Abstract
The initiation of human immunodeficiency virus type 1 (HIV-1) reverse transcription occurs at the primer binding site (PBS) that is complementary to the 3′-terminal nucleotides of tRNA3Lys. Why all known strains of HIV-1 select tRNA3Lys for replication is unknown. Previous studies on the effect of altering the PBS of HIV-1 on replication identified an HIV-1 with a PBS complementary to tRNAGlu. Since the virus was not initially designed to use tRNAGlu, the virus had selected tRNAGlu from the intracellular pool of tRNA for use in replication. Further characterization of HIV-1 that uses tRNAGlu may provide new insights into the preference for tRNA3Lys. HIV-1 constructed with the PBS complementary to tRNAGlu was more stable than HIV-1 with the PBS complementary to tRNAMet or tRNAHis; however, all of these viruses eventually reverted back to using tRNA3Lys following growth in SupT1 cells or peripheral blood mononuclear cells (PBMCs). New HIV-1 mutants with nucleotides in U5 complementary to the anticodon of tRNAGlu remained stable when grown in SupT1 cells or PBMCs, although the mutants grew more slowly than the wild-type virus. Sequence analysis of the U5 region and the PBS revealed additional mutations predicted to further promote tRNA-viral genome interaction. The results support the importance of the tRNA anticodon-genome interaction in the selection of the tRNA primer and highlight the fact that unique features of tRNA3Lys are exploited by HIV-1 for selection as the reverse transcription primer.
Human immunodeficiency virus type 1 (HIV-1), like all retroviruses, utilizes a cellular tRNA as the primer for reverse transcription (1, 25). The tRNA primer binds to a region of the HIV-1 genome that is complementary to the 3′-terminal 18 nucleotides of the tRNA, termed the primer binding site (PBS) (7). HIV-1 has evolved to selectively use 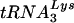 for replication. Previous studies have shown that modifying the PBS to correspond to other tRNAs results in the use of these tRNAs for reverse transcription (6, 15, 27). However, a hallmark of all of these studies is that the virus reverts back to using
for replication. Previous studies have shown that modifying the PBS to correspond to other tRNAs results in the use of these tRNAs for reverse transcription (6, 15, 27). However, a hallmark of all of these studies is that the virus reverts back to using 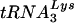 after limited time in culture.
after limited time in culture.
The exclusive use of 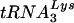 by HIV-1 as the primer for replication and the propensity of mutant HIV-1 viruses with altered PBS regions to revert back to using
by HIV-1 as the primer for replication and the propensity of mutant HIV-1 viruses with altered PBS regions to revert back to using 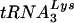 suggest that unique features of
suggest that unique features of 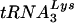 and the HIV-1 genome may be involved in primer selection. Our laboratory has taken a genetic approach to understand the mechanism of primer selection. HIV-1 mutants have been created in which the PBSs have been modified to be complementary to tRNAs other than
and the HIV-1 genome may be involved in primer selection. Our laboratory has taken a genetic approach to understand the mechanism of primer selection. HIV-1 mutants have been created in which the PBSs have been modified to be complementary to tRNAs other than 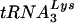 (12-14, 26, 27). RNA modeling of the U5-PBS region suggests that a stem-loop structure exists in which nucleotides that are complementary to the anticodon of
(12-14, 26, 27). RNA modeling of the U5-PBS region suggests that a stem-loop structure exists in which nucleotides that are complementary to the anticodon of 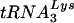 (the A-loop or anticodon binding loop) are displayed on a loop. Both chemical and enzymatic analyses of U5-PBS-tRNA interactions support the idea that the A-loop is in a complex with the anticodon of
(the A-loop or anticodon binding loop) are displayed on a loop. Both chemical and enzymatic analyses of U5-PBS-tRNA interactions support the idea that the A-loop is in a complex with the anticodon of 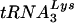 (9). Support for a role of the anticodon binding loop in the selection of the tRNA has come from previous studies in which HIV-1 mutants that could use tRNAs other than
(9). Support for a role of the anticodon binding loop in the selection of the tRNA has come from previous studies in which HIV-1 mutants that could use tRNAs other than 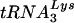 for replication were created (11-14, 26). Viruses in which both the anticodon binding regions and PBSs were altered to correspond to the new tRNAs were able to stably use these tRNAs for replication.
for replication were created (11-14, 26). Viruses in which both the anticodon binding regions and PBSs were altered to correspond to the new tRNAs were able to stably use these tRNAs for replication.
Although in the majority of instances the PBS reverted back to being complementary to 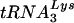 , with some U5-PBS combinations HIV-1 selected a new, unexpected tRNA from the intracellular milieu (12, 14, 29). In one of these instances, several PBS sequences that were complementary to tRNAGlu were identified (16). Since the wild-type PBS complementary to
, with some U5-PBS combinations HIV-1 selected a new, unexpected tRNA from the intracellular milieu (12, 14, 29). In one of these instances, several PBS sequences that were complementary to tRNAGlu were identified (16). Since the wild-type PBS complementary to 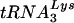 was not detected, it was possible that virus with a PBS complementary to tRNAGlu might have had an advantage over the viruses that used
was not detected, it was possible that virus with a PBS complementary to tRNAGlu might have had an advantage over the viruses that used 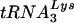 . Analysis of HIV-1 with both U5 and the PBS complementary to tRNAGlu, then, may provide insights into the selection process and even the cause of specificity for
. Analysis of HIV-1 with both U5 and the PBS complementary to tRNAGlu, then, may provide insights into the selection process and even the cause of specificity for 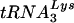 . HIV-1 mutants were created in which either the PBS alone or the PBS and the anticodon binding region were altered to be complementary to tRNAGlu. Surprisingly, we found that HIV-1 with a PBS complementary to tRNAGlu was more stable than HIV-1 with a PBS complementary to tRNAMet or tRNAHis; however, the HIV-1 with a PBS complementary to tRNAGlu eventually reverted to using
. HIV-1 mutants were created in which either the PBS alone or the PBS and the anticodon binding region were altered to be complementary to tRNAGlu. Surprisingly, we found that HIV-1 with a PBS complementary to tRNAGlu was more stable than HIV-1 with a PBS complementary to tRNAMet or tRNAHis; however, the HIV-1 with a PBS complementary to tRNAGlu eventually reverted to using 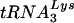 . Viruses with both PBS and A-loop complementarity to tRNAGlu were stable following extended growth in SupT1 cells or peripheral blood mononuclear cells (PBMCs). Even though tRNAGlu was present in cells at levels greater than those of
. Viruses with both PBS and A-loop complementarity to tRNAGlu were stable following extended growth in SupT1 cells or peripheral blood mononuclear cells (PBMCs). Even though tRNAGlu was present in cells at levels greater than those of 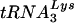 and virions contained more tRNAGlu than
and virions contained more tRNAGlu than 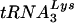 , HIV-1 that used tRNAGlu grew more slowly in both SupT1 cells and PBMCs. The results of these studies further support a role for U5 in the selection of this tRNA primer and suggest that additional unique features of
, HIV-1 that used tRNAGlu grew more slowly in both SupT1 cells and PBMCs. The results of these studies further support a role for U5 in the selection of this tRNA primer and suggest that additional unique features of  support high-level HIV-1 replication.
support high-level HIV-1 replication.
MATERIALS AND METHODS
Construction of mutant proviral genomes.
