesis of RNA. As shown in Fig. 4, the Km values for ATP and GTP utilized for elongation are 2.25 ± 0.84 and 0.37 ± 0.07 μM, respectively. For comparison, the Km values for ATP and GTP utilized by HCV NS5B in elongation are 2.34 ± 0.07 and 1.85 ± 0.28 μM, respectively, on the HCV-derived, D(+) template (48). In the HCV study, only total RNA synthesis, which represents both the hairpin and template-sized products, was measured in the assays. Furthermore, GTP utilization exhibited a biphasic profile, and a second Km of 39 μM was obtained, which led the authors to suggest that this low-affinity value for GTP is for de novo synthesis of RNA (48). In our study, the Km for ATP utilization by the DEN2 polymerase for de novo synthesis is 5.43 ± 2.50 μM, which is about 2.5-fold higher than that required for the 3′ elongation reaction (see above). For comparison, the Km of BMV polymerase for ATP in de novo synthesis from the BMV RNA template is 13.53 ± 2.65 μM (65). However, we could not determine accurately the Km for GTP required for de novo initiation in this study, because as stated above, the threshold of GTP concentration below which no appreciable de novo product could be detected was ∼150 μM. The effects of gradually increasing the GTP concentration to 1 mM did not follow Michaelis-Menten kinetics (data not shown). Moreover, the N-terminal methyltransferase domain was reported to have a novel GTP binding site with a dissociation constant (Kd) of 58 ± 14 μM, and the 7MeGpppA also binds to the same site with a Kd of 255 ± 5 μM (18). The HCV NS5B does not have an RNA methyltransferase domain analogous to that of DEN2 NS5. Thus, the effect of GTP binding on DEN2 polymerase is more complex, and further study is needed to differentiate between the novel GTP binding to the N-terminal domain and the site in the polymerase domain responsible for de novo RNA synthesis. The Vmaxs for ATP and GTP were calculated, from the results of experiments shown in Fig. 4, to be 2.3 ± 1.0 and 1.26 ± 0.6 pmol of NMP/μg (10 pmol) of DEN2 polymerase enzyme/h, respectively, for elongation; the Vmax for ATP in de novo synthesis was calculated to be 0.37 ± 0.015 pmol of AMP/μg of enzyme (10 pmol)/h. For HCV NS5B polymerase, Vmax values for ATP and GTP were reported to be 4.8 and 5.1 pmol of NMP/μg of enzyme/h for total RNA synthesis (47).
FIG. 4.
Effects of varying amounts of ATP and GTP on RdRP activity. RdRP assays were carried out as described in Materials and Methods except that the concentrations of ATP (A and B) or GTP (C) were varied. The amounts of the hairpin product produced by 3′ elongation and the template-sized product produced by de novo synthesis of RNA were determined by counting the radioactivity by use of a scintillation counter. Data are presented as double-reciprocal plots (1/V versus 1/[S]) generated with Microsoft Excel. The plot for GTP utilization in de novo synthesis showed a complex kinetics and therefore could not be accurately determined (see the text).
Template primer-directed RNA synthesis at the 3′ end of DEN2 subgenomic RNA.
Viral RdRPs, in addition to being able to initiate RNA synthesis de novo, are also able to utilize short oligonucleotides as primers for the elongation reaction (15, 20, 39, 54, 65, 78). Many RdRPs are able to catalyze synthesis of RNA by a template-primed reaction resulting from a fold-back structure of the template RNA (1, 4, 75). In this study, we sought to determine whether the DEN2 polymerase could utilize exogenously added RNA primers of various lengths that are complementary to the template RNA.
The 3′-terminal sequence of DEN2 viral RNA is -GGUUCU-3′. Since either ATP or GTP alone is sufficient to form the de novo preinitiation complex, we surmised that the DEN2 polymerase synthesizes a tetranucleotide RNA primer, AGAA, and remains in a heparin-resistant initiation complex prior to the addition of CTP and UTP for the elongation reaction. Therefore, we investigated the efficiency with which the polymerase can utilize exogenously added RNA primers of varying lengths complementary to the 3′ end of the template RNA. We carried out RdRP assays in the presence of primers AG, AGA, AGAA, and AGAACC. The reaction temperature was kept at 35°C. At this temperature, the polymerase essentially produced the hairpin product from the template RNA by 3′-end elongation in the absence of any primers (1). Thus, if the exogenously added primer is utilized by the polymerase under these conditions, we would be able to detect the template-sized product synthesized from the primer by elongation.
