Abstract
Cottontail rabbit papillomavirus (CRPV) genomes mutated in the trans-activation domain of the E2 protein, which stimulates both viral DNA replication and transcription, are severely impaired in their ability to induce tumors in New Zealand White rabbits. A number of papillomaviruses encode, in addition to full-length E2, a shortened E2 protein or an E2 protein fused to a short stretch of amino acids derived from the small E8 open reading frame that counteract the activities of E2. We identified and cloned the novel cDNA E9^E2C of CRPV from papillomas of New Zealand White and cottontail rabbits and characterized the functions of the encoded gene product. E9^E2C was shown to be a bona fide repressor of minimal viral promoters, with the E9 domain being essential for this activity, and to repress E1/E2-dependent replication of a CRPV origin construct. In addition, E9^E2C counteracted the transactivation effect of the full-length E2 on minimal promoters containing several E2 binding sites. To investigate the role of E9^E2C in tumorigenesis, we constructed two CRPV genomes mutated in E9^E2C. One, designated CRPV-E9atgmut-pLAII, contained a mutation in the unique start codon in the E9 open reading frame, and the second E9^E2C mutant was constructed by the introduction of a stop codon close to the splice donor site at nucleotide 3714 that additionally prevented the correct splicing of the transcript. When we infected New Zealand White rabbits with these constructs, we surprisingly noted no differences in tumor induction efficiency, viral genome copy number, and viral transcription in comparison to wild-type CRPV.
Papillomaviruses have attracted special attention because persistent infection with a subset of human papillomaviruses (HPV) is a necessary factor for the development of anogenital cancer (4, 43). In contrast, the role of HPV in skin cancer, which is the most common cancer of the Caucasian race, is still undefined, although a very first link was obtained in 1935, when Rous et al. (30) described the development of squamous cell carcinomas in rabbits after experimental infection with cottontail rabbit papillomavirus (CRPV). Skin cancer in humans has been recognized as a common side effect in long-term immunosuppressed patients, who have a 65-fold increased risk compared to immunocompetent individuals. Shortly after the onset of immunosuppressive therapy, HPV-induced warts develop in these individuals, and 20 years after the beginning of immunosuppression nonmelanocytic skin cancer can be found in up to 70% of these patients (6). A similar situation has been described in the rare genetic disorder epidermodysplasia verruciformis, but the mechanisms leading to the development of skin cancer and especially the role of papillomaviruses are not yet fully understood (3, 12, 38).
The only model system for studying tumor induction and progression after infection of the skin with papillomaviruses is the domestic rabbit infected with CRPV, in which local tumors develop within 3 to 6 weeks postinfection and progress within 6 to 14 months in more than 80% of the cases into invasive growing and finally metastasizing carcinomas (39, 40). Recently, we demonstrated with CRPV genomes mutated within the E2 open reading frame (ORF) that a transactivator function of the viral protein E2 plays a major role in tumor induction and growth extension (16). The E2 protein is known to be involved in DNA replication and early gene expression of papillomaviruses by forming dimers, which specifically bind to the palindromic DNA sequence ACCN6GGT (E2 binding site [E2BS]) found in multiple copies in the upstream region of early and late promoters of papillomaviruses (23). In transient reporter assays, E2 is capable of stimulating transcription from artificial promoter constructs in an E2BS-dependent manner as well as from several early bovine papillomavirus type 1 (BPV1) promoters and from the E7 promoter of HPV 6a (23, 27).
During DNA replication, E2 forms a complex with the viral E1 protein and thereby increases the recognition of the replication origin by E1 and subsequently DNA replication efficiency (34). This transactivating and replication-stimulating property resides in the N terminus of E2, whereas the C terminus is responsible for DNA binding, dimerization, and transcriptional repression from the promoter-proximal E2BS (23). Aside from the E2 activator protein, two different E2 repressor proteins have been identified in BPV1-infected cells (10, 15, 17, 19). One is an N-terminally truncated E2 protein generated from a promoter located within the E2 gene, which has been designated E2C or E2TR (17), and the other one is generated from an alternatively spliced transcript, in which the E8 gene, overlapping the ORF of E1, is linked to the splice acceptor in the hinge region of the full-length E2 (10) and was designated E8/E2. Both proteins have the hinge region and the C terminus of the full-length E2 with its ability to bind DNA and form dimers. These proteins are in principle able to form homo- and heterodimers with the full-length E2 protein and to bind specifically to the E2 binding sites (2, 21, 24).
In tissue culture systems, both truncated E2 proteins proved to be repressors of the full-length E2 protein (10, 17, 18, 21). Moreover, they repressed the promoters within the noncoding region of BPV1 that are activated by the full-length E2 protein (10, 17). Similar spliced viral transcripts comparable to E8/E2 of BPV1 have also been described for HPV-11-, HPV-16-, HPV-31-, and HPV-33-infected cells or tissues. The proteins have been designated E8^E2C, sE2, and E2C, depending on the virus type (13, 29, 33, 36). All sE2, E2C, and E8^E2C proteins examined have been demonstrated to inhibit the transactivation activity of the full-length E2 protein in reporter assays and its stimulatory effect on the replication of the viral origin (5, 7, 8, 9, 22, 36, 37).
Recent findings furthermore suggested that E8^E2C is not only a counteractor of the full-length E2 protein but might be a general repressor of the viral promoters (36, 37). These data support the model that E2 repressor proteins of papillomaviruses might play an important role in the regulation of the viral life cycle, but nothing is known so far about their involvement in tumor induction and progression in vivo. In the case of CRPV, no E2 repressor proteins have been described so far. Transcript mapping experiments do not support the existence of a truncated E2 transcript, as described for BPV1, but they indicated the presence of a splice donor site that could give rise to a possible E8^E2C transcript (25).
To identify potential transcripts encoding E2 repressors, we analyzed a number of CRPV-induced tumors from New Zealand White and cottontail rabbits and successfully cloned an E9^E2C cDNA, whose encoded protein was functionally characterized with regard to transcriptional regulation and repression of DNA replication and for its role in tumor induction and progression with rabbits as the animal model (39, 40). Our results show that CRPV E9^E2C is a potent repressor of transcription and viral DNA replication in tissue culture but is dispensable for papilloma induction in New Zealand White rabbits.
MATERIALS AND METHODS
Animal system.
New Zealand White rabbits were anesthetized with 25 mg of ketamine hydrochloride and 5 mg of xylazine per kg; the backs of the rabbits were shaved with electric clippers and a razor blade. Sites of infection were marked with tattoo ink. Wild-type and mutant CRPV DNAs were injected into the rabbit skin with a Helios Gene Gun (Bio-Rad). For this purpose, 1 μg of DNA was precipitated on 0.5 mg of gold as described in the manufacturer's instructions. Each marked injection site on the rabbit skin was bombarded seven times with 1 μg of DNA with the help of the gene gun and helium gas at 350 to 400 lb/in2 pressure (42). Each rabbit had an equal number of injection sites for wild-type CRPV-pLAII DNA and either CRPV-E9atgmut-pLAII or CRPV-E9stop-pLAII DNA. Three and 6 months later, the number of papillomas that developed was determined, and the maximum diameter of each papilloma was measured. Biopsies were removed surgically, snap-frozen, and stored at −80°C.