The wild-type PBS of the shuttle vector pUC119PBS, which contains a HpaI-to-PstI DNA fragment containing the 5′ long terminal repeat, PBS, and leader region of gag from wild-type pHXB2, was mutated to have a sequence complementary to the 18 3′-terminal nucleotides of tRNAGlu, forming pUC119PBS(Glu). Mutagenesis was performed by using the Gene Editor in vitro mutagenesis system (Promega, Madison, Wis.) along with the primer 5′-CGCTTTCAATTCCCGGTCAGGGAACCACTGCTAGAGATTTTCCAC-3′. The pUC119PBS(Glu) vector was then used as the substrate to form the A-loop mutant vectors pUC119PBS(Glu Loop 1) and pUC119PBS(Glu Loop 2). The primers used for the A-loop mutagenesis were 5′-CCACTGCTAGAGACTCAACTCTACACTGACTA AAAGGGTC-3′ and 5′-CCACTGCTAGAGACTCAACTCTAGCCTGACTAAAAGG GTC-3′, respectively. All mutagenesis was performed according to the manufacturer's instructions, and the resulting mutant sequences were verified by DNA sequencing. The 868-bp HpaI-BssHII fragments of pUC119PBS(Glu), pUC119PBS(Glu Loop 1), and pUC119PBS(Glu Loop 2) containing the U5 and PBS regions were then subcloned between the HpaI and BssHII sites of pHXB2 to form the complete proviral clones pHXB2(Glu), pHXB2(Glu Loop 1), and pHXB2(Glu Loop 2). The proviral clones were screened by restriction digestion, and the proper sequences were verified by DNA sequencing.
Tissue culture.
All cultures were maintained at 37°C and 5% CO2. COS-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), and SupT1 cells were grown in RPMI 1640 medium containing 15% FBS. PBMCs were isolated from HIV-1-seronegative whole blood by Ficoll-Paque Plus (Amersham Pharmacia Biotech, Uppsala, Sweden) density gradient centrifugation. The PBMCs were stimulated for 2 days in RPMI 1640 medium containing 15% FBS and 3 μg of phytohemagglutinin (Sigma, St. Louis, Mo.)/ml. After stimulation, the medium was removed and replaced with RPMI 1640 medium supplemented with 15% FBS and 30 U of recombinant human interleukin-2 (Roche Diagnostics, Indianapolis, Ind.)/ml.
Virus production and analysis of viral infectivity.
COS-1 cells at 75% confluency were transfected with 5 μg of the appropriate pHXB2 proviral DNA using DEAE-dextran. After 48 h, the virus supernatants were removed and filtered through 0.45-μm-pore-size syringe filters (Gelman Sciences, Ann Arbor, Mich.) and the p24 concentration of each was determined by immunoassay (Coulter, Miami, Fla.). Virus supernatant containing 100 ng of p24 was then used to infect 10 × 106 SupT1 cells or 20 × 106 PBMCs. The virus was allowed to adsorb for 24 h, and the infected cells were pelleted by centrifugation at 1,000 × g, washed with fresh medium, and further cultured. The infected SupT1 cells were passaged 1:6 every 3 days, and the PBMCs were passaged 1:2 every 3 days. Supernatants were collected from both SupT1 and PBMC infected cultures every 6 days. The infected SupT1 cultures were visually monitored for the formation of syncytia and clearance of the cells. Once the infected SupT1 cultures were found to be cleared of cells, 106 new cells were added to continue the culture and p24 collections were ceased. For the infected PBMC cultures, 5 × 106 new cells were added every 12 days and supernatant collection was continued for the duration of the culture. The concentrations of p24 antigen in all collected supernatants were determined by using the p24 immunoassay.
DNA sequence analysis of proviral U5 and PBS regions.
High-molecular-weight DNA was obtained from a 1-ml aliquot of infected SupT1 cells or PBMCs every 12 days of culture by using the Wizard Genomic DNA isolation kit (Promega) according to the manufacturer's instructions. A fragment containing the U5 and PBS regions of the integrated provirus was amplified from the high-molecular-weight template DNA by PCR with the primers 5′-CGGAATTCTCTCCTTCTAGCCTCCGCTAGTC-3′ and 5′-CCTTGAGCATGCGATCTACCACACACAAGGC-3′. The resulting PCR products were gel purified and ligated to the cloning vector pGem-T Easy (Promega), and the ligation mixes were used to transform competent Escherichia coli DH5α cells (Invitrogen, Carlsbad, Calif.). Plasmid DNA obtained from individual colonies was screened for inserts by using restriction digestion and then subjected to automated sequence analysis with the primer 5′-CGGAATTCTCTCCTTCTAGCCTCCGCTAGTC-3′.
Analysis of tRNA in cells and HIV-1 virions.
Total cellular RNA was isolated from uninfected SupT1 cells by using Tri-reagent (Sigma) according to the manufacturer's instructions. Northern blotting was performed by using the NorthernMax-Gly kit (Ambion, Houston, Tex.) according to the manufacturer's instructions. The cellular RNA samples contained 5 μg of total cellular SupT1 RNA, and the control samples each consisted of 1 ng of in vitro transcripts of 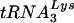 or tRNAGlu created by using the MEGAshortscript transcription kit (Ambion) according to the manufacturer's instructions. Oligonucleotides complementary to human
or tRNAGlu created by using the MEGAshortscript transcription kit (Ambion) according to the manufacturer's instructions. Oligonucleotides complementary to human 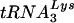 (5′-CGCCCGAACAGGGACTTGAACCCTGGACCCTCAGATTAAAAGTCTGATGCTCTACCGACTGAGCTATCC-3′)and tRNAGlu (5′-TTCCCTGACCGGGAATCGAACCCGGGCCGCGGCGGTGAGAGCGCCGAATCCTAACCACTA-3′) served as probes. Equal amounts of the probes (15 pmol) were 5′-end labeled with [γ-32P]ATP by using the Ready-To-Go T4 polynucleotide kinase kit (Amersham Pharmacia Biotech, Piscataway, N.J.), and free nucleotides were removed by centrifugation by using ProbeQuant G-50 micro columns (Amersham Pharmacia Biotech). Equal amounts of labeled probe (106 cpm/pmol) were added to the membranes. The signals of the hybridized and washed membranes were detected with a phosphorimager, and the relative amounts were determined by volume analysis by using ImageQuant version 1.2 (Molecular Dynamics, Piscataway, N.J.).
(5′-CGCCCGAACAGGGACTTGAACCCTGGACCCTCAGATTAAAAGTCTGATGCTCTACCGACTGAGCTATCC-3′)and tRNAGlu (5′-TTCCCTGACCGGGAATCGAACCCGGGCCGCGGCGGTGAGAGCGCCGAATCCTAACCACTA-3′) served as probes. Equal amounts of the probes (15 pmol) were 5′-end labeled with [γ-32P]ATP by using the Ready-To-Go T4 polynucleotide kinase kit (Amersham Pharmacia Biotech, Piscataway, N.J.), and free nucleotides were removed by centrifugation by using ProbeQuant G-50 micro columns (Amersham Pharmacia Biotech). Equal amounts of labeled probe (106 cpm/pmol) were added to the membranes. The signals of the hybridized and washed membranes were detected with a phosphorimager, and the relative amounts were determined by volume analysis by using ImageQuant version 1.2 (Molecular Dynamics, Piscataway, N.J.).
To analyze 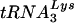 or tRNAGlu in virions, virus from SupT1 cultures infected with wild-type HIV-1 HXB2 or HXB2(Glu Loop 2) was collected and filtered through a 0.45-μm-pore-size low-protein binding filter (Pall Gelman Laboratory, Ann Arbor, Mich.). Virion-associated RNA was isolated via phenol-chloroform extraction followed by ethanol precipitation. Equal quantities of virion-associated RNA were blotted and probed for
or tRNAGlu in virions, virus from SupT1 cultures infected with wild-type HIV-1 HXB2 or HXB2(Glu Loop 2) was collected and filtered through a 0.45-μm-pore-size low-protein binding filter (Pall Gelman Laboratory, Ann Arbor, Mich.). Virion-associated RNA was isolated via phenol-chloroform extraction followed by ethanol precipitation. Equal quantities of virion-associated RNA were blotted and probed for 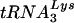 or tRNAGlu.
or tRNAGlu.
RESULTS
Characterization of HIV proviral genome with PBS complementary to tRNAGlu.
Previous studies have described HIV-1 proviral genomes in which the 18-nucleotide PBS has been altered to be complementary to a variety of tRNAs (11, 12, 27). Analysis of HIV-1 with different U5-PBS combinations revealed the presence of a PBS complementary to tRNAGlu in integrated proviruses from a long-term culture. Since we did not know if this PBS would be from a full-length provirus, in our first set of experiments we created an HIV-1 that has a PBS complementary to tRNAGlu (Fig. 1).