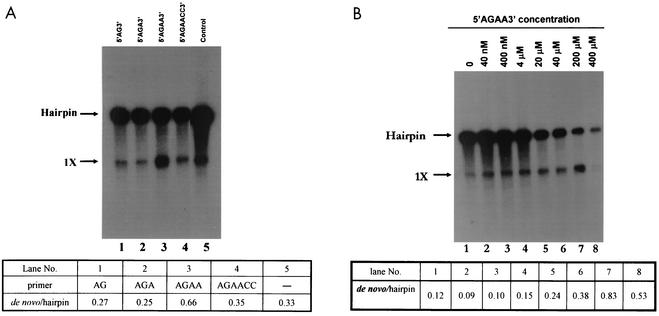
The results shown in Fig. 5A indicate that in the absence of any exogenously added primer, the template was utilized by the polymerase to yield predominantly a hairpin product by a 3′-end elongation reaction (lane 5), in agreement with our previous observation (1). The di- and trinucleotide primers (40 μM) did not significantly change the proportion of the hairpin product produced in the control assay (Fig. 5A; compare lanes 1 and 2 with lane 5). However, when AGAA (40 μM) was used as a primer, there was a distinct shift toward synthesis of the template-sized product (Fig. 5A, lane 3). This experiment has been repeated twice, with similar results (see also Fig. 4B). One possible cause for these differences in the efficiency of utilization of these primers by the DEN2 polymerase could be differences in the stability of the ternary complex consisting of the template, primer, and enzyme. Next, we tested the utilization of primer AGAACC, which is complementary to the 3′-terminal sequence of DEN2 RNA (GGUUCU-3′). As shown in Fig. 5A, lane 4, the efficiency of utilization of this hexanucleotide primer was so low that results were comparable to those for the control without any primer. These results suggested that the tetranucleotide primer AGAA binds optimally at the active site of the enzyme to initiate RNA synthesis. For HCV polymerase, di- and trinucleotide primers were the most efficient, whereas increasing the length further had a drastic effect on efficiency (78).
FIG. 5.
Analysis of primer utilization by the DEN2 polymerase reveals that the optimal primer for DEN2 polymerase is AGAA. (A) The oligoribonucleotide primer AG, AGA, AGAA, or AGAACC (40 μM) (lanes 1 to 4) was simply mixed with the subgenomic RNA template (0.1 μg) in a standard RdRP assay. The temperature of incubation of the RdRP assays was kept at 35°C, at which predominantly a hairpin product was formed in the absence of any primer (lane 5). (B) The concentration of AGAA was varied while all other components were held at constant levels. Products were analyzed by formaldehyde-agarose gel electrophoresis and autoradiography as described in Materials and Methods. The products were quantified by densitometric analysis as described in the legend to Fig. 3.
Next, we sought to determine the optimum concentration of this primer in the RdRP assay. As shown in Fig. 5B, increasing the concentration of primer AGAA up to 200 μM progressively increased the ratio of the template-sized product to the hairpin product. There was also a progressive decrease in 3′-end elongation, suggesting that once the productive template-primer-enzyme ternary complex is formed, excess primer competes with the fold-back structure of the template RNA for binding with the enzyme, inhibiting RNA synthesis by 3′-end elongation and thus decreasing overall RNA synthesis.
Mutational analysis of the subgenomic RNA template efficiency in RdRP assays.