Recombinant plasmids.
The E2-dependent luciferase reporter plasmids p6xE2BS-luc and pC18-Sp1-luc have been described previously (35, 37). The construct CRPV-PLP1-pGL3 was derived from plasmid CRPV-pLAII (26) by PCR with the upstream primer P7328 (5′-CGCAAGAGAGGTACCTAATTG-3′) and the downstream primer P116 (5′-GGCAATGGGTACCGATCTCAGATAC-3′), each with a KpnI restriction site and subsequent cloning of the amplified fragment into the pGL3-Basic vector (Promega, Heidelberg, Germany). The noncoding region (NCR) of CRPV (CRPV nucleotides 7339 to 151) was amplified by PCR with plasmid CRPV-pLAII (26) as the template and an upstream primer containing an SstI restriction site (5′-CTAAACGCAAGAGCTCTACTTAATTG-3′) and a downstream primer with an NcoI restriction site (5′-GCAGTTCTCCATGGGCTCTAAGTTTC-3′). The PCR-generated fragment was cloned into the pGL3-Basic vector and designated CRPV-pGL3-NCR.
The coding region of CRPV-E2 (CRPV nucleotides 3111 to 4284) was amplified by PCR with plasmid CRPV-pLAII as the template and an upstream (5′-GAAGACGAGGGATCCGATGGAGGCTCTC-3′) and a downstream (5′-GGGTTACATGCATACAGGATCCTAAAGCCC-3′) primer, each containing a BamHI restriction site. To amplify the CRPV E1 gene (CRPV nucleotides 1362 to 3171), an upstream primer containing an EcoRI restriction site (5′-CCCGGAGTGTTGTAAGAATTCATGGCTG-3′) and a downstream primer with a BamHI restriction site (5′-CCAAACTCGTGCTCGGATCCTCATAGAG-3′) were used. E1 and E2 were cloned into the expression vector pSG5 (Stratagene, Amsterdam, The Netherlands) and designated E1-pSG5 and E2-pSG5, respectively.
To generate an E9^E2C expression vector, the cloned cDNA was amplified with a forward primer containing an EcoRI restriction site (5′-CGCCCTTGTACTGGAATTCATGAAGCTGC-3′) and p3883, digested with EcoRI and MluI, and used to replace the EcoRI-MluI fragment from CRPV E2-pSG5, giving rise to E9^E2C-pSG5. The E9 deletion mutant E9K2A(d3-10)^E2C was constructed by PCR with primer pMA3702 (5′-CAAAACATGCGCCTCTCTGAATTCATGGCCAGCTCCCC-3′) and the downstream primer p3883. The amplicon was digested with EcoRI and MluI and used to replace the EcoRI-MluI fragment in E9^E2C-pSG5.
Mutation of the E9 ATG codon or the change of amino acid 10 of E9 to a stop codon in the context of the whole CRPV genome was performed by overlap extension PCR with primers E9atgmut (5′-GTACTGACGGAGACGAAGCTGCTTC-3′ and reverse complement sequence) or E9 stop (5′-CTGGTGCTGAGTCGTTAGAGGTAGAATC-3′ and reverse complement sequence) and primers p685 (5′-CTGGAGGAACTTGGATTATATCC-3′) and p1920rev (5′-CCCAATCCCAACTCATAGTCTTG-3′) and CRPV-pLAII as a template. The amplicons were digested with AvrII and SstI and used to replace the AvrII-SstI fragment in CRPV-pLAII. The plasmids obtained were designated CRPV-E9atgmut-pLAII and CRPV-E9stop-pLAII (see Fig. 6). All PCR-amplified fragments were confirmed by DNA sequence analysis. Expression vectors for H-ras (pT24ras) and for HPV16 E7 (pE7) have been described earlier (20, 32).
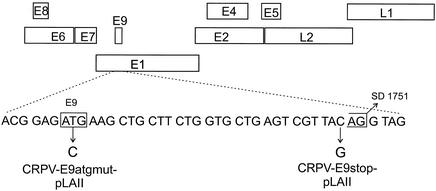
FIG. 6.
Construction of E9^E2C mutants in the context of the CRPV genome. The nucleotide sequence of the CRPV E9 gene is shown below the genome of CRPV. Positions of the ATG codon within the ORF E9 and the splice donor (SD) consensus site at nucleotide 1751 are indicated. Mutated nucleotides leading to the disruption of the ATG codon (CRPV-E9atgmut-pLAII) or to the introduction of a stop codon in E9 (CRPV-E9stop-pLAII) are indicated by arrows.
Cell culture.
The SCC13 cell line was maintained as described (35). The CRL 6502 cell line (American Type Culture Collection), an epithelial cottontail rabbit cell line, was maintained in medium consisting of a 1:1 mixture of M199 (Invitrogen, Karlsruhe, Germany) and F12 (Invitrogen, Karlsruhe, Germany) supplemented with 1 nM dexamethasone, 100 pM 17-β-estradiol, 1 nM T3, 10 ng of platelet-derived growth factor 2α per ml, 5 μg of insulin per ml, 100 IU of penicillin-streptomycin (Invitrogen, Karlsruhe, Germany) per ml, 7 mM sodium bicarbonate, 50 mM HEPES, 20 mM NaOH, 10 ng of mouse epidermal growth factor (Sigma Aldrich, Taufkirchen, Germany) per ml, and 5% (vol/vol) fetal calf serum (Seromed, Berlin, Germany).
To generate an immortalized keratinocyte cell line from New Zealand White rabbits, primary keratinocytes were isolated from skin epithelium as described previously for the isolation of human keratinocytes from human foreskin (31). Primary cultures were maintained in supplemented keratinocyte- and serum-free medium (Invitrogen, Karlsruhe, Germany). To immortalize primary rabbit keratinocytes, cells were grown to 70% confluency on a 35-mm dish and cotransfected with 300 ng each of pT24ras, pE7, and pSV2neo in the presence of 5 μl of Lipofectamine and OptiMEM (Invitrogen, Karlsruhe, Germany). Transfected cells were selected in keratinocyte- and serum-free medium (Invitrogen, Karlsruhe, Germany) supplemented with G418 (100 μg/ml) for 14 days. Pooled cultures of G418-resistent clones were expanded and designated RK1-16E7/ras.
Transient luciferase expression assays.
Approximately 105 SCC13, CRL6502, or RK1-16E7/ras cells were seeded into 35-mm-diameter dishes. The next day cells were cotransfected in the presence of 5 μl of Lipofectamine (Invitrogen, Karlsruhe, Germany) with 200 ng of luciferase reporter plasmids and CRPV expression vectors as indicated in the legends to Fig. 2, 3, and 4. The total amount of transfected DNA was kept constant within each experiment by adding parental pSG5 expression plasmid DNA. Luciferase assays were carried out 48 h after transfection. The cells were washed twice with cold phosphate-buffered saline and then lysed by adding 150 μl of cold luciferase extraction buffer (0.1 M potassium phosphate [pH 7.8], 1% Triton X-100, 1 mM dithiothreitol). Lysates were cleared by centrifugation (20,000 × g, 5 min, 4°C), and 20- to 40-μl aliquots of extract were subjected to luminometer analysis as described in the manufacturer's manual. Luciferase expression assays were carried out in duplicate and repeated two to four times to ensure reproducibility.