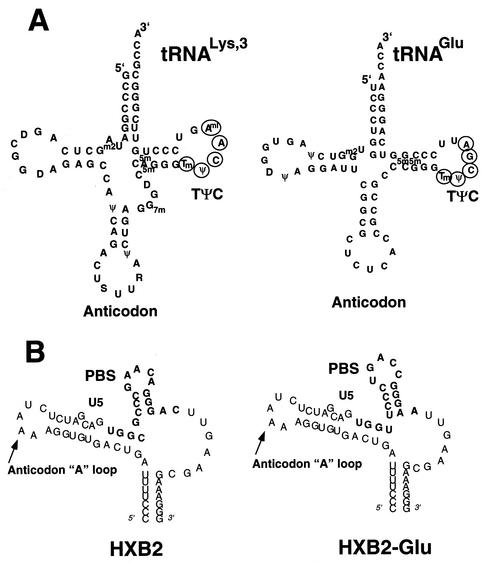
FIG. 1.
Diagrams of tRNAs and U5-PBS RNA. (A) 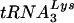 and tRNAGlu. The cloverleaf diagrams of
and tRNAGlu. The cloverleaf diagrams of 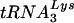 and tRNAGlu are presented with the modified bases in both tRNAs noted. The PBS of HIV is complementary to the 3′-terminal nucleotides of
and tRNAGlu are presented with the modified bases in both tRNAs noted. The PBS of HIV is complementary to the 3′-terminal nucleotides of 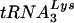 ; a second HIV genome was constructed in which the PBS was complementary to the 3′-terminal 18 nucleotides of tRNAGlu. Those nucleotides circled in the TφC-loop of
; a second HIV genome was constructed in which the PBS was complementary to the 3′-terminal 18 nucleotides of tRNAGlu. Those nucleotides circled in the TφC-loop of 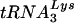 and tRNAGlu are complementary to regions downstream of the PBS. Abbreviations: S, 5-methoxycarbonylmethyl-2-thiouridine; ψ, pseudouridine; D, dihydrouridine; Tm, 2′-O-methyl-5-methyluridine; Aml, 1-methyladenosine; C5m, 5-methylcytidine; Gm2, N2-methylguanosine; G7m, 7-methylguanosine. (B) The U5-PBS region from HXB2 and that from HXB2(Glu) in which the PBS has been altered to be complementary to tRNAGlu are shown. The PBS is part of a stem-loop structure in which adenines are displayed at the crown of the stem-loop.
and tRNAGlu are complementary to regions downstream of the PBS. Abbreviations: S, 5-methoxycarbonylmethyl-2-thiouridine; ψ, pseudouridine; D, dihydrouridine; Tm, 2′-O-methyl-5-methyluridine; Aml, 1-methyladenosine; C5m, 5-methylcytidine; Gm2, N2-methylguanosine; G7m, 7-methylguanosine. (B) The U5-PBS region from HXB2 and that from HXB2(Glu) in which the PBS has been altered to be complementary to tRNAGlu are shown. The PBS is part of a stem-loop structure in which adenines are displayed at the crown of the stem-loop.
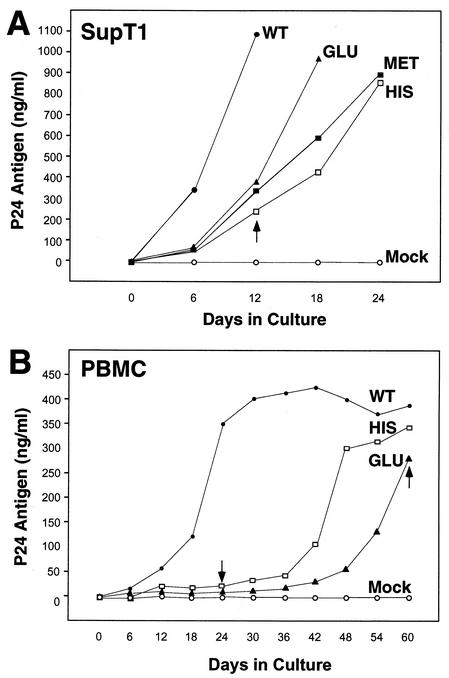
We first examined the replication of HIV-1 in which the PBS was altered to be complementary to tRNAGlu in both SupT1 cells and PBMCs (Fig. 2). Consistent with previous studies, we first generated virus by transfection of COS-1 cells, and an infection was initiated in SupT1 cells by using identical amounts of virus measured by a p24 antigen capture assay. Following initiation of the infection, culture supernatants were analyzed for p24 antigen at various times postinfection. Analysis of the virus production in these cultures during the first 24 days revealed rapid growth of the wild-type virus, as previously described (11, 12, 26). The virus with the PBS complementary to tRNAGlu exhibited a delay in replication compared to the wild type, followed by a rapid increase during the next 12 days to reach a level similar to that of the wild-type virus. We also compared the replication of the virus with a PBS complementary to tRNAGlu to that of viruses in which the PBSs were complementary to tRNAHis or tRNAMet. In this case, we found that the virus with the PBS complementary to tRNAGlu appeared to grow faster than these viruses, as measured by this p24 antigen capture assay. However, the viruses with the PBSs complementary to tRNAHis or tRNAMet eventually reached the same level of replication, as judged by a p24 antigen capture assay at 24 days postinitiation of the culture. Previous studies that characterized these viruses have shown that the rapid rise in p24 antigen correlated with the reversion of these viruses to the utilization of 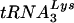 for replication (12, 26). To address this issue, we analyzed the PBSs of these viruses at several time points during the culture (Table 1). As expected, by day 24 all of the viruses with the PBSs complementary to tRNAMet or tRNAHis had reverted back to the wild type. To our surprise, we found that the viruses with the PBSs complementary to tRNAGlu remained stable for up to 48 days in culture. Thus, in contrast to the viruses which use tRNAHis or tRNAMet, HIV-1 that uses tRNAGlu was able to grow to substantial levels during the first 24 days in culture and remained stable during this time.
for replication (12, 26). To address this issue, we analyzed the PBSs of these viruses at several time points during the culture (Table 1). As expected, by day 24 all of the viruses with the PBSs complementary to tRNAMet or tRNAHis had reverted back to the wild type. To our surprise, we found that the viruses with the PBSs complementary to tRNAGlu remained stable for up to 48 days in culture. Thus, in contrast to the viruses which use tRNAHis or tRNAMet, HIV-1 that uses tRNAGlu was able to grow to substantial levels during the first 24 days in culture and remained stable during this time.
FIG. 2.
Replication of HIV-1 with altered PBS in SupT1 cells and PBMCs. (A) The replication of the wild-type virus (WT) and viruses with PBSs complementary to tRNAGlu (Glu), tRNAMet (Met), and tRNAHis (His) was analyzed in SupT1 cells. Cultures were initiated as described in Materials and Methods, and the supernatants were analyzed for p24 antigen production during a 24-day culture period. The values represented are from a single experiment; repeat experiments gave similar replication profiles. The arrow denotes the sample time at which PBSs from both viruses which used tRNAMet and tRNAHis reverted to PBSs complementary to 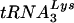 (Table 1). (B) Replication of HIV-1 with the PBS complementary to tRNAHis or tRNAGlu in PBMCs. Cultures were initiated as described in Materials and Methods, and virus replication was monitored by analysis of p24 antigen in culture supernatants. The arrow denotes the sample time at which the PBSs complementary to tRNAHis and tRNAGlu were found to revert to PBSs complementary to
(Table 1). (B) Replication of HIV-1 with the PBS complementary to tRNAHis or tRNAGlu in PBMCs. Cultures were initiated as described in Materials and Methods, and virus replication was monitored by analysis of p24 antigen in culture supernatants. The arrow denotes the sample time at which the PBSs complementary to tRNAHis and tRNAGlu were found to revert to PBSs complementary to 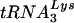 (Table 2). Results presented are from a single experiment; repeat experiments gave similar replication profiles for each virus.
(Table 2). Results presented are from a single experiment; repeat experiments gave similar replication profiles for each virus.