The results of our experiments so far indicated that ATP and GTP are required for the formation of the de novo preinitiation complex and that high concentrations of GTP are required for de novo initiation. Two possibilities could explain the requirement for a high concentration of GTP for de novo initiation: either (i) with increasing concentrations of GTP, the polymerase changes its conformation, which is then able to bind to the 3′ end of the template RNA and catalyze de novo synthesis of negative-strand RNA, or (ii) GTP serves as the initiating nucleotide complementary to the penultimate C of the template RNA. For example, minus-strand synthesis by BMV polymerase has been reported to begin at the penultimate cytidylate residue on the template, and thus a guanylate is the first nucleotide incorporated (51). In this case then, the terminal nucleotide is added by other mechanisms.
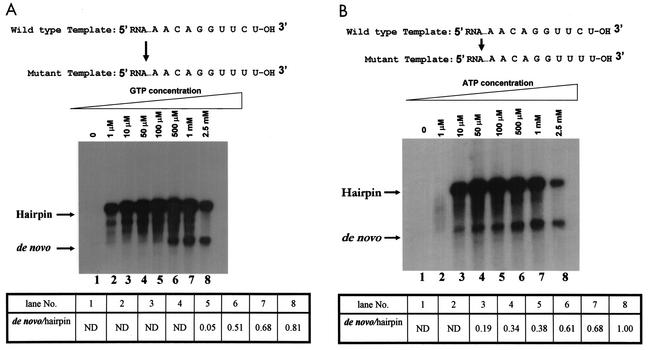
To examine these possibilities in detail, the 3′-penultimate nucleotide of the subgenomic RNA template was mutated from C to U (UUCU-3′ to UUUU-3′). First, we determined the ATP and GTP concentration requirements of this mutant template for the de novo and 3′-end elongation reactions. As shown in Fig. 6A, between 1 and 100 μM GTP, no significant de novo synthesis occurred; under these conditions, GTP was essentially used for the 3′-end elongation reaction to yield the hairpin product. However, at 500 μM GTP, de novo synthesis of RNA was stimulated about 10-fold, even though the first 6 nucleotides of the template RNA (-GGUUUU-3′) did not require incorporation of a guanylate residue. These results suggested that the high GTP requirement in de novo synthesis of RNA plays a conformational role rather than being necessary for incorporation. By varying the ATP concentrations, we observed that even at 10 μM ATP, both 3′-end elongation and de novo synthesis occurred (1×/2× ratio, 0.19); increasing the ATP concentration to 100 μM stimulated de novo synthesis about twofold, and increasing it from 100 to 500 μM brought about a further increase of 1.6-fold (Fig. 6B). This result was different from that for GTP, in which case no appreciable de novo synthesis occurred below a threshold of 100 μM (Fig. 6A).
FIG. 6.
High GTP concentration requirement for de novo initiation on a mutant RNA template. The positive-strand RNA template containing the 3′-end sequence GGUUCU-3′ in the wild-type RNA was mutated to GGUUUU-3′ by PCR as described in Materials and Methods. RdRP assays were performed at 30°C by using fixed amounts of all components except GTP (A) or ATP (B). Products were analyzed by formaldehyde-agarose gel electrophoresis followed by autoradiography. The intensities of bands were quantified by using the imagej program as described in the Fig. 3 legend. ND, not done.
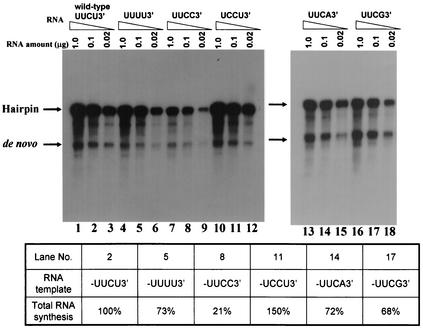
The 3′-terminal dinucleotide sequence CU is highly conserved in all mosquito-borne flaviviruses and therefore is likely to play an important role in viral replication. Therefore, we asked whether the 3′-terminal nucleotide of subgenomic RNA of positive polarity is important for negative-strand RNA synthesis in vitro. To address this question, subgenomic mutant RNA templates in which the UUCU-3′ (wild type) sequence was mutated to UUCC-3′, UUCA-3′, UUCG-3′, or UCCU-3′ were generated as described in Materials and Methods. We determined the template efficiencies of these mutants along with those of the UUUU-3′ mutant and the wild-type template in RdRP assays. The results shown in Fig. 7 indicate that mutation of the 3′-terminal nucleotide from U to C reduced the template efficiency about fivefold, whereas mutation of the penultimate nucleotide from C to U (UUCU-3′ to UUUU-3′) or of the third-position nucleotide from U to C (UUCU-3′ to UCCU-3′) did not affect template efficiency to a significant extent. Similarly, mutation of the 3′-terminal U to A or G also did not affect template efficiency significantly (Fig. 7). From these results, we conclude that the 3′-terminal nucleotide, U, plays a critical role in polymerase binding and that the 3′-terminal nucleotide has a preferred order in efficient minus-strand RNA synthesis.