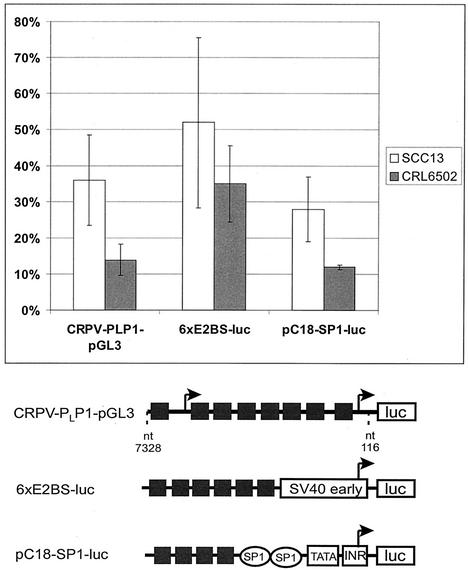
FIG. 2.
E9^E2C represses the basal activity of different promoters in epithelial cells. Reporter plasmids CRPV-PLP1-pGL3, 6×E2BS-luc, and pC18-SP1-luc, each containing several E2 binding sites (black boxes) upstream of the viral promoters PL and P1 of CRPV, the simian virus 40 (SV40) early promoter, or the minimal adenoviral promoter (INR) that drive the expression of the luciferase gene (luc) were cotransfected in SCC13 or CRL6502 cells with either 10 ng of CRPV-E9^E2C-pSG5 or 10 ng of the empty backbone expression vector pSG5. Luciferase activities are given relative to the activity in pSG5-transfected cells, which was set to 1. Standard deviations are indicated by error bars. Each measurement presents the average of 6 to 10 independent experiments.
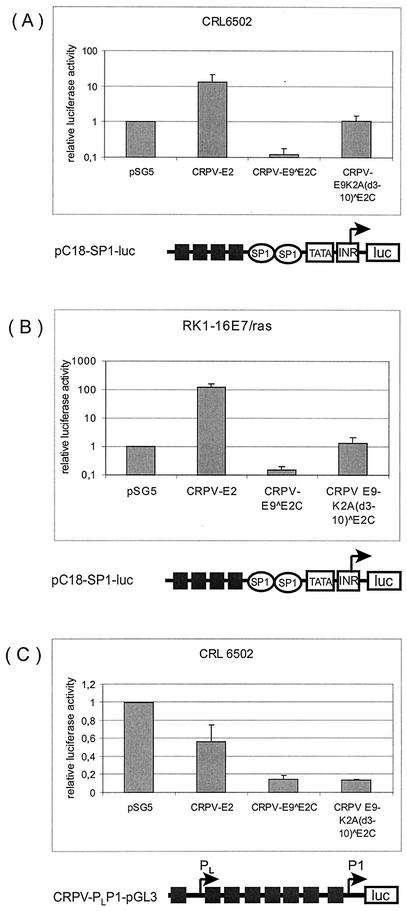
FIG. 3.
E9 domain is responsible for repression of minimal viral promoters. CRL6502 (A and C) or RK1-16E7/ras (B) cells were transfected with 200 ng of the reporter plasmid pC18SP1-luc (A and B) or 200 ng of the reporter plasmid CRPV-PLP1-pGL3 (C) and 10 ng of the empty expression vector pSG5 or expression vectors for CRPV-E2, wild-type CRPV-E9^E2C, or the E9 deletion mutant CRPV-E9K2A(d3-10)^E2C. Luciferase activities are given relative to the activity in pSG5-transfected cells, which was set to 1. Standard deviations are indicated by error bars. Note logarithmic scale on y axis (A and B).
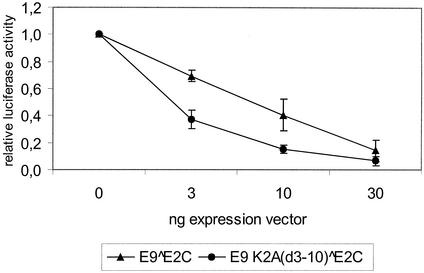
FIG. 4.
E9^E2C counteracts the transactivation mediated by full-length E2. SCC13 cells were cotransfected with 200 ng of the reporter plasmid pC18-SP1-luc (see Fig. 2 legend), 10 ng of CRPV-E2-pSG5, and an increasing amount of CRPV-E9^E2C-pSG5 or CRPV-E9K2A(d3-10)^E2C. Luciferase activities are shown relative to that of cells not transfected with E9^E2C-pSG5, which was set to 1. Standard deviations are indicated by error bars.
RNA isolation and analysis.
Total RNA was extracted from papillomas and carcinomas of rabbits by the guanidinium thiocyanate method (11) with Trizol reagent (Invitrogen, Karlsruhe, Germany) as recommended by the manufacturer. Five hundred nanograms of each RNA was used for reverse transcription-PCR with the SuperScript one-step system (Invitrogen, Karlsruhe, Germany) and the forward primer P1708 (5′-GTACTGACGGAGATGAAGCTGCTTC-3′) and either the reverse primer P3883 (5′-GTTTTCGTTGCTTTGCCGGCCTTC-3′) or the reverse primer P4289 (5′-CGTCACTAAAGCCCATAAAAATTCC-3′) with the following conditions: reverse transcription for 30 min at 42°C, denaturation for 10 min at 95°C, and then 35 cycles consisting of denaturation at 94°C for 20 s, annealing at 55°C for 20 s, and extension for 40 s at 72°C. The amplification products were cloned into the PCR 2.1Topo vector with the Topo TA cloning kit (Invitrogen, Karlsruhe, Germany), and then sequenced. RNA used for the quantitative reverse transcription-PCR was treated with DNase I (amplification grade; Invitrogen, Karlsruhe, Germany). After ethanol precipitation, RNA was reverse transcribed with the Superscript II reverse transcription kit (Invitrogen, Karlsruhe, Germany) with oligo(dT) as the primer for 50 min at 42°C. For each real-time PCR, 60 ng of cDNA and a 0.4 μM concentration of each primer were used with a final MgCl2 concentration of 4 mM.