TABLE 1.
Summary of PBS stability during culture in SupT1 cells
| Initial provirusa | No. of viruses with wild-type PBSs/total no. of PBSs sequenced on day:b
|
|||
|---|---|---|---|---|
| 12 | 24 | 36 | 48 | |
| His | 9/13 | 17/17 | ND | ND |
| Met | 8/12 | 12/12 | ND | ND |
| Glu | 0/13 | 0/13 | 0/12 | 12/12 |
His, HIV-1 with a PBS complementary to tRNAHis; Met, HIV-1 with a PBS complementary to tRNAMet; Glu, HIV-1 with α PBS complementary to tRNAGlu.
ND, not done.
Up to this point, our studies have used a continuously dividing T-cell line, SupT1, to assess the effects that altering the PBS has on replication. Since the availability of tRNA could be influenced by the metabolic activity of the cell, we next wanted to assess the replication of HIV-1 with an altered PBS in primary PBMCs (Fig. 2). Equal amounts of virus generated from the transfection of COS-1 cells were used to initiate PBMC infections. A rapid increase in p24 antigen production, peaking at approximately 30 days postinitiation of culture, was observed for the wild-type virus. In contrast, viruses in which the PBSs were complementary to tRNAHis or tRNAGlu grew slowly compared to the wild type. By 42 days postinitiation of culture, we found a slight increase in the growth of virus with the PBS complementary to tRNAHis; we observed a rapid growth of this virus in the subsequent 12 days of culture, with the p24 values peaking at days 54 to 60. In contrast, the virus that used tRNAGlu began a slow and steady rise of virus production starting at approximately day 42. By day 60, this virus had reached a plateau of p24 antigen production similar to that of the wild-type virus and virus which used tRNAHis.
The PBSs of these viruses were analyzed at various times postinitiation of culture. By day 24 of culture, we detected reversion of the PBS complementary to tRNAHis back to the wild-type PBS; the numbers of clones with wild-type PBSs increased at days 36 and 48 of culture (Table 2). The reversion back to the wild-type PBS correlated with the rapid increase in levels of p24 antigen in the culture. In contrast, the virus with the PBS complementary to tRNAGlu remained stable for up to 48 days postinitiation of culture. DNA sequence analysis of the PBSs from viruses with PBSs complementary to tRNAGlu from both SupT1 cells (Tables 3, 4, and 5) and PBMCs (Tables 6 and 7) following reversion revealed that the PBSs had indeed reverted to being complementary to 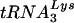 . In addition, we found few sequence changes in the initial input sequence in the U5 region; the replacement of a G immediately 5′ of the PBS with an A or C was found in the viruses grown in both SupT1 cells and PBMCs (Tables 3, 4, 5, 6, and 7).
. In addition, we found few sequence changes in the initial input sequence in the U5 region; the replacement of a G immediately 5′ of the PBS with an A or C was found in the viruses grown in both SupT1 cells and PBMCs (Tables 3, 4, 5, 6, and 7).
TABLE 2.
Summary of PBS stability during growth in PBMCs
| Initial provirusa | No. of viruses with wild-type PBSs/total no. of PBSs sequenced on day:
|
||||
|---|---|---|---|---|---|
| 12 | 24 | 36 | 48 | 60 | |
| His | 0/10 | 4/10 | 7/10 | 10/10 | NDb |
| Glu | 0/10 | 0/10 | 0/10 | 0/10 | 3/10 |
| HXB2(Glu Loop 2) | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
His, HIV-1 with a PBS complementary to tRNAHis, Glu, HIV-1 with a PBS complementary to tRNAGlu.
ND, not done.
TABLE 3.
DNA sequences of U5 and PBS regions of proviruses derived from pHXB2(Glu) on day 48 of culture in SupT1 cells
| Frequencya | Changes in the following input DNA sequenceb (5′-3′): |
|---|---|
| CCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGTGGTTCCCTGACCGGGAATTGAAAGCGAAAGGGAA | |
| 9/12 | ******************************************CG***GA**A****C***************** |
| 1/12 | ******************************************CG***GA**A****CG**************** |
| 1/12 | **************************************A***CG***GA**A****C***************** |
| 1/12 | **************************************C***CG***GA**A****C***************** |
Number with DNA sequence/total number sequenced.
The PBS complementary to tRNAGlu is shown in bold in the input sequence. In each case the changes resulted in a PBS complementary to 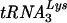 . Asterisks represent conserved nucleotides.
. Asterisks represent conserved nucleotides.
TABLE 4.
DNA sequences of U5 and PBS regions of proviruses derived from pHXB2(Glu Loop 1) on day 48 of culture in SupT1 cells
| Frequencya | Changes in the following input DNA sequenceb (5′-3′): |
|---|---|
| CAGACCCTTTTAGTCAGTGTAGAGTTGAGTCTCTAGCAGTGGTTCCCTGACCGGGAATTGAAAGCGAAAGGGAA | |
| 1/12 | ************************************************************************** |
| 1/12 | **************************************A*********************************** |
| 1/12 | **************************************A*******************C*************** |
| 1/12 | *********************A******A*********A*********************************** |
| 1/12 | ********C******T*****A******A*********A*********************************** |
| 3/12 | ****************AGC******************************************************* |
| 1/12 | ***************************************************A****C***************** |
| 1/12 | ****************************A**********************A****C***************** |
| 1/12 | ****************************A*********A************A****C***************** |
| 1/12 | ***********************************************GA**A****C***************** |
Number with DNA sequence/total number sequenced.
The PBS and anticodon region complementary to tRNAGlu are shown in bold in the input sequence. Asterisks represent conserved nucleotides.
TABLE 5.
DNA sequences of U5 and PBS regions of proviruses derived from pHXB2(Glu Loop 2) on day 48 of culture in SupT1 cells
| Frequencya | Changes in the following input DNA sequenceb (5′-3′): |
|---|---|
| CAGACCCTTTTAGTCAGGCTAGAGTTGAGTCTCTAGCAGTGGTTCCCTGACCGGGAATTGAAAGCGAAAGGGAA | |
| 2/12 | ************************************************************************** |
| 1/12 | ****************A********************************************************* |
| 3/12 | ****************A************A******************************************** |
| 1/12 | ****************A************A*******************************************G |
| 4/12 | *****************TG******************************************************* |
| 1/12 | *****************TG********************************A****C***************** |
Number with DNA sequence/total number sequenced.
The PBS and anticodon region complementary to tRNAGlu are shown in bold in the input sequence. Asterisks represent conserved nucleotides.
TABLE 6.
DNA sequences of U5 and PBS regions of proviruses derived from pHXB2(Glu) on day 60 of culture in PBMCs
| Frequencya | Changes in the following input DNA sequenceb (5′-3′): |
|---|---|
| CCTCAGACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGTGGTTCCCTGACCGGGAATTGAAAGCGAAAGGGAA | |
| 3/10 | ************************************************************************** |
| 1/10 | **************************************C*********************************** |
| 1/10 | *******************************G****************************************** |
| 2/10 | ***********************************************GA**A****C***************** |
| 2/10 | ******************************************CG***GA**A****C*****************c |
| 1/10 | **************************************A***CG***GA**A****C*****************c |
Number with DNA sequence/total number sequenced.
The PBS complementary to tRNAGlu is shown in bold in the input sequence. Asterisks represent conserved nucleotides.
Changes resulted in a wild-type PBS complementary to tRNA3Lys.
TABLE 7.
DNA sequences of U5 and PBS regions of proviruses derived from pHXB2(Glu Loop 2) on day 60 of culture in PBMC.
| Frequencya | Changes in the following input DNA sequenceb (5′-3′): |
|---|---|
| CAGACCCTTTTAGTCAGGCTAGAGTTGAGTCTCTAGCAGTGGTTCCCTGACCGGGAATTGAAAGCGAAAGGGAA | |
| 6/10 | ************************************************************************** |
| 1/10 | **************************************A*********************************** |
| 1/10 | ***********************************************************A************** |
| 1/10 | ****************A********************************************************* |
| 1/10 | ****************A************A******************************************** |
Number with DNA sequence/total number sequenced.