FIG. 7.
Mutational analysis of the 3′-terminal sequences of the template RNA. The wild-type sequence, UUCU-3′, was mutated to UUUU-3′ (see Fig. 6), UCCU-3′, UUCA-3′, UUCG-3′, or UUCC-3′ as described in Materials and Methods. The mutant template RNAs in three different concentrations (0.02, 0.1, and 1.0 μg) were used in the RdRP assays, and the products were analyzed by formaldehyde-agarose gel electrophoresis and autoradiography. The radioactivities for the 1× and 2× products were quantified by liquid scintillation counting. The sum of the radioactivity, representing total RNA synthesis, was determined by using 0.1 μg each of wild-type and mutant RNA templates. The activity of each mutant relative to that of wild-type RNA, taken as 100%, is given below the gel.
DISCUSSION
In previous studies, an in vitro RdRP assay system was established using cytoplasmic extracts from DEN2-infected C6/36 or monkey kidney cells and subgenomic 770-nucleotide RNA templates of positive-sense polarity. RNA synthesis required both terminal regions of the template RNA, which revealed that there is a functional interaction between these two regions (74, 75). The template RNAs used in our assays contained highly conserved 3′ and 5′ stem-loop structures (9, 52, 62) and CYC motifs (26; for a review see reference 12). Two products were formed in the in vitro assays: a template-sized RNA and a hairpin RNA twice the size of the template (75). In a later study, it was shown that DEN2 RdRP expressed and purified from E. coli was active on exogenous subgenomic 770-nucleotide RNA templates and also produced two products similar to those of the cell lysate system described above; the template-sized product was shown to be the product of de novo synthesis, and the hairpin RNA resulted from 3′-end elongation of a fold-back structure of template RNA (1).
In that study, using the purified DEN2 NS5 RdRP, it was shown that the viral polymerase switches the mode of RNA synthesis from synthesis of a de novo product at low temperatures to that of a hairpin product by 3′-end elongation of a fold-back RNA structure at higher temperatures. The ratio of the de novo to the hairpin product decreased from 2 to 0.25 by a temperature shift from 20 to 40°C (1). A heparin trap protocol was used to isolate de novo initiation events from elongation in a single RNA synthesis event by the enzyme. The results showed that de novo initiation events required incubation at low temperatures and that once a stable de novo preinitiation complex was formed, the elongation step was insensitive to temperature variations. However, in the context of viral infection, only the synthesis of the de novo initiation product by the polymerase is physiologically important for faithful reproduction of progeny viral RNA molecules. The polymerase is likely to be regulated by its interaction with other viral proteins as well as host proteins in the viral replicase complex and the viral RNA template and by its membrane localization. Our in vitro assay will be useful for dissection of the components of the viral replicase complex and analysis of the contribution of the membrane association of template RNA to de novo synthesis of the RNA product.
Using this in vitro RdRP assay system, it was shown that RNA synthesis required both 5′- and 3′-terminal regions including the highly conserved CYC motifs in the subgenomic RNA template (1). The 3′ stem-loop structure and the CYC motifs have also been shown to be important for RNA synthesis in vitro using infected cell lysates (74) as well as for viral RNA replication in cultured cells, as shown by mutagenesis of infectious DEN2 RNA (76) or either Kunjin virus viral replicon (36) or yellow fever virus replicon RNA (16).