To determine the CRPV transcript number, sequence-specific primers within the E4 ORF of CRPV were used (p3776, 5′-CAGTGCCCGCCGCTCAAAAGA-3′; p4016rev, 5′-GTTATAAGCTCGCGAAGCC-3′). PCR was carried out under the following conditions: denaturation at 95°C for 10 min, 45 cycles of amplification with denaturation at 95°C for 10 s, annealing at 52°C for 10 s, and extension at 72°C for 20 s. To confirm the detection of the correct amplicon, a melting curve analysis was performed following the amplification. Transcript numbers were standardized for β-actin transcripts determined in the LightCycler with the help of the following sequence specific primers: forward, 5′-GACTCGTCGTACTCCTGCTTG-3′; and reverse, 5′-CATTGCCGACAGGATCCAGA-3′. For the Northern blot analysis, polyadenylated RNA was selected from the total RNA isolated from the tumors by the Oligotex mRNA mini kit (Qiagen, Hilden, Germany). Polyadenylated RNA (600 ng of each) was used for the Northern blot analysis performed with the Northern Max-Gly Northern blotting kit (Ambion, Huntingdon, United Kingdom) as recommended by the manufacturer. As a probe, 32P-labeled CRPV-pLAII DNA was used.
Transient replication analysis.
Approximately 5 × 105 SCC13 cells were seeded into 60-mm-diameter dishes. The next day cells were cotransfected in the presence of 15 μl of Lipofectamine with 1 μg of E1-pSG5, 500 ng of CRPV-pGL3-NCR, 200 ng of E2-pSG5, and various amounts of E9^E2C-pSG5 or E9K2A(d3-10)^E2C-pSG5. The total amount of transfected DNA was kept constant by adding the parental pSG5 expression plasmid DNA. Cells were harvested 72 h after transfection, and low-molecular-weight DNA was purified as described previously (35). DNA samples were incubated with restriction enzymes DpnI and HpaI at 37°C for 5 h and then analyzed by Southern blotting.
DNA analysis.
DNA was isolated from papillomas by proteinase K digestion in melting buffer (0.1 M EDTA [pH 8.0], 0.05 M Tris [pH 8.0], 0.5% [vol/vol] sodium dodecyl sulfate) followed by phenol-chloroform extraction and ethanol precipitation. The E9 gene was amplified by PCR (35 cycles) with the primers P685 and P1920rev (see above) from total cellular DNA isolated from papillomas. The amplicon was sequenced with primer P1920rev and the BigDye Terminator DNA sequencing kit (ABI Prism) and analyzed in an ABI Prism 310 genetic analyzer. Viral copy number was determined by real-time quantitative PCR with 30 ng of DNA, 0.25 μM sequence-specific hybrid probes CRPV FL (5′-CTATCAGCTTCGTCTGCGTCTGTGCX-3′) and CRPV-LC (5′-LightCyc-ler-Red640-CCAGAAGCCATAAGAACCTTGAATCG-3′) and a 0.5 μM concentration (each) of sequence-specific primers CRPV-E7 F (5′-AGAAAGGCCCTATGCAGTGTC-3′) and CRPV-E7 R (5′-AGGGAAAGCGATGCGGATA-3′) in a final MgCl2 concentration of 3 mM.
The real-time PCR was carried out with LightCycler equipment (Roche Molecular Diagnostics, Penzberg, Germany) under the following conditions: denaturation at 95°C for 10 min, amplification for 40 cycles with 10 s of denaturation at 95°C, 10 s of annealing at 55°C, and 20 s of extension at 72°C. CRPV copy numbers are expressed relative to the cellular α-tubulin gene copy numbers. The number of α-tubulin genes was determined by real-time quantitative PCR with SYBR Green I and primers atub-forward (5′-CTGGAGCACTCTGATTGTGC-3′) and atub-reverse (5′-CTGTGGTGGTTGCTCAGCATG-3′).
Southern hybridization analysis.
DNA was transferred onto a nylon membrane (Gene Screen Plus; NEN Life Science Products) for 5 h by upward capillary transfer with 0.4 M NaOH as the transfer buffer. The 32P-labeled full-length CRPV genome served as a hybridization probe and was generated with Ready-To-Go DNA labeling beads (without dCTP) (Amersham Pharmacia, Freiburg, Germany) and purified by NucTrapProbe columns (Stratagene, Amsterdam, The Netherlands). Hybridization was carried out with 107 cpm of the probe in 10 ml of hybridization buffer (4× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}]), 5× Denhardt’s, 50% formamide, 1% sodium dodecyl sulfate, 10% dextran sulfate, and 0.2 mg of salmon sperm (Invitrogen, Karlsruhe, Germany) per ml overnight at 42°C. Then the membrane was washed at room temperature twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate, twice in 0.1× SSC-0.1% sodium dodecyl sulfate, and twice in 0.1× SSC-1% sodium dodecyl sulfate at 50°C. Afterwards the blot was autoradiographed with an intensifying screen or exposed to storage screens and visualized with a Fuji BAS 1800 phosphorimager and analyzed with AIDA software.
RESULTS
Identification of a novel ORF termed E9 involved in generation of a spliced E9^E2C transcript and cloning of its cDNA.
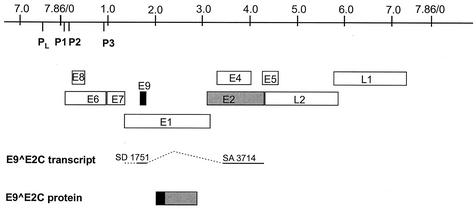
E8^E2C transcripts encode fusion proteins which have as a common feature 10 to 12 amino acids of the small E8 gene which are linked through a splice event to the hinge region and the C terminus of the E2 protein. To investigate whether a similar transcript is also present in CRPV-induced tumors, we isolated RNA from papillomas and carcinomas of different domestic rabbits and from a papilloma of a cottontail rabbit. These RNAs were used as templates in a reverse transcription-PCR with primers P1708 and P3883, which are located at the putative E9 start codon and downstream of the putative splice acceptor site within the E2 ORF. With this approach we amplified a cDNA of about 200 bp. Cloning and sequencing revealed a splice donor site at nucleotide 1751 that was linked to the acceptor site at nucleotide 3714 (Fig. 1). A subsequent reverse transcription-PCR with primer P1708 and a primer located at the very 3′ end of E2 (P4289) resulted in a cDNA of 600 bp, and sequencing confirmed that the spliced transcript encoded the complete C terminus of E2. This novel CRPV transcript encodes an E9^E2C fusion protein similar to E8^E2C, but as an E8 ORF in the CRPV genome overlapping the E6 ORF (Fig. 1) has already been described, the new ORF was labeled E9 (14).
FIG. 1.
CRPV genome organization and structure of the E9^E2C transcript. The linearized genome of CRPV with the various ORFs (E1 to E9, L1, and L2), the late viral promoter (PL), and the early viral promoters (P1, P2, and P3) are shown at the top. Below, structure of the E9^E2C transcript with the splice donor (SD) site at nucleotide 1751 linked to the splice acceptor (SA) site at nucleotide 3714, giving rise to a fusion protein consisting of 10 amino acids derived from ORF E9 (black box) linked to the hinge and C terminus of E2 (grey box).
E9^E2C is a negative regulator of transcription and DNA replication in tissue culture.