The PBS and anticodon sequence complementary to tRNAGlu are shown in bold. Asterisks represent conserved nucleotides.
Characterization of HIV-1 with the PBS and anticodon binding domains in U5 complementary to tRNAGlu.
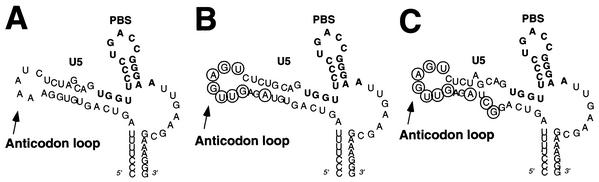
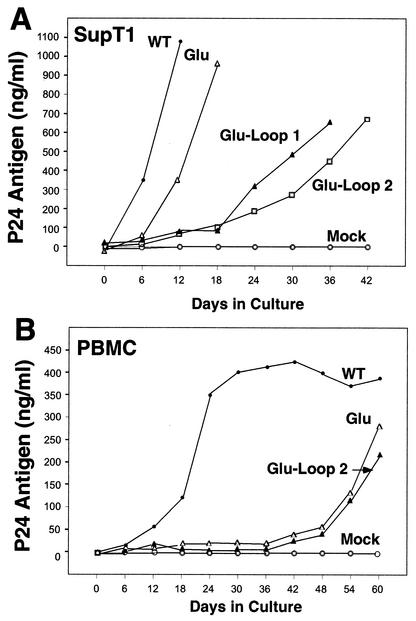
Previous studies have found that the alteration of a region in U5 (designated the A-loop) to correspond to the anticodon domains of certain tRNAs allows stable use of these tRNAs for replication by HIV-1. In an attempt to further stabilize the use of tRNAGlu as the primer for HIV replication, we made additional mutations in the U5 region in which the nucleotides in the A-loop were replaced with those complementary to the anticodon of tRNAGlu (Fig. 3). Additional nucleotide substitutions in the stem were made to facilitate a predicted U5-PBS stem-loop (31, 32). Two RNA structures (Glu loop 1 and Glu loop 2) were chosen that had similar free energy values for the U5-PBS regions; modeling of these U5-PBS regions resulted in a stem-loop structure that was very similar to that of the wild-type HIV genome. We chose to analyze both mutants since previous studies analyzing HIV-1 mutants that used tRNAMet or tRNAHis had suggested that bulges within the RNA stem might be important for tRNA selection and replication (14, 29). The replication of two mutant HIV genomes in which the anticodon binding regions and PBSs had been altered to correspond to tRNAGlu was first analyzed in SupT1 cells (Fig. 4). Both viruses demonstrated a delay in replication compared to the wild-type virus (or even the virus with a PBS complementary to tRNAGlu). By 42 days postinitiation of culture, these viruses still had not produced p24 antigen to the level observed for the wild-type virus at day 12 of culture. These virus cultures were subsequently grown for an additional 6 days in culture (to day 48), followed by analysis of the PBSs at various times of culture. In this case, we found that the PBSs of all samples analyzed were still complementary to tRNAGlu (Tables 4 and 5). We also noted additional mutations in the U5 region; in some instances, common mutations were found between different PCR clones that were analyzed.
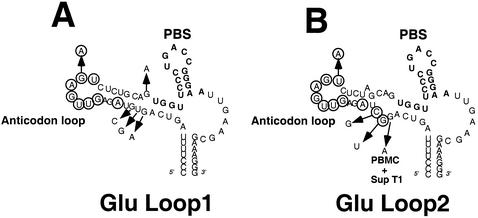
FIG. 3.
HIV-1 with the U5-PBS region complementary to tRNAGlu. HIV-1 genomes in which the PBS and A-loop regions have been altered to be complementary to the 3′-terminal nucleotides and anticodon of tRNAGlu are presented. The PBS region is noted in bold. The circled nucleotides are those which were changed to be complementary to the anticodon of tRNAGlu (CUC). (A) Wild-type HIV-1 genome in which the PBS has been altered to be complementary to tRNAGlu. (B) HIV-1 genome with additional nucleotides making it complementary to the anticodon of tRNAGlu [HXB2(Glu Loop 1)]. An additional nucleotide change was added to enhance the stem of the U5-PBS stem-loop (A, circled). (C) HIV-1 genome in which the PBS and A-loop have been altered to correspond to the 3′-terminal nucleotides of tRNAGlu and the anticodon region. Additional nucleotides inserted within the stem destabilized the stem-loop, creating a bulge region; virus with this genome is designated HXB2(Glu Loop 2).
FIG. 4.
Replication of HIV genomes with U5-PBS regions optimized for use of tRNAGlu. (A) Replication in SupT1 cells. Virus cultures were initiated as described in Materials and Methods. The supernatants from the cultures were analyzed for p24 antigen production at various days postinitiation of culture. For comparison, results for viruses in which only the PBS was altered to correspond to tRNAGlu (Glu) are shown. Results for two additional viruses with both the U5 and the PBS altered to correspond to tRNAGlu (Glu-Loop 1 and Glu-Loop 2) are presented. WT, wild type. (B) Replication of viruses in PBMCs. The replication of one of the viruses represented in panel A (Glu-Loop 2) was analyzed in PBMCs. For comparison, results for the virus with only the PBS altered to be complementary to tRNAGlu (Glu) are presented. The arrow denotes the sample time at which the virus with only the PBS complementary to tRNAGlu had a PBS complementary to 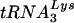 . Results are from a single experiment; the cultures were performed in duplicate with similar replication profiles.
. Results are from a single experiment; the cultures were performed in duplicate with similar replication profiles.
The replication of these viruses was also analyzed in PBMCs (Fig. 4). We again found a severe delay in the production of infectious virus in PBMC cultures compared to that of the wild-type virus. In this case, replication of the virus with a PBS and anticodon binding loop complementary to tRNAGlu was slow, reaching approximately 50% of that of the wild type by 60 days postinitiation of culture. The replication of this virus paralleled that seen for the virus in which only the PBS was made complementary to tRNAGlu. The PBSs of these viruses were analyzed to determine whether they had reverted back to the wild type. Sequence analysis of these viruses at day 60 revealed a remarkable conservation of the initial DNA sequence; in 60% of the sequences analyzed, we obtained the input sequence (Table 7). In the remaining 40%, we observed only single nucleotide changes mostly confined to the U5 region. Interestingly, similar changes were noted following the growth of this same virus in SupT1 cells.
Comparison of tRNALys and tRNAGlu in cells and HIV-1 virions.
One simple explanation for the reversion of the mutant PBS back to one complementary to 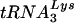 is that this tRNA is predominant in the host cells. To address this issue, we compared the relative levels of
is that this tRNA is predominant in the host cells. To address this issue, we compared the relative levels of 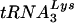 with those of tRNAGlu in SupT1 cells, which allow high-level replication of HIV-1, prior to analysis of HIV-1 growth in these cells (Fig. 5). Identical amounts of total RNA from SupT1 cells were probed for tRNAGlu or
with those of tRNAGlu in SupT1 cells, which allow high-level replication of HIV-1, prior to analysis of HIV-1 growth in these cells (Fig. 5). Identical amounts of total RNA from SupT1 cells were probed for tRNAGlu or 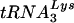 ; in vitro-transcribed tRNAs were used to confirm specificity of the probes. Under these experimental conditions, we found that the amounts of tRNAGlu were larger than those of
; in vitro-transcribed tRNAs were used to confirm specificity of the probes. Under these experimental conditions, we found that the amounts of tRNAGlu were larger than those of 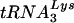 in SupT1 cells. Quantitation from three independent samples revealed that the cellular tRNAGlu/
in SupT1 cells. Quantitation from three independent samples revealed that the cellular tRNAGlu/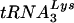 ratio was approximately 10:1. Analysis of mitogen-activated PBMCs demonstrated a similar pattern in which the level of
ratio was approximately 10:1. Analysis of mitogen-activated PBMCs demonstrated a similar pattern in which the level of 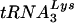 was less than that of tRNAGlu (data not shown).
was less than that of tRNAGlu (data not shown).