In this study, we further define the components of the preinitiation complex. Our results indicate that incubation of polymerase, template, ATP, and GTP at low temperatures is sufficient for preferred synthesis of the de novo product. CTP and UTP can be added subsequent to the addition of heparin and are therefore required only for the elongation reaction. If any of the required initiation components were omitted in the preinitiation complex prior to the addition of heparin but added subsequently, there was a significant reduction in the proportion of the de novo product. Moreover, the concentration of GTP in the preinitiation complex should be high, although varying the ATP concentration did not have much effect in de novo synthesis. A high GTP concentration is not required, however, for the elongation reaction from the 3′ end of a fold-back structure of the template RNA, a result which is similar to the property of BMV RdRP described previously (33). De novo initiation by BMV polymerase (33), Qβ RdRP (5), and HCV NS5B (48, 78, 79) also requires high GTP concentrations, whereas the influenza virus polymerase requires high concentrations of ATP (39). Luo et al. (48) observed a biphasic kinetics in the utilization of GTP by the HCV NS5B RdRP, which had two Km values for GTP; the lower Km corresponded to those of the other NTPs whereas the higher Km was attributed to de novo initiation.
Since a high GTP concentration is required for de novo initiation but not for elongation, it is possible that GTP is the initiating nucleotide for negative-strand RNA synthesis. This has been shown for BMV RdRP, which initiates with the guanylate complementary to the penultimate cytosine (51). Recent crystal soaking experiments with the φ6 RdRP have demonstrated that the initial NTP binding site is complementary to the second nucleotide from the 3′ end of the template strand (10). After the initial Watson-Crick binding of GTP to the second nucleotide from the 3′ end of the template, the φ6 polymerase steps back. The “stepping back” then allows the binding of a second GTP to the 3′-terminal nucleotide of the template, followed by the formation of the first phosphodiester bond (10). Similarly, in vitro RNA synthesis by short oligoribonucleotide primers for the Tacaribe arenavirus RNA polymerase indicated that stimulation of polymerase activity by GpC (complementary to positions +2 and +3 of the template) was greater than that by CpG (complementary to positions +1 and +2). Analysis of the 5′ ends of the in vitro transcripts was consistent with a model in which genome replication begins with pppGpC on the +2 and +3 nucleotide positions on the template; the enzyme steps back on the template so that the 5′ end of the newly synthesized transcript is at position −1 prior to elongation (20). A similar explanation was also put forward as a possible mechanism for RNA synthesis by the respiratory syncytial virus polymerase (42).
The stepping-back mechanism for initiation of DNA replication by viral DNA polymerases to ensure copying of the terminal sequences on the template has been well studied (38, 50). In the case of DEN2, the second nucleotide in the negative-strand RNA is a guanylate residue. If a high GTP concentration is required to initiate at the penultimate nucleotide, then mutation of this residue from C to U in the template RNA (from the wild-type UUCU-3′ to UUUU-3′) would be expected to shift this requirement for a high concentration of the initiating nucleotide from GTP to ATP. On the other hand, a high GTP concentration was still required for de novo synthesis of RNA, and the de novo initiation complex was formed only when the template, polymerase, ATP, and high concentrations of GTP were preincubated prior to the addition of heparin (data not shown). This finding still does not rule out the possibility that the initiating nucleotide is GTP, which can form a wobble base pair with the 3′-penultimate uridylate on the mutant template RNA. Under these conditions, the same primer, AGAA (see below), may still be made during the initiation step. Alternatively, GTP binding to a specific site on the DEN2 polymerase changes its conformation such that the enzyme is able to bind productively at the 3′ end of either the wild-type or the mutant template RNA for de novo initiation.