In order to characterize the functions of CRPV-E9^E2C, we constructed a eukaryotic expression vector (E9^E2C-pSG5) by cloning the E9^E2C cDNA into the pSG5 vector. First the effect of E9^E2C on different E2-responsive promoter constructs was investigated in SCC13 cells, a human squamous carcinoma cell line, as well as in CRL 6502 cells, an epithelial cottontail rabbit cell line. Both cell lines were transfected either with 10 ng of E9^E2C-pSG5 or with the parental expression vector pSG5 and 200 ng of reporter plasmids containing E2 binding sites upstream of a variety of different promoters driving the luciferase gene (Fig. 2). E9^E2C repressed the basal activity of all reporter constructs in both cell lines. A reporter construct containing both the late (PL) and early (P1) E6 promoters of CRPV in their natural context within the NCR (CRPV-PLP1-pGL3) was repressed by E9^E2C to a similar extent as synthetic promoter constructs containing multiple E2BS and a minimal adenovirus major late promoter (pC18-Sp1-luc) or a minimal simian virus 40 early promoter (6xE2BS-luc) in SCC13 cells. Interestingly, all reporter constructs were more efficiently repressed in CRL 6502 cells (Fig. 2). E9^E2C was also able to strongly repress the heterologous P97 promoter of HPV-31 in normal human keratinocytes (data not shown).
To evaluate the contribution of the E9 domain of E9^E2C to this general transcription repression property, a deletion mutant was constructed, in which amino acid residues 3 to 10 of E9 were deleted in the E9^E2C-pSG5 expression vector. In addition, the lysine at position 2 was exchanged to alanine, as lysines were shown to be important for the repression activity of the HPV-31 E8^E2C protein (37). The mutant designated E9K2A(d3-10)^E2C-pSG5 was transfected into CRL6502 cells together with 200 ng of CRPV-PLP1-pGL3 or pC18-Sp1-luc and compared to the activity of wild-type E9^E2C and also full-length E2 (Fig. 3A and C). E2 strongly activated whereas E9^E2C repressed pC18-Sp1-luc (Fig. 3A). These experiments were also carried out with immortalized domestic rabbit keratinocytes (RK1-16E7/ras), which demonstrated that the E9 domain was also required for promoter repression by E9^E2C in keratinocytes from New Zealand White rabbits (Fig. 3B).
In both cell lines, E9K2A(d3-10)^E2C showed a complete loss of repression activity, demonstrating that the E9 part is essential for this function (Fig. 3A and B). All E2 proteins inhibited the basal activity of the homologous promoter construct CRPV-PLP1-pGL3, but to different extents (Fig. 3C). E2 showed a weaker repression activity than E9^E2C (56% versus 14%, Fig. 3C). Interestingly, E9K2A(d3-10)^E2C displayed the same repression activity as E9^E2C, indicating that the E2 C terminus is sufficient for this function (Fig. 3C).
Next, the influence of E9^E2C on the transactivation by CRPV-E2 was examined by cotransfecting 200 ng of the reporter plasmid pC18-SP1-luc with 10 ng of CRPV E2-pSG5 expression vector and increasing amounts of expression vectors (0, 3, 10, or 30 ng) for E9^E2C-pSG5 or the deletion mutant E9K2A(d3-10)^E2C into RK1 cells. As a result, we observed a concentration-dependent repression of full-length E2-mediated promoter activation by E9^E2C similar to that observed earlier with HPV-31 E8^E2C (36, 37) as well as with the deletion mutant, indicating that the C terminus of E2 itself can counteract transactivation by E2 and that this does not require the E9 part of the fusion protein (Fig. 4).
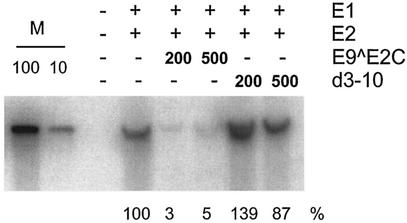
To investigate the influence of E9^E2C on the E2-dependent replication of the CRPV origin in a transient replication assay, SCC13 cells were transfected with a replication reporter construct (CRPV-pGL3-NCR) (16) together with an expression plasmid for CRPV-E1 (E1-pSG5) or CRPV-E2 (E2-pSG5) and with increasing amounts of E9^E2C-pSG5 or E9K2A(d3-10)^E2C (0 ng, 200 ng, or 500 ng). Low-molecular-weight DNA was isolated, digested with the restriction enzyme DpnI in order to distinguish replicated DNA from transfected DNA, and analyzed by Southern blot analysis (Fig. 5). Cotransfection of increasing amounts of E9^E2C-pSG5 resulted in a decrease of newly replicated origin plasmid, whereas the E9K2A(d3-10)^E2C mutant showed no effect on replication efficiency. Taken together, these results suggested that the CRPV E9^E2C protein is a bona fide E2 repressor protein that can counteract the ability of E2 to stimulate transcription and DNA replication and is also a transcriptional repressor on its own. Moreover, the E9 domain is required for the repression of a synthetic reporter construct and for the repression of the E1/E2-dependent viral DNA replication but not for the inhibition of the promoter activity of the CRPV regulatory region or the inhibition of E2 in transcriptional activation assays.
FIG. 5.
E9^E2C represses the E1- and E2-dependent replication of the CRPV origin. Autoradiography of a transient DNA replication assay. SCC13 cells were transfected with 500 ng of the CRPV origin plasmid CRPV-pGL3-NCR and with the expression vectors CRPV-E1-pSG5 (E1) and CRPV-E2-pSG5 (E2) and increasing amounts (in nanograms) of CRPV-E9^E2C-pSG5 (E9^E2C). Low-molecular-weight DNA was extracted, digested with DpnI and HpaI, and analyzed by Southern blot hybridization. Lane M, 100 and 10 pg of CRPV-pGL3-NCR plasmid linearized by HpaI. Quantitative analysis of the amount of replicated DNA is shown below the autoradiograph. The replication efficiencies are expressed relative to the replication level in cells not transfected with E9^E2C-pSG5, which was set to 100%.
E9 knockout mutations show no phenotype in New Zealand White rabbits.
The transactivation domain of CRPV E2 is required for efficient tumor induction by CRPV in New Zealand White rabbits, suggesting a role for E2 proteins in the process of tumor development and progression (16). To investigate the role of the E9^E2C repressor protein in these processes, we constructed a mutant CRPV genome designated CRPV-E9atgmut-pLAII, in which the unique start codon in the E9 ORF was mutated to threonine (Fig. 6). This mutation is silent within the E1 ORF. CRPV-E9atgmut-pLAII as well as wild-type CRPV-pLAII DNA were injected into 77 sites in the skin of a total of six rabbits with the help of a Helios gene gun. After 3 months we observed the appearance of papillomas with a comparable average maximum diameter in 47 of the 77 injected sites (61%) with both constructs (Table 1).
TABLE 1.