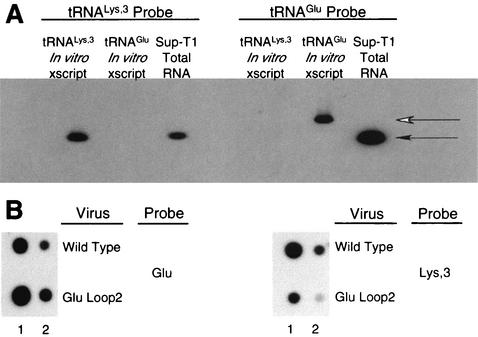
FIG. 5.
Analysis of intracellular and HIV-1 virion-associated 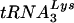 and tRNAGlu. (A) Analysis of tRNA from SupT1 cells. Total RNA from SupT1 cells was isolated as described in Materials and Methods. Following electrophoresis and transfer onto nitrocellulose membrane, equal amounts were probed with oligonucleotides specific for
and tRNAGlu. (A) Analysis of tRNA from SupT1 cells. Total RNA from SupT1 cells was isolated as described in Materials and Methods. Following electrophoresis and transfer onto nitrocellulose membrane, equal amounts were probed with oligonucleotides specific for 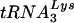 or tRNAGlu. Specificity controls included equal amounts (1 ng) of in vitro RNA transcription products; note that the in vitro transcript (xscript) for tRNAGlu contains extra nucleotides that retard the migration in the agarose gels used for this analysis. The densities of the probes reacting with the cognate tRNA for both
or tRNAGlu. Specificity controls included equal amounts (1 ng) of in vitro RNA transcription products; note that the in vitro transcript (xscript) for tRNAGlu contains extra nucleotides that retard the migration in the agarose gels used for this analysis. The densities of the probes reacting with the cognate tRNA for both 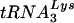 and tRNAGlu were similar (i.e., the density of the
and tRNAGlu were similar (i.e., the density of the 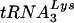 probe with 1 ng of in vitro-transcribed
probe with 1 ng of in vitro-transcribed 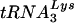 versus the density of the tRNAGlu probe with 1 ng of in vitro-transcribed tRNAGlu). The use of scanning densitometry to determine the cellular amounts of tRNAGlu and
versus the density of the tRNAGlu probe with 1 ng of in vitro-transcribed tRNAGlu). The use of scanning densitometry to determine the cellular amounts of tRNAGlu and 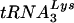 revealed an approximate 10:1 ratio. This analysis was repeated three times, and a representative blot is shown. The closed arrow denotes migration of full-length tRNALys or tRNAGlu. The open arrow denotes migration of the tRNAGlu in vitro transcript containing additional nucleotides from cloning of the tRNA gene into the transcription plasmid. (B) Analysis of tRNA from HIV-1 virions. Total virion RNA collected from viruses isolated from supernatant of SupT1 cells infected with HXB2 or HXB2(Glu Loop 2). HIV-1 virions were isolated by ultracentrifugation, and RNA was extracted and prepared as described in Materials and Methods. The blot was probed for tRNAGlu and
revealed an approximate 10:1 ratio. This analysis was repeated three times, and a representative blot is shown. The closed arrow denotes migration of full-length tRNALys or tRNAGlu. The open arrow denotes migration of the tRNAGlu in vitro transcript containing additional nucleotides from cloning of the tRNA gene into the transcription plasmid. (B) Analysis of tRNA from HIV-1 virions. Total virion RNA collected from viruses isolated from supernatant of SupT1 cells infected with HXB2 or HXB2(Glu Loop 2). HIV-1 virions were isolated by ultracentrifugation, and RNA was extracted and prepared as described in Materials and Methods. The blot was probed for tRNAGlu and 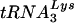 as indicated. Lane 1, 1 μg of RNA isolated from virions; lane 2, 0.1 μg of RNA isolated from virions.
as indicated. Lane 1, 1 μg of RNA isolated from virions; lane 2, 0.1 μg of RNA isolated from virions.
We next compared the levels of 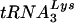 and tRNAGlu found in wild-type HIV-1 virions (HXB2) and virions with U5-PBS regions complementary to tRNAGlu [HXB2(Glu Loop 2)] (Fig. 5). RNA from virions from infected SupT1 cultures were isolated and normalized and dilutions were analyzed for
and tRNAGlu found in wild-type HIV-1 virions (HXB2) and virions with U5-PBS regions complementary to tRNAGlu [HXB2(Glu Loop 2)] (Fig. 5). RNA from virions from infected SupT1 cultures were isolated and normalized and dilutions were analyzed for 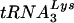 or tRNAGlu by using specific oligonucleotide probes. tRNALys and tRNAGlu were detected in both wild-type virus and the virus with a U5-PBS region complementary to tRNAGlu. The proportion of
or tRNAGlu by using specific oligonucleotide probes. tRNALys and tRNAGlu were detected in both wild-type virus and the virus with a U5-PBS region complementary to tRNAGlu. The proportion of 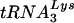 varied depending on the virus. In the wild type, the proportion of
varied depending on the virus. In the wild type, the proportion of 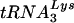 was greater than that in the virus with a U5-PBS region complementary to tRNAGlu, while slightly more tRNAGlu was detected in viruses that used tRNAGlu than in the wild-type virus.
was greater than that in the virus with a U5-PBS region complementary to tRNAGlu, while slightly more tRNAGlu was detected in viruses that used tRNAGlu than in the wild-type virus.
DISCUSSION
In this study, we have investigated the selection of the tRNA primer required by HIV-1 for replication by constructing a virus in which the PBS has been made complementary to tRNAGlu. Analysis of virus replication in both SupT1 cells and PBMCs revealed that the HIV-1 with a PBS complementary to tRNAGlu grew more slowly than the wild-type virus but more rapidly than viruses with PBSs complementary to tRNAMet or tRNAHis. Most importantly, the PBSs of viruses that used tRNAGlu were more stable, as judged by a reduced propensity to mutate back to utilizing 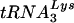 , than PBSs complementary to tRNAMet or tRNAHis. We further engineered the HIV-1 genome to include a region within the U5 that was complementary to the anticodon nucleotides of tRNAGlu. This additional mutation resulted in HIV-1 that stably utilized tRNAGlu when grown in SupT1 cells or PBMCs. Thus, the selective engineering of the HIV-1 genome with mutations in both U5 and the PBS has resulted in a virus in which the tRNA primer has been successfully altered from
, than PBSs complementary to tRNAMet or tRNAHis. We further engineered the HIV-1 genome to include a region within the U5 that was complementary to the anticodon nucleotides of tRNAGlu. This additional mutation resulted in HIV-1 that stably utilized tRNAGlu when grown in SupT1 cells or PBMCs. Thus, the selective engineering of the HIV-1 genome with mutations in both U5 and the PBS has resulted in a virus in which the tRNA primer has been successfully altered from 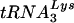 to tRNAGlu.
to tRNAGlu.