Recent structural analysis of HCV NS5B showed that GTP binds to a specific low-affinity surface binding site that cannot bind to other NTPs, 30 Å away from the catalytic site, adjacent to a region where thumb and finger domains interact (7). It appears that other nucleotides can bind to the catalytic site of HCV NS5B through their triphosphate moieties even in the absence of the template. At present, it has not been conclusively established whether this surface binding site is responsible for de novo initiation by the HCV NS5B. It has also been suggested that this surface site may be involved in oligomerization of the polymerase, which may be required for optimal initiation of RNA synthesis, as proposed for the poliovirus polymerase, 3Dpol (8, 27, 28, 58). A recent study has indicated that the N-terminal region consisting of 296 amino acid residues has a 2′-O-methyltransferase activity, and the crystal structure of this domain has been determined (18). This domain bound to GTP, as shown by UV cross-linking, and the Km for GTP was around 58 μM. It is not clear if this GTP binding site plays any role in the initiation of RNA synthesis.
The 3′-terminal dinucleotide CU is highly conserved in all mosquito-borne flaviviruses. Our results indicate that mutation of the 3′-terminal nucleotide from U to C reduced the template activity about fivefold, whereas mutation of the penultimate nucleotide C to U was well tolerated (as seen from the activity of the UUUU-3′ template, which was similar to that of the wild type). Recently, Shim et al. (64), using a synthetic short RNA for purified HCV NS5B polymerase in the in vitro RdRP assays, showed that U was preferred, followed by G, A, and C. However, Yi and Lemon reported that the order for replication of HCV replicon RNA in cultured cells (from most to least preferred) was U (wild type), C, A, and G (73). These results from experiments performed in vitro using purified polymerase and in cultured mammalian cells using replicon RNA are different in the order of preference at the 3′-terminal nucleotide, and in both studies, the U residue was still the preferred nucleotide at the 3′ end. Interestingly, purified DEN2 NS5 and HCV NS5B (64) exhibit similar orders of preference for the 3′-terminal nucleotide (U, A∼G, C). As Shim et al. (64) suggested, the 3′-terminal base is not the sole determinant for template specificity; that specificity is collectively determined by the 3′-terminal nucleotide, incoming NTP, and the active-site structure of the polymerase.
We tested primer utilization by the polymerase by using synthetic AG, AGA, AGAA, and AGAACC primers and the subgenomic RNA template in the RdRP assays at 35°C. This temperature was chosen because in the absence of any primer, the product formed is predominantly a hairpin RNA (1). Therefore, if the primer is utilized by the polymerase for 3′-end elongation to yield a template-sized product, it could easily be detected by electrophoresis on a denaturing gel and autoradiography. Our results indicated that AGAA was the optimal length of the primer utilized by the polymerase and that neither a shorter nor a longer (AGAAGG) primer was efficient. These results suggest that primer AGAA may fit into the active site of the polymerase optimally at the temperature that favors an open conformation (see below). Furthermore, AGAA is likely to be a stable product formed by the polymerase during the de novo initiation step which is then elongated to yield the template-sized product. Moreover, RNA synthesis by elongation at the 3′ end of the AGAA primer required a reduced concentration of GTP (10 μM) (data not shown). This observation is consistent with the finding of a previous study that primer GpG could substitute for the high GTP requirement for stimulation of negative-strand RNA synthesis by BMV polymerase (33). In addition, the influenza virus polymerase exhibited a 10-fold-higher Km for ATP than for the other NTPs for initiation, but during elongation the Km for ATP was same as that for the other NTPs (39).
Ackermann and Padmanabhan proposed a model to explain the temperature-dependent shift from de novo synthesis to 3′-end elongation of the fold-back structure on the template RNA (1). This model was based on a previous study that analyzed template-primer utilization by the poliovirus polymerase 3Dpol and the HCV polymerase NS5B and correlated the results to differences in the conformation of the active sites of these viruses (29). The active site of the poliovirus polymerase 3Dpol is in an open conformation and is active on double-stranded RNA templates. However, the HCV RdRP has an extra β-hairpin of 12 amino acid residues in the thumb subdomain which occludes the active site (and which is absent in 3Dpol). HCV NS5B RdRP is thus unable to bind to the double-stranded template RNA-primer structure productively (29). In support of this model for active-site occlusion, deletion of 4 amino acid residues on either side of the β-hairpin in the active site of HCV NS5B conferred on the polymerase the ability to initiate synthesis on a double-stranded template primer as efficiently as the poliovirus 3Dpol.