Tumor development in New Zealand White rabbits after injection with wild-type CRPV-pLAII or E9 mutant CRPV-pLAII
| Rabbit no. | DNA injected | No. of papillomas/no. of infection sites (% of sites) | Avg diam at 3 mo postinfection (cm) | Avg CRPV copy no.a ± SD | Ratio of CRPV transcripts per β-actin copy ± SD |
|---|---|---|---|---|---|
| 1-6 | CRPV-E9atg mut-pLAII | 47/77 (61%) | 0.74 | 15.7 ± 13.2 | 4.8 ± 1.7 |
| CRPV-pLAII | 47/77 (61%) | 0.88 | 22.7 ± 16.6 | 5.8 ± 1.4 | |
| 7-11 | CRPV-E9stop-pLAII | 39/40 (98%) | 2.7 | 18.0 ± 7.1 | 5.0 ± 0.8 |
| CRPV-pLAII | 40/40 (100%) | 3.1 | 24.4 ± 7.6 | 3.8 ± 0.7 |
Expressed in relation to α-tubulin copy number.
To confirm this phenotype, a second E9^E2C knockout CRPV genome was constructed by introduction of a stop codon into the E9 gene downstream from the putative start codon. This mutation causes, in addition to the disruption of E9, an amino acid change from glutamine to glutamic acid in the overlapping E1 ORF. Injection of CRPV-E9stop-pLAII into 40 sites on five domestic rabbits induced papillomas within 3 months with 98% efficiency, in comparison to 100% efficiency with wild-type CRPV-pLAII. Biopsies were taken 3 and 6 months postinjection, and DNA and RNA were isolated. The presence of the E9 mutations in the papillomas could be demonstrated by repeated sequencing analysis of the corresponding DNA fragments amplified by PCR.
To investigate if the loss of E9^E2C function shown to be a potent repressor of transcription and replication in vitro influences CRPV genome copy numbers or viral transcription in the lesions, we performed quantitative real-time PCR experiments. The relative CRPV genome copy number in the biopsies was determined by amplification of a fragment of the E7 gene and of the α-tubulin gene in the same DNA. No significant difference in viral genome copy numbers per cell between wild-type CRPV-induced or E9 mutant-induced papillomas could be detected (Table 1). To investigate the physical status of the viral DNA in the different biopsies, a Southern blot was performed with 32P-labeled CRPV DNA as a probe. Isolated biopsy DNA (10 μg each) digested with EcoRI, which cuts once within CRPV and once within the pLAII vector, or with XhoI, which is a noncutting enzyme, was loaded.
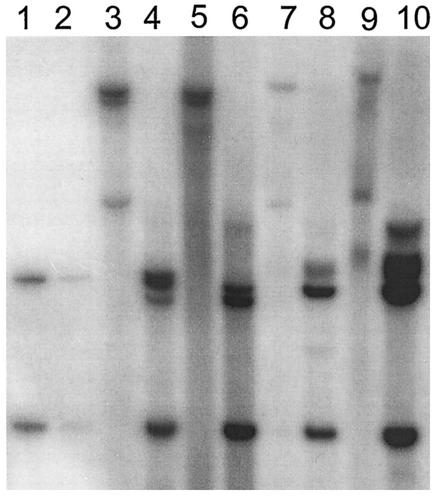
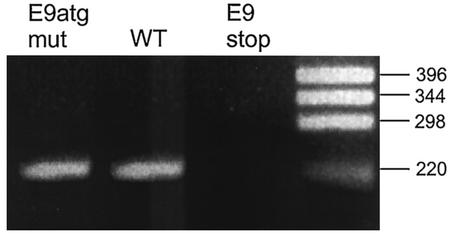
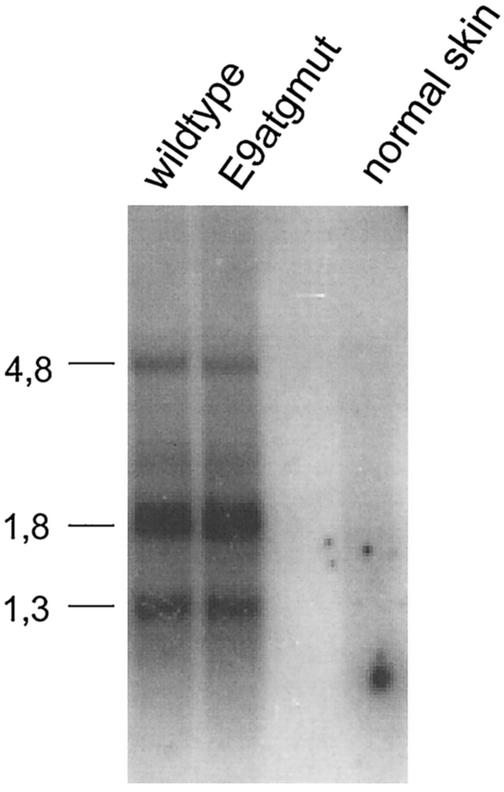
As shown in Fig. 7, the restriction pattern of the DNA isolated from wild-type CRPV-pLAII induced papillomas and the one from either CRPV-E9atgmut-pLAII or CRPV-E9stop-pLAII are similar. The slower-migrating band indicative of high-molecular-weight DNA within the XhoI-digested lanes 3, 5, 7, and 9 of Fig. 7 suggests the existence of some integrated CRPV DNA copies, whereas the faster migrating band reveals open circle forms that point in addition to the presence of episomal DNA. Next, RNA was isolated from the papilloma biopsies and reverse transcription-PCR was performed in order to investigate if the E9^E2C transcript was present within the tumors. E9^E2C messages were detectable in wild-type- and CRPV-E9atgmut-pLAII-induced tumors, but were repeatedly not found in CRPV-E9stop-pLAII-induced tumors (Fig. 8). This indicated that the E9stop mutation, which is in the immediate vicinity of the splice donor site, additionally prevented the formation of the correctly spliced E9^E2C transcript.
FIG. 7.
Southern blot analysis of DNA isolated from papillomas induced by wild-type CRPV-pLAII (lanes 5, 6, 9, and 10), CRPV-E9atgmut-pLAII (lanes 7 and 8), or CRPV-E9stop-pLAII (lanes 3 and 4). Biopsy DNA (10 μg) was either digested with XhoI (lanes 3, 5, 7, and 9), which does not cut within CRPV-pLAII, or with EcoRI (lanes 4, 6, 8, and 10), which cuts twice within CRPV-pLAII. As marker fragments, we used 20 pg (lane 2) and 200 pg (lane 1) of CRPV-pLAII plasmid DNA digested with EcoRI.
FIG. 8.
Detection of CRPV-E9^E2C transcript by reverse transcription-PCR. A total of 500 ng of RNA isolated from papillomas induced by wild-type CRPV-pLAII (WT), CRPV-E9atgmut-pLAII (E9atgmut), or CRPV-E9stop-pLAII (E9stop) was used in a reverse transcription-PCR with primers P1708 (upstream of SD1751) and P3883 (downstream of SA3714) and Ready-To-Go reverse transcription-PCR beads. The correct size of the cDNA fragment derived from the spliced E9^E2C transcript is shown on a 2% agarose gel. The sizes of molecular size markers are given on the right (in base pairs).