Similar to previous studies, the present study has taken a genetic approach to investigate the mechanism of primer tRNA selection by HIV-1. In particular, we were interested in determining why HIV-1 has evolved to select 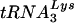 over other tRNAs, including
over other tRNAs, including 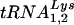 that is present in greater amounts in the virion (10, 30). To address this question, we analyzed sequences of U5-PBS regions from previous studies for the presence of new, unexpected tRNAs that were selected by HIV-1. The use of these other tRNAs by HIV-1 may indicate a preference for the other tRNAs over
that is present in greater amounts in the virion (10, 30). To address this question, we analyzed sequences of U5-PBS regions from previous studies for the presence of new, unexpected tRNAs that were selected by HIV-1. The use of these other tRNAs by HIV-1 may indicate a preference for the other tRNAs over 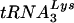 . A previous study identified several PBS clones that were complementary to tRNAGlu (16). As a first step in the study, we created HIV-1 in which only the PBS was complementary to tRNAGlu and analyzed the replication and stability of the PBS following extended culture in SupT1 cells and PBMCs. To our surprise, the PBS of this virus demonstrated considerable stability following growth of the virus in both cell types. In fact, the HIV-1 with a PBS complementary to tRNAGlu was more stable than any of the other viruses we have created thus far with only the PBSs changed. One explanation for the stability could be that the amounts of tRNAGlu detected in SupT1 cells were greater than those of
. A previous study identified several PBS clones that were complementary to tRNAGlu (16). As a first step in the study, we created HIV-1 in which only the PBS was complementary to tRNAGlu and analyzed the replication and stability of the PBS following extended culture in SupT1 cells and PBMCs. To our surprise, the PBS of this virus demonstrated considerable stability following growth of the virus in both cell types. In fact, the HIV-1 with a PBS complementary to tRNAGlu was more stable than any of the other viruses we have created thus far with only the PBSs changed. One explanation for the stability could be that the amounts of tRNAGlu detected in SupT1 cells were greater than those of 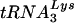 . However, if the amounts of tRNA in the cytoplasm were the sole determinant for selection, we would have expected that HIV-1 with a PBS complementary to tRNAGlu would replicate as well as the wild-type virus, which was not observed in SupT1 cells or PBMCs. This discrepancy highlights the complicated nature of the HIV-1 primer selection process. The mechanism underlying the selection of
. However, if the amounts of tRNA in the cytoplasm were the sole determinant for selection, we would have expected that HIV-1 with a PBS complementary to tRNAGlu would replicate as well as the wild-type virus, which was not observed in SupT1 cells or PBMCs. This discrepancy highlights the complicated nature of the HIV-1 primer selection process. The mechanism underlying the selection of 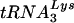 by HIV-1 for use as the primer in replication is unknown. Previous studies have suggested that the viral proteins nucleocapsid and reverse transcriptase or the Gag-Pol protein are involved in selection (3, 8, 18, 19). Recent studies have demonstrated that the lysyl-tRNA synthetase is present in HIV-1 virions and may act as a chaperone for tRNA (4). In addition, studies have found that elongation factor 1α (EF1A), known to interact with tRNAs, is also found in HIV virions (5, 21, 22). What is not clear from these studies is whether the tRNA entering the virions in complex with these proteins is actually the
by HIV-1 for use as the primer in replication is unknown. Previous studies have suggested that the viral proteins nucleocapsid and reverse transcriptase or the Gag-Pol protein are involved in selection (3, 8, 18, 19). Recent studies have demonstrated that the lysyl-tRNA synthetase is present in HIV-1 virions and may act as a chaperone for tRNA (4). In addition, studies have found that elongation factor 1α (EF1A), known to interact with tRNAs, is also found in HIV virions (5, 21, 22). What is not clear from these studies is whether the tRNA entering the virions in complex with these proteins is actually the 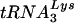 that is selected for use as the primer. A previous study found that the tRNA composition of a virus that uses tRNAHis had the same ratios of
that is selected for use as the primer. A previous study found that the tRNA composition of a virus that uses tRNAHis had the same ratios of 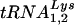 and
and 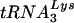 as the wild-type virus; no enrichment of tRNAHis was found (30). In contrast, HXB2(Glu Loop 2) had an enrichment of tRNAGlu in the virions compared to the wild-type virus, suggesting that in this case, the alteration of the U5-PBS region was sufficient to change the tRNA composition in the virion. However, even with the increased level of tRNAGlu in the virion, the virus did not replicate in a fashion similar to that of the wild type. Thus, the amounts of tRNA that we detected in the cell and virion may not necessarily reflect the amount available for HIV-1 to select as a primer. The tRNA in the cell is probably not free but in complex with EF1A prior to use in translation or with the aminoacyl-tRNA synthetase after ejection from the E site of the ribosome (20, 24). Interestingly, a previous study reported that EFTu (the E. coli equivalent of EF1A)-GTP-aminoacylated tRNA can interact efficiently with other tRNAs that have complementary anticodons (28). Nuclear magnetic resonance studies of the A-loop region of HIV-1 suggest that the U5 stem-loop is similar to that of a tRNA anticodon (23), making it possible that the A-loop-
as the wild-type virus; no enrichment of tRNAHis was found (30). In contrast, HXB2(Glu Loop 2) had an enrichment of tRNAGlu in the virions compared to the wild-type virus, suggesting that in this case, the alteration of the U5-PBS region was sufficient to change the tRNA composition in the virion. However, even with the increased level of tRNAGlu in the virion, the virus did not replicate in a fashion similar to that of the wild type. Thus, the amounts of tRNA that we detected in the cell and virion may not necessarily reflect the amount available for HIV-1 to select as a primer. The tRNA in the cell is probably not free but in complex with EF1A prior to use in translation or with the aminoacyl-tRNA synthetase after ejection from the E site of the ribosome (20, 24). Interestingly, a previous study reported that EFTu (the E. coli equivalent of EF1A)-GTP-aminoacylated tRNA can interact efficiently with other tRNAs that have complementary anticodons (28). Nuclear magnetic resonance studies of the A-loop region of HIV-1 suggest that the U5 stem-loop is similar to that of a tRNA anticodon (23), making it possible that the A-loop-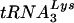 interaction could have similarities to codon-anticodon interactions that occur during translation. An EF1A-GTP-aminoacylated tRNA complex may then play a role in tRNA selection, possibly by chaperoning the tRNA to the complementary region in U5. After the EF1A-tRNA complex interacts with the U5 loop region, the tRNA would need to be released from the complex to associate with the PBS. It is interesting that a previous study found that the affinities of EFTu-GTP for tRNAGlu and tRNALys were two of the weakest among those for tRNAs (17). Additional studies then will be needed to address the role of EF1A in selection of the HIV-1 primer tRNA.
interaction could have similarities to codon-anticodon interactions that occur during translation. An EF1A-GTP-aminoacylated tRNA complex may then play a role in tRNA selection, possibly by chaperoning the tRNA to the complementary region in U5. After the EF1A-tRNA complex interacts with the U5 loop region, the tRNA would need to be released from the complex to associate with the PBS. It is interesting that a previous study found that the affinities of EFTu-GTP for tRNAGlu and tRNALys were two of the weakest among those for tRNAs (17). Additional studies then will be needed to address the role of EF1A in selection of the HIV-1 primer tRNA.
The development of HIV-1 that uses tRNAGlu rather than 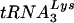 now brings the number of viruses that can use alternative tRNAs for replication to four. Previous studies have characterized HIVs that use tRNAHis, tRNAMet, or
now brings the number of viruses that can use alternative tRNAs for replication to four. Previous studies have characterized HIVs that use tRNAHis, tRNAMet, or 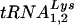 for replication rather than
for replication rather than 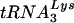 (11, 13, 26). A common feature of all of these viruses is that the U5 was mutated to display nucleotides complementary to the anticodon of the tRNA on the loop of an RNA stem-loop. Analysis of the U5 regions from HIV-1 that use tRNAGlu following long-term growth in SupT1 cells or PBMCs lends further support for a role of the U5-PBS RNA stem-loop in primer selection. We found that the U5 region from HIV-1 that grew in SupT1 cells and PBMCs contained mutations that could result in a stem region even more similar to that of the wild-type virus; the substituted nucleotides would be predicted to form bulges within the stem of the U5-PBS stem-loop (Fig. 6). It is possible that the location of mismatched nucleotides, creating bulges in the stem, or specific nucleotides in the U5-PBS are required for high-level replication. The enhanced growth of the HIV-1 in which only the PBS was complementary to tRNAGlu in SupT1 cells compared to that of viruses with mutations in U5 [e.g., HXB2(Glu Loop 1) and HXB2(Glu Loop 2)] might have been due in part to the predicted conservation of the wild-type U5-PBS RNA stem-loop, since previous studies have shown that mutations within the RNA stem-loop in the wild-type genome can affect viral replication (2). Additional study of these viruses will be needed in order to refine our understanding of the importance of the U5-PBS RNA stem-loop structure in the selection of the tRNA primer.