If the DEN2 polymerase structurally resembles the HCV enzyme, the active site of the enzyme is likely to be in equilibrium between a more rigid, closed conformation at low temperatures and a more flexible, open conformation at higher temperatures; in the former state, the active site of the polymerase is positioned for de novo initiation, whereas in the latter state, the polymerase is able to bind to double-stranded template-primer structures and catalyze 3′-end elongation of the primers or of a 3′ fold-back structure of the template RNA to yield a template-sized or a hairpin product, respectively. There appears to be some structural similarity among flaviviruses, hepaciviruses, and pestiviruses, as a sequence akin to the HCV β-loop is expected in the proximity of the conserved GDD motif. Moreover, the positions of aromatic amino acid residues within the putative β-loop also seem to be conserved (32). Further work including determination of the crystal structure of the DEN2 polymerase is necessary to clarify the role of the GTP binding site(s) on the polymerase and its relationship to de novo initiation of RNA synthesis.
Acknowledgments
M. Nomaguchi and M. Ackermann contributed equally to this study.
This work was supported by Public Health Service grant AI-32078.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Bartelma, G., and R. Padmanabhan. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122-132. [DOI] [PubMed] [Google Scholar]
- 3.Bazan, J. F., and R. J. Fletterick. 1989. Comparative analysis of viral cysteine protease structural models. FEBS Lett. 249:5-7. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal, T. 1980. Q beta replicase template specificity: different templates require different GTP concentrations for initiation. Proc. Natl. Acad. Sci. USA 77:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowski, P., A. Niebuhr, O. Mueller, M. Bretner, K. Felczak, T. Kulikowski, and H. Schmitz. 2001. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75:3220-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 10.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 11.Cahour, A., B. Falgout, and C.-J. Lai. 1992. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J. Virol. 66:1535-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 13.Chambers, T. J., D. W. McCourt, and C. M. Rice. 1989. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology 169:100-109. [DOI] [PubMed] [Google Scholar]
- 14.Chambers, T. J., R. C. Weir, A. Grakoui, D. W. McCourt, J. F. Bazan, R. J. Fletterick, and C. M. Rice. 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc. Natl. Acad. Sci. USA 87:8898-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, D., and J. T. Patton. 2000. De novo synthesis of minus strand RNA by the rotavirus RNA polymerase in a cell-free system involves a novel mechanism of initiation. RNA 6:1455-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui, T., R. J. Sugrue, Q. Xu, A. K. Lee, Y. C. Chan, and J. Fu. 1998. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology 246:409-417. [DOI] [PubMed] [Google Scholar]
- 18.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falgout, B., M. Pethel, Y. M. Zhang, and C. J. Lai. 1991. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J. Virol. 65:2467-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcin, D., and D. Kolakofsky. 1992. Tacaribe arenavirus RNA synthesis in vitro is primer dependent and suggests an unusual model for the initiation of genome replication. J. Virol. 66:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., A. P. Donchenko, E. V. Koonin, and V. M. Blinov. 1989. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 17:3889-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubler, D. J., and G. G. Clark. 1995. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg. Infect. Dis. 1:55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 26.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 27.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 28.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 30.Irie, K., P. M. Mohan, Y. Sasaguri, R. Putnak, and R. Padmanabhan. 1989. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain). Gene 75:197-211. [DOI] [PubMed] [Google Scholar]
- 31.Kadare, G., and A. L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, C. C., P. Singh, and D. J. Eckert. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 33.Kao, C. C., and J. H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaicvirus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapoor, M., L. Zhang, M. Ramachandra, J. Kusukawa, K. E. Ebner, and R. Padmanabhan. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100-19106. [DOI] [PubMed] [Google Scholar]
- 35.Kautner, I., M. J. Robinson, and U. Kuhnle. 1997. Dengue virus infection: epidemiology, pathogenesis, clinical presentation, diagnosis, and prevention. J. Pediatr. 131:516-524. [DOI] [PubMed] [Google Scholar]