To quantitate CRPV gene expression, the RNA isolated from the tumors was digested with DNase I, reverse transcribed, and then analyzed by quantitative real-time PCR. This analysis was carried out with primers E4for and E4rev, located within the E4 gene downstream of the splice acceptor at site 3714, as this sequence is present in all known early CRPV transcripts. The absolute CRPV transcript numbers determined were standardized against the number of β-actin transcripts. No significant differences in relative transcript numbers between RNA from wild-type CRPV-pLAII- and CRPV-E9atgmut- or CRPV-E9stop-pLAII-induced papillomas were observed (Table 1). To look for qualitative differences in the viral transcription pattern, a Northern blot analysis was performed with polyadenylated RNA from wild-type CRPV-pLAII- and CRPV-E9atgmut-pLAII-induced papillomas and 32P-labeled CRPV DNA as a probe (Fig. 9). Again no obvious difference was detectable between the RNAs tested. We surprisingly found no phenotype of knockout mutations of the E9^E2C repressor when infecting New Zealand White rabbits with CRPV DNA.
FIG. 9.
Northern blot analysis of polyadenylated RNA isolated from papillomas induced by wild-type CRPV-pLAII (wild type) or CRPV-E9atgmut-pLAII (E9atgmut) or from normal rabbit skin (normal skin). A total of 600 ng of RNA per lane was hybridized to 32P-labeled CRPV DNA as the probe. The sizes of molecular size markers are given on the left (in kilobases).
DISCUSSION
In this study we identified the novel E9 ORF in the CRPV genome, which is part of a spliced transcript giving rise to an E9^E2C fusion protein. This transcript was found in tumors of both domestic and cottontail rabbits. The transcript structure is comparable to that of E8^E2C transcripts described for BPV1, HPV-11, HPV-16, HPV-31, and HPV-33 (10, 13, 29, 33, 36). The E9^E2C protein behaves as a bona fide E2 repressor protein. It inhibits the transactivating effect of the full-length E2 protein and the E1/E2-dependent transient replication of the CRPV origin. It also represses the basal promoter activity of the CRPV regulatory region. Furthermore, E9^E2C represses the activity of minimal viral promoters when E2BS are present.
Transcriptional repression of artificial promoters is different from the repression of transcription driven by the CRPV NCR promoters, as it is dependent on the E9 domain, which is similar to the behavior of HPV-31 E8^E2C (37). However, a comparison of the amino acid sequences of HPV-31 E8 and CRPV E9 revealed only a moderate degree of sequence homology between these domains. A KWK amino acid motif that is especially highly conserved among different HPVs is absent from CRPV E9^E2C. Both the CRPV E9 and the HPV-31 E8 domain have a high content of hydrophobic and basic amino acids, making it likely that not the primary amino acid sequence but structural features are important for the repression activity. Interestingly, the E9 domain is required for the repression of DNA replication by E9^E2C but is not needed to interfere with E2's ability to transactivate a synthetic reporter construct. The inability of the isolated E2 C terminus to interfere with DNA replication, although it can counteract E2-mediated transactivation, probably by binding site competition and/or heterodimer formation, could be due to the large number of E2BS present in the NCR of CRPV, which can possibly not all be saturated. Taken together, these data suggest that the E9 domain is involved both in the inhibition of transcription from artificial promoters and in the repression of viral DNA replication.
When we introduced mutations into the E9 gene in the context of the whole CRPV genome either by mutating the ATG start codon or by introducing a stop codon into E9, that additionally resulted in the complete disappearance of the spliced E9^E2C transcript in infected cells, and when we examined their activity in domestic rabbits, we surprisingly found no phenotype for E9^E2C knockout viruses.
A comparable situation is seen with BPV1 E8/E2 knockout genomes, which display no phenotype in tissue culture cells with respect to viral genome copy number, viral transcription, E2-mediated transactivation, and transformation of the host cell (18). In concordance, CRPV E9 mutants showed no difference in viral copy number or viral transcription compared to wild-type CRPV. However, in vitro transformation assays cannot be performed with full-length CRPV genomes because of the lack of a tissue culture system and the lack of immortalizing activity of the E6 and E7 genes in keratinocytes (32). Interestingly, comparable mutations in HPV-31 genomes that disrupt E8^E2C expression have been reported to strongly increase transient DNA replication in tissue culture but subsequently prevent long-term maintenance of HPV-31 episomes in normal human keratinocytes (36). A major difference to the results obtained with BPV1 and HPV-31 is that these data were generated in tissue culture systems that may not be reflected by the situation in CRPV-infected domestic rabbits. Vice versa, we have been unable, despite numerous attempts and long-term experience with HPV-31, to establish a replication system for genomic CRPV DNA in cultured cells.
In agreement with our results obtained for CRPV-E9^E2C, E8/E2 of BPV1 has been demonstrated to be a repressor of E2-mediated transactivation. Indeed, CRPV E9^E2C may fulfill a function comparable to that of BPV1 E8/E2 in the viral life cycle, although the exact nature of this function is still unclear (10). BPV1 encodes a second E2 repressor protein, termed E2C, which represents the major E2 species in BPV1-positive cells (15). Although BPV1 E2C shows activities similar to those of BPV1 E8/E2, knockout mutations in E2C caused a strong increase in viral copy number, viral transcript numbers, and enhanced soft agar colony formation and focus-forming activity (18, 28).
An apparent phenotype for BPV1 E8/E2 was only visible after the construction of an E8/E2-E2C double mutant BPV1 genome and resulted in complex behavior, including lack of focus-forming activity (18). It is therefore tempting to speculate about the existence of a second E2 repressor in CRPV as well. However, earlier mapping experiments of CRPV transcripts by S1-analysis do not support this possibility, in spite of the fact that the E9 splice donor site was found in the course of these experiments, although attached to another exon (25, 41). Furthermore, the analysis of E2 proteins translated from different polycistronic RNAs revealed the existence of a single 49-kDa protein, corresponding to the full-length E2 protein (1).
The function of E9^E2C in the CRPV life cycle remains elusive. According to the activities of E9^E2C detected in tissue culture experiments, one would have expected a change in viral genome copy number in papillomas induced by CRPV E9^E2C knockout genomes in comparison to wild-type virus. However, New Zealand White rabbits, the animals employed here, are distinguished from the natural host, cottontail rabbits, by an abortive viral life cycle characterized by low levels of viral DNA amplification and the lack of virus production. Viral copy numbers are much lower in domestic rabbit papillomas than in cottontail rabbit papillomas (10 versus 400 genome copies/cell) (40; S. Jeckel, unpublished data). This difference between these species with regard to productive DNA amplification might be explained by the existence of a cellular repressor protein in keratinocytes of New Zealand White rabbits whose presence might completely overshadow the effects caused by the loss of E9^E2C function.
Acknowledgments
This work was supported by a grant from the Wilhelm Sander-Stiftung (no. 99.111.1) to T.I.
We thank Felix O. Wettstein for providing papilloma biopsies from cottontail rabbits.
REFERENCES
- 1.Barbosa, M. S., and F. O. Wettstein. 1988. E2 of cottontail rabbit papillomavirus is a nuclear phosphoprotein translated from an mRNA encoding multiple open reading frames. J. Virol. 62:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsoum, J., S. S. Prakash, P. Han, and E. J. Androphy. 1992. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J. Virol. 66:3941-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout, R. J., L. M. Tieben, H. L. Smits, J. N. Bavinck, B. J. Vermeer, and J. ter Schegget. 1995. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J. Clin. Microbiol. 33:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvard, V., A. Storey, D. Pim, and L. Banks. 1994. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J. 13:5451-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouwes Bavinck, J. N., D. R. Hardie, A. Green, S. Cutmore, A. MacNaught, B. O'Sullivan, V. Siskind, F. J. Van Der Woude, and I. R. Hardie. 1996. The risk of skin cancer in renal transplant recipients in Queensland, Australia. A follow-up study. Transplantation 15:715-721. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, C.-M., G. Dong, T. R. Broker, and L. T. Chow. 1992. Control of human papillomavirus type 11 origin of replication by the E2 family of transcription regulatory proteins. J. Virol. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, C.-M., T. R. Broker, and L. T. Chow. 1991. An E1M^E2C fusion protein encoded by human papillomavirus type 11 is a sequence-specific transcription repressor. J. Virol. 65:3317-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin, M. T., R. Hirochika, H. Hirochika, T. R. Broker, and L. T. Chow. 1988. Regulation of human papillomavirus type 11 enhancer and E6 promoter by activating and repressing proteins from the E2 open reading frame: functional and biochemical studies. J. Virol. 62:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe, J., P. Vaillancourt, A. Stenlund, and M. Botchan. 1989. Bovine papillomavirus type 1 encodes two forms of a transcriptional repressor: structural and functional analysis of new viral cDNAs. J. Virol. 63:1743-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 12.de Jong-Tieben, L. M., R. J. Berkhout, H. L. Smits, J. N. Bouwes Bavinck, B. J. Vermeer, F. J. van der Woude, and J. ter Schegget. 1995. High frequency of detection of epidermodysplasia verruciformis-associated human papillomavirus DNA in biopsies from malignant and premalignant skin lesions from renal transplant recipients. J. Investig. Dermatol. 105:367-371. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 14.Giri, I., O. Danos, and M. Yaniv. 1985. Genomic structure of the cottontail rabbit (Shope) papillomavirus. Proc. Natl. Acad. Sci. USA 82:1580-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbert, N. L., J. T. Schiller, D. R. Lowy, and E. J. Androphy. 1988. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc. Natl. Acad. Sci. USA 85:5864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeckel, S., E. Huber, F. Stubenrauch, and T. Iftner. 2002. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. J. Virol. 76:11209-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert, P. F., B. A. Spalholz, and P. M. Howley. 1987. A transcriptional repressor encoded by BPV-1 shares a common carboxy-terminal domain with the E2 transactivator. Cell 50:69-78. [DOI] [PubMed] [Google Scholar]
- 18.Lambert, P. F., B. C. Monk, and P. M. Howley. 1990. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 64:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert, P. F., N. L. Hubbert, P. M. Howley, and J. T. Schiller. 1989. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J. Virol. 63:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Land, H., L. F. Parada, and R. A. Weinberg. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304:596-602. [DOI] [PubMed] [Google Scholar]
- 21.Lim, D. A., M. Gossen, C. W. Lehman, and M. R. Botchan. 1998. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J. Virol. 72:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, J. S., S. R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 23.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 24.McBride, A. A., J. C. Byrne, and P. M. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasseri, M., and F. O. Wettstein. 1984. Differences exist between viral transcripts in cottontail rabbit papillomavirus-induced benign and malignant tumors as well as non-virus-producing and virus-producing tumors. J. Virol. 51:706-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasseri, M., C. Meyers, and F. O. Wettstein. 1989. Genetic analysis of CRPV pathogenesis: the L1 open reading frame is dispensable for cellular transformation but is required for papilloma formation. Virology 170:321-325. [DOI] [PubMed] [Google Scholar]
- 27.Rapp, B., A. Pawellek, F. Kraetzer, M. Schaefer, C. May, K. Purdie, K. Grassmann, and T. Iftner. 1997. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J. Virol. 71:6956-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riese, D. J., II, J. Settleman, K. Neary, and D. DiMaio. 1990. Bovine papillomavirus E2 repressor mutant displays a high-copy-number phenotype and enhanced transforming activity. J. Virol. 64:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotenberg, M. O., L. T. Chow, and T. R. Broker. 1989. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, with the polymerase chain reaction. Virology 172:489-497. [DOI] [PubMed] [Google Scholar]
- 30.Rous, P., and J. W. Beard. 1935. The progression to carcinoma of virus-induced rabbit papillomas (Shope). J. Exp. Med. 62:523-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt, A., J. B. Harry, B. Rapp, F. O. Wettstein, and T. Iftner. 1994. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J. Virol. 68:7051-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snijders, P. J., A. J. van den Brule, H. F. Schrijnemakers, P. M. Raaphorst, C. J. Meijer, and J. M. Walboomers. 1992. Human papillomavirus type 33 in a tonsillar carcinoma generates its putative E7 mRNA via two E6* transcript species which are terminated at different early region poly(A) sites. J. Virol. 66:3172-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenlund, A. 1996. Papillomavirus DNA replication, p. 679-698. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8∧E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stubenrauch, F., T. Zobel, and T. Iftner. 2001. The E8 domain confers a novel long-distance transcriptional repression activity on the E8^E2C protein of high-risk human papillomavirus type 31. J. Virol. 75:4139-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surentheran, T., C. A. Harwood, P. J. Spink, A. L. Sinclair, I. M. Leigh, C. M. Proby, J. M. McGregor, and J. Breuer. 1998. Detection and typing of human papillomaviruses in mucosal and cutaneous biopsies from immunosuppressed and immunocompetent patients and patients with epidermodysplasia verruciformis: a unified diagnostic approach. J. Clin. Pathol. 51:606-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syverton, J. T. 1952. The pathogenesis of the rabbit papilloma to carcinoma sequence. Ann. N.Y. Acad. Sci. 54:1126-1140. [DOI] [PubMed] [Google Scholar]
- 40.Wettstein, F. O. 1987. Papillomaviruses and carcinogenic progression I — cottontail rabbit (Shope) papillomavirus, p. 167-186. In N. P. Salzman and P. M. Howley (ed.), The Papovaviridae, vol. 2. Plenum Publishing Corp., New York, N.Y.
- 41.Wettstein, F. O., M. S. Barbosa, and M. Nasseri. 1987. Identification of the major cottontail rabbit papillomavirus late RNA cap site and mapping and quantitation of an E2 and minor E6 coding mRNA in papillomas and carcinomas. Virology 159:321-328. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, W., and J. L. Brandsma. 1996. High efficiency, long-term clinical expression of cottontail rabbit papillomavirus (CRPV) DNA in rabbit skin following particle-mediated DNA transfer. Nucleic Acids Res. 24:2620-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.zur Hausen, H. 1991. Viruses in human cancers. Science 254:1167-1173. [DOI] [PubMed] [Google Scholar]