(11, 13, 26). A common feature of all of these viruses is that the U5 was mutated to display nucleotides complementary to the anticodon of the tRNA on the loop of an RNA stem-loop. Analysis of the U5 regions from HIV-1 that use tRNAGlu following long-term growth in SupT1 cells or PBMCs lends further support for a role of the U5-PBS RNA stem-loop in primer selection. We found that the U5 region from HIV-1 that grew in SupT1 cells and PBMCs contained mutations that could result in a stem region even more similar to that of the wild-type virus; the substituted nucleotides would be predicted to form bulges within the stem of the U5-PBS stem-loop (Fig. 6). It is possible that the location of mismatched nucleotides, creating bulges in the stem, or specific nucleotides in the U5-PBS are required for high-level replication. The enhanced growth of the HIV-1 in which only the PBS was complementary to tRNAGlu in SupT1 cells compared to that of viruses with mutations in U5 [e.g., HXB2(Glu Loop 1) and HXB2(Glu Loop 2)] might have been due in part to the predicted conservation of the wild-type U5-PBS RNA stem-loop, since previous studies have shown that mutations within the RNA stem-loop in the wild-type genome can affect viral replication (2). Additional study of these viruses will be needed in order to refine our understanding of the importance of the U5-PBS RNA stem-loop structure in the selection of the tRNA primer.
FIG. 6.
Summary of sequence changes found in U5-PBS regions following virus growth in SupT1 cells and PBMCs. (A) Changes found in the U5 region of the virus HXB2(Glu Loop 1) following replication in SupT1 cells. The arrows denote the major changes observed following replication. An additional change of G to A within the A-loop region (circled) was also found. Note that we did not see reversion of the PBS back to the wild-type PBS complementary to 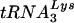 . (B) Changes found in the virus HXB2(Glu Loop 2) following in vitro culture in SupT1 cells or PBMCs. The major changes were noted within the stem region of the U5 PBS stem-loop. In this case, C-to-G, G-to-U, and G-to-A changes were noted in viruses grown in SupT1 cells; a G-to-A change was noted in viruses grown in PBMCs and SupT1 cells (PBMC + SupT1). These changes will be predicted to maintain a bulge region reduced from that of the initial HXB2(Glu Loop 2). Note again that reversion of the PBS to the wild-type PBS complementary to
. (B) Changes found in the virus HXB2(Glu Loop 2) following in vitro culture in SupT1 cells or PBMCs. The major changes were noted within the stem region of the U5 PBS stem-loop. In this case, C-to-G, G-to-U, and G-to-A changes were noted in viruses grown in SupT1 cells; a G-to-A change was noted in viruses grown in PBMCs and SupT1 cells (PBMC + SupT1). These changes will be predicted to maintain a bulge region reduced from that of the initial HXB2(Glu Loop 2). Note again that reversion of the PBS to the wild-type PBS complementary to 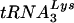 was found.
was found.
Acknowledgments
We thank Kenda Moore for helpful comments. Sylvia McPherson of the UAB CFAR Molecular Biology Core is acknowledged for help with construction of HIV-1 proviral genomes (A27727). All DNA sequencing was carried out by the UAB CFAR DNA Sequencing Core (A27727). We thank Barry Kosloff for preparation of PBMCs, and we thank Adrienne Ellis for preparation of the manuscript. C.D.M. acknowledges help from M.A.R.
L.C.D., N.J.K., and T.E.E. were supported by training grant AI 07493. This research was supported by grants GM56544 (C.D.M. and S.C.H.) and AI34749 (C.D.M.).
REFERENCES
- 1.Baltimore, D. 1970. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 226:1209-1211. [DOI] [PubMed] [Google Scholar]
- 2.Beerens, N., B. Klaver, and B. Berkhout. 2000. A structured RNA motif is involved in correct placement of the tRNA3Lys primer onto the human immunodeficiency virus genome. J. Virol. 74:2227-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 4.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 75:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, A. T., B. Klaver, and B. Berkhout. 1995. Reduced replication of human immunodeficiency virus type 1 mutants that use reverse transcription primers other than the natural tRNA3Lys. J. Virol. 69:3090-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilboa, E., S. W. Mitra, S. Goff, and D. Baltimore. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93-100. [DOI] [PubMed] [Google Scholar]
- 8.Harrich, D., and B. Hooker. 2002. Mechanistic aspects of HIV-1 reverse transcription initiation. Rev. Med. Virol. 12:31-45. [DOI] [PubMed] [Google Scholar]
- 9.Isel, C., C. Ehresmann, G. Keith, B. Ehresmann, and R. Marquet. 1995. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer). J. Mol. Biol. 247:236-250. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 67:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang, S.-M., and C. D. Morrow. 1999. Genetic analysis of a unique human immunodeficiency virus type 1 (HIV-1) with a primer binding site complementary to tRNAMet supports a role for U5-PBS stem-loop RNA structures in initiation of HIV-1 reverse transcription. J. Virol. 73:1818-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang, S.-M., J. K. Wakefield, and C. D. Morrow. 1996. Mutations in both the U5 region and the primer-binding site influence the selection of the tRNA used for the initiation of HIV-1 reverse transcription. Virology 222:401-414. [DOI] [PubMed] [Google Scholar]
- 13.Kang, S.-M., Z. Zhang, and C. D. Morrow. 1999. Identification of a human immunodeficiency virus type 1 that stably uses tRNALys1,2 rather than tRNALys,3 for initiation of reverse transcription. Virology 257:95-105. [DOI] [PubMed] [Google Scholar]
- 14.Kang, S.-M., Z. Zhang, and C. D. Morrow. 1997. Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J. Virol. 71:207-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X., J. Mak, E. J. Arts, Z. Gu, L. Kleiman, M. A. Wainberg, and M. A. Parniak. 1994. Effects of alterations of primer-binding site sequences on human immunodeficiency virus type 1 replication. J. Virol. 68:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., Z. Zhang, S.-M. Kang, J. L. Buescher, and C. D. Morrow. 1997. Insights into the interaction between tRNA and primer binding site from characterization of a unique HIV-1 virus which stably maintains dual PBS complementary to tRNAGly and tRNAHis. Virology 238:273-282. [DOI] [PubMed] [Google Scholar]
- 17.Louie, A., N. S. Ribeiro, B. R. Reid, and F. Jurnak. 1984. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259:5010-5016. [PubMed] [Google Scholar]
- 18.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mak, J., A. Khorchid, Q. Cao, Y. Huang, I. Lowy, M. A. Parniak, V. R. Prasad, M. A. Wainberg, and L. Kleiman. 1997. Effects of mutations in Pr160gag-pol upon tRNALys,3 and Pr160gag-pol incorporation into HIV-1. J. Mol. Biol. 265:419-431. [DOI] [PubMed] [Google Scholar]
- 20.Negrutskii, B. S., and M. P. Deutscher. 1991. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. USA 88:4991-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 22.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. I. Sowder, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 23.Puglisi, E. V., and J. D. Puglisi. 1998. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 5:1033-1036. [DOI] [PubMed] [Google Scholar]
- 24.Stapulionis, R., and M. P. Deutscher. 1995. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 92:7158-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temin, H. M., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226:1211-1213. [DOI] [PubMed] [Google Scholar]
- 26.Wakefield, J. K., S.-M. Kang, and C. D. Morrow. 1996. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol. 70:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield, J. K., A. G. Wolf, and C. D. Morrow. 1995. Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA3Lys. J. Virol. 69:6021-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamane, T., D. L. Miller, and J. J. Hopfield. 1981. Interaction of elongation factor Tu with the aminoacyl transfer ribonucleic acid dimer Phe-tRNA-Glu-tRNA. Biochemistry 20:449-452. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Z., S.-M. Kang, Y. Li, and C. D. Morrow. 1998. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA 4:394-406. [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Z., S. M. Kang, A. LeBlanc, S. L. Hajduk, and C. D. Morrow. 1996. Nucleotide sequences within the U5 region of the viral RNA genome are the major determinants for a human immunodeficiency virus type 1 to maintain a primer binding site complementary to tRNAHis. Virology 226:306-317. [DOI] [PubMed] [Google Scholar]
- 31.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]
- 32.Zuker, M., and P. Stiegler. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9:133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]