- 36.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1997. Towards defining a minimal functional domain for NTPase and RNA helicase activities of the hepatitis C virus NS3 protein. Virus Res. 49:17-25. [DOI] [PubMed] [Google Scholar]
- 38.King, A. J., and P. C. Van der Vliet. 1994. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 13:5786-5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klumpp, K., M. J. Ford, and R. W. Ruigrok. 1998. Variation in ATP requirement during influenza virus transcription. J. Gen. Virol. 79:1033-1045. [DOI] [PubMed] [Google Scholar]
- 40.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 41.Koonin, E. V. 1992. A new group of putative RNA helicases. Trends Biochem. Sci. 17:495-497. [DOI] [PubMed] [Google Scholar]
- 42.Kuo, L., R. Fearns, and P. L. Collins. 1997. Analysis of the gene start and gene end signals of human respiratory syncytial virus: quasi-templated initiation at position 1 of the encoded mRNA. J. Virol. 71:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo, M. D., C. Chin, S. L. Hsu, J. Y. Shiao, T. M. Wang, and J. H. Lin. 1996. Characterization of the NTPase activity of Japanese encephalitis virus NS3 protein. J. Gen. Virol. 77:2077-2084. [DOI] [PubMed] [Google Scholar]
- 44.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and the RNA-stimulated NTPase domains of dengue virus type 2 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 47.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 48.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Markoff, L. 1989. In vitro processing of dengue virus structural proteins: cleavage of the pre-membrane protein. J. Virol. 63:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendez, J., L. Blanco, J. A. Esteban, A. Bernad, and M. Salas. 1992. Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. USA 89:9579-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller, W. A., J. J. Bujarski, T. W. Dreher, and T. C. Hall. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537-546. [DOI] [PubMed] [Google Scholar]
- 52.Mohan, P. M., and R. Padmanabhan. 1991. Detection of stable secondary structure at the 3′ terminus of dengue virus type 2 RNA. Gene 108:185-191. [DOI] [PubMed] [Google Scholar]
- 53.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagy, P. D., C. D. Carpenter, and A. E. Simon. 1997. A novel 3′-end repair mechanism in an RNA virus. Proc. Natl. Acad. Sci. USA 94:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowak, T., P. M. Farber, G. Wengler, and G. Wengler. 1989. Analyses of the terminal sequences of West Nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology 169:365-376. [DOI] [PubMed] [Google Scholar]
- 56.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Reilly, E. K., and C. C. Kao. 1998. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 252:287-303. [DOI] [PubMed] [Google Scholar]
- 58.Pata, J. D., S. C. Schulz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 59.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preugschat, F., C. W. Yao, and J. H. Strauss. 1990. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J. Virol. 64:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. T. Sarisky, and C. C. Kao. 2001. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 63.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 73:2129-2134. [DOI] [PubMed] [Google Scholar]
- 64.Shim, J. H., G. Larson, J. Z. Wu, and Z. Hong. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J. Virol. 76:7030-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, J. H., S. Adkins, G. Faurote, and C. C. Kao. 1996. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligonucleotides. Virology 226:1-12. [DOI] [PubMed] [Google Scholar]
- 66.Svitkin, Y. V., V. N. Lyapustin, V. A. Lashkevich, and V. I. Agol. 1984. Differences between translation products of tick-borne encephalitis virus RNA in cell-free systems from Krebs-2 cells and rabbit reticulocytes: involvement of membranes in the processing of nascent precursors of flavivirus structural proteins. Virology 135:536-541. [DOI] [PubMed] [Google Scholar]
- 67.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 68.Warrener, P., J. K. Tamura, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wengler, G., G. Czaya, P. M. Farber, and J. H. Hegemann. 1991. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 72:851-858. [DOI] [PubMed] [Google Scholar]
- 70.Wengler, G., and G. Wengler. 1991. The carboxy-terminal part of the NS3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 71.Wengler, G., and G. Wengler. 1993. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 72.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 73.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 75.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 76.Zeng, L., B. Falgout, and L. Markoff. 1998. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J. Virol. 72:7510-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, L., P. M. Mohan, and R. Padmanabhan. 1992. Processing and localization of dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J. Virol. 66:7549-